Abstract
The safe and effective delivery of RNA interference (RNAi) therapeutics remains an important challenge for clinical development. The diversity of current delivery materials remains limited, in part because of their slow, multi-step syntheses. Here we describe a new class of lipid-like delivery molecules, termed lipidoids, as delivery agents for RNAi therapeutics. Chemical methods were developed to allow the rapid synthesis of a large library of over 1,200 structurally diverse lipidoids. From this library, we identified lipidoids that facilitate high levels of specific silencing of endogenous gene transcripts when formulated with either double-stranded small interfering RNA (siRNA) or single-stranded antisense 2′-O-methyl (2′-O Me) oligoribonucleotides targeting microRNA (miRNA). The safety and efficacy of lipidoids were evaluated in three animal models: mice, rats and nonhuman primates. The studies reported here suggest that these materials may have broad utility for both local and systemic delivery of RNA therapeutics.
The specific reduction, or silencing, of gene expression through RNAi has considerable potential to create a new class of therapeutics that addresses previously untreatable diseases1,2. High doses of cholesterol-modified siRNA3 and cholesterol-modified 2′-OMe oligoribonucleotides (antagomirs)4 have been demonstrated to reduce gene expression specifically in vivo in liver and other tissues. Certain viruses, such as respiratory syncytial virus can be inhibited locally after administration of naked siRNA5,6. Recently, systemic delivery of siRNA to the liver of mice and nonhuman primates was demonstrated using a lipid formulation7,8. Despite these advances, there are few published reports demonstrating efficacy of systemically delivered siRNA in primates8,9. Although significant efforts have been dedicated to cationic lipid delivery systems, the development of safe and effective methods both in vitro and in vivo has proven challenging.
To date, the delivery of siRNA has been mediated by direct conjugation of delivery agents to the RNA moiety3,10–12, formulation using lipid-7,8,13–16, polymer-9,17–20 or peptide-based delivery systems21–23 and, more recently, complexation with antibody fusion proteins24,25. A key barrier to the exploration of delivery material space is new material synthesis. Conventional lipid synthesis typically requires individually optimized, multiple-step synthesis, including time-intensive procedures such as chemical protection and deprotection, use and removal of catalysts, solvent exchanges and purification26. The customization of each synthetic reaction and the multiple steps required limit throughput and, correspondingly, limit the ability to generate substantial library size and diversity. To address these problems, we developed chemical methods capable of rapid, parallel generation of lipid-like molecules (Fig. 1 and Supplementary Methods online).
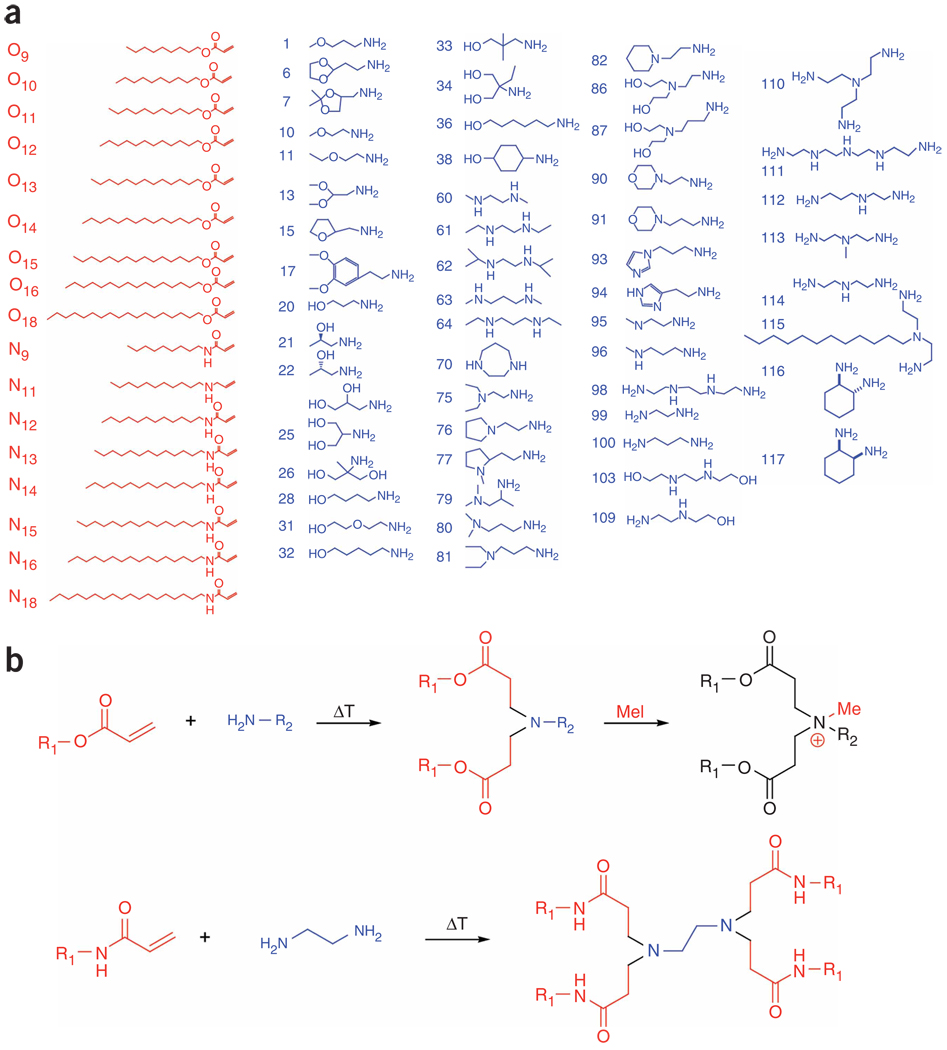
Figure 1.
Synthesis of lipidoids. (a) Alkyl-acrylate, alkyl-acrylamide and amino molecules were used to synthesize a combinatorial library of lipidoids. (b) Synthesis occurs through the conjugate addition of amines to an acrylate or acrylamide. Depending on the number of addition sites in the amino monomer, lipidoids can be formed with anywhere from 1 to 7 tails. Amino groups in the lipidoid can be quaternized by treatment with methyl iodide. For ease of nomenclature, lipidoids are named as follows: (amine number)(acrylate or acrylamide name)-(number of tails)(“+” if quaternized).
RESULTS
Synthesis of lipidoid library
The synthesis scheme is based upon the conjugate addition of alkyl-acrylates or alkyl-acrylamides to primary or secondary amines (Fig. 1b). This particular chemistry, unlike many traditional lipid synthesis chemistries, allows for reactions in the absence of solvent or catalysts, results only in lipidoid product, and thereby eliminates the need for protection and deprotection steps, purification or concentration steps (Supplementary Methods and Supplementary Table 1 online for details on synthesis and characterization). Using these methods, we first synthesized a pilot library of nearly 700 lipidoid members, in which a number of parameters were systematically varied, including (i) alkyl chain length from C10 to C18, (ii) linkage between the alkyl chain and the amine through the degradable ester or the more stable amide, (iii) primary R group on the amine and (iv) the post-synthetic introduction of a constitutive positive charge to certain lipidoids by quaternization of the amine with the alkylating agent methyl iodide.
Characterization of representative lipidoids demonstrates nearly complete conjugation for acrylate-based lipidoids under the conditions used (Supplementary Methods). Acrylamide conjugation was slower, but after one week at elevated temperature the majority of material was conjugated (Supplementary Methods). This combinatorial approach has the advantages of both simple and rapid synthesis, as well as the potential to generate a large and diverse collection of materials.
Lipidoid-mediated delivery in vitro
Once synthesized, this first library was screened for the ability to deliver siRNA to a HeLa cell line that stably expresses both firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferase (Fig. 2a). Efficacy of siRNA delivery by lipidoids was determined by treating cells with siRNA-lipidoid complexes, prepared using a firefly luciferase-targeting siRNA (siLuc), and then measuring the ratio of firefly to Renilla luciferase expression. In this assay, toxic or other nonspecific effects result in reduction of expression of both luciferase proteins, whereas noncytotoxic, specific silencing results in reduction of only firefly luciferase. To facilitate screening throughput, we formed siRNA-lipid complexes by simple mixing of siRNA-lipidoid solutions in microtiter plates.
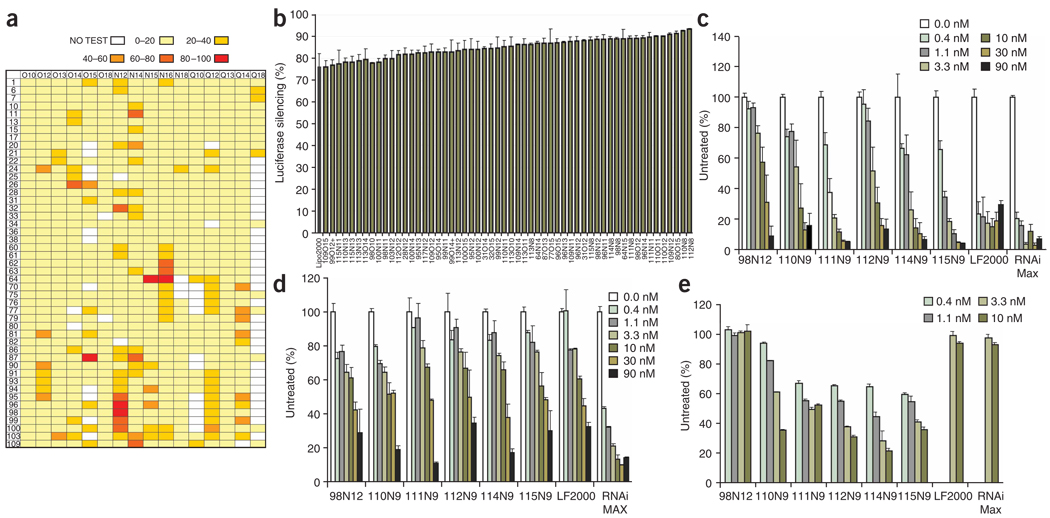
Figure 2.
In vitro screening of lipidoids for siRNA delivery. (a) HeLa cells expressing both firefly and Renilla luciferase were treated with firefly luciferase targeting siRNA-lipidoid complexes. The average percent reduction in firefly luciferase activity after treatment with siRNA-lipidoid complexes at a 5:1 (wt/wt) ratio in quadruplicate is shown. For ease of analysis, data are grouped as follows: no test, 0–20% knockdown, 20–40%, 40–60%, 60–80%, 80–100%. (b) Optimized in vitro knockdown by lipidoids in HeLa cells. Lipidoids were optimized for delivery using four lipidoid/siRNA ratios: 5:1, 10:1, 15:1 and 20:1. Materials were tested in quadruplicate. Data are presented for the optimal siRNA/lipidoid wt/wt ratio for each lipidoid, including s.d. This data set includes only lipidoids with no significant cytoxicity, as assessed by no significant change in Renilla luciferase expression relative to untreated cells. (c–e) Dose response of silencing in HeLa (c), HepG2 (d) and primary macrophage cultures (e). Data were generated in quadruplicate, as a function of siRNA molarity, at ratio of 5:1 wt/wt. s.d. is shown. Day 5 GFP-expressing bone marrow-derived macrophage cultures were incubated with siRNA-lipidoid complexes composed of the indicated lipidoids or commercial transfection reagents (Lipofectamine 2000 and Lipofectamine RNAiMAX) and siGFP or siCD45 for 6 h. GFP expression was quantified by flow cytometry. Silencing is expressed as the percentage of untreated cultures performed in parallel.
Analysis of these results revealed several trends in lipidoids capable of delivering siRNA to HeLa cells. Overall, enhanced delivery performance was achieved using lipidoids containing more than two amines per head unit (e.g., monomers 61–64, 95–103). Furthermore, effective materials often had either two long amide tails (e.g., N16) or several smaller amide tails (e.g., 98N12). Based on this data set, we synthesized a second-generation library of 500 lipidoids, expanding the structures tested based on these trends. In particular, we hypothesized that a number of these design features could enrich the library with effective materials. Specifically, we expanded the library to include lipidoids with even shorter amide tails (e.g., N11, N9), and more head units (e.g., monomers 109–117). Because many of these head units have the potential to form more than two tails, lipidoids were resynthesized with varied reaction stoichiometry to generate lipidoids with a diverse number of tails.
The ratio of delivery material to nucleic acid is known to affect delivery potential of formulations. Therefore, this second generation library of 500 lipidoids was screened for delivery to HeLa cells at four different lipidoid/siRNA ratios, in quadruplicate. Using these screening methods, we identified 56 lipidoids that induced gene silencing at levels similar to the commercially available in vitro transfection agent, Lipofectamine 2000 (Fig. 2b). Notably, the top performing lipidoids in this second library contained several structural similarities: (i) amide linkages, (ii) more than two alkyl tails, (iii) tail length in the range of 8–12 carbons and (iv) one tail short of total substitution of the amine reactants, therefore containing one secondary amine. These relatively short tails are surprising in comparison to typical gene delivery lipids such as DOTAP and DOTMA, which contain 18 aliphatic carbon atoms26. Notably, many of the effective lipidoids are structurally unlike either conventional lipids or cationic polymers (e.g., 100N9). This synthesized set of effective siRNA delivery materials substantially expands the collection and chemical diversity of materials known to facilitate siRNA delivery to cells.
To more closely examine in vitro performance, we tested a collection of purified lipidoids selected from those showing efficacy in HeLa cells at different doses, with three cell types. HeLa, human hepatocellular liver carcinoma cell line HepG2 and primary bone marrow–derived murine macrophages (Fig. 2c–e). In vitro analysis shows that, in general, both lipidoids and commercial lipids are relatively noncytotoxic at the concentration ranges in which they are efficacious (Supplementary Fig. 1a–d online). Notably, these lipidoids performed differently in the different cell types. Whereas all materials were able to facilitate good silencing at the highest siRNA levels, the commercial reagents Lipofectamine RNAiMAX and Lipofectamine 2000 were able to facilitate greater silencing at the lowest siRNA levels in HeLa and HepG2 cells. In contrast, with primary bone marrow–derived macrophages, lipidoids were more effective delivery vehicles at low siRNA concentrations. Dose-dependent silencing of green fluorescent protein (GFP) expression was observed for 6 of the 7 lipidoids tested with 50% silencing observed even at a 1-nM siRNA concentration (Fig. 2e). In contrast Lipofectamine 2000 and Lipofectamine RNAiMAX were not effective at silencing GFP expression even at higher siRNA concentrations (Fig. 2e). Whereas the overall silencing at low complex concentrations in HeLa and HepG2 cells lines is less than with commercial reagents, these lipidoids provide for more effective silencing in primary bone marrow–derived macrophages. This library approach provides a substantial number of new materials that may be useful for transfecting cells that have thus far proven refractory to transfection by currently available reagents.
Lipidoid-mediated delivery of siRNA to mouse and rat liver
To examine the utility of this library, we first tested the lipidoids for the in vivo systemic delivery of siRNA to liver. The liver represents an attractive organ for therapeutic intervention, because of the number of potential hepatic targets and the highly perfused nature of the organ, which may render it more amenable to delivery of exogenous siRNAs. Using a liver-directed in vivo screen of the lipidoid libraries, we identified a series of compounds that facilitate high levels of siRNA-mediated gene silencing in hepatocytes, the cells comprising the liver parenchyma. Factor VII, a blood clotting factor, is an ideal target gene for assaying functional siRNA delivery to liver. Because this factor is produced specifically in hepatocytes, gene silencing indicates successful delivery to parenchyma, as opposed to delivery to the cells of the reticulo-endothelial system (e.g., Kupffer cells). Furthermore, Factor VII is a secreted protein that can be readily measured in serum, obviating the need to euthanize animals. Finally, because of the protein’s short half-life (2–5 h), silencing at the mRNA level is manifest as silencing at the protein level with minimal lag.
Seventeen top-performing lipidoids, identified through in vitro screens, were formulated for in vivo use. The complexes contained lipidoid, cholesterol and PEG-lipid. Generally, simple ionic complexes are not suitable for in vivo systemic administration owing to poor serum stability, tendency to form aggregates and poor in vivo tolerability. At least seven lipidoid formulations were identified that mediated significant reduction of serum Factor VII protein levels, with the largest reduction observed for 98N12 (>90%) (Fig. 3a). Five variants of 98N12 containing different tail numbers were purified and tested individually (Supplementary Fig. 2 online). 98N12-5 (5-tail) was found to be optimal for in vivo delivery. To verify the specificity of gene silencing, liver mRNA levels were measured for both Factor VII and another hepatocyte-expressed gene, apolipoprotein B (Apob) (Fig. 3b). In animals treated with formulations containing siFVII, only silencing of the Factor VII mRNA was observed. Conversely, in animals treated with formulations containing Apob-specific siRNA (siApoB), only silencing of the Apob mRNA was observed. Further, administration of a lipidoid-formulated 1:1 (wt/wt) mixture of the two siRNAs resulted in silencing of both Factor VII and Apob genes with no detectable loss in potency or competition between siRNAs. No silencing of Factor VII was observed when a mismatch Factor VII siRNA was used (data not shown). These data indicate that the observed gene silencing is a direct result of the specific effects of lipidoid-siRNA on mRNA levels in the liver and that these effects are applicable to multiple hepatocyte-expressed genes.
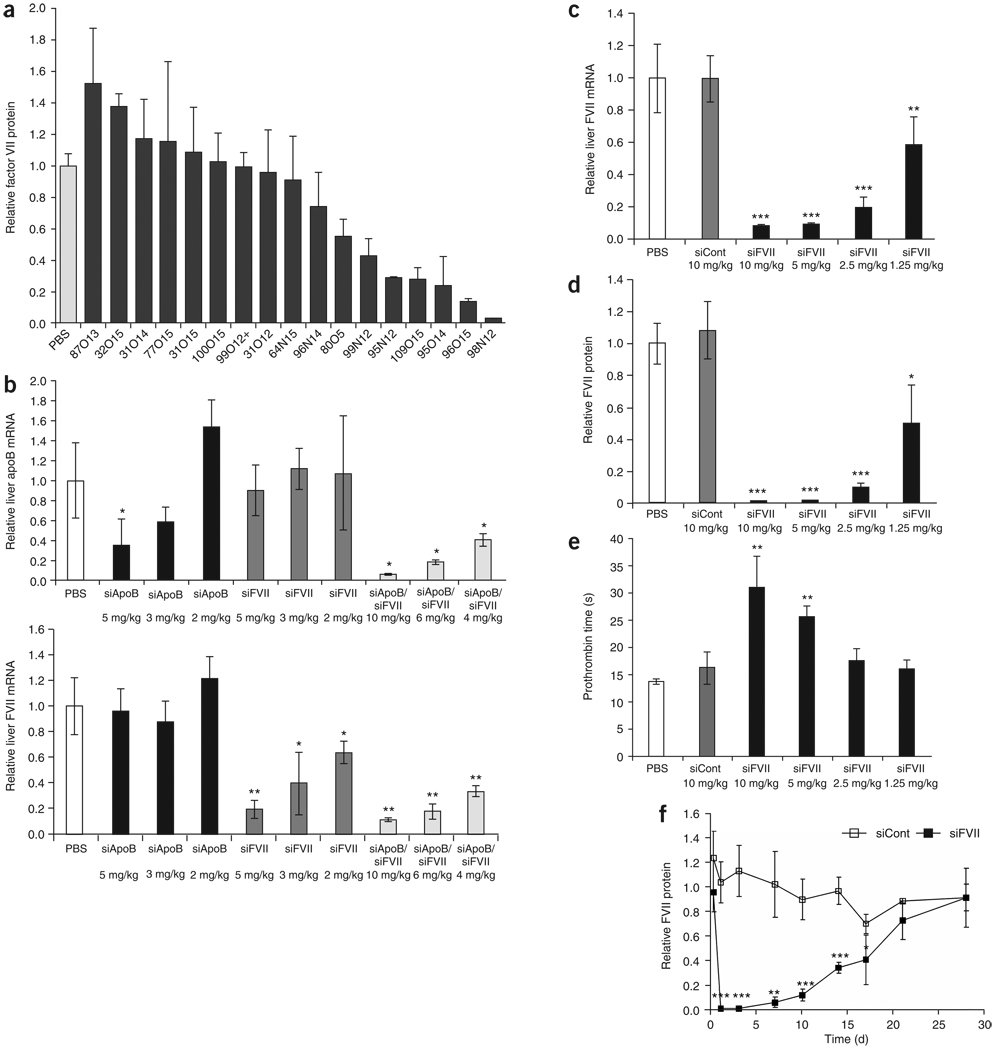
Figure 3.
In vivo delivery of siRNA to liver in rodents. (a) Mice (n = 3) received two daily i.v. injections of different lipidoid formulations of siRNA at a dose of 2.5 mg/kg. Factor VII protein levels were quantified 24 h after the second administration. (b) Simultaneous silencing of two genes in vivo in mice. Mice (n = 3) received a single i.v. bolus injection of a 98N12-5(1)-formulated 1:1 (wt/wt) mixture of siFVII and siApoB at 10, 6 or 4 mg/kg (5, 3 or 2 mg/kg of each siRNA). For comparison, control animals received PBS or lipidoid-formulated siFVII alone or lipidoid-formulated siApoB alone at 5, 3 or 2 mg/kg. Forty-eight hours after administration animals were killed and livers were harvested. Liver mRNA levels of Factor VII or ApoB (normalized to GAPDH) were determined by branched DNA assay. (c–e) Rats (n = 4) were injected with lipidoid-formulated siRNA at 1.25, 2.5, 5 and 10 mg/kg. Animals were bled and killed 48 h after administration; shown are liver mRNA levels (c), serum Factor VII protein levels (d) and prothrombin time (e). (f) Durability of silencing in rats. Rats (n = 5) received a single i.v. administration of lipidoid-formulated siRNA at 5 mg/kg. Animals were bled at various time points after administration and serum Factor VII protein levels were quantified. Data points represent group mean ± s.d. Data points marked with asterisks are statistically significant relative to control treated groups (*, P < 0.05; **, P < 0.005; ***, P < 0.001; t-test, single-tailed).
To further explore the in vivo activity of 98N12-5 observed in mice, we conducted efficacy and tolerability studies in rats. Rats were given a single intravenous (i.v.) injection of siFVII formulated in 98N12-5 at doses of 1.25, 2.5, 5 and 10 mg/kg. Significant, dose-dependent reductions in liver Factor VII mRNA levels were observed, with 40%, 80% and >90% silencing at 1.25, 2.5 and 5 mg/kg, respectively (Fig. 3c and Supplementary Table 2 online). No silencing was observed using a formulated control siRNA (siCont), demonstrating specificity of silencing. The reduction in liver Factor VII mRNA levels produced a concomitant dose-dependent reduction in serum Factor VII protein levels, with nearly complete silencing at the highest dose levels (Fig. 3d).
As would be expected, significantly reduced serum Factor VII levels produced a phenotypic effect in the treated animals. As Factor VII is part of the extrinsic coagulation pathway, treated animals had impaired clotting through this pathway as measured by prolongation in prothrombin time (Fig. 3e). The phenotypic effect was found to be specific and not attributable to the delivery vehicle, as the formulated control group demonstrated no perturbations in prothrombin time. The resultant gene silencing was highly sustainable. Single injections of 98N12-5 formulated Factor VII-targeting siRNA (siFVII) were capable of mediating silencing that persisted for nearly 4 weeks (Fig. 3f).
Next, we investigated the tolerability of the 98N12-5 formulation in rats. Animals received four once-per-week i.v. bolus injections of a formulated siRNA at doses as high as 10 mg/kg/week. The non-physiological siRNA siCont was used to eliminate any potential target silencing–related toxicities. The formulation was generally well-tolerated at the dose levels tested, as determined by cage-side observations and measures of clinical chemistry and hematology parameters (Table 1). However, enlargement of the spleen—a major clearance organ for nanoparticles—was observed at the highest doses. The appearance and weight of all other organs were normal.
Table 1.
Clinical chemistry and hematology parameters for lipidoid-siRNA treated rats
| Group | ALT (U/L) | AST (U/L) | Bilirubin (mg/dl) | BUN (mg/dl) | RBC (× 106/µl) | Hemoglobin (g/dl) | WBC (× 103/µl) | PLT (× 103/µl) | Spleen (g) |
|---|---|---|---|---|---|---|---|---|---|
| 0 mg/kg | 34 ± 3 | 134 ± 11 | 0.1 ± 0.0 | 13.5 ± 1.3 | 6.9 ± 0.4 | 14.7 ± 0.3 | 7.6 ± 3.0 | 1,019 ± 91 | 0.67 ± 0.13 |
| 2 mg/kg | 37 ± 5 | 118 ± 16 | 0.1 ± 0.0 | 15.3 ± 0.5 | 6.4 ± 0.4 | 13.8 ± 0.8 | 6.5 ± 1.5 | 999 ± 16 | 0.79 ± 0.11 |
| 5 mg/kg | 39 ± 6 | 114 ± 27 | 0.1 ± 0.0 | 15.0 ± 1.8 | 6.6 ± 0.4 | 13.9 ± 0.7 | 5.4 ± 2.1 | 866 ± 187 | 1.10 ± 0.39 |
| 7.5 mg/kg | 41 ± 8 | 98 ± 30 | 0.1 ± 0.0 | 16.5 ± 2.1 | 6.3 ± 0.5 | 13.5 ± 0.8 | 9.1 ± 1.8 | 1,106 ± 156 | 1.04 ± 0.13 |
| 10 mg/kg | 43 ± 9 | 86 ± 8 | 0.1 ± 0.1 | 16.5 ± 2.9 | 6.2 ± 0.4 | 13.5 ± 0.6 | 9.2 ± 0.7 | 871 ± 83 | 1.12 ± 0.09 |
Sprague-Dawley rats (n = 8) were given four once-per-week bolus i.v. injections of formulated siCont at different dose levels. Blood samples were taken 2 d after the 4th and final dose (day 24). ALT, alanine aminotransferase; AST, aspartate aminotransfersase; BUN, blood urea nitrogen; RBC, red blood cells; WBC, white blood cells; PLT, platelets.
siRNA delivery to lung and peritoneal macrophages in vivo
We examined an additional disease model to explore the versatility of the lipidoid approach. Local siRNA delivery to the lung after intranasal administration was tested in a mouse model of respiratory syncytial virus infection (Fig. 4a)5. In separate experiments, we confirmed the absence of gene silencing in the liver and kidney after local pulmonary administration of the formulation (data not shown). Whereas ‘naked’ siRNA provided roughly one log reduction in viral plaques, 98N12-5-formulated siRNA at the same dose provided greater than two log reduction in viral plaques. These data demonstrate that lipidoids can be used in nonsystemic applications of RNAi technology and are capable of delivering siRNA to nonhepatic cell types.
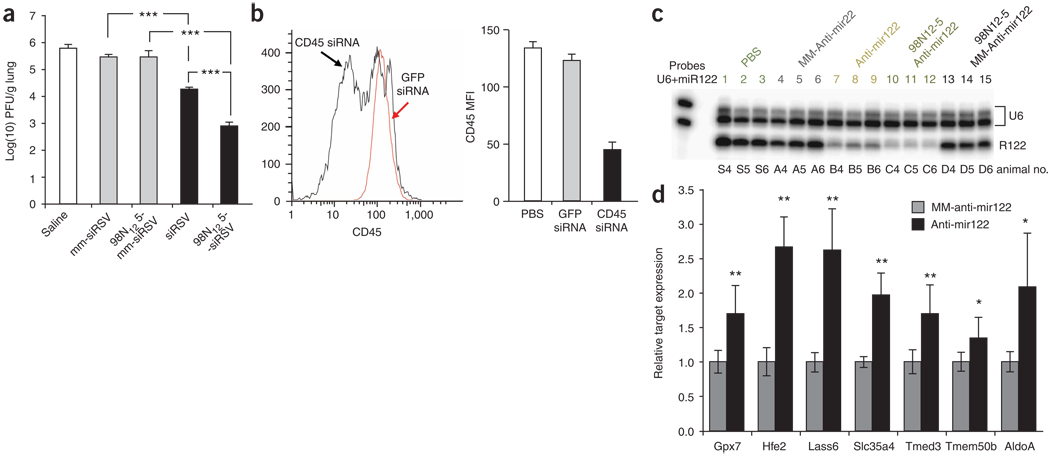
Figure 4.
In vivo delivery of siRNA to lung and peritoneal macrophages and delivery of anti-miRs to liver. (a) Inhibition of RSV/A2 in BALB/c mouse lungs. Mice (n = 5) were intranasally administered naked siRNA or lipidoid-siRNA (siRSV or mm-siRSV) formulation at a dose of 2 mg/kg. Lungs were harvested at day 4 post-infection and assayed by immunostaining plaque assay. (b) Inhibition of CD45 protein in thioglycollate elicited mouse peritoneal macrophages. Mice (n = 4) were injected with thioglycollate (i.p.) 3 d before treatment with 10 mg/kg of lipodoid-formulated siCD45 or siGFP administered via i.p. injection. We analyzed 4 d post administration CD45 expression on macrophages by flow cytometry. Macrophage cells were gated and median fluorescence intensity of the CD45 staining is plotted. MFI, mean fluorescence intensity. (c) Lipidoid delivery of anti-miR122 in vivo. Nuclease protection assay and miRNA detection. Liver RNA samples of three representative animals per treatment group are shown. The U6 signal serves as RNA input control. Animals treated with antagomir122 show a marked decrease of miR-122 signal (lanes 7–9). Even further reduction of miR-122 signal is observed for lipidoid-formulated anti-miR122–treated animals (lanes 10–12). The mismatched control antagomir and anti-miR had little effect on miR-122 signals (lanes 4–6 and 13–15). (d) Derepression of miR-122 target genes after miR-122 inhibition in mice. Mice (n = 6) received i.v. injections of either lipidoid-anti-miR122 (black bars) or lipidoid-mm-anti-miR122 (gray bars) at a daily dose of 5 mg/kg for 3 consecutive days. Twenty-four hours after the last injection, expression levels of seven reported miR-122 target genes were analyzed. Data points represent group mean ± s.d. Data points marked with asterisks are statistically significant relative to control treated groups (* P < 0.05, ** P < 0.005, *** P < 0.001; t-test, single-tailed).
The macrophage is a cell type frequently implicated in the pathology of inflammatory diseases. Following our success in transfecting siRNAs into primary macrophages in in vitro cultures (Fig. 2e), we tested whether lipidoid-formulated siRNAs can mediate silencing in macrophages in vivo. Mice were injected intraperitoneally with thioglycollate as a sterile inflammation stimulus followed by injection of 98N12-5-formulated siCD45. A 65% reduction of CD45 protein expression was observed in the peritoneal macrophage population (Fig. 4b). Although the intraperitoneal administration circumvents the localization challenge associated with systemic delivery to peritoneal macrophages, these results indicate that lipidoid formulations can potentially be used for the delivery of siRNA to macrophages in vivo.
Delivery of single-stranded oligoribonucleotides targeting miRNAs in vivo
To examine the utility of lipidoid materials in the delivery of other nucleic acid therapeutic drugs, we tested the potential of 98N12-5 to facilitate the delivery of single-stranded 2′-OMe oligoribonucleotides targeting miRNAs (anti-miRs). In vivo delivery of anti-miR results in silencing of a specific target miRNA, and consequently, the upregulation of specific genes regulated by the target miRNA4. 98N12-5-formulated anti-miR122 dosed at 5 mg/kg on 3 consecutive days in mice resulted in greater miR-122 repression than the cholesterol-conjugated version of the same oligoribonucleotide (antagomir122) dosed at 80 mg/kg on 3 consecutive days (Fig. 4c). Further, this effect was shown to be specific, as mismatched control anti-miR122 (mm-anti-miR122) did not result in appreciable effects on miR-122. Consistent with miR-122 downregulation, gene targets of miR-122 were shown to be derepressed in anti-miR122–treated mice relative to mismatched controls (Fig. 4d).
Lipidoid-mediated gene silencing in nonhuman primates
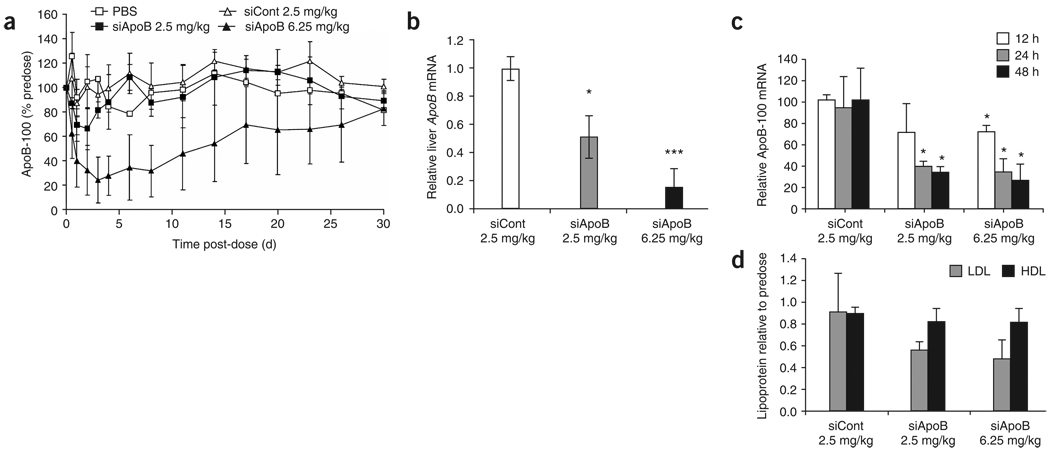
To determine the effects of 98N12-5 lipidoid in a third animal species, we initiated studies in nonhuman primates. Cynomolgus monkeys were treated with a single i.v. injection of lipidoid-formulated siApoB at siRNA doses of 2.5 and 6.25 mg/kg. Separate control animals were treated with either saline or lipidoid-formulated siCont. Serum samples were taken from animals for up to 30 d after administration to determine both the extent and duration of serum ApoB protein silencing. Silencing of ApoB was observed in a dose-dependent manner, with maximal serum ApoB reduction of up to 75% relative to pre-dose levels (Fig. 5a). In contrast, no significant silencing was observed in the saline or formulated control groups, indicating specific activity of the formulated ApoB siRNA. Silencing was remarkably persistent for animals in the high-dose group after a single i.v. bolus injection, with the lowest levels of ApoB achieved rapidly by day 3, >50% silencing still maintained at 2 weeks and full recovery of expression achieved only 1 month later. The data herein regarding durability for ApoB silencing extends the previous reports for systemic RNAi in nonhuman primates reported by Zimmermann et al.8, where the study terminated at 11 d. Consistent with ApoB silencing, therapeutic efficacy as measured by specific and persistent reductions in low-density lipoprotein cholesterol (LDL-C) of up to 60% was also observed (Supplementary Fig. 3 online).
Figure 5.
In vivo delivery in primates. (a) Extent and duration of serum ApoB-100 protein reduction in cynomolgus monkeys after single bolus i.v. administration of lipidoid-formulated siRNA For all groups except saline control; n = 6 for data points up to and including 2 d and n = 3 for data points after 2 d. For saline control, n = 4 for data points up to and including 2 d, and n = 2 for data points beyond 2 d. Data points represent group mean ± s.d. No error bars shown for saline group where n = 2. (b–d) Animals (n = 3) were treated with either formulated control siRNA at 2.5 mg/kg, formulated Apob targeting siRNA at 2.5 mg/kg, or formulated Apob targeting siRNA at 6.25 mg/kg. (b) Liver Apob mRNA levels normalized to GAPDH mRNA 48 h after administration. (c) Serum ApoB-100 protein reduction at 12, 24 and 48 h after administration as percentage of predose levels. (d) LDL-C and high-density lipoprotein (HDL) cholesterol levels at 48 h after administration, normalized to predose levels. Data points represent group mean ± s.d. Data points marked with asterisks are statistically significant relative to control treated groups (* P < 0.05, ** P < 0.005, *** P < 0.001; t-test, single-tailed).
A subsequent primate study was performed with a next-generation 98N12-5 lipidoid-based formulation in cynomolgus monkeys. This formulation was optimized in part by maximizing the siRNA loading in the lipidoid complex. Notably, the total mass of delivery material relative to siRNA in this formulation is roughly one-third that of the previously published stable nucleic acid lipid particles (SNALP) delivery system8. Also, unlike the previously published SNALP system, this formulation contains fewer total components: three different agents plus siRNA instead of four. Animals were treated with formulated ApoB siRNA at siRNA doses of 2.5 and 6.25 mg/kg, or formulated control siRNA at 2.5 mg/kg administered via single i.v. injection. Tissues were harvested 48 h after administration for liver mRNA determination. Silencing of ApoB liver mRNA of up to 85% was observed (Fig. 5b), corresponding to a maximal reduction in serum ApoB protein of up to 74% (Fig. 5c). As early as 2 d after administration, LDL-C was reduced by >50% (Fig. 5d). Toxicological analysis indicated that the formulations were well tolerated at the dose levels tested, with no observed treatment-related changes in appearance or behavior. No clinically relevant changes in coagulation or hematological parameters were observed other than a mild reduction in platelets at the highest dose. No significant changes in clinical chemistry parameters were observed except for a slight increase in alanine aminotransferase (ALT) and aspartate aminotransferase (AST). These elevations were less than those observed with the previously published SNALP formulations8. (Table 2 and Supplementary Table 3 online).
Table 2.
ALT and AST levels for lipidoid-siRNA treated cynomolgus monkeys
| Group | Time | ALT (U/L) | AST (U/L) |
|---|---|---|---|
| siCont, 2.5 mg/kg | Pre-dose | 50 ± 10 | 53 ± 14 |
| 24 h | 76 ± 5 | 100 ± 5 | |
| 48 h | 64 ± 27 | 61 ± 19 | |
| siApoB, 2.5 mg/kg | Pre-dose | 40 ± 21 | 45 ± 42 |
| 24 h | 61 ± 10 | 65 ± 13 | |
| 48 h | 53 ± 38 | 48 ± 45 | |
| siApoB, 6.25 mg/kg | Pre-dose | 44 ± 19 | 53 ± 27 |
| 24 h | 70 ± 6 | 127 ± 10 | |
| 48 h | 67 ± 32 | 81 ± 38 |
DISCUSSION
RNAi technology is poised to form the basis for the next major class of pharmaceutical drugs. However, effective delivery of RNAi therapeutics remains the key hurdle for advancement of this technology. We believe the development of lipidoids could extend the scale and diversity of available delivery solutions. Lipidoid formulations of siRNA and anti-miR demonstrated potent, specific and durable effects on gene expression in three distinct species, including nonhuman primates. Therapeutic efficacy was observed in vivo in liver, lung and peritoneal macrophages. Prior studies have demonstrated successful silencing in lung models using delivery reagents; however, in contrast to our results, these cases did not demonstrate substantial improvement in silencing over that achieved with naked siRNA5,6. We note that gene silencing in lung continues to be difficult to achieve, even with the best available lung delivery formulations27. In addition, to our knowledge, there are currently no other reagents available that have demonstrated specific, endogenous gene silencing in peritoneal macrophages. Previous reports, based on delivery of Cy3-labeled siRNA, suggest Transit-TKO may have this ability, but data demonstrating in vivo silencing have not been published28.
We believe the development of this library of lipid-like materials represents an important expansion of the diversity and collection of intracellular delivery materials. This one-step synthetic scheme enables the straightforward parallel generation of large libraries of delivery material. Notably, a number of materials were identified with both in vitro and in vivo utility and the common structural features of these materials suggest certain design criteria for creating future intracellular delivery agents, including (i) amide linkages, (ii) more than two alkyl tails, (iii) tail length in the range of 8–12 carbons and (iv) a secondary amine. Finally, further studies are warranted to investigate lipidoid-based delivery of RNA and other drugs to extend this technology for the broadest applications of RNAi therapy and drug delivery.
METHODS
Library synthesis
Lipidoids were synthesized by addition of acrylamides or acrylates to amines. Amines were purchased from Sigma-Aldrich and TCI America. Acrylates were purchased from Sigma-Aldrich, Dajac Monomer- Polymer, Hampford Research, Scientific Polymer and TCI America. Acrylamides were synthesized by the drop-wise addition of acryloyl chloride to the appropriate 1-aminoalkane (see Supplementary Methods for details). The ester portion of the lipidoid library was synthesized at a ratio of 2:1 acrylate/amine, with no solvent, unless otherwise specified. The amide portion of the lipidoid library was synthesized at the maximal ratio of acylamide/amine for each amine (e.g., 6 acrylamide/amine for amine monomer 98). All library reactions were carried out in 5-ml Teflon-lined glass screw-top vials. 200 mg of amine was added to the corresponding amount of acrylate or acrylamide. The mixture was stirred at 90 °C for 1 or 7 d for acrylate or acrylamide monomers, respectively. After cooling, the lipid mixtures were used without purification unless otherwise specified.
Nucleic acids
All siRNAs and 2′-OMe oligoribonucleotides were synthesized by Alnylam. Oligonucleotides were characterized by electrospray mass spectrometry and anion-exchange high-performance liquid chromatography. The sequences for the sense and antisense strands of siRNAs are as follows:
siLuc sense: 5′-CUUACGCUGAGUACUUCGATT-3′, antisense: 5′-UCGAAGUACUCAGCGUAAGTT-3′.
siFVII sense: 5′-GGAucAucucAAGucuuAcT*T-3′, antisense: 5′-GuAAGAcuuGAGAuGAuccT*T-3′.
siApoB sense: 5′-GGAAUCuuAuAuuuGAUCcA*A-3′, antisense: 5′-uuGGAUcAAAuAuAAGAuUCc*c*U-3′.
siCont sense: 5′-cuuAcGcuGAGuAcuucGAT*T-3′, antisense: 5′-UCGAAGuACUcAGCGuAAGT*T-3′.
siGFP sense: 5′-CcAcAuGAAGcAGcACGACu*U-3′, antisense: 5′-AAGUCGUGCUGCUUCAUGUGg*u*C-3′.
siCD45 sense: 5′-cuGGcuGAAuuucAGAGcAT*T-3′, antisense: 5′-UGCUCUGAAAUUcAGCcAGT*T-3′.
mm-siRSV sense: 5′-CGAUUAUAUUACAGGAUGAT*T-3′, antisense: 5′- UCAUCCUGUAAUAUAAUCGT*T-3′.
siRSV sense: 5′-UCCUAGAAUCAAUAAAGGGTT-3′, antisense: 5′- CCCUUUAUUGAUUCUAGGATT-3′.
- 2′-OMe oligoribonucleotides:
- antagomir122: 5′-a*c*aaacaccauugucacacu*c*c*a-Cholesterol-3′
- mm-antagomir122: 5′- a*c*acacaacacugucacauu*c*c*a-Cholesterol-3′
- anti-miR122: 5′-a*c*aaacaccauugucacacu*c*c*a-3′
- mm-anti-miR122: 5′- a*c*acacaacacugucacauu*c*c*a-3′
2′-OMe modified nucleotides are in lower case, 2′-fluoro modified nucleotides are in bold lower case, and phosphorothioate linkages are represented by asterisks. siRNAs were generated by annealing equimolar amounts of complementary sense and antisense strands.
In vitro siRNA transfection assay
The protocol was adapted from ref. 29. HeLa cells, stably expressing firefly luciferase and Renilla luciferase were seeded (14,000 cells/well) into each well of an opaque white 96-well plate (Corning-Costar) and allowed to attach overnight in growth medium. Growth medium was composed of 90% phenol red-free DMEM, 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin (Invitrogen). Cells were transfected with 50 ng of firefly-specific siLuc complexed with lipidoid at lipidoid/siRNA ratios of 2.5:1, 5:1, 10:1 and 15:1 (wt/wt) to determine the optimum for transfection efficiency. Transfections were performed in quadruplicate.
Working dilutions of each lipid were prepared (at concentrations necessary to yield the different lipid/siRNA weight ratios) in 25-mM sodium acetate buffer (pH 5).
We added 25 µl of the diluted lipid to 25 µl of 60 µg/ml siRNA in a well of a 96-well plate. The mixtures were incubated for 20 min to allow for complex formation, and then 30 µl of each of the lipidoid/siRNA solutions was added to 200 µl of medium in 96-well polystyrene plates. The growth medium was removed from the cells using a 12-channel aspirating wand (V&P Scientific) after which 150 µl of the DMEM/lipidoid/siRNA solution was immediately added. Cells were allowed to grow for 1 d at 37 °C, 5% CO2 and were then analyzed for luciferase expression. Control experiments were performed with Lipofectamine 2000, as described by the vendor (Invitrogen).
Firefly and Renilla luciferase expression was analyzed using Dual-Glo assay kits (Promega). Luminescence was measured using a Victor3 luminometer (Perkin Elmer). A standard curve for luciferase was generated by titration of luciferase enzyme (Promega) into growth medium in an opaque white 96-well plate.
Bone marrow–derived macrophage transfection
Murine bone marrow–derived macrophages were cultures according to standard protocol30. C57Bl/6 mice expressing GFP under the control of the RAGE locus promoter were used as a source of bone marrow31. Cells were cultured in 12-well dishes for 5 d in the presence of 8 ng/ml of macrophage colony stimulating factor (M-CSF). The optimal siRNA/lipidoid ratio was determined for each lipidoid (a ratio of either 5 or 10 wt/wt was used). Mixtures of siGFP or control siCD45 with lipidoids were prepared as described above. Lipofectamine 2000 and Lipofectamine RNAiMAX (Invitrogen) were complexed with siRNA according to manufacturer’s instruction. siRNA-lipidoid mixtures were added to macrophage cultures at the desired concentrations for 6 h. Medium was exchanged and GFP expression was analyzed by flow cytometry 5 d later.
Lipidoid-siRNA formulation
Lipidoid-based siRNA formulations comprised lipidoid, cholesterol, polyethylene glycol-lipid (PEG-lipid) and siRNA. Formulations were prepared using a protocol similar to that described in refs. 32,33. Stock solutions of 98N12-5(1)·4HCl MW 1489, mPEG2000-Ceramide C16 (Avanti Polar Lipids) MW 2634 or mPEG2000-DMG MW 2660 (synthesized by Alnylam, see Supplementary Methods), and cholesterol MW 387 (Sigma-Aldrich) were prepared in ethanol and mixed to yield a molar ratio of 42:10:48. Mixed lipids were added to 125 mM sodium acetate buffer pH 5.2 to yield a solution containing 35% ethanol, resulting in spontaneous formation of empty lipidoid nanoparticles. Resulting nanoparticles were extruded through a 0.08 µm membrane (two passes). siRNA in 35% ethanol and 50 mM sodium acetate pH 5.2 was added to the nanoparticles at 1:7.5 (wt/wt) siRNA/total lipids and incubated at 37 °C for 30 min. Ethanol removal and buffer exchange of siRNA-containing lipidoid nanoparticles was achieved by tangential flow filtration against PBS using a 100,000 MWCO membrane. Finally, the formulation was filtered through a 0.2 µm sterile filter. Particle size was determined using a Malvern Zetasizer NanoZS (Malvern). siRNA content was determined by UV absorption at 260 nm and siRNA entrapment efficiency was determined by Ribogreen assay34. Resulting particles had a mean particle diameter of ~50 nm, with peak width of 20 nm, and siRNA entrapment efficiency >95%.
In vivo rodent Factor VII and Apob silencing experiments
All procedures used in animal studies conducted at Alnylam were approved by the Institutional Animal Care and Use Committee (IACUC) and were consistent with local, state and federal regulations as applicable. C57BL/6 mice (Charles River Labs) and Sprague-Dawley rats (Charles River Labs) received either saline or siRNA in lipidoid formulations via tail vein injection at a volume of 0.01 ml/g. At various time points after administration, animals were anesthetized by isofluorane inhalation and blood was collected into serum separator tubes by retroorbital bleed. Serum levels of Factor VII protein were determined in samples using a chromogenic assay (Coaset Factor VII, DiaPharma Group; or Biophen FVII, Aniara Corporation) according to manufacturers’ protocols. A standard curve was generated using serum collected from saline-treated animals. In experiments where liver mRNA levels were assessed, at various time points after administration, animals were killed and livers were harvested and snap frozen in liquid nitrogen. Frozen liver tissue was ground into powder. Tissue lysates were prepared and liver mRNA levels of Factor VII and Apob were determined using a branched DNA assay (QuantiGene Assay, Panomics)8.
In vivo mouse RSV silencing experiments
BALB/c mice (Harlan Sprague-Dawley Laboratories) were anesthetized by intraperitoneal (i.p.) administration of 2,2,2-tribromoethanol (Avertin) and instilled intranasally (i.n.) with lipidoid-siRNA formulations in a total volume of 50 µl. At 4 h after siRNA instillation, the mice were anesthetized and infected i.n. with 106 plaque-forming units (PFU) of respiratory syncytial virus (RSV)/A2 or RSV/B1. Before removal of lungs at day 4 after infection, anesthetized mice were exsanguinated by severing the right caudal artery. Lung tissue was collected on ice in PBS (Invitrogen) to determine virus titers. RSV titers from lungs were measured by immunostaining plaque assay. Lungs were homogenized with a hand-held Tissumiser homogenizer (Fisher Scientific). The lung homogenates were placed on ice for 5–10 min to allow debris to settle. Clarified lung lysates were diluted tenfold in serum-free DMEM, added to 95% confluent Vero E6 cells cultured in DMEM in 24-well plates, and incubated for 1 h at 37 °C, followed by 2% methylcellulose overlay. At 5 d post-infection, the medium was removed and the cells were fixed with acetone/methanol (60:40) and immunostained. Plaques were counted and log (10) PFU/g lung versus PBS or siRNA mismatch control was determined.
Silencing in peritoneal macrophages
C57Bl/6J mice (Jackson Labs) were injected intraperitoneally with 1 ml of 4% Brewers thioglycollate medium (Difco) 3 d before injecting 10 mg/kg of siCD45, or siGFP i.p. (four mice per group). Peritoneal lavage was collected 4 d later and stained with fluorophore-conjugated antibodies to CD11b, Gr1 and CD45 (BD Biosciences). Flow cytometry samples were run on the LSRII flowcytometer (BD Bioscience) and FlowJo software (Treestar) was used to identify the CD11bhighGr1low macrophage population and quantify CD45 expression.
In vivo miRNA silencing experiments
C57BL/6NCRL mice (Charles River) received lipidoid formulations of antagomir or anti-miR via tail vein injection at 5 mg/kg (0.5 mg/ml) on 3 consecutive days. Livers were taken at day 4 and expression levels of miR-122 were determined. Liver tissue was dissolved in proteinase K-containing cell and tissue lysis buffer (EPICENTRE) and subjected to sonication. Total RNA was extracted with TE-saturated phenol (Roth) and subsequent precipitation in ethanol. Synthetic DNA probes complementary to mouse miR-122, as well as mouse U6 RNA were 5′-end labeled using polynucleotide kinase (New England Biolabs) and γ-32P ATP (GE Healthcare). Probe sequences were: miR-122, 5′- AAACACCATTGTCACACTCCACAGCTCTCTCTTCT -3′; U6, 5′ -CACGAATTTGCGTGTCATCCTTGCGCAGGGGCCATGTTCTTCTTCTTCTTC - 3.
Total liver RNA was simultaneously hybridized in solution to a miR-122-specific probe and the U6 probe. The hybridization conditions allowed detection of U6 RNA and mature miRNA, but not pre-miRNA. After treatment with S1 nuclease, samples were loaded on denaturing 10% acrylamide gels. Gels were exposed to a phosphoimager screen and analyzed on a Typhoon 9200 instrument (GE Healthcare). Relative signal intensities of miR-122 versus U6 were calculated for each sample.
Expression level analysis of miR-122 target genes by branched DNA assay
Assay was performed as described before4. Briefly, 30–50mg of frozen liver tissue was lysed in 1 ml tissue and cell lysis buffer (EPICENTRE) by sonication. 10–40 µl lysate was used for branched DNA assay, depending on signal strength of target gene. Probe sets were designed using QuantiGene ProbeDesigner software. Target gene expression was assayed according to QuantiGene Detection Assay recommendations and normalized to corresponding glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeper expression from same liver tissue lysate.
In vivo nonhuman primate experiments
All procedures using cynomolgus monkeys were conducted by a certified contract research organization using protocols consistent with local, state and federal regulations as applicable and approved by the Institutional Animal Care and Use Committee (IACUC). Cynomolgus monkeys (n = 6 per group) received either 5 ml/kg PBS, 2.5 mg/kg formulated siCont (1.25 ml/kg), 2.5 mg/kg (1.25 ml/kg) formulated siApoB or 6.25 mg/kg (3.125 ml/kg) formulated siApoB as bolus i.v. injections via the brachial vein. For apoB-100 protein measurements, serum was collected pre-dose and at 0.5, 1, 2, 3, 4, 6, 8, 11, 14, 17, 20, 23, 26 and 30 d after administration. In a subsequent experiment, cynomolgus monkeys (n = 3 per group) received either 2.5 mg/kg formulated siCont or 2.5 or 6.25 mg/kg of formulated siApoB as bolus i.v. injections via the saphenous vein. For apoB-100 protein measurements, serum was collected pre-dose and at 12, 24 and 48 h after administration. ApoB-100 protein was determined by using an enzyme-linked immunosorbent assay as previously described8. Clinical chemistries were analyzed at pre-dose and 24 and 48 h after administration. Hematology and coagulation parameters were analyzed at pre-dose and 48 h after administration. Animals were killed at 48 h. Liver Apob mRNA levels were determined in liver samples using a branched DNA assay (QuantiGene Assay)8.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank funding by the National Institutes of Health grant R01 EB00244. A.Z. would like to thank the Swiss National Science Foundation for his postdoctoral fellowship. We would also like to thank John Maraganore for helpful comments and Maryellen Duckman for assistance with manuscript preparation.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 3.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 4.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 5.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 6.Li BJ, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrissey DV, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 9.Heidel JD, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc. Natl. Acad. Sci. USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNamara JO, II, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 12.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 13.Pal A, et al. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. Int. J. Oncol. 2005;26:1087–1091. [PubMed] [Google Scholar]
- 14.Palliser D, et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 15.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Engineering mucosal RNA interference in vivo. Mol. Ther. 2006;14:336–342. doi: 10.1016/j.ymthe.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Ge Q, et al. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. USA. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard KA, et al. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol. Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 20.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 21.Davidson TJ, et al. Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation. J. Neurosci. 2004;24:10040–10046. doi: 10.1523/JNEUROSCI.3643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim WJ, et al. Cholesteryl oligoarginine delivering vascular endothelial growth factor siRNA effectively inhibits tumor growth in colon adenocarcinoma. Mol. Ther. 2006;14:343–350. doi: 10.1016/j.ymthe.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 23.Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc. Natl. Acad. Sci. USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 26.Miller A. Cationic Liposomes for Gene Therapy. Angew. Chem. Int. Ed. 1998;37:1769–1785. [Google Scholar]
- 27.Griesenbach U, et al. Inefficient cationic lipid-mediated siRNA and antisense oligonucleotide transfer to airway epithelial cells in vivo. Respir. Res. 2006;7:26. doi: 10.1186/1465-9921-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amarzguioui M, et al. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat. Protocols. 2006;1:508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- 29.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol. Ther. 2005;11:426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Cunnick J, Kaur P, Cho Y, Groffen J, Heisterkamp N. Use of bone marrow-derived macrophages to model murine innate immune responses. J. Immunol. Methods. 2006;311:96–105. doi: 10.1016/j.jim.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Constien R, et al. Characterization of a novel EGFP reporter mouse to monitor Cre recombination as demonstrated by a Tie2 Cre mouse line. Genesis. 2001;30:36–44. doi: 10.1002/gene.1030. [DOI] [PubMed] [Google Scholar]
- 32.Maurer N, et al. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys. J. 2001;80:2310–2326. doi: 10.1016/S0006-3495(01)76202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semple SC, et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta. 2001;1510:152–166. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 34.Heyes J, Palmer L, Bremner K, Maclachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.