Abstract
Noroviruses are the principal cause of epidemic gastroenteritis worldwide. Multiple reports have concluded that the major capsid proteins of GII.4 strains, which cause 80% of norovirus infections worldwide, are evolving rapidly, resulting in new epidemic strains. Surrogate neutralization assays using sera from outbreaks and from immunized mice suggest that, as with influenza virus, antigenic variation maintains GII.4 persistence in the face of human population herd immunity. To test this hypothesis, mice were hyperimmunized with virus-like particles (VLPs) representing an early (GII.4-1987) and a contemporary (GII.4-2006) GII.4 strain. Anti-GII.4-1987 IgG monoclonal antibodies (MAbs) strongly reacted with GII.4 VLPs derived between only 1987 and 2002. Ligand binding blockade was more efficient with GII.4-1987 and GII.4-1997 VLPs than with GII.4-2002. Anti-GII.4-2006 IgG MAbs recognized either a broad panel of GII.4 VLPs (1987 to 2006) or a subset of contemporary (2004 to 2006) VLPs. Most 2006 antibodies did not recognize or only poorly recognized GII.4 VLPs of 2007 or 2008, documenting rapid antigenic evolution of GII.4 capsids. Generally, 2006 MAbs blocked homotypic VLP-ligand binding but were unable to block VLPs representing strains primarily circulating during or earlier than 2002. These analyses demonstrate that both subtle and significant evolutionary change has occurred within antibody epitopes between epidemic strains, providing direct evidence that the GII.4 noroviruses are undergoing antigenic variation, likely in response to herd immunity. As with influenza virus, HIV, and hepatitis C virus, norovirus antigenic variation will significantly influence the design of efficacious vaccines and immunotherapeutics against these important human pathogens.
Noroviruses are the leading cause of severe viral gastroenteritis. Although the severity of disease is usually moderate, infection can be especially virulent in young children and the elderly (10, 16, 25, 27, 32, 48). It is estimated that 200,000 people die each year from norovirus infections, especially in the developing world (50). An effective vaccine would be particularly advantageous to young and aged populations, military personnel, food handlers, and child and health care providers and in the developing world. The major obstacle to successful norovirus vaccine development is the lack of understanding of the extensive antigenic relationships between norovirus strains and the complex relationship between host protective immunity and antigenic heterogeneity.
Genetically, noroviruses are grouped by the major capsid protein amino acid sequence. Viruses with less than 14.3% difference are classified as the same strain, those with 14.3 to 43.8% difference are classified as the same genotype, and those with 45 to 61.4% difference are classified as the same genogroup (68). Currently, noroviruses are grouped into five genogroups (GI to GV). Genogroups GI and GII are responsible for most human infections, and these genogroups are further subdivided into more than 25 different genotypes (68). The majority of norovirus outbreaks are caused by the GII.4 genotype. Between 1995 and 2006 four major GII.4 strain pandemics have been identified. The first was recognized in the mid-1990s (46). During this time, strain US95/96 was responsible for ∼55% of the norovirus outbreaks in the United States and 85% of the outbreaks in the Netherlands (63). In 2002, the US95/96 strain was replaced by the Farmington Hills strain (66), which was associated with ∼80% of norovirus outbreaks (17) in the United States. Simultaneously in Europe, the GII.4b variant emerged and caused outbreaks during the winter, spring, and summer (42, 44, 51). In 2004, the Hunter GII.4 variant was detected in Australia, Europe, and Asia (7, 33, 51). This strain was subsequently replaced in 2006 by two new cocirculating GII.4 variants in the United States and Europe, Laurens (2006a) and Minerva (2006b) (10, 33, 57).
Structurally, noroviruses are ∼38-nm icosahedral viruses with an ∼7.5-kb single-stranded, positive-sense RNA genome that encodes three large open reading frames (ORFs). ORF1 encodes the replicase polyprotein, while ORFs 2 and 3 encode the major and minor capsid proteins, respectively. Expression of the major capsid protein (ORF2) in baculovirus and Venezuelan equine encephalitis (VEE) virus results in formation of virus-like particles (VLPs) composed of 180 copies of the monomeric protein (53). The monomer is structurally divided into the shell domain (S) that forms the core of the particle and the protruding domain (P) that extends away from the core. The P domain is further subdivided into the P1 subdomain (residues 226 to 278 and 406 to 520) and the P2 subdomain (residues 279 to 405) (53). P2 represents the most exposed surface of the viral particle and determines interaction with both potential neutralizing antibody and histo-blood group antigens (HBGAs) (9, 12, 39, 41).
Multiple recent reports have concluded that the major capsid proteins of GII.4 strains are evolving rapidly, resulting in new epidemic strains with altered antigenicity (4, 6, 39, 59). The majority of these changes are occurring within the surface-exposed P2 subdomain. Surrogate neutralization assays using both sera collected from human GII.4 outbreaks and from norovirus-immunized mice suggest that potential neutralizing epitopes are not conserved among GII.4 noroviruses. This antigenic variation and accompanying host immune evasion may contribute to GII.4 persistence in human populations (8, 39). Additional compelling evidence for long-term protective immunity to norovirus infection also comes from reports indicating that periods of “high norovirus activity” correlated with the emergence of new GII.4 strains (1, 5, 29, 47, 55, 62) and are followed by years characterized by decreased numbers of outbreaks. These data suggest that herd immunity may be an important regulator of GII.4 norovirus evolution and persistence in human populations (8, 39).
Successful RNA viruses have been shown to evade host immunity via several modes, including antigenic variation (43, 65). Influenza virus hemagglutinin (HA) and HIV-1 gp120 Env are highly antigenic, well-characterized viral proteins documented to change neutralizing epitopes with passage (26, 34, 43, 45). Similarly, there is evidence of antigenic variants in immunocompromised individuals chronically infected with norovirus (56). How viruses evolve under pressure from host protective immunity to escape neutralizing antibody is controversial. Historically, the paradigm has been that herd immunity causes antigenic drift, the slow accumulation of mutations of small effect that eventually result in significant antigenic change. This hypothesis has been challenged by arguments for epochal evolution in which mutations of large effect are admixed within neutral mutations and by the idea that these combined small- and large-effect mutations actually drive antigenic change (31). This hypothesis argues that a complex relationship exists between mutation and antigenic variation, with specific sites differentially driving escape from herd immunity.
In the absence of a small-animal model or a cultivatable virus, we prepared panels of both time-ordered GII.4 VLPs and monoclonal antibodies (MAbs) to study the mechanisms of GII.4 persistence in human populations, including antigenic variation. Previous studies have demonstrated that VLPs are antigenically very similar to native noroviruses (28, 36), and mouse MAbs can be used to compare epitopes between different VLPs. Therefore, we constructed VLPs representing each of the pandemic strains [GII.4-1997 (US95/96), GII.4-2002 (Farmington Hills), GII.4-2004 (Hunter), and GII.4-2006 (Minerva)] in addition to an ancestral strain that circulated prior to any known pandemic outbreak (GII.4-1987). An additional VLP (GII.4-2005) representing Sakai was used to represent a strain circulating during the brief window between the 2004 and 2006 pandemic strains. Sakai is a neoteric GII.4 outbreak strain associated with outbreaks in health care facilities in Southeast Asia (48) and sporadically detected in the United States and the Netherlands (10). Two new VLPs, GII.4-2007 and GII.4-2008, were constructed to represent strains circulating within communities after the Minerva pandemic peaked. Previously, we reported the histo-blood group antigen binding patterns (putative cellular receptors) and antigenic relationships between these GII.4 strains circulating between 1987 and 2006 (8, 39). In these studies, VLP GII.4-1987, GII.4-1997, and GII.4-2006 bound to multiple HBGAs but preferentially to H type 3. VLP GII.4-2002 also bound H type 3 but showed preferential binding to Lewis y antigen (Ley). Under our conditions of treatment, a strong affinity of HGBA ligand for VLP GII.4-2004 and GII.4-2005 was not identified. Using enzyme immunoassays (EIAs) and surrogate neutralization assays based on these VLP-HBGA interactions, GII.4-1987 and -1997 were antigenically indistinguishable from each other with both human norovirus outbreak sera and VLP-immunized mouse sera. VLPs of strains circulating in 2002 or later had significantly less reactivity with sera directed against VLPs of earlier strains. Based upon these results, five putative antibody epitopes were predicted based upon evolving surface-exposed residues in the GII.4 capsid (14). These observations provide empirical support for bioinformatic predications that GII.4 noroviruses are undergoing antigenic variation (2, 39, 59).
In this study we demonstrate that the antibody epitopes of GII.4 norovirus major capsid proteins are changing by comparing the reactivity of a panel of monoclonal antibodies generated independently against an early GII.4 strain (GII.4-1987) and a contemporary GII.4 strain (GII.4-2006) against a panel of time-ordered GII.4 VLPs representing major evolutionary steps and consequently major outbreak strains ranging between 1987 and 2008. These results not only support the hypothesis that noroviruses are undergoing rapid antigenic variation but also provide a model platform to evaluate whether change is mediated by classical antigenic drift or by epochal evolution.
MATERIALS AND METHODS
Phylogenetic analysis of GII.4 ORF2 sequences.
Thirty-three genotype GII.4 capsid P2 subdomain amino acid sequences, including 11 representative sequences and 22 reference sequences, were aligned by ClustalX, version 1.83 (13), using default parameters. The alignment was refined manually, and sites of variation, defined as any site with a quality score of less than 100, were exported in table format and ordered by genotype and date. To eliminate potential sequencing errors, positions that were different in only one representative sequence were removed. Variable sites that occurred in the P2 subdomain were exported using Microsoft Excel. Since the residues within predicted epitopes (14) appear to reuse the same residues from previous outbreak strains at various times and because different portions of the protein evolve at different rates (39), molecular clock analysis was not conducted on these data. Further, molecular clock analysis has been previously reported for the GII.4 noroviruses (4, 58).
A maximum-likelihood phylogenetic tree was generated using the PhyML program (20) as implemented in the Geneious package (Biomatters, Auckland, New Zealand), using the MtREV substitution model. Boot-strapping was conducted generating 100 bootstrapped data sets, and a consensus tree was generated using Consensus from the Phylip package (18). Trees were visualized in the Geneious tree viewer, and the Seaview tool (19) was used for editing and rearranging branches.
Virus-like particles.
A panel of time-ordered GII.4 VLPs representing GII.4 strains circulating in 1987, 1997, 2002, 2004, 2005 (39), 2006 (8), 2007 (AB496912), and 2008 (AB492092) was assembled as previously described. Briefly, all ORF2 constructs were inserted directly into the VEE replicon vector for the production of virus replicon particles (VRPs), and VLPs were expressed in VRP-infected BHK cells. Subsequently, VLPs were purified by velocity sedimentation in sucrose, and approximately 35-nm particles were visualized by negative-staining electron microscopy (EM) (3, 40). VLP protein concentrations were determined by a Bio-Rad protein assay (Bio-Rad, Hercules, CA).
Mouse immunization, hybridoma production, and IgG purification.
Monoclonal antibodies were produced and purified by the University of North Carolina (UNC)—Chapel Hill, Immunology Core (http://mabs.unc.edu). Briefly, to create the UNC50 anti-GII.4-1987 monoclonal antibodies, Swiss Webster mice were given oral immunizations with 5 μg of VLP/100 μl on days 0, 2, 4, 16, and 18. On day 21 mice were boosted with 5 μg of VLP/100 μl and 2 μg/10 μl intravenously (i.v.) before splenocyte fusion. UNC56 anti-GII.4-1987 hybridomas were produced from mice immunized as above with an additional boost intraperitoneally (i.p.) at 5 months with 50 μg/200 μl plus Gerbu adjuvant and subsequent fusion. UNC67 anti-GII.4-2006 hybridomas were produced from mice immunized with 50 μg/100 μl plus Gerbu adjuvant on days 0, 20, 41, 57, and 84, and splenocytes fused on day 88. Resulting hybridomas were subcloned by limiting dilution, isotyped, and purified by protein G chromatography.
EIAs.
Antibody reactivity was determined by EIA. Plates were coated at 1 μg/ml of VLP in phosphate-buffered saline (PBS) for 4 h at room temperature and blocked overnight at 4°C in 5% Carnation dry milk in PBS-0.05% Tween 20 before the addition of either serially diluted IgM-containing tissue culture supernatant or 2 μg/ml purified IgG; samples were incubated for 1 h at 37°C, followed by anti-mouse-IgM or IgG-alkaline phosphatase (Sigma Chemicals) for 30 min at 37°C and color development with para-nitrophenyl phosphate (pNPP; Sigma Chemicals) at room temperature for 15 min. Each step was followed by washing with PBS-0.05% Tween 20, and all antibodies were diluted in 5% Carnation dry milk in PBS-0.05% Tween 20. All samples were assayed in triplicate as both tissue culture supernatants and purified antibodies. Establishment of EIAs using new MAbs included PBS and genogroup I VLP-coated wells as negative controls and polyclonal anti-VLP mouse or human serum as a positive control. Antibodies were considered positive for reactivity if the mean optical density (OD) for VLP-coated wells was greater than three times the mean optical density for PBS-coated wells. Assay plates were considered valid if negative controls were below 0.08 OD units and positive controls were above 0.5 OD units.
Carbohydrate binding blockade assays.
VLP-HBGA binding blockade experiments were done as reported previously by our group (38, 39) with minor modification. Synthetic HBGAs were bound to NeutriAvidin-coated plates (Pierce, Rockford, IL) at 10 μg/ml (Glycotech, Gaithersburg, MD) for 1 h before the addition of MAb-pretreated VLP at 1 μg/ml for 1.5 h. VLP binding was detected with rabbit anti-GII VLP polyclonal antiserum, followed by anti-rabbit IgG-alkaline phosphatase (Sigma Aldrich, St. Louis, MO) and pNPP (Sigma Aldrich). The percent control binding was defined as the binding level in the presence of antibody pretreatment compared to the binding level in the absence of antibody pretreatment, multiplied by 100. HBGA ligands were H type 3 for GII.4-1987, -1997, and -2006 and Ley for GII.4-2002 (38, 39). All incubations were done at room temperature. Each step was followed by washing with PBS-0.05% Tween 20, and all reagents were diluted in 5% Carnation dry milk in PBS-0.05% Tween 20. All samples were assayed in triplicate as both tissue culture supernatants and purified antibodies. Two criteria were used to designate an antibody as blockade competent: (i) a positive dose response between antibody treatment and mean percent control binding, as indicated by a negative slope significantly different from zero by linear regression analysis, and (ii) at least a 50% block of the VLP-HBGA interaction within the dilution series tested. The exceptions to these criteria were GII.4-2006-G3, -G4, and -G7. These antibodies completely blocked homotypic VLP-HBGA interaction at all dilutions tested and were determined to be blockade competent even though they did not meet the first criterion. Establishment of blockade assays using new MAbs included PBS and genogroup I VLP as negative controls and polyclonal anti-VLP mouse or human serum as a positive control.
RESULTS
GII.4 sequence variation and phylogeny.
A phylogenetic tree was generated using maximum-likelihood analysis from an alignment of the P2 subdomain of 33 GII.4 sequences, including 11 representative sequences (Fig. 1). The tree shows that distinct clusters arose over time, starting with the earliest known GII.4 strain from 1974 through 2008, with distinct clusters correlating with major outbreaks.
FIG. 1.
Maximum-likelihood analysis of the P2 subdomain of the GII.4 noroviruses. A phylogenetic tree was generated using maximum-likelihood analysis from an alignment of the P2 subdomain of 33 GII.4 sequences, including 11 representative sequences. The tree shows that distinct clusters arose over time, starting with the ancestral GII.4-1974 cluster. Evolution appears to have proceeded linearly over time, with new strains emerging from previous strains, often using recycled residues within important surface-exposed epitopes..
Informative sites from the P2 subdomain of 11 representative sequences were exported and ordered by time (Fig. 2). Analysis indicated that significant changes occurred in the P2 subdomain over time, consistent with results previously reported for the GII.4 genotype (39). In general, it appears that the GII.4 genotype underwent epochal evolution whereby periods of evolutionary stasis were followed by bursts of adaptation that gave rise to novel strains that emerged from previous circulating strains, often via recycling of residues within important surface-exposed epitopes. Moreover, two extant clusters, Hunter (GII.4-2004) and Minerva (GII.4-2006), appear to be persisting in human populations by evolving novel capsid variations. The Hunter cluster has two representative strains (2004 and 2007) that are significantly different in the P2 subdomain, with amino acid differences at five positions (296, 298, 364, 365, and 407). Positions 296, 298, and 407 are surface-exposed residues that lie in predicted antigenic epitopes (14). This observation suggests that these two distinct capsids from the Hunter cluster likely have different antigenic properties. In addition, within the Minerva cluster there are 16 amino acid differences in the P2 subdomain between GII.4-2006 and GII.4-2008, most of which map to the surface of the capsid and lie within predicted antigenic epitopes (Fig. 2). Taken together, these observations suggest that two extant strains are currently competing for persistence in human populations by evolving novel genotypes capable of evading existing herd immunity.
FIG. 2.
GII.4 P2 subdomain antigenic variation. Eleven representative sequences were selected from each major phylogenetic cluster, beginning with strains originating in 1974 through extant strains that emerged in 2008. The 11 representative capsid sequences were aligned with ClustalX, and informative sites in the P2 subdomain were exported to this table and ordered by time. Each color represents amino acid changes that occurred within the subcluster. Ancestral GII.4, light yellow; GII.4-1987 of Camberwell, yellow; GII.4-1997 of Grimsby, red; GII-2002 and GII-2002a of Farmington Hills, blue; GII-2004 of Hunter, green; GII-2005 of Sakai, orange; and GII-2006 of Minerva, purple; GII.5-2007, light green; GII.4-2008 of Stockholm, tan; and GII.4-2008a of Appledorn, brown.
Characterization of broadly reactive anti-GII.4-1987 IgM MAbs.
Supporting earlier bioinformatic analysis that indicated that GII.4 strain persistence and outbreak potential corresponded with the emergence of antigenic variants, antibody cross-reactivity patterns (39) and surrogate neutralization assays using polyclonal sera from GII.4 outbreaks and immunized mice suggested that GII.4 noroviruses were undergoing antigenic variation and using immune evasion to escape protective herd immunity targeting ancestral strains in human populations. Antigenic variation is proposed to mediate GII.4 strain persistence in human populations. To directly test this hypothesis, we prepared panels of monoclonal antibodies against an early GII.4 strain, GII.4-1987, and a contemporary strain, GII.4-2006, and then compared the reactivity of these antibodies against a panel of GII.4 VLPs presenting GII.4 outbreak strains circulating globally from 1987 until 2008.
Immunization with GII.4-1987 VLPs resulted in seven MAbs, two IgM and five IgG isotypes (Table 1). Hybridoma tissue culture supernatants containing each of the IgM MAbs had a broad degree of recognition across a diverse panel of norovirus VLPs, as measured by EIA (see Fig. S1a in the supplemental material). GII.4-1987-M1 and -M2 reacted not only across the panel of time-ordered GII.4 VLPs but also with GII-1, -2, -3, and -14 strains. Contrary to IgG findings (21, 37, 61), the IgM MAb reactivity extended across genogroups, recognizing VLPs from GI-1, -2, -3, and -4.
TABLE 1.
Antibody reactivity to immunization with GII.4 strains
| Clone name | Clone no. | Immunogen | Isotype |
|---|---|---|---|
| GII.4-1987-M1 | 44.14.1 | GII.4-1987 | IgM |
| GII.4-1987-M2 | 42.12.1 | GII.4-1987 | IgM |
| GII.4-1987-G1 | 159.1.1 | GII.4-1987 | IgG2b |
| GII.4-1987-G2 | 65.5.1 | GII.4-1987 | IgG1 |
| GII.4-1987-G3 | 163.5.9 | GII.4-1987 | IgG1 |
| GII.4-1987-G4 | 116.10.12 | GII.4-1987 | IgG1 |
| GII.4-1987-G5 | 174.5.4 | GII.4-1987 | IgG1 |
| GII.4-2006-G1 | 70.12.6 | GII.4-2006 | IgG2b |
| GII.4-2006-G2 | 95.11.1 | GII.4-2006 | IgG2b |
| GII.4-2006-G3 | 274.12.1 | GII.4-2006 | IgG2a |
| GII.4-2006-G4 | 351.12.1 | GII.4-2006 | IgG1 |
| GII.4-2006-G6 | 4.4.10 | GII.4-2006 | IgG1 |
| GII.4-2006-G7 | 416.9.1 | GII.4-2006 | IgG1 |
To assess if these highly reactive IgM MAbs could be potentially neutralizing, we evaluated the ability of each IgM antibody to block interaction of the immunizing VLP, GII.4-1987, with its HBGA binding partner, H type 3 (39), in a surrogate neutralization assay. GII.4-1987 VLPs were pretreated with serially diluted IgM-containing hybridoma tissue culture supernatant before addition to H type 3-coated plates and determination of the amount of VLP bound in the presence or absence of the MAb. Neither monoclonal IgM inhibited VLP interaction with HBGA ligand at any of the dilutions tested (see Fig. S1b in the supplemental material). These data suggest that both GI and GII norovirus strains share common epitopes within the intact particle that are antibody accessible, but these epitopes are likely conserved outside the carbohydrate binding domain. Cross-genogroup reactivity is in contrast to observations comparing IgG cross-reactivity (see below).
Characterization of anti-GII.4-1987 IgG MAbs against a panel of time-ordered GII.4 VLPs.
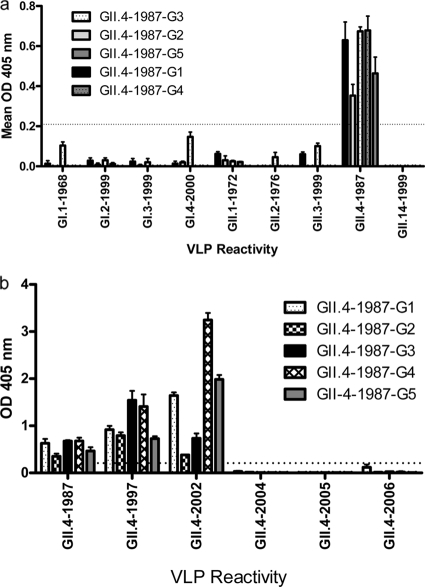
In contrast to the broad reactivity of the IgM MAbs, the anti-GII.4-1987 IgG MAbs were genotype specific. The five IgG MAbs were purified, and the reactivity of each across the panel of GI and GII VLPs was measured by EIA. All five antibodies preferentially reacted to the GII.4-1987 VLP with low or no reactivity to VLPs tested from other GII or any GI genotypes (Fig. 3a). Expansion of the GII.4 genotype to include the complete panel of time-ordered GII.4 VLPs identified a clear demarcation of anti-GII.4 MAb reactivity limited to strains that circulated primarily before 2004. Each IgG recognized GII.4-1987, -1997, and -2002 but did not recognize GII.4-2004, -2005, -2006 (Fig. 3b), -2007, or -2008 (data not shown) VLPs by EIA. As the five IgG MAb epitopes changed over time, these data are consistent with the hypothesis that encoded epitopes within the GII.4 noroviruses are clearly undergoing antigenic variation over time.
FIG. 3.
Anti-GII.4-1987 monoclonal IgG antibodies are genotype specific. Anti-GII.4-1987 monoclonal IgG antibodies were purified by protein G affinity chromatography, diluted to 2 μg/ml, and assayed in triplicate by EIA for reactivity to a panel of GI and GII VLPs (a) and an expanded panel of time-ordered GII.4 VLPs (b). Bars represent mean optical density values with standard errors of the mean. The dashed line indicates the value equal to three times the background level.
The neutralization potential of each of these MAbs was evaluated using a surrogate neutralization assay, which provides a biochemical measure of the capacity of each antibody to specifically block ligand HBGA-VLP interaction (23, 39). Serial dilutions of purified IgG were incubated with GII.4-1987, -1997, -2002, or -2006 before VLPs were added to HBGA-coated plates, and the VLP binding with antibody pretreatment was compared to the binding without antibody pretreatment (Fig. 4). All of the IgG antibodies had similar effects on GII.4-1987 and -1997 interaction with H type 3, supporting previous findings that these two strains are, so far, antigenically indistinguishable (39). Monoclonal antibody GII.4-1987-G1, -G4, and -G5 blockade of GII.4-1987, -1997, and -2002 VLPs was dose dependent (see Materials and Methods) and nearly complete at the highest doses tested. However, blockade of GII.4-2002 required ∼4-fold or more MAb to block at least 50% binding compared to the concentrations needed to block GII.4-1987 (Fig. 4a, c, and e). MAbs GII.4-1987-G2 and -G3 did not significantly affect HBGA binding of any of the VLPs (Fig. 4b and c). These surrogate neutralization assays combined with EIA data for the anti-GII.4-1987 monoclonal IgG (mIgG) antibodies indicate that some of the time-sensitive GII.4 epitopes are likely located in or near the carbohydrate binding site.
FIG. 4.
Anti-GII.4-1987 mIgG can block VLP-HBGA interaction of early GII.4 strains but not contemporary GII.4 strains. Purified antibodies were assayed for blockade of the interaction between GII.4-1987, GII.4-1997, and GII.4-2006 strains and H type 3 and between GII.4-2002 and Ley, and the mean percent control binding was calculated compared to the no-antibody pretreatment control binding. Error bars represent standard errors of the means of samples run in triplicate.
Characterization of a panel of anti-GII.4-2006 IgG MAbs.
Monoclonal antibodies directed against the early GII.4-1987 strain support the hypothesis that GII.4 epitopes are changing over time via antigenic drift and/or by selection driven mutation. To study this time-sensitive antigenic variation more thoroughly, we prepared a second set of monoclonal antibodies against a contemporary GII.4 epidemic strain, GII.4-2006, and compared antibody reactivity across our panel of norovirus VLPs. Six stable IgG-producing hybridoma lines were established (Table 1). Among the six MAbs, two cross-reactivity groups were identified by EIA (Fig. 5). The first group (GII.4-2006-G1, -G2, -G3, and -G4) was genogroup specific, reacting only with the immunizing GII.4-2006 VLP (Fig. 5a). The second group (GII.4-2006-G6 and -G7) reacted more broadly, including with other GII VLPs (GII.1-1972, GII.2-1976, GII.3-1999, and GII.14-2000), but not with any GI VLPs tested (Fig. 5b). Expansion of the GII.4 genotype to include the complete panel of time-ordered GII.4 VLPs again supported two reactivity patterns for this set of MAbs. The genocluster-specific MAbs had limited reactivity across the GII.4 time-ordered VLPs. All reacted with the immunizing strain, GII.4-2006 (Fig. 6). Among the four genotype-specific IgG MAbs, three (GII.4-2006-G1, -G3, and -G4) cross-reacted with GII.4-2005, two (GII.4-2006-G2 and -G3) cross-reacted with GII.4-2004, and one (GII.4-2006-G3) cross-reacted with GII.4-2002. None of the genotype-specific MAbs cross-reacted with the VLPs representing strains circulating primarily before 2002 (Fig. 6a). The two cross-GII genotype-reactive IgG MAbs (GII.4-2006-G6 and -G7) also recognized GII.4-2005, -2004, -2002, and -1997. Only GII.4-2006-G6 had a robust reactivity to GII.4-1987 VLPs under these treatment conditions (Fig. 6b).
FIG. 5.
Cross-genogroup reactivity of anti-GII.4-2006 monoclonal IgG antibodies. Anti-GII.4-2006 mIgG antibodies were purified by protein G affinity chromatography, diluted to 2 μg/ml, and assayed by EIA for reactivity to a panel of GI and GII VLPs. (a) MAbs without GII cross-genotype reactivity. (b) MAbs with GII cross-genotype reactivity. Bars represent mean optical density values with standard errors of the means. The dashed line indicates the value equal to three times the background level.
FIG. 6.
GII.4 VLP reactivity of anti-GII.4-2006 monoclonal IgG antibodies. Anti-GII.4-2006 mIgG antibodies were purified by protein G affinity chromatography, diluted to 2 μg/ml, and assayed by EIA for reactivity to an expanded panel of time-ordered GII.4 VLPs. (a) MAbs without cross-genotype reactivity. (b) MAbs with GII cross-genotype reactivity. Bars represent mean optical density values with standard errors of the means. The dashed line indicates the value equal to three times the background level.
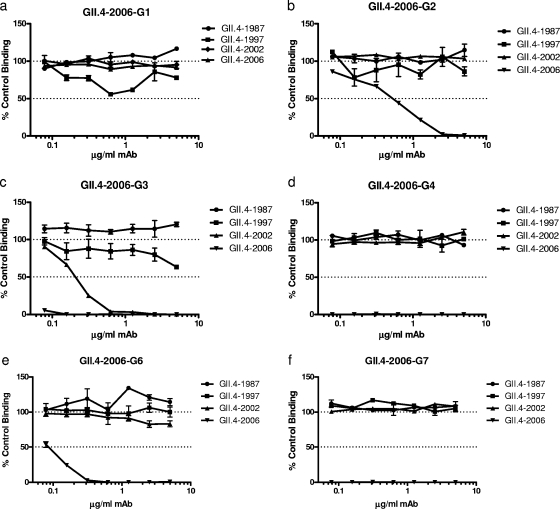
Each MAb was evaluated for its ability to disrupt ligand HBGA-VLP interaction using the surrogate neutralization assay, as described above. In contrast to the EIA data, the blockade assay did not differentiate the anti-GII.4-2006 MAbs into two different phenotype groups. Five of the six MAbs were able to block the immunizing strain VLP-HBGA interaction at least 50% with dose dependency (Fig. 7). Only GII.4-2006-G1 was unable to block binding. Despite a broad homotypic blockade, only one MAb (GII.4-2006-G3) (Fig. 7c) was able to block GII.4-2002-Ley interaction. None of the anti-GII.4-2006 MAbs inhibited GII.4-1987 or -1997 VLP interaction with H type 3, despite EIA recognition of these VLPs by two of the MAbs (Fig. 6b). Further, despite EIA recognition by GII.4-2006-G6 and -G7, neither GII.2-1976 nor GII.3-1999 VLP interaction with H type 3 was affected by antibody pretreatment (data not shown). As carbohydrate binding ligands for GII.1-1972 and GII.14-1999 have not been identified, blockade potential for these VLPs could not be determined. As observed with MAbs against GII.4-1987, these EIA and surrogate neutralization assays using anti-GII.4-2006 MAbs indicate that some of the GII.4 epitopes are changing over time and that some of these time-sensitive epitopes are located in or near the carbohydrate binding site.
FIG. 7.
Anti-GII.4-2006 MAb can block VLPs from contemporary GII.4 strains but not early GII.4 strains. Purified antibodies were assayed for blockade of the interaction between GII.4-1987, GII.4-1997, and GII.4-2006 strains and H type 3 and between GII.4-2002 and Ley, and the mean percent control binding was calculated compared to the no-antibody pretreatment control binding. Error bars represent standard errors of the means. (a to d) MAbs without cross-genotype reactivity. (e and f) MAbs with GII cross-genotype reactivity. Error bars represent standard errors of the means of samples run in triplicate.
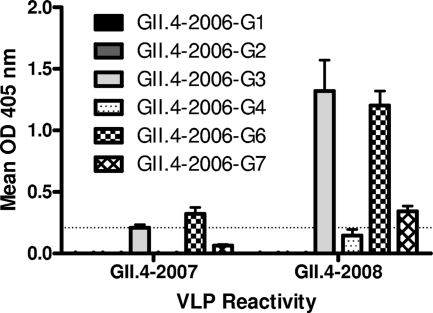
Since 2006, several new norovirus strains have been detected in outbreak investigations although none has circulated at pandemic levels. To test if further genetic change reported in these later strains may be leading to antigenic changes, we built two more VLPs. The first, GII.4-2007, represents Cruise Ship virus, a strain with high homology to GII.4-2004 Hunter (Fig. 1). The second, GII.4-2008, represents Stockholm virus, a strain closely related to GII.4-2006 Minerva (Fig. 1). Neither of these strains, which fall within previously defined genoclusters, was pandemic in circulation. VLPs for these new strains were produced as before, and the structure of particles was verified by electron microscopy before antigenic characterization. Both panels of MAbs were assayed for reactivity to GII.4-2007 and GII.4-2008 by EIA (Fig. 8). As expected from previous results, none of the anti-GII.4-1987 MAbs recognized either GII.4-2007 or GII.4-2008 (data not shown). The Hunter-like GII.4-2007 VLP reacted weakly with only one MAb, GII.4-2006-G6. Supporting bioinformatic predictions (Fig. 2), GII.4-2008 was antigenically distinct from GII.4-2006. Three of six GII.4-2006 MAbs cross-reacted with GII.4-2008. GII.4-2006-G3 and -G6 demonstrated strong binding signals while GII.4-2006-G7 reacted only modestly, and GII.4-2006-G1, -G2, and -G4 did not react. These data support a continued antigenic divergence even within genoclusters, as previously described for GII.4-2002 variants (39). The Hunter-like GII.4-2007 VLP was recognized by only one GII.4-2006 MAb compared to recognition by four MAbs of GII.4-2004 Hunter VLPs, suggesting that herd immunity against GII.4 Minerva-like strains may also be shaping Hunter capsid protein evolution (Fig. 6 and 8). Similarly, only three of the six anti-GII.4-2006 MAbs recognized GII.4-2008, a Minerva-like strain found in the same genocluster as GII.4-2006 (Fig. 8). Further emphasizing the degree of antigenic change in these later strains, we were unable to evaluate blockade potential of the MAbs for GII.4-2007 and -2008 due to insufficient cross-reactivity of these VLPs with the rabbit polyclonal sera used for blockade detection.
FIG. 8.
GII.4-2007 and -2008 are antigenically diverse from GII.4-2006. Anti-GII.4-2006 monoclonal IgG antibodies were purified by protein G affinity chromatography, diluted to 2 μg/ml, and assayed by EIA for reactivity to GII.4-2007 and -2008 VLPs. Bars represent mean optical density values with standard errors of the means. The dashed line indicates the value equal to three times the background level.
DISCUSSION
GII.4 noroviruses have been recognized as a leading cause of severe viral gastroenteritis worldwide for more than 20 years. The mechanism of continued persistence of this specific genocluster among a large family of genetically and antigenically diverse strains represents a fundamental question in norovirus biology as it combines questions of viral fitness with questions of host genetic susceptibility and immunity. These questions remained largely unanswered until recently, and all of them impact future vaccine design. The major obstacle to successful norovirus vaccine design is the lack of understanding of the extensive antigenic relationships between norovirus strains and how this heterogeneity affects host protective immunity. Despite this, we developed the first multivalent vaccines with broad cross-reactive and protective responses against noroviruses in mice (40). To address these issues and inform vaccine design, we prepared a panel of MAbs generated independently against an early GII.4 strain (GII.4-1987) and a contemporary GII.4 strain (GII.4-2006). By comparing the EIA reactivity of these MAbs against a panel of time-ordered (1987 to 2008) GII.4 VLPs, we have identified MAbs that recognize epitopes present only in strains chronologically near to the immunizing strain. More importantly, the inability of the 1987 monoclonal antibodies to bind contemporary 2006 VLPs confirmed the existence of evolving epitopes, epitopes which escaped antibody recognition most likely from mutations in and around surface-exposed sites which varied over time. These data provide the first direct evidence that epitopes in the GII.4 norovirus major capsid protein are undergoing antigenic variation over time, most likely in response to human herd immunity, much like other RNA viruses including influenza virus and HIV but in contrast to genogroup I noroviruses (15, 38). Our data support a biphasic model of antigenic variation that emerges in GII.4 viruses in response to host herd immunity over time. First, we describe the emergence of GII.4 variation that differentially regulates the ability of ancestral strain monoclonal antibodies to block carbohydrate binding by the new emergent outbreak strain. Then over time, additional GII.4 variation eventually allows for complete or nearly complete escape from ancestral humoral immune response recognition/neutralization. These data also support earlier arguments that blockade responses are indicative of protective immunity (23, 39, 54). Larger monoclonal antibody panels, including human monoclonal antibodies and robust tissue culture models, would certainly enrich support for this model of GII.4 evolution.
Monoclonal antibodies to a limited assortment of norovirus VLPs have been produced previously to evaluate the serologic relationships between different genotypes and genogroups (22, 30, 41, 49). Most groups developing norovirus monoclonal antibodies have used VLPs for antibody reactivity characterization, which preferentially selects for antibodies that target surface-exposed conformation-retained epitopes. This is in contrast to immunizing and screening with P particles or P dimers as antigens (67), which likely externally display undefined combinations of surface and some internal epitopes that are normally buried in native virions. Further, the use of Western blotting as reported in Yang et al. (67) failed to detect significant strain-antigenic differences. This approach may be experimentally flawed, as only 1 out of the 11 mIgGs characterized here bound to denatured VLP by Western blotting. One anti-GII.4-1987 MAb bound to denatured VLPs of GII.4 strains from 1987 to 2006 (data not shown), which is in sharp contrast to EIA data which demonstrated more specific MAb interaction patterns when incubated with native conformation-intact VLPs (Fig. 3b). Allan et al. (2) have compared reactivity of five monoclonal antibodies against a pre- and post-2002 epidemic GII.4 strain. These data also identified a conformational epitope composed of residues 294 to 296 and 393 to 395. Unfortunately, none of our VLPs allowed for direct comparison between the two studies, but the finding that residues 393 to 395 were antigenically important supported our previously published work identifying amino acid 395 as an antigenic determinant in the GII.4-2002 Farmington Hills strain (39). Nevertheless, our study is the first to examine the extent of antigenic variations within a genotype over decades, including variations occurring through four epidemic strains, and is consistent with earlier findings using polyclonal antinorovirus human and mouse sera (8, 39). The panel of MAbs was not able to distinguish clear antigenic differences between GII.4-1987 and GII.4-1997 VLPs by either EIA (Fig. 3 and 6) or surrogate neutralization assays (Fig. 4 and 7), indicating that these two strains are antigenically very similar. These data are compelling because they support an earlier hypothesis (39) that the emergence of the pandemic strain US95/96 may have been driven by altered host range, primarily mediated by enhanced affinity for an expanded group of HBGAs in these strains that allowed for more efficient infection of some human populations rather than immune evasion. In comparison, MAb reactivity across the time-ordered panel of GII.4 VLPs strongly suggests that GII.4-2002 (Farmington Hills) strains signaled a key shift in the antigenic milieu of the GII.4 noroviruses. By EIA, all five anti-GII.4-1987 mIgGs recognized GII.4-1987, -1997, and -2002 but had no reactivity to -2004, -2005, -2006, -2007, and -2008 VLPs (Fig. 3; also data not shown). Moreover, this sharp leading edge in cross-reactivity was also associated with a rapid loss in the ability of antibodies to block GII.4-2002 carbohydrate interactions (Fig. 4). If the in vitro blockade assay faithfully captures a neutralization response associated with preventing virus particle-receptor interaction and entry, then it seems likely that GII.4-2002 viruses may be highly resistant to ancestral GII.4 immune responses. Clearly, GII.4 strains of 2004 to 2008 are completely resistant to blockade, allowing for escape from antibody-mediated host immunity. These data likely explain, in part, the ability of new contemporary strains to out-compete ancestral strains by circumventing host herd immune responses. Importantly, one of the five MAbs that blocked GII.4-2006 VLP interaction with ligand also blocked GII.4-2002-ligand interaction (Fig. 7). These data suggest an intriguing possibility that evolution selects for a successful contemporary GII.4 strain that retains or evolves one epitope or a few epitopes which simultaneously allow for the induction of cross-blockade antibody responses against ancestral isolates. By inducing cross-blockade responses against the more recent ancestral strains, successful new GII.4 isolates may have evolved a second strategy that specifically reinforces protective population-level herd immune responses against recent ancestral strains that are competing with new strains for naive hosts.
While GII.4-1987 antibodies were GII.4 specific; GII.4-2006 antibodies displayed very different binding patterns within and across genogroups. Minerva strains caused a global pandemic in 2006 and are still circulating in human populations as the predominant epidemic strain. Phylogenetic analyses demonstrate the emergence of significant evolution in the 2008 GII.4 Minerva-like strains, demonstrating that this lineage is evolving rapidly (Fig. 1). As most of the GII.4-2006 monoclonal antibodies did not recognize GII.4-2008, it seems likely that much of this evolution is in response to host herd immunity and that a major new pandemic strain may be emerging in the near future. In addition, the extant Minerva strain contains the same Asn393 and Ala395 residues found in the GII.4-2002 pandemic strain, and the Ala395 change was sufficient to alter HBGA binding and antigenicity in this subcluster. This provides further evidence that a subset of residues is tolerated at specific positions that allow for innovations and that these residues are likely recycled over time. It further suggests that the extant Minerva strain may exhibit novel antigenic properties compared to the 2006 Minerva strain.
Five of the seven anti-GII.4-2006 MAbs blocked homotypic VLP-HBGA interaction, and blockade potential did not correlate with either broad EIA reactivity or contemporary-strain EIA reactivity (Fig. 6 and 7). Four of the seven MAbs reacted with only contemporary (2002 to 2006) GII.4 VLPs (Fig. 6a). Of these, one epitope was maintained from 2002 to 2006, one was found in only 2004 and 2006, and two were found in 2005 and 2006 only, indicating that the epitopes are changing more quickly post-2002 and that some epitopes may be recycling in and out of strains, as previously predicted (14, 15, 39). These data are in agreement with phylogenetic data demonstrating that the GII.4 strains are evolving at an accelerated rate post-2002, with epidemic strains emerging approximately every 2 to 3 years (39, 59). Contrary to data demonstrating that the GI strains share common epitopes between the genogroups that may lead to cross-protection and/or original antigenic sin (15, 38), relatively little cross-reactivity exists between the GII genoclusters with this group of GII.4-2006 MAbs, with no detectable cross-genocluster VLP-HBGA blockade. The remaining two anti-GII.4-2006 MAbs recognized not only the entire GII.4 panel but also other GII strains, although with various levels of sensitivity (Fig. 6a). These MAbs presumably recognize well-conserved genogroup epitopes which could subtly influence pathogenic outcomes and/or also serve as potential diagnostic or therapeutic reagents. Previous studies using outbreak samples have suggested that GII.3 outbreaks in the early 1990s may have induced immune responses that influenced GII.4-1997 evolution, suggesting that complex cross-epitope recognition patterns may exist across GII strains (8). It also seems likely that GII.4-2006 immune responses are driving GII.4-2004 evolution. Surprisingly, two of the broad-spectrum GII.4 2006 MAbs are potentially neutralizing as they specifically blocked GII.4-2006-HBGA interaction. Despite broad GII.4 VLP recognition by EIA, these MAbs did not block GII.4-1987, -1997, or -2002 HBGA binding (Fig. 7) or block cross-genogroup GII.2-1976 or GII.3-1999 ligand binding. It is intriguing that GII.4-1987 VLPs did not elicit similar broadly reactive monoclonal antibodies. While speculative, these data suggest that very complex cross-reactive epitope patterns may exist among time-ordered GII VLPs, which could potentially influence disease susceptibility patterns and clinical disease. Such a hypothesis is also in agreement with previous findings studying polyclonal antisera from infected humans and immunized mice indicating that anti-GII.4 antibody responses are broadly reactive against GII.4 VLPs by EIA and that blockade assays provide a powerful biochemical assay to define unique biological differences in antibody-VLP interactions with carbohydrate binding partners (8, 39). Using a modified version of the blockade assay designed and utilized extensively by our group (23, 38-40), Reeck et al. (54) recently reported an association between blockade antibody titers, but not EIA titers, and protection from illness in GI.1-1968-infected volunteers. These data support use of the blockade assay as a surrogate neutralization assay for noroviruses. Similar findings have been reported for influenza virus H3 hemagglutinin, where anti-H3 MAbs were shown to cross-react with other HA molecules by immunofluorescence assay (IFA), but to neutralize only select H3-containing viruses (64). Importantly, molecular evolution and antigenic differences between GII.4-2008 (Minerva-like) and GII.4-2007 (Hunter-like) support the notion that each is evolving/coevolving rapidly, most likely in response to human herd immunity, and that either/both may lead to future emergent pandemic strains.
Eight out of 11 MAbs blocked homotypic VLP-HBGA interaction, demonstrating potential neutralization activity. Currently, the only means to test actual neutralization would be to test the ability of these MAbs to protect gnotobiotic pigs from GII.4 challenge (11). It is not surprising that the majority of MAbs blocked ligand binding as the P2 domain of the capsid protein is both the site of HBGA interaction and the most surface-exposed portion of the VLP, i.e., available for antibody interaction. Fine-mapping studies with peptides or mutant capsids will be necessary to map key interaction residues. Further, cocrystalization of blocking MAbs with the panel of VLPs may provide structural information important for predicting modes of carbohydrate blockade and mapping epitopes onto the structure (52, 60). We have shown here by comparing blockade of GII.4-2002 with GII.4-1987 and −1997 that regardless of mechanism, change in the binding site sequence can lead to significant changes in neutralization potential (8, 39). It is important to note that protection is not always dependent upon binding near or within the receptor-binding site. Three MAbs recognized GII.4 VLPs but did not block VLP-HBGA binding, indicating that they may recognize epitopes outside the binding site. While not potentially neutralizing, these MAbs could be protective by interfering with a nonreceptor-binding step or by causing viral aggregation. In concert, mutations outside the capsid sequence may also have influenced GII.4 emergence as a recent report demonstrated that the RNA-dependent RNA polymerase of pandemic strains acquired a key mutation after 2001, resulting in an increased mutation rate and subsequent evolution rate (6).
Determination of the mechanisms of antibody neutralization and any corresponding protection from norovirus infection is seriously limited by the lack of a cell culture model or small-animal model for norovirus propagation. We have developed a large pool of anti-GII.4 MAbs from immunized mice, and these have yielded useful information on the changing antigenic relationship between the evolving GII.4 strains. The correlation between mouse and human norovirus epitopes is unknown and constitutes an important limiting factor in our study, as our approach has a bias toward mouse and not human antigenicity. However, multiple studies have shown that mice mount robust antibody responses to the capsid protein and that these mouse antibodies have similar reactivities to and blockade capacities for norovirus VLPs (24, 40). In addition to diagnostic reagents, these mouse antibodies could have potential vaccine use in the GII.4-infected gnotobiotic pig model and possibly in human therapeutics as the process of “humanizing” mouse antibodies becomes more accessible although combination cocktails and/or reformulation will likely be necessary to counter virus selection by antibody escape. Importantly, study of the GII.4 noroviruses with strain-matched VLPs and panels of MAbs is a useful model system for dissecting complex molecular mechanisms governing virus-ligand interaction and escape from herd immunity by antigenic variation in noncultivatable viruses.
As with HIV, hepatitis C virus (HCV), and influenza virus, antigenic variation will complicate the design of an effective GII.4 norovirus vaccine. It is likely that any vaccine will require frequent reformulation and need to include multiple components, as is the case for successful influenza vaccination. Preliminary data of multicomponent vaccination with both norovirus (40) and influenza virus (64) suggest that this vaccination approach can result in potentially neutralizing antibody production both to new strains not included in the cocktail and to multiple versions of the receptor-binding protein undergoing antigenic variation. Finally, the antigenic variation noted here is likely being driven by protective herd immunity. Contrary to early norovirus studies (35), these data continue to support the argument that norovirus infection does illicit protective immune responses in many individuals, making an effective multivalent vaccine design possible (40).
Supplementary Material
Acknowledgments
We thank Drew Trevor Scobey and Victoria Madden and C. Robert Bagnell, Jr., of Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, University of North Carolina—Chapel Hill, for expert technical support and Martha Collier and Robert Johnston of the Carolina Vaccine Institute for VRP production.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (grant AI056351).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print on 27 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adamson, W. E., R. N. Gunson, A. Maclean, and W. F. Carman. 2007. Emergence of a new norovirus variant in Scotland in 2006. J. Clin. Microbiol. 45:4058-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, D. J., J. J. Gray, C. I. Gallimore, J. Xerry, and M. Iturriza-Gomara. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., B. Yount, L. Lindesmith, P. R. Harrington, S. R. Greene, F. C. Tseng, N. Davis, R. E. Johnston, D. G. Klapper, and C. L. Moe. 2002. Expression and self-assembly of Norwalk virus capsid protein from Venezuelan equine encephalitis virus replicons. J. Virol. 76:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok, K., E. J. Abente, M. Realpe-Quintero, T. Mitra, S. V. Sosnovtsev, A. Z. Kapikian, and K. Y. Green. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buesa, J., B. Collado, P. Lopez-Andujar, R. Abu-Mallouh, J. Rodriguez Diaz, A. Garcia Diaz, J. Prat, S. Guix, T. Llovet, G. Prats, and A. Bosch. 2002. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 40:2854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull, R. A., J. S. Eden, W. D. Rawlinson, and P. A. White. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon, J. L., L. C. Lindesmith, E. F. Donaldson, L. Saxe, R. S. Baric, and J. Vinje. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2007. Norovirus activity-United States, 2006-2007. MMWR Morb. Mortal. Wkly. Rep. 56:842-846. [PubMed] [Google Scholar]
- 11.Cheetham, S., M. Souza, T. Meulia, S. Grimes, M. G. Han, and L. J. Saif. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80:10372-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, R., J. D. Neill, M. K. Estes, and B. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donaldson, E. F., L. C. Lindesmith, A. D. Lobue, and R. S. Baric. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190-211. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, E. F., L. C. Lindesmith, A. D. Lobue, and R. S. Baric. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467-474. [DOI] [PubMed] [Google Scholar]
- 17.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1989. Mathematics vs. evolution: mathematical evolutionary theory. Science 246:941-942. [DOI] [PubMed] [Google Scholar]
- 19.Gouy, M., S. Guindon, and O. Gascuel. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221-224. [DOI] [PubMed] [Google Scholar]
- 20.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 21.Hale, A. D., D. C. Lewis, X. Jiang, and D. W. Brown. 1998. Homotypic and heterotypic IgG and IgM antibody responses in adults infected with small round structured viruses. J. Med. Virol. 54:305-312. [PubMed] [Google Scholar]
- 22.Hale, A. D., T. N. Tanaka, N. Kitamoto, M. Ciarlet, X. Jiang, N. Takeda, D. W. Brown, and M. K. Estes. 2000. Identification of an epitope common to genogroup 1 “Norwalk-like viruses.” J. Clin. Microbiol. 38:1656-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington, P. R., L. Lindesmith, B. Yount, C. L. Moe, and R. S. Baric. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington, P. R., B. Yount, R. E. Johnston, N. Davis, C. Moe, and R. S. Baric. 2002. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J. Virol. 76:730-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, J. P., W. J. Edmunds, R. Pebody, D. W. Brown, and B. A. Lopman. 2008. Deaths from norovirus among the elderly, England and Wales. Emerg. Infect. Dis. 14:1546-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hensley, S. E., S. R. Das, A. L. Bailey, L. M. Schmidt, H. D. Hickman, A. Jayaraman, K. Viswanathan, R. Raman, R. Sasisekharan, J. R. Bennink, and J. W. Yewdell. 2009. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326:734-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutson, A. M., R. L. Atmar, and M. K. Estes. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, X., M. Wang, D. Graham, and M. Estes. 1992. Expression, self-assembly and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansen, K., K. Mannerqvist, A. Allard, Y. Andersson, L. G. Burman, L. Dillner, K. O. Hedlund, K. Jonsson, U. Kumlin, T. Leitner, M. Lysen, M. Thorhagen, A. Tiveljung-Lindell, C. Wahlstrom, B. Zweygberg-Wirgart, and A. Widell. 2008. Norovirus strains belonging to the GII.4 genotype dominate as a cause of nosocomial outbreaks of viral gastroenteritis in Sweden 1997-2005. Arrival of new variants is associated with large nation-wide epidemics. J. Clin. Virol. 42:129-134. [DOI] [PubMed] [Google Scholar]
- 30.Kitamoto, N., T. Tanaka, K. Natori, N. Takeda, S. Nakata, X. Jiang, and M. K. Estes. 2002. Cross-reactivity among several recombinant calicivirus virus-like particles (VLPs) with monoclonal antibodies obtained from mice immunized orally with one type of VLP. J. Clin. Microbiol. 40:2459-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koelle, K., S. Cobey, B. Grenfell, and M. Pascual. 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314:1898-1903. [DOI] [PubMed] [Google Scholar]
- 32.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181(Suppl. 2):S262-S269. [DOI] [PubMed] [Google Scholar]
- 33.Kroneman, A., H. Vennema, J. Harris, G. Reuter, C. H. von Bonsdorff, K. O. Hedlund, K. Vainio, V. Jackson, P. Pothier, J. Koch, E. Schreier, B. E. Bottiger, and M. Koopmans. 2006. Increase in norovirus activity reported in Europe. Euro. Surveill. 11:E061214.1. [DOI] [PubMed] [Google Scholar]
- 34.Kwong, P. D., and I. A. Wilson. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leon, J. S., S. Menira, Q. Wang, E. R. Smith, L. J. Saif, and C. L. Moe. 2008. Immunology of norovirus infection, p. 219-262. In M. Vajdy (ed.), Immunity against mucosal pathogens. Springer Science, Boston, MA.
- 36.Li, X., R. Zhou, Y. Wang, H. Sheng, X. Tian, H. Li, and H. Qiu. 2009. Identification and characterization of a native epitope common to norovirus strains GII/4, GII/7 and GII/8. Virus Res. 140:188-193. [DOI] [PubMed] [Google Scholar]
- 37.Lindesmith, L., C. Moe, J. Lependu, J. A. Frelinger, J. Treanor, and R. S. Baric. 2005. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 79:2900-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindesmith, L. C., E. Donaldson, J. Leon, C. L. Moe, J. A. Frelinger, R. E. Johnston, D. J. Weber, and R. S. Baric. 2010. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J. Virol. 84:1800-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LoBue, A. D., L. Lindesmith, B. Yount, P. R. Harrington, J. M. Thompson, R. E. Johnston, C. L. Moe, and R. S. Baric. 2006. Multivalent norovirus vaccines induce strong mucosal and systemic blocking antibodies against multiple strains. Vaccine 24:5220-5234. [DOI] [PubMed] [Google Scholar]
- 41.Lochridge, V. P., K. L. Jutila, J. W. Graff, and M. E. Hardy. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799-2806. [DOI] [PubMed] [Google Scholar]
- 42.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682-688. [DOI] [PubMed] [Google Scholar]
- 43.McHardy, A. C., and B. Adams. 2009. The role of genomics in tracking the evolution of influenza A virus. PLoS Pathog. 5:e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medici, M. C., M. Martinelli, L. A. Abelli, F. M. Ruggeri, I. Di Bartolo, M. C. Arcangeletti, F. Pinardi, F. De Conto, G. Izzi, S. Bernasconi, C. Chezzi, and G. Dettori. 2006. Molecular epidemiology of norovirus infections in sporadic cases of viral gastroenteritis among children in Northern Italy. J. Med. Virol. 78:1486-1492. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, M. I., and E. C. Holmes. 2007. The evolution of epidemic influenza. Nat. Rev. Genet. 8:196-205. [DOI] [PubMed] [Google Scholar]
- 46.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 47.Okada, M., T. Ogawa, H. Yoshizumi, H. Kubonoya, and K. Shinozaki. 2007. Genetic variation of the norovirus GII-4 genotype associated with a large number of outbreaks in Chiba prefecture, Japan. Arch. Virol. 152:2249-2252. [DOI] [PubMed] [Google Scholar]
- 48.Okada, M., T. Tanaka, M. Oseto, N. Takeda, and K. Shinozaki. 2006. Genetic analysis of noroviruses associated with fatalities in healthcare facilities. Arch. Virol. 151:1635-1641. [DOI] [PubMed] [Google Scholar]
- 49.Parker, T. D., N. Kitamoto, T. Tanaka, A. M. Hutson, and M. K. Estes. 2005. Identification of genogroup I and genogroup II broadly reactive epitopes on the norovirus capsid. J. Virol. 79:7402-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel, M. M., M. A. Widdowson, R. I. Glass, K. Akazawa, J. Vinje, and U. D. Parashar. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phan, T. G., T. Kuroiwa, K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, A. Yamamoto, K. Sugita, T. Nishimura, F. Yagyu, S. Okitsu, W. E. Muller, N. Maneekarn, and H. Ushijima. 2006. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 78:971-978. [DOI] [PubMed] [Google Scholar]
- 52.Prabakaran, P., J. Gan, Y. Feng, Z. Zhu, V. Choudhry, X. Xiao, X. Ji, and D. S. Dimitrov. 2006. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 281:15829-15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 54.Reeck, A., O. Kavanagh, M. K. Estes, A. R. Opekun, M. A. Gilger, D. Y. Graham, and R. L. Atmar. 2010. Serological correlate of protection against norovirus-induced gastroenteritis. J. Infect. Dis. 202:1212-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reuter, G., P. Pankovics, and G. Szucs. 2008. Genetic drift of norovirus genotype GII-4 in seven consecutive epidemic seasons in Hungary. J. Clin. Virol. 42:135-140. [DOI] [PubMed] [Google Scholar]
- 56.Schorn, R., M. Hohne, A. Meerbach, W. Bossart, R. P. Wuthrich, E. Schreier, N. J. Muller, and T. Fehr. 2010. Chronic norovirus infection after kidney transplantation: molecular evidence for immune-driven viral evolution. Clin. Infect. Dis. 51:307-314. [DOI] [PubMed] [Google Scholar]
- 57.Siebenga, J., A. Kroneman, H. Vennema, E. Duizer, and M. Koopmans. 2008. Food-borne viruses in Europe network report: the norovirus GII.4 2006b (for US named Minerva-like, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Euro Surveill. 13:8009. [PubMed] [Google Scholar]
- 58.Siebenga, J. J., P. Lemey, S. L. Kosakovsky Pond, A. Rambaut, H. Vennema, and M. Koopmans. 2010. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 6:e1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su, H. P., J. W. Golden, A. G. Gittis, J. W. Hooper, and D. N. Garboczi. 2007. Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein. Virology 368:331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Treanor, J. J., X. Jiang, H. P. Madore, and M. K. Estes. 1993. Subclass-specific serum antibody responses to recombinant Norwalk virus capsid antigen (rNV) in adults infected with Norwalk, Snow Mountain, or Hawaii virus. J. Clin. Microbiol. 31:1630-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu, E. T., R. A. Bull, G. E. Greening, J. Hewitt, M. J. Lyon, J. A. Marshall, C. J. McIver, W. D. Rawlinson, and P. A. White. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin. Infect. Dis. 46:413-420. [DOI] [PubMed] [Google Scholar]
- 63.Vinje, J., S. Altena, and M. Koopmans. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]
- 64.Wang, T. T., G. S. Tan, R. Hai, N. Pica, E. Petersen, T. M. Moran, and P. Palese. 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 6:e1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welsh, R. M., J. W. Che, M. A. Brehm, and L. K. Selin. 2010. Heterologous immunity between viruses. Immunol. Rev. 235:244-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 190:27-36. [DOI] [PubMed] [Google Scholar]
- 67.Yang, Y., M. Xia, M. Tan, P. Huang, W. Zhong, X. L. Pang, B. E. Lee, J. Meller, T. Wang, and X. Jiang. 2010. Genetic and phenotypic characterization of GII-4 noroviruses that circulated during 1987 to 2008. J. Virol. 84:9595-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.