Abstract
Transgenic expression of the RNA-dependent RNA polymerase 3Dpol inhibited infection of Theiler's murine encephalitis virus (TMEV), a picornavirus from which it was derived. Here, we infected 3Dpol transgenic mice with another picornavirus, as well as an alphaherpesvirus and a rhabdovirus. 3Dpol transgenic FVB mice had significantly lower viral loads and survived longer after infection with all three types of viruses than nontransgenic FVB mice. Viral inhibition among three different types of virus by transgenic 3Dpol suggests that the mechanism of action is not the direct interference with picornaviral 3Dpol but instead may be the changing of host cells to an antiviral state before or after viral infection occurs, as basal interferon levels were higher in 3Dpol transgenic mice before infection. Further study of this mechanism may open new possibilities for future antiviral therapy.
Viral infections cause many chronic devastating diseases, and acute viral infections can be life threatening. Currently, vaccinations are the main strategy for preventing viral epidemics. Although vaccination regimens are effective, we do not have vaccines for some common viral illnesses. Furthermore, an effective vaccine may protect 80 to 90% of the population, which still leaves 10 to 20% vulnerable to the illness. Therefore, much research has focused on finding novel antiviral therapies. Antiviral drugs have been and are presently being developed to inhibit viral infection. These include synthetic molecules that block viral entry or release, nucleoside analogs which inhibit viral RNA or DNA polymerases, and small interfering RNA (siRNA) strategies (4-6, 14). However, as is also the case for vaccinations, many of these drugs are highly virus specific. Furthermore, most antivirals developed to date have side effects or induce the development of mutated viruses that become resistant to treatment.
We showed that RNA-dependent RNA polymerase (3Dpol) from Theiler's murine encephalitis virus (TMEV) inhibited TMEV infection and also the pathological injury associated with persistent TMEV infection when expressed transgenically in mice via the human ubiquitin promoter (3). We further investigated other viral infections by using the 3Dpol transgenic mice to test whether viruses other than picornaviruses may be inhibited by this RNA polymerase. In this study, we infected 3Dpol transgenic and nontransgenic FVB mice with a picornavirus, encephalomyocarditis virus (EMCV), a alphaherpesvirus, pseudorabies virus (PRV), and a rhabdovirus, vesicular stomatitis virus (VSV).
EMCV (ATCC VR129B) is a cardiovirus in the picornavirus family. It is closely related to TMEV and contains a viral RNA-dependent RNA polymerase similar, although not identical, to that of TMEV. Therefore, we predicted that EMCV would be influenced by 3Dpol transgenic mice and would serve as a positive control. PRV 152 is an attenuated live vaccine strain of PRV (PRV Bartha) that contains an enhanced green fluorescent protein (EGFP)-expressing cassette cloned into the middle of the PRV gG gene (11). Despite its name, PRV has no relationship to rabies virus. Unlike rabies virus, which is a rhabdovirus, PRV is an alphaherpesvirus and has a large double-stranded DNA genome. PRV does not contain RNA-dependent RNA polymers, such as 3Dpol from TMEV. VSV (Indiana subtype) is an enveloped negative-strand RNA virus of the rhabdovirus family, and its 11.2-kb genome encodes only seven viral proteins (N, P, M, G, L, C, and C′) (8). This virus also expresses a green fluorescent protein that resides between the G and L genes (1). VSV does contain an RNA-dependent RNA polymerase; however, it is not structurally related to 3Dpol from picornaviruses.
Mice were infected with 40 PFU of EMCV intraperitoneally, 2 × 107 PFU of PRV 152 intramuscularly, and 2 × 103 PFU of VSV intracerebrally. All viruses chosen infect the central nervous system (CNS) by the route of infection mentioned above. We concentrated on quantifying viral load and injury in the CNS to utilize the strengths of established techniques in our laboratory. At a specified time point, mouse tissues were harvested for analysis of viral load or pathology, or the survival days for the mice were recorded. All experiments were done in accordance with the Mayo Clinic Institutional Animal Care and Use Committee guidelines.
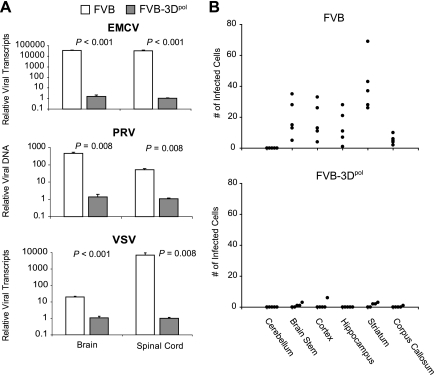
To determine whether infected transgenic 3Dpol mice had reduced viral loads, we harvested the brain and spinal cord at the indicated time points after infection with EMCV, PRV, and VSV (Fig. 1A) and quantified viral RNA or DNA by real-time quantitative PCR using SYBR green. All samples were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and relative viral RNA or DNA was calculated using the Livak method (10). Three days after infection with EMCV, 3Dpol transgenic mice had 10,000-fold fewer viral copies in both the brain (P = 0.008, rank sum) and spinal cord (P = 0.008, rank sum) than the wild-type mice. Six days after infection with PRV, 3Dpol transgenic mice contained 500-fold less viral DNA in the brain (P = 0.008, rank sum) and approximately 100-fold less viral DNA in the spinal cord (P = 0.008, rank sum) than wild-type mice. Likewise, after infection with the negative-stranded RNA virus VSV, we detected significantly less viral RNA in the brain (P < 0.001, Student's t test) and spinal cord (P = 0.008, rank sum) of 3Dpol transgenic mice than in those of wild-type mice 3 days after infection, with the brain containing 20-fold fewer viral transcripts in 3Dpol transgenic mice and the spinal cord containing >1,000-fold fewer transcripts than in wild-type FVB mice.
FIG. 1.
Viral load in 3Dpol transgenic and nontransgenic FVB mice after infection with EMCV, PRV, or VSV. (A) Relative viral RNA transcripts (EMCV and VSV) or DNA copies (PRV). Mice were harvested 3 days after infection with EMCV or VSV and 6 days after infection with PRV. Data were normalized to GAPDH for RNA quantitation and to genomic IL-2 for quantitation of PRV. All groups contained five mice. (B) Quantification of cells in areas of the brain expressing GFP from VSV infection after immunoperoxidase staining. Data are expressed as the number of positively stained cells in the ×40 magnification area of maximal pathology. One dot represents an individual infected mouse. FVB, nontransgenic mice; FVB-3Dpol, FVB 3Dpol transgenic mice.
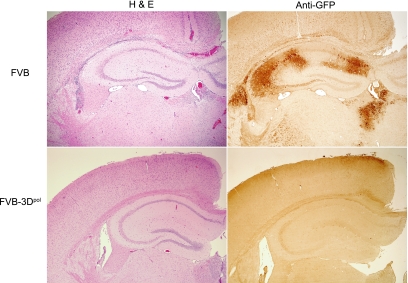
To confirm the reduction of viral load observed for infected 3Dpol transgenic mice by real-time quantitative PCR, we counted cells containing GFP in the brains of VSV-infected mice. Because the vesicular stomatitis virus expressed a green fluorescent protein, we stained tissue sections from paraffin-embedded sections of infected brain by immunoperoxidase using anti-GFP. Each area of the brain was scored, and data were expressed as the number of antigen-positive cells per 40× high-power field in areas of maximal pathology (Fig. 1B). 3Dpol transgenic mice contained little or no infected GFP-expressing cells 3 days after infection in all areas of the brain examined, whereas nontransgenic mice had significantly more virus-positive cells in the brain (P < 0.001, rank sum), with the majority of infected cells in the brain stem, cortex, hippocampus, and striatum. This is visually observed in the hippocampus from a representative VSV-infected nontransgenic compared to that of a 3Dpol transgenic mouse 3 days after infection (Fig. 2).
FIG. 2.
Hippocampus sections from a VSV-infected 3Dpol transgenic mouse versus a nontransgenic FVB mouse show reduced inflammatory infiltrates in 3Dpol transgenic mice (hematoxylin and eosin stain [H & E]) and reduced numbers of VSV-infected cells (anti-GFP immunoperoxidase stain).
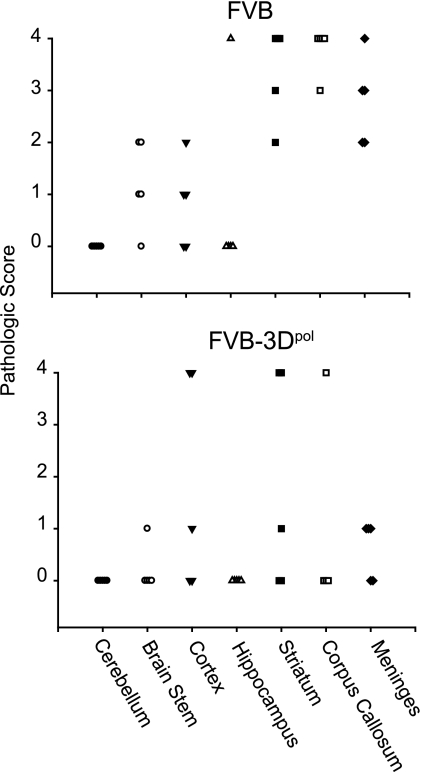
To determine if lower viral loads in VSV-infected 3Dpol transgenic mice also correlated with less inflammation and overall pathology in the brain, we scored day 3 infected mouse brains of 3Dpol transgenic and nontransgenic FVB mice. The methods have been previously described (7). Following fixation by intracardiac perfusion, each area of the brain was graded on a four-point scale as follows: 0, no pathology; 1, no tissue destruction and only minimal inflammation; 2, early tissue destruction (loss of architecture) and moderate inflammation; 3, definite tissue destruction (demyelination, parenchymal damage, cell death, neurophagia, and neuronal vacuolation); and 4, necrosis (complete loss of all tissue elements with associated cellular debris). The area with maximal tissue damage was used for the assessment of each brain region. 3Dpol transgenic mice had significantly less pathological injury, overall, in the brain (P = 0.006, rank sum) than that seen for nontransgenic mice (Fig. 3).
FIG. 3.
Pathological brain scores in mice 3 days after infection with VSV. Seven areas of the brain (cerebellum, brain stem, cortex, hippocampus, striatum, corpus callosum, and meninges) were graded based on a four-point scale as described in the text. Each point represents the score from an individual mouse.
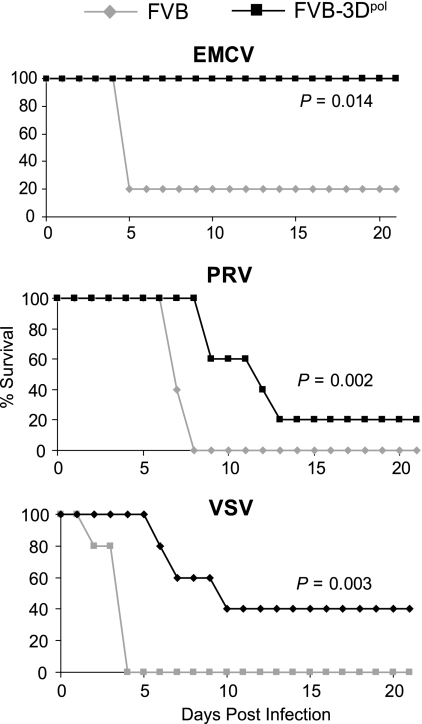
Ultimately, the best indication of transgenic 3Dpol's antiviral function is survival after injection of lethal amounts of virus. Therefore, mice were injected with each of the three viruses, and survival was recorded over a period of 21 days after infection. 3Dpol transgenic mice survived significantly longer after infection with each of the three viruses (Fig. 4). All five 3Dpol transgenic mice survived up to 21 days after infection with EMCV, whereas 4 of 5 nontransgenic mice died by day 5. Nontransgenic mice had a mean survival time of 7.4 ± 0.2 days after infection with the alphaherpesvirus PRV, whereas 3Dpol transgenic mice survived significantly longer (P = 0.002, log rank test), with a mean survival time of 12.8 ± 2.3 days. Likewise, 3Dpol transgenic mice, which had a mean survival time of 13.0 ± 3.7 days after infection with the rhabdovirus VSV, survived significantly longer (P = 0.003) than the nontransgenic FVB mice, which had a mean survival time of 3.6 ± 0.4 days.
FIG. 4.
Survival of 3Dpol transgenic mice versus nontransgenic mice after infection with EMCV, PRV, or VSV.
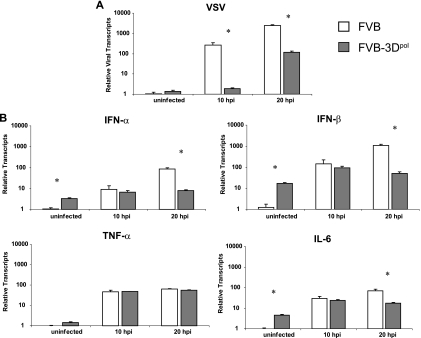
To observe the early innate immune responses of 3Dpol transgenic mice, we examined the RNA transcript levels of important early immune mediators of viral infection, alpha interferon (IFN-α), IFN-β, tumor necrosis factor alpha (TNF-α), and interleukin-6 (IL-6), before VSV infection and 10 and 20 h after infection from the brains of nontransgenic and 3Dpol transgenic mice (Fig. 5). Viral transcripts in 3Dpol transgenic mice show viral inhibition as early as 10 h after infection, with a P value of <0.001 (Fig. 5A). Of interest is the observation that type I interferon and IL-6 levels were significantly higher in 3Dpol transgenic mice before infection (Fig. 5B). After 10 h, levels of all cytokines measured were similar between nontransgenic and 3Dpol transgenic mice even though levels of viral transcripts were already significantly higher in nontransgenic FVB mice. It was not until 20 h after infection that the cytokine levels, with the exception of TNF-α, for nontransgenic mice eclipsed those for 3Dpol transgenic mice, likely because of higher viral loads in the nontransgenic mice. Further studies will be done to determine whether the higher levels of type I interferon transcripts in 3Dpol transgenic mice before infection is the main mediator of the antiviral effects in 3Dpol transgenic mice.
FIG. 5.
Viral transcripts and corresponding cytokine levels in 3Dpol transgenic and nontransgenic FVB mice before infection and 10 or 20 h after infection with VSV. (A) Relative viral RNA transcripts in the brain after VSV infection. (B) Relative cytokine transcripts in the brain after VSV infection. Data were normalized to GAPDH and are relative to the background threshold cycle (CT) values obtained for brains of uninfected nontransgenic mice. The numbers of mice used were as follows: uninfected, 4 nontransgenic mice (FVB) and 4 uninfected FVB 3Dpol transgenic mice (FVB-3Dpol); 10 hours postinfection (hpi), 4 FVB and 3 FVB-3Dpol; 20 hpi, 3 FVB and 4 FVB-3Dpol. *, statistical significance, with a P value of <0.05 by Student's t test (unequal variance).
It was of interest that transgenic plants expressing viral RNA-dependent RNA polymerases have been shown to inhibit specific viral infections (2, 9). Viral inhibition in replicase transgenic plants is thought to occur from protein-mediated (dominant negative effects) or RNA-mediated (RNA interference) mechanisms (12, 13). In effect, the replicase transgenic plants inhibit viral infection only from the virus from which the transgene was derived or from closely related viruses.
Previously, we showed that transgenic expression of 3Dpol polymerase in mice inhibited infection of the virus from which this polymerase was derived (TMEV). We also provided evidence that this inhibitory effect on TMEV was not the consequence of a change in the adaptive immune response to TMEV, as Rag knockout of 3Dpol transgenic mice still resulted in decreased viral loads and increased survival after TMEV infection. Furthermore, we determined that this effect was not due to an “insertional effect” of the transgene, as two separate embryonic mouse lines showed a similar lack of disease pathology in the spinal cord after infection with TMEV. In a previous study, we hypothesized that this effect may be seen only for picornaviruses or viruses containing a similar 3Dpol RNA-dependent RNA polymerase due to a possibly less functional transgenic 3Dpol competing for viral 3Dpol in the viral replication complex (3). Our study directly tested this hypothesis by using one picornavirus and two nonpicornaviruses for infection. We showed that the hypothesis is not true, as 3Dpol transgenic mice have a significantly lower viral load and live longer after infection with three different types of viruses. We now propose that the mechanism by which 3Dpol polymerase acts is through changing some or all host cell types to an antiviral state, possibly by stimulating some aspect of the nonadaptive immune response before or soon after viral infection. Finding this mechanism may open new avenues for antiviral research and potential strategies for broad antiviral therapies in the future.
Footnotes
Published ahead of print on 20 October 2010.
REFERENCES
- 1.Fernandez, M., M. Porosnicu, D. Markovic, and G. N. Barber. 2002. Genetically engineered vesicular stomatitis virus in gene therapy: application for treatment of malignant disease. J. Virol. 76:895-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golemboski, D. B., G. P. Lomonossoff, and M. Zaitlin. 1990. Plants transformed with a tobacco mosaic virus nonstructural gene sequence are resistant to the virus. Proc. Natl. Acad. Sci. U. S. A. 87:6311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerkvliet, J., L. Zoecklein, L. Papke, A. Denic, A. J. Bieber, L. R. Pease, C. S. David, and M. Rodriguez. 2009. Transgenic expression of the 3D polymerase inhibits Theiler's virus infection and demyelination. J. Virol. 83:12279-12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonard, J. N., and D. V. Schaffer. 2006. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 13:532-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lew, W., X. Chen, and C. U. Kim. 2000. Discovery and development of GS 4104 (oseltamivir): an orally active influenza neuraminidase inhibitor. Curr. Med. Chem. 7:663-672. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien, J. J., and D. M. Campoli-Richards. 1989. Acyclovir: an updated review of its antiviral activity, pharmacokinetic properties and therapeutic efficacy. Drugs 37:233-309. [DOI] [PubMed] [Google Scholar]
- 7.Pavelko, K. D., C. L. Howe, K. M. Drescher, J. D. Gamez, A. J. Johnson, T. Wei, R. M. Ransohoff, and M. Rodriguez. 2003. Interleukin-6 protects anterior horn neurons from lethal virus-induced injury. J. Neurosci. 23:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peluso, R. W., J. C. Richardson, J. Talon, and M. Lock. 1996. Identification of a set of proteins (C′ and C) encoded by the bicistronic P gene of the Indiana serotype of vesicular stomatitis virus and analysis of their effect on transcription by the viral RNA polymerase. Virology 218:335-342. [DOI] [PubMed] [Google Scholar]
- 9.Prins, M., M. Laimer, E. Noris, J. Schubert, M. Wassenegger, and M. Tepfer. 2008. Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9:73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmittgen, T. D., and K. J. Livak. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101-1108. [DOI] [PubMed] [Google Scholar]
- 11.Smith, B. N., B. W. Banfield, C. A. Smeraski, C. L. Wilcox, F. E. Dudek, L. W. Enquist, and G. E. Pickard. 2000. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. U. S. A. 97:9264-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenllado, F., I. Garcia-Luque, M. T. Serra, and J. R. Diaz-Ruiz. 1995. Nicotiana benthamiana plants transformed with the 54-kDa region of the pepper mild mottle tobamovirus replicase gene exhibit two types of resistance responses against viral infection. Virology 211:170-183. [DOI] [PubMed] [Google Scholar]
- 13.Tenllado, F., I. Garcia-Luque, M. T. Serra, and J. R. Diaz-Ruiz. 1996. Resistance to pepper mild mottle tobamovirus conferred by the 54-kDa gene sequence in transgenic plants does not require expression of the wild-type 54-kDa protein. Virology 219:330-335. [DOI] [PubMed] [Google Scholar]
- 14.Yin, Z., Y. L. Chen, W. Schul, Q. Y. Wang, F. Gu, J. Duraiswamy, R. R. Kondreddi, P. Niyomrattanakit, S. B. Lakshminarayana, A. Goh, H. Y. Xu, W. Liu, B. Liu, J. Y. Lim, C. Y. Ng, M. Qing, C. C. Lim, A. Yip, G. Wang, W. L. Chan, H. P. Tan, K. Lin, B. Zhang, G. Zou, K. A. Bernard, C. Garrett, K. Beltz, M. Dong, M. Weaver, H. He, A. Pichota, V. Dartois, T. H. Keller, and P. Y. Shi. 2009. An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 106:20435-20439. [DOI] [PMC free article] [PubMed] [Google Scholar]