Abstract
Human immunodeficiency virus (HIV)-positive individuals can be superinfected with different virus strains. Individuals who control an initial HIV infection are therefore still at risk for subsequent infection with divergent viruses, but the barriers to such superinfection remain unclear. Here we tested long-term nonprogressors' (LTNPs') susceptibility to superinfection using Indian rhesus macaques that express the major histocompatibility complex class I (MHC-I) allele Mamu-B*17, which is associated with control of the pathogenic AIDS virus SIVmac239. The Mamu-B*17-restricted CD8+ T cell repertoire is focused almost entirely on 5 epitopes. We engineered a series of SIVmac239 variants bearing mutations in 3, 4, or all 5 of these epitopes and used them to serially challenge 2 Mamu-B*17-positive LTNPs. None of the escape variants caused breakthrough replication in LTNPs, although they readily infected Mamu-B*17-negative naive macaques. In vitro competing coculture assays and examination of viral evolution in hosts lacking Mamu-B*17 suggested that the mutant viruses had negligible defects in replicative fitness. Both LTNPs maintained robust immune responses, including simian immunodeficiency virus (SIV)-specific CD8+ and CD4+ T cells and neutralizing antibodies. Our results suggest that escape mutations in epitopes bound by “protective” MHC-I molecules may not be sufficient to establish superinfection in LTNPs.
The correlates of protection against human immunodeficiency virus (HIV) remain unknown, despite 25 years of research. For this reason there has been increasing interest in individuals who effectively control virus replication. Long-term nonprogressors (LTNPs) are HIV-infected individuals who remain clinically healthy for years and even decades. A subset of LTNPs, “elite controllers” (ECs), spontaneously control HIV replication in plasma to levels below the threshold of detection in commercial assays, currently 50 viral genomic RNA (vRNA) copies/ml plasma (12). Defining the mechanisms by which LTNPs and ECs establish and maintain effective control over virus replication, as well as understanding potential limits to this control, may provide critical insights into the types of immune responses that successful HIV vaccines should elicit.
Several lines of evidence suggest that virus-specific CD8+ T cell responses play a key role in the effective control of HIV replication. Resolution of acute viremia is temporally associated with the appearance of CD8+ T cell responses in most subjects (5, 27). Elite control is associated with expression of certain human leukocyte antigen (HLA) class I alleles, in particular HLA-B27 and -B57 (7, 8, 13, 18, 20, 38). CD8+ T cell populations restricted by these molecules are immunodominant during acute infection (1) and frequently select for escape mutant viruses (14, 25, 28, 46). Notably, when such escape mutant viruses are transmitted to HLA-B27- or HLA-B57-positive individuals, these individuals are frequently unable to control virus replication, despite their expression of “protective” HLA alleles (10, 19).
An animal model of HIV control can complement studies of human LTNPs/ECs in several key ways, including unequivocal knowledge of the infecting virus sequence and the timing of infection, as well as providing opportunities to perform interventions that cannot be done in humans. It is therefore striking that a subset of Indian-origin rhesus macaques (Macaca mulatta) are able to durably control replication of pathogenic simian immunodeficiency viruses (SIVs) in a fashion similar to that of human LTNPs/ECs. Control of SIV replication is associated with the major histocompatibility complex class I (MHC-I) alleles Mamu-B*17 (59) and -B*08 (30). Transient depletion of CD8+ cells in ECs resulted in a loss of containment of virus replication, and control was reestablished when CD8+ cells repopulated the periphery (17). This animal model has thus provided further evidence that ongoing CD8+ T cell responses are critical for maintaining durable control over AIDS virus replication.
Studies of both humans and macaques have suggested that individuals who maintain low viremia after an initial immunodeficiency virus challenge can be superinfected with viruses whose sequences diverge from that of the initial infecting virus. Macaques vaccinated with a live attenuated SIV initially controlled challenge with a divergent pathogenic virus isolate but later experienced breakthrough viremia and progressed to AIDS. The breakthrough viruses had mosaic genomes resulting from multiple recombination events between the vaccine and challenge strains, which yielded viruses capable of persistent high-titer replication (50). A human subject who maintained virus loads below 5,000 copies/ml plasma following structured therapy interruption in the acute phase of HIV infection was later superinfected with a second clade B virus with sequence differences in multiple epitopes recognized by his CD8+ T cells, which caused a marked increase in viremia (2). A subsequent study of breakthrough virus replication showed that loss of control over HIV replication was the result of superinfection and subsequent selection for recombinant viruses bearing escape mutations in immunodominant CD8+ T cell epitopes (52).
Here we tested the hypothesis that ECs and LTNPs are susceptible to challenge with viruses bearing mutations in CD8+ T cell epitopes bound by “protective” MHC-I molecules. We reasoned that challenge with viruses harboring consensus escape mutations in Mamu-B*17-restricted epitopes could “dissect out” the CD8+ T cell populations responsible for durable control of SIVmac239, resulting in superinfection. The Mamu-B*17-restricted CD8+ T cell repertoire is focused on 5 epitopes in most LTNPs, ECs, and normal progressors expressing this molecule (34). We therefore constructed a series of SIVmac239 variants encoding escape mutations in Mamu-B*17-restricted epitopes and used them to challenge Mamu-B*17-positive macaques, one an LTNP and one an EC, that had controlled an initial infection with SIVmac239.
MATERIALS AND METHODS
Animals.
Rhesus macaques (Macaca mulatta) of Indian descent were housed at the Wisconsin National Primate Research Center (WNPRC), assigned to institutional animal care and use committee (IACUC)-approved protocols and cared for according to the National Research Council's Guide for the Care and Use of Laboratory Animals (39). Animals were screened for the presence of a panel of MHC-I alleles by PCR with sequence-specific primers (PCR-SSP) as described previously (24). Mamu-B*17-positive elite controllers rhAJ11 and r95071 were infected intrarectally with wild-type SIVmac239 nefopen (49) as part of previous studies (37, 59).
Viruses and inoculations.
For this study, animals were inoculated intravenously with 100 50% tissue culture infective doses (TCID50) of SIVmac239 variant viruses. After infection, plasma virus loads were measured using a quantitative reverse transcription (QRT)-PCR assay targeting the gag gene as described previously (9, 54).
To produce variant viruses bearing escape mutations in Mamu-B*17-restricted CD8+ T cell epitopes, we first identified nonsynonymous substitutions that commonly occurred in Mamu-B*17-positive progressor macaques with high-level chronic viremia (15, 34, 41; G. E. May, E. J. León, N. J. Maness, and T. C. Friedrich, unpublished data). We reasoned that these mutations would be most likely to confer effective escape from Mamu-B*17-restricted CD8+ T cells while minimizing costs to viral replicative fitness. The wild-type and mutant sequences of Mamu-B*17-restricted epitopes are presented in Table 1. We introduced targeted substitutions into the SIVmac239 molecular clone by the use of mutagenic primers with the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX). Cycling conditions for mutagenesis reactions were as follows: 95° for 1 min; then 15 cycles of 95° for 50 s, 60° for 50 s, and 68° for 10 min; and a final extension of 68° for 7 min. To rescue infectious SIV viruses, we transfected Vero cells with plasmid DNA, cocultured these cells with CEMx174 cells for 2 days, and then harvested viruses in cell-free supernatant approximately 6 days later. This procedure has previously been described in detail (15, 54).
TABLE 1.
Sequences and Mamu-B*17 binding efficiencies of wild-type and mutant epitope peptides
| SIV strain | Epitope | Sequencea | IC50 (nM) | Reference |
|---|---|---|---|---|
| Wild type | Vif HW8 | HLEVQGYW | 29 | 35 |
| 3×, 4×, 5× | Vif HW8 H1Y | YLEVQGYW | 434 | 35 |
| Wild type | Env FW9 | FHEAVQAVW | 7 | 35 |
| 3×, 4×, 5× | Env FW9 H2Y | FYEAVQAVW | 1,313 | 35 |
| Wild type | Nef IW9 | IRYPKTFGW | 7.6 | 16 |
| 3×, 4×, 5× | Nef IW9 I1T, T6I | TRYPKIFGW | 339 | 16 |
| Wild type | Nef MW9 | MHPAQTSQW | 3.7 | 35 |
| 4×, 5× | Nef MW9 M1V | VHPAQTSQW | 294 | 35 |
| Wild type | Cryptic RW9 | RHLAFKCLW | 32 | 34 |
| 5× | Cryptic RW9 W9R | RHLAFKCLR | 948 | 34 |
Bold type indicates positions of sequence substitution.
Competition assays to measure viral replicative fitness.
We assessed the replicative fitness of cloned mutant viruses using a newly developed in vitro competing coculture assay. Briefly, we produced a reference virus bearing a genetic “barcode” of synonymous substitutions in gag that abrogated binding of the primers and probes used in our standard QRT-PCR diagnostic assay. Similarly, primers and probes specific for the “barcoded” virus, which we termed SIVmac239-10s, do not amplify wild-type SIVmac239 sequences, allowing for specific quantitation of wild-type and reference viruses in the same sample. In 7-day competing coculture assays, SIVmac239-10s showed no detectable fitness defect in comparison with wild-type SIVmac239, as we recently described (54).
To compare the growth levels of mutant viruses and SIVmac239-10s, fresh peripheral blood mononuclear cells (PBMC) were isolated from an SIV-naive rhesus macaque and incubated overnight at 37°C in RPMI containing 10% fetal bovine serum (FBS) and 5 μg/ml phytohemagglutinin (PHA). Approximately 18 h later, PBMC were washed twice with phosphate-buffered saline (PBS) and resuspended at 4 million cells/ml in R10 containing 20 units interleukin-2 (IL-2) per ml (R10-20). Two million PBMC were then infected with a total amount of virus equivalent to of 50 million vRNA copies by adding the appropriate volume of virus and incubating at 37°C for 4 h. PBMC were then washed twice with R10 to remove the virus and plated in duplicate in a 24-well plate at one million PBMC/well in 2 ml R10-20. Viral supernatant (1 ml) was harvested at days 1, 3, 5, and 7 postinfection. Absolute quantification of SIVmac239-10s and mutant viruses was performed using duplicate samples in a QRT-PCR as described previously (54).
Tracking evolution of viruses in vivo.
vRNA was extracted from plasma using the Qiagen MinElute kit (Qiagen, Valencia, CA) or by guanidium thiocyanate extraction. Overlapping amplicons representing all SIVmac239 open reading frames (ORF) were amplified using the Qiagen one-step RT-PCR kit and SIVmac239-specific primer pairs as previously described (17, 29). cDNA was sequenced on a 3730 DNA analyzer (Applied Biosystems, Foster City, CA), and sequences were assembled using CodonCode Aligner (CodonCode, Deadham, MA).
Tracking cellular immune responses: intracellular cytokine staining.
We used intracellular cytokine staining as previously described (17) to enumerate lymphocytes secreting cytokines to SIV antigens in elite controllers. Briefly, freshly isolated PBMC were incubated with 5 μM peptide(s) and antibodies against CD28 (clone L293) and CD49d (clone 9F10) for 1.5 h at 37°C, after which 10 μg brefeldin A was added to halt extracellular transport. Cells were incubated a further 5 h, washed and stained for surface expression of CD4 (allophycocyanin [APC]-labeled clone SK3) and CD8 (peridinin chlorophyll protein [PerCP]-labeled clone SK1), and fixed overnight in 1% paraformaldehyde at 4°C. The following day, cells were permeabilized in buffer containing 0.1% saponin and stained for expression of the cytokines gamma interferon (IFN-γ; fluorescein isothiocyanate [FITC]-labeled clone 4S.B3) and IL-2 (phycoerythrin [PE]-labeled clone MQ1-17H12). All antibodies were obtained from BD Biosciences and were titrated for optimal staining. After intracellular staining, PBMC were fixed in 1% paraformaldehyde for 2 h at 4°C. Events were then collected on a FACSCalibur flow cytometer and analyzed with FlowJo software (Tree Star, Ashland, OR).
Tracking cellular immune responses: ELISPOT.
We used an IFN-γ enzyme-linked immunospot (ELISPOT) assay to measure responses to Mamu-B*17-restricted CD8+ T cell epitopes. Assays were performed as described previously (57), using 100,000 PBMC and 10 μM synthetic peptide (or 10-fold serial dilutions of peptide) per well in precoated ELISpotPLUS kits (Mabtech, Cincinnati, OH). All samples were run in duplicate, and negative (no peptide) and positive (concanavalin A) controls were included on each plate.
Neutralization assays.
Pseudoviruses were generated in 293T cells, and neutralization with single-round infectious SIVmac239 pseudovirus was performed using TZM-bl cells as targets for infection as described previously (60). Briefly, pseudovirus was previously titrated on TZM-bl cells, and a predetermined amount of virus that produced ∼300,000 relative light units (RLUs) was incubated for 1 h at 37°C with serially diluted sera and control CD4 IgG2. TZM-bl cells were resuspended in medium containing a final concentration of dextran at 20 μg/ml, washed, counted, and plated at 1 × 105 cells per well over the incubated solution of virus and sera for an additional 48 h. The degree of virus neutralization by macaque serum was calculated by measuring luciferase activity. The wells were aspirated and washed once with PBS, and 60 μl of luciferase cell culture lysis reagent (Promega, Madison, WI) was added. The lysate was mixed by pipetting, 50 μl was transferred to a round-bottom plate (Corning, Corning, NY), and the plate was centrifuged at 1,800 × g for 10 min at 4°C. Twenty μl was transferred to an opaque assay plate (Corning), and the luciferase activity was measured on a luminometer (EG&G Berthold LB 96V; Perkin Elmer, Gaithersburg, MD) by using a luciferase assay reagent (Promega, Madison, WI).
Enzyme-linked immunosorbent assays (ELISAs) for detection of Env-binding antibodies.
Recombinant gp130 SIVmac239 was coated onto the wells of microtiter plates (Corning) at a concentration of 5 μg/ml by incubation overnight at 4°C. The plates were washed four times with PBS-0.05% Tween 20 and blocked with 4% milk solution (Bio-Rad, Hercules, CA). After washing, serial dilutions of serum samples were applied to the plate and incubated for 2 h at room temperature. A CD4 IgG2 antibody standard curve was run on each plate. The plates were washed again, goat anti-human IgG Fc fragment-specific antibody coupled to alkaline phosphatase (Pierce/Thermo Fisher Scientific, Rockford, IL) was added, and the plates were incubated for 1 h at room temperature. The plates were washed, and bound conjugate was detected with p-nitrophenyl phosphate substrate (Sigma, St. Louis, MO).
RESULTS
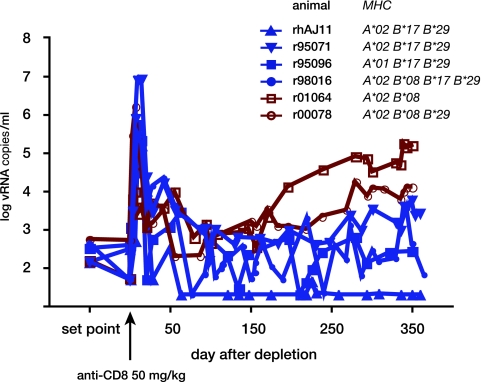
In a previous study, we transiently depleted CD8+ cells from the periphery in a cohort of rhesus macaques of Indian descent that had exhibited elite control of pathogenic SIVmac239 (17). The treatment interrupted the animals' ability to control SIV replication, indicating that ongoing CD8+ cell responses were required to maintain EC status. For the present study, we monitored long-term trends in viremia in CD8+ cell-depleted ECs. Control over SIV replication appeared to be more stable following transient CD8+ cell depletion in the Mamu-B*17-positive ECs than in ECs that did not express this allele (Fig. 1). We therefore further examined the mechanism of MHC-I-associated elite control after CD8+ cell depletion in Mamu-B*17-positive ECs. For this study, we used 2 Mamu-B*17-positive ECs that also expressed the high-frequency MHC-I allele Mamu-A*02. Both of these animals maintained extremely low plasma viremia prior to CD8+ cell depletion (geometric mean chronic phase virus load of <200 copies/ml) (17). Following CD8+ cell depletion, one animal, rhAJ11, controlled virus replication to a level below the limit of detection. In the other animal, r95071, a new steady-state level of viremia of ∼10,000 copies/ml was established. Therefore, although it maintained low viremia after CD8+ cell depletion, r95071 could no longer be considered an EC. For this study we will refer to both animals using the broader terms “LTNPs” or “controllers.”
FIG. 1.
SIVmac239 replication in elite controller macaques during and after transient depletion of CD8+ cells. Six SIV elite controllers were transiently depleted of CD8+ cells in a previous study (17) and were followed for a year afterwards for the present study. Plasma viremia was measured using a quantitative RT-PCR assay targeting SIV gag. Mamu-B*17-positive ECs are shown in blue traces with solid symbols. Mamu-B*17-negative animals are depicted with red traces and open symbols. Known MHC-I alleles expressed by each EC are listed. “Set point” represents the geometric mean plasma virus load for each animal from 12 weeks postinfection until 4 weeks prior to CD8+ cell depletion. Anti-CD8 antibody was administered on study day 0.
Few fixed amino acid replacements in virus replicating in Mamu-B*17-positive ECs.
We previously determined that the SIV-specific Mamu-B*17-restricted CD8+ T cell repertoire is restricted to 5 epitopes in the majority of animals infected with SIVmac239 (34), so we focused our analysis on regions of the viral genome encoding these epitopes. We found few complete substitutions in Mamu-B*17-restricted epitope sequences in ECs expressing this allele in recrudescent virus replicating during the period of CD8+ cell depletion: only the Nef IW9 epitope, which is immunodominant in acute infection of Mamu-B*17-positive animals, harbored substitutions in all 4 Mamu-B*17-positive ECs in our study (Fig. 2). In addition, a histidine-to-glutamine (H-to-Q) substitution at position 2 of the Env FW9 epitope appeared to be fixed in the virus of one animal, r95096, at both 2 weeks and 1 year following CD8+ cell depletion (Fig. 2). Although we observed apparently complete substitutions in other epitopes 2 weeks after CD8+ cell depletion, they were lost over time in replicating virus. These results are consistent with an earlier study in which we observed infrequent amino acid substitutions in Mamu-B*17-positive ECs (34). They also suggest a paradox: Mamu-B*17-restricted CD8+ T cell responses appear to be associated with effective control of SIV replication, are focused on a relatively narrow repertoire, and yet do not seem to exert strong selective pressure on replicating viruses. Since Mamu-B*17-restricted CD8+ T cell epitope sequences do accumulate mutations in Mamu-B*17-positive progressors, we next asked whether viruses encoding such mutations might establish breakthrough infection in Mamu-B*17-positive controllers.
FIG. 2.
Few amino acid substitutions in Mamu-B*17-restricted epitopes become fixed in controllers. We directly sequenced plasma virus amplicons from Mamu-B*17-positive and -negative controllers at 2 weeks and 1 year after CD8+ cell depletion to identify nonsynonymous substitutions in the 5 epitopes constituting the Mamu-B*17-restricted repertoire. Codons matching the SIVmac239 sequence are represented with dots. Nonsynonymous substitutions are shown by the corresponding single-letter amino acid codes. Capital letters indicate complete replacement as detected by Sanger sequencing, while lowercase letters indicate sites of mixed-base heterogeneity. “na” indicates that we were unable to amplify viral cDNA from animal rhAJ11, whose virus load was undetectable throughout most of the study period.
Mamu-B*17-positive LTNPs resist rechallenge with escape mutant SIV.
We used site-directed mutagenesis to produce variants of the SIVmac239 molecular clone encoding epitope substitutions that had been observed in Mamu-B*17-positive macaques with persistently high viremia (Table 1). When multiple escape variants had been identified for a particular epitope, we introduced substitutions that reduced binding to Mamu-B*17 by the greatest degree in in vitro assays.
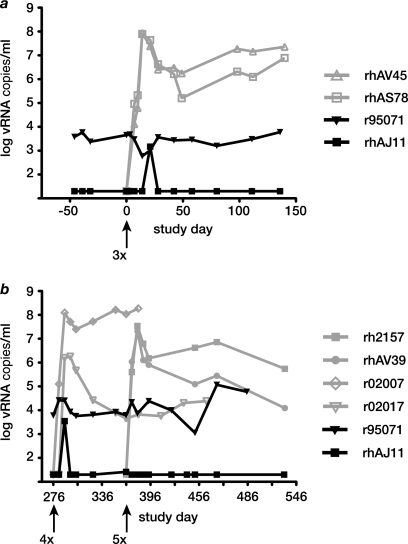
In total, we produced 3 mutant SIV virus species bearing mutations in 3, 4, or all 5 CD8+ T cell epitopes that constitute the Mamu-B*17-restricted repertoire (called 3×SIV, 4×SIV, or 5×SIV). We then serially rechallenged 2 Mamu-B*17-positive controllers with each of these viruses, each time also challenging 2 Mamu-B*17-negative control animals that had not previously been infected with SIV. To ensure a stringent challenge, we administered 100 TCID50 of virus intravenously. 3×SIV encoded putative escape substitutions in Vif HW8, Env FW9, and Nef IW9 (Table 1). This virus caused persistent high viremia in 2 Mamu-B*17-negative control animals. In contrast, virus loads in the controllers remained steady for more than 6 months following 3×SIV challenge, except for a single “blip” of 1,500 copies/ml plasma 21 days after 3×SIV challenge in animal rhAJ11 (Fig. 3a).
FIG. 3.
Engineered escape mutant viruses replicate in Mamu-B*17-negative control animals but do not cause breakthrough viremia in Mamu-B*17-positive LTNPs. We inoculated 2 Mamu-B*17-positive LTNPs with a series of engineered escape variants of SIVmac239. Each inoculation was 100 TCID50 given intravenously. Two previously uninfected Mamu-B*17-negative control animals were also inoculated with each mutant virus. Mutant viruses were inoculated at time points indicated with arrows. (a) 3×SIV productively infected and caused high-level persistent viremia in 2 control animals (gray traces and open symbols), while virus loads in the 2 LTNPs (black traces and solid symbols) were unchanged. (b) Viremia remained steady in LTNPs (black) after challenge with 4×SIV and 5×SIV, while both of these viruses caused persistent infection of previously uninfected controls (gray).
We next challenged the LTNPs with 4×SIV, which harbored the same mutations as 3×SIV, as well as a substitution in the subdominant Nef MW9 epitope. Animal rhAJ11's CD8+ T cell repertoire had been focused exclusively on this epitope immediately following CD8+ cell depletion treatment (17), so we reasoned that 4×SIV might be more likely than 3×SIV to cause breakthrough virus replication in LTNPs. However, this virus also failed to disrupt the LTNPs' control over SIV replication, although it replicated in Mamu-B*17-negative control animals (Fig. 3b). 5×SIV, which encodes all escape mutations in 4×SIV as well as a substitution in the cRW9 epitope encoded by a cryptic open reading frame overlapping the env ORF (33), was our final challenge virus. We inoculated the 2 Mamu-B*17-positive controllers with 5×SIV 90 days after the 4×SIV challenge. 5×SIV also failed to cause breakthrough viremia in the 2 Mamu-B*17-positive controllers, despite its ability to replicate to high titer in vivo in previously uninfected Mamu-B*17-negative animals (Fig. 3b).
Animal r95071 experienced increasing viremia beginning approximately 3 months after 5×SIV challenge (Fig. 3b). Interestingly, however, we did not detect sequences representing 3×SIV, 4×SIV, or 5×SIV in either controller at any time point after inoculation (data not shown). Together, our data indicate that viruses bearing escape substitutions in Mamu-B*17-restricted epitopes are replication competent in vivo, but these substitutions do not confer the ability to superinfect Mamu-B*17-positive LTNPs. Furthermore, mutant virus challenge does not appear to have influenced the eventual failure of immune control in the viremic LTNP animal r95071.
Substitutions in Mamu-B*17-restricted epitopes have negligible effects on viral fitness.
We next assessed the replicative abilities of the mutant viruses in competing coculture with a reference virus using an assay we recently described (54). When 3×SIV, 4×SIV, or 5×SIV was inoculated either in equal proportions to the reference virus (SIVmac239-10s) or in 10-fold excess, we detected a small downward trend in the proportion of epitope mutant viruses in the culture over the 7-day assay, although these differences were not significant (Fig. 4a). These results suggest that 3×SIV, 4×SIV, and 5×SIV are not seriously impaired in their growth capacities, but it is possible that more subtle fitness differences would have been detectable in competing coculture assays lasting longer than 7 days.
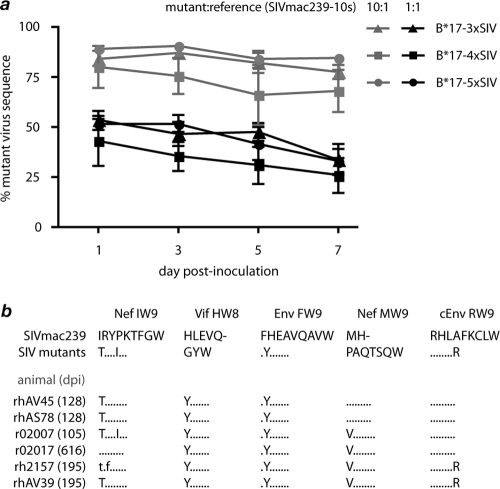
FIG. 4.
Mutations in Mamu-B*17-restricted CD8+ T cell epitopes exact negligible costs to viral fitness. We assessed the fitness of engineered escape mutant viruses in vitro using a novel competing coculture assay that we recently described (54) and in vivo by sequencing plasma virus replicating in chronically infected Mamu-B*17-negative control animals. (a) Mutant viruses were mixed at defined ratios with the reference strain SIVmac239-10s and used to infect macaque PBMC, and each viral species was quantified in separate QRT-PCRs at the indicated time points after inoculation as described in Materials and Methods. We calculated the proportion of mutant virus species present in each sample by dividing the number of mutant virus genome copies by the number of reference virus copies. Shown are data from four independent experiments with inoculum ratios of mutant to reference of 10:1 (gray) or 1:1 (black). Error bars indicate standard error of the mean for each time point. (b) We sequenced regions encoding the 5 Mamu-B*17-restricted CD8+ T cell epitopes in plasma virus from Mamu-B*17-negative animals infected with 3×SIV, 4×SIV, or 5×SIV at the indicated number of days postinfection (dpi).
Mutations in HIV and SIV epitopes that are under evolutionary constraints frequently revert to wild type upon transmission to individuals that lack the MHC-I allele that restricts the selecting CD8+ T cell response (6, 15, 28, 32). We therefore sequenced SIV genomic regions encoding Mamu-B*17-restricted epitopes in plasma from the 6 previously uninfected animals during chronic infection with 3×SIV, 4×SIV, or 5×SIV. The engineered escape mutations were completely maintained in 4 of 5 epitopes in the absence of Mamu-B*17-mediated selection pressure (Fig. 4b). The only epitope in which we found evidence of reversion to the wild type was Nef IW9. One of the engineered substitutions (epitope position 1 threonine to isoleucine; T1I) was lost over time in 5 of 6 Mamu-B*17-negative animals, while the other, I6T, reverted completely to wild type only in r02017, the animal that survived the longest following SIV infection (Fig. 4b). Strikingly, all engineered viral mutations were completely preserved in animal r02007, which was euthanized 105 days after infection with extremely high virus loads (>1.8 × 108 copies/ml) (Fig. 3b) and signs of rapidly progressive disease. These mutations in Mamu-B*17-restricted CD8+ T cells thus do not prevent high-titer virus replication and do not frequently revert in the absence of Mamu-B*17-mediated selection in vivo. Together, our results suggest that the putative escape mutations used in our study, including those in Nef IW9, exact negligible costs to viral replicative fitness.
Cellular immune response breadth and magnitude following serial rechallenge of LTNPs.
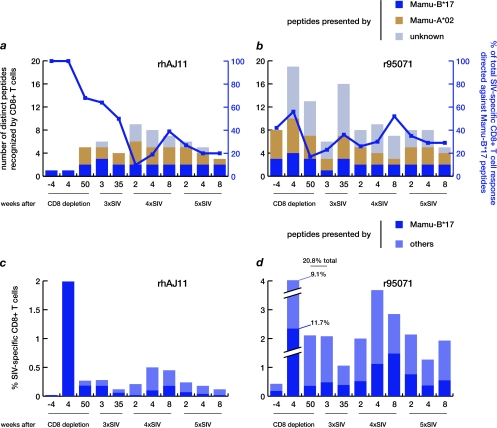
When CD8+ T cells repopulated the periphery after transient depletion in our earlier study, we observed high-frequency responses in ECs against epitopes that had been subdominant in acute infection (17). In animal rhAJ11, the returning CD8+ T cell response was focused on a single epitope, the Mamu-B*17-restricted Nef MW9 (Fig. 5a). In contrast, animal r95071 recognized at least 19 different SIVmac239 epitopes (4 presented by Mamu-B*17, 6 by Mamu-A*02, and 9 by unknown MHC-I molecules) (Fig. 5b). In this study, intracellular cytokine staining (ICS) assays detecting IFN-γ revealed that prior to challenge with the first mutant virus, animal rhAJ11's CD8+ T cell repertoire had broadened to include 5 distinct epitopes (2 Mamu-B*17 and 3 Mamu-A*02) (Fig. 5a), while r95071's repertoire had narrowed to focus on 13 (3 Mamu-B*17, 4 Mamu-A*02, and 6 of unknown restriction) (Fig. 5b). Interestingly, both the epitopic breadth (Fig. 5a and b) and the total magnitude (Fig. 5c and d) of CD8+ T cell responses remained relatively constant following challenge with each mutant virus. Strikingly, however, the contribution of Mamu-B*17-restricted CD8+ T cells to the total SIV-specific cellular immune response waned dramatically in rhAJ11 over the course of the study. Before challenge with 3×SIV, Mamu-B*17-restricted populations accounted for over 60% of the total SIV peptide-specific CD8+ T cell response by magnitude in this animal, but by 8 weeks after challenge with 5×SIV, Mamu-B*17-restricted cells made up only 20% of the total CD8+ T cell response (Fig. 5a and c). In contrast, the proportion of Mamu-B*17-restricted SIV-specific CD8+ T cells in r95071 oscillated around a mean of 35% of the total response by magnitude throughout the rechallenge study (Fig. 5b and d).
FIG. 5.
Breadth and magnitude of SIV-specific CD8+ T cell responses in LTNPs following serial challenge with escape mutant SIV. SIV-specific CD8+ T cell responses were assessed by stimulating PBMC with pooled synthetic peptides representing the entire SIVmac239 proteome as well as individual peptides bound by Mamu-B*17 and Mamu-A*02, which were both expressed by both LTNPs. Expression of IFN-γ was assessed by intracellular cytokine staining (ICS) at the indicated time points after CD8 depletion treatment and challenge with 3×SIV, 4×SIV, and 5×SIV. (a and b) Bars are plotted against the left y axis and indicate the number of distinct peptides recognized by each animal that were presented by Mamu-B*17, Mamu-A*02, or undetermined MHC-I molecules. Lines are plotted against the right y axis and indicate the proportion of the total SIV-specific response accounted for by Mamu-B*17-restricted CD8+ T cells (calculated as the frequency of all Mamu-B*17-restricted CD8+ T cell populations divided by the total frequency of all SIV-specific CD8+ T cells). (c and d) Bars depict the frequency of CD8+ T cells recognizing peptides presented by Mamu-B*17 or other MHC-I molecules. Note that only Mamu-B*17- and -A*02-restricted peptides were tested in rhAJ11 at 4 weeks prior to CD8+ cell depletion due to the limited number of available PBMC.
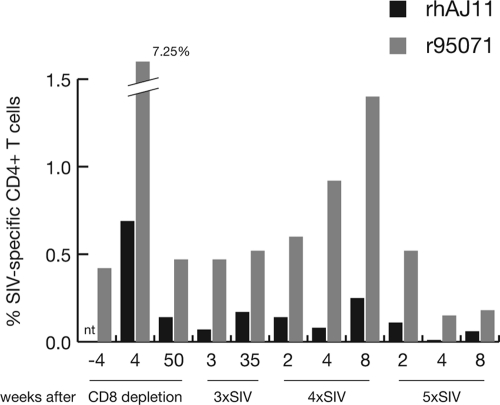
We also consistently detected SIV-specific CD4+ T cell responses in both LTNPs. The total magnitude of these responses was strongest in both animals immediately after CD8+ cell depletion, but throughout the study, frequencies of SIV peptide-specific CD4+ T cells ranged between 0.01% and 0.25% in rhAJ11 and 0.15% and 1.4% in r95071 (Fig. 6a). There was no apparent correlation between the timing of challenges with mutant SIV constructs and fluctuations in the magnitude of the CD4+ T cell response. CD4+ T cells from both animals consistently recognized multiple distinct SIV peptide pools (rhAJ11, 1 to 9 pools; r95071, 5 to 12 pools) (data not shown). Neither animal's total CD4+ T cell count showed a significant decline over the course of the study (data not shown).
FIG. 6.
Breadth and magnitude of SIV-specific CD4+ T cell responses in LTNPs following serial challenges with escape mutant SIV. Frequencies of CD4+ T cells responding to SIVmac239 peptides were measured by ICS as described in the legend to Fig. 5. Bars indicate the sum of the frequencies of all CD4+ T cells responding to SIV peptides in rhAJ11 or r95071. “nt” indicates that, due to limited availability of PBMC, CD4+ T cell responses to the entire viral proteome were not measured prior to CD8+ cell depletion in rhAJ11.
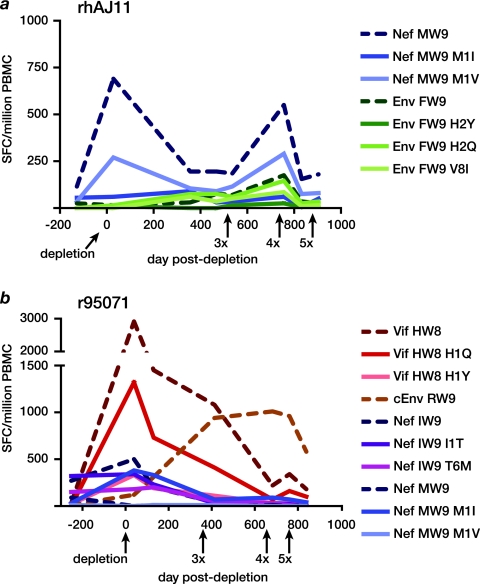
LTNPs' CD8+ T cell repertoires evolve with time and include responses against epitope variants.
Since the LTNPs maintained detectable CD4+ T cell responses, we reasoned that they could have made CD8+ T cell responses to the mutant epitopes encoded by 3×SIV, 4×SIV, and 5×SIV that played a role in their ability to resist superinfection. Indeed, IFN-γ ELISPOT analysis revealed that rhAJ11's strong Nef MW9-specific population included cells that recognized the position 1 methionine-to-valine (M1V) variant carried by the 4×SIV and 5×SIV mutant viruses (Fig. 7a), although this epitope sequence was never detected in the animal's autologous virus (data not shown). Interestingly, populations specific for a different Mamu-B*17-restricted epitope, Env FW9, and 2 variants became detectable in rhAJ11 prior to the rechallenge study. These responses increased in magnitude after challenge with 3×SIV, despite the fact that the epitope variant encoded by the mutant viruses, Env FW9 H2Y, is predicted to bind only weakly if at all to Mamu-B*17 (50% inhibitory concentration [IC50] of 1,313 nM) (Table 1).
FIG. 7.
Evolution of responses to variant Mamu-B*17-restricted CD8+ T cell epitopes in LTNPs. We assessed the ability of cryopreserved PBMC from LTNPs to recognize variant Mamu-B*17 peptides in an IFN-γ ELISPOT assay. Results are displayed as the frequency of spot-forming cells (SFC) per million PBMC. Peptides that elicited a response at at least one time point are shown. Broken lines indicate wild-type epitope sequences; solid lines indicate epitope variants. CD8+ cell depletion and challenges with mutant viruses occurred at the indicated time points. (a) rhAJ11 responded to variants of Nef MW9 and Env FW9. (b) r95071 responded to variants of Vif HW8, Nef IW9, and Nef MW9. In addition, a response to the cryptic epitope cEnv RW9 emerged beginning after CD8+ cell depletion.
In contrast, the ability of CD8+ T cells from r95071 to recognize variant Mamu-B*17-restricted peptides decreased over the course of the study. CD8+ T cell populations in this animal recognized multiple variants of Vif HW8 and Nef IW9 around the time of CD8+ cell depletion, but these populations, together with the previously immunodominant response against wild-type Vif HW8, waned in frequency during the serial rechallenge study. Instead, we detected a response against the cryptic epitope cEnv RW9 that increased in frequency after challenge with 3×SIV and 4×SIV; the Env cRW9 sequence is intact in each of these viruses. However, cEnv RW9-specific CD8+ T cells declined in frequency following challenge with 5×SIV, which harbored a potent escape mutation in this epitope (Fig. 7b), even though we could not detect the emergence of variation in this epitope in r95071's autologous virus (data not shown).
Neutralizing and Env-binding antibody responses are detected in chronic infection of both controllers and progressors.
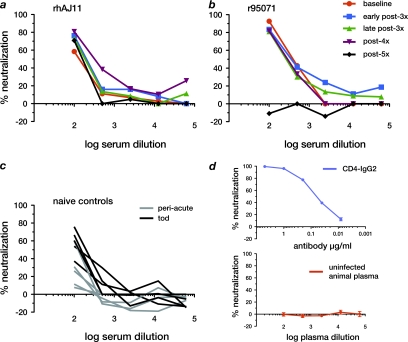
Since particular CD8+ T cell responses did not appear to be associated with the LTNPs' ability to control superinfection with escape mutant viruses, we measured both neutralizing and Env gp130-binding antibody responses to SIVmac239 in the controllers and previously uninfected animals. Both LTNPs had detectable neutralizing antibodies prior to the serial rechallenge experiments, which were maintained throughout the study period at similar titers; serum from both LTNPs had 50% neutralizing activity at dilutions between 1:100 and 1:500 (Fig. 8a and b). By the time of necropsy, all Mamu-B*17-negative control animals had also developed detectable neutralizing antibody responses, with titers comparable to those observed with the LTNPs (Fig. 8c). Measurements of serum antibodies capable of binding SIVmac239 Env gp130 revealed that the LTNPs had total serum antibody titers similar to those of 3 of 6 previously uninfected animals, while the other 3 previously uninfected animals made only weak total antibody responses (data not shown). Our results therefore show that controllers' antibody responses in chronic infection are similar to those observed with progressors. Importantly, although this level of neutralizing activity was not capable of effectively controlling virus replication in the chronic phase of an initial infection in the Mamu-B*17-negative control animals, neutralizing antibodies may have played an important role in protecting the LTNPs against superinfection.
FIG. 8.
Neutralizing antibody responses to SIVmac239 in LTNPs and Mamu-B*17-negative control animals. We measured neutralizing antibody levels against SIVmac239 in macaques' plasma or serum in a pseudotyped-virion assay. We tested samples taken from Mamu-B*17-positive LTNPs rhAJ11 (a) and r95071 (b) prior to challenge with 3×SIV (baseline) and at the indicated times after challenge with mutant viruses. We also measured neutralizing antibody titers in the six Mamu-B*17-negative control animals (c) at 7 to 12 weeks postinfection (peri-acute) and at the time of sacrifice (tod). (d) The chimeric monoclonal antibody CD4-IgG2 was used as a positive control, while plasma from an uninfected macaque served as a negative control. Positive- and negative-control samples were tested in triplicate; error bars represent standard error of the mean computed for each antibody/serum concentration.
DISCUSSION
The SIV-infected macaque model offers an important complement to the study of human control of HIV replication, since it allows for the control of key parameters, such as the strain of the infecting virus as well as the timing and dose of infection. Strikingly, Asian macaques exhibit MHC-associated control of pathogenic SIV that is very similar to that observed with humans (30, 42, 59). Transient depletion of CD8+ cells in ECs interrupts this control (17). The protective MHC-I molecules Mamu-B*08 and HLA-B27 even bind similar peptides (31). In the present study, we sought to identify particular epitope-specific CD8+ T cell populations restricted by the protective MHC-I molecule Mamu-B*17 that were required for long-term control of the molecularly cloned virus SIVmac239 by serially challenging Mamu-B*17-positive controllers with SIVmac239 variants encoding putative escape mutations in increasing numbers of Mamu-B*17-restricted epitopes. These engineered escape mutant viruses appeared replicatively fit in vitro and in vivo, with the potential exception of a small cost to viral fitness associated with one of the substitutions in Nef IW9. Nonetheless, they failed to cause superinfection in the two LTNPs in this study.
Exposure to viruses bearing amino acid substitutions in CD8+ T cell epitopes was thought to play a key role in the superinfection of some HIV-positive subjects that maintained low viremia after a first infection (2, 52). Indeed, recent studies suggest that HIV superinfection may be more common than was initially believed (47, 48), so it is perhaps surprising that these animals control serial challenges with viruses bearing mutations in epitopes that are targeted by an apparently protective cellular immune response. However, the envelope sequences of the variant viruses used to challenge the LTNPs in this study were identical to those of SIVmac239 except for the single amino acid substitution in the FW9 CD8+ T cell epitope located in gp41. It is therefore possible that the neutralizing antibody responses that we detected in the LTNPs made an important contribution to their immunity to superinfection. Early passive transfer studies in macaques suggested that relatively high serum concentrations of neutralizing antibody might be required to achieve protection against intravenous or mucosal challenge (35, 44, 51), but more recent investigations suggest that substantially lower antibody titers can protect macaques against acquisition of simian-human immunodeficiency viruses (SHIVs) administered in a multiple-low-dose mucosal challenge regimen (21-23). Moreover, in one study, even very low titers of passively transferred monoclonal antibody 2G12 provided sterilizing protection in 2 of 4 animals against vaginal challenge with a dose of SHIV that productively infected 5 of 5 animals after a single challenge (36). While it is difficult to directly compare data from these previous studies to our own due to the different routes, doses, and strains of challenge viruses used, as well as the different assays used to measure antibody activity, together they suggest that virus-specific neutralizing antibody responses do indeed play an important role in the LTNPs' resistance to superinfection.
Studies aimed at identifying the mechanisms tying “protective” genotypes to the outcome of successful control of HIV have yet to yield conclusive results. In perhaps the best-characterized system, it appears that recognition of the Gag KK10 epitope is critical for the control of HIV replication associated with HLA-B27 (19, 20, 25). There are some indications that HLA-B57+ ECs can make de novo responses to epitope variants (3) and can maintain control of emerging escape viruses (40), though the extent to which variant-specific responses are present in HLA-B57+ progressors remains uncertain. In our study, it is possible that the brief viral recrudescence during CD8+ cell depletion acted to “boost” particular SIV-specific CD8+ T cell responses, stimulating a more diverse CD8+ T cell repertoire that helped to control subsequent challenges. In support of this interpretation, responses to variants of Nef MW9 and Env FW9 emerged over time in animal rhAJ11, peaking after challenge with 4×SIV, even as viremia remained undetectable in this animal. In contrast, the frequencies of almost all variant-specific CD8+ T cells dwindled with time in r95071 and were barely detectable by the time it was challenged with 5×SIV. r95071's virus load remained steady at around 5,000 copies/ml until 3 months after 5×SIV challenge, when it began to steadily increase. Interestingly, this increasing viremia was not associated with a loss of SIV-specific CD4+ T cell responses or a decline in the CD4+ T cell count. We speculate that the continuing coevolution of the virus and CD8+ T cell response could have played a role in rhAJ11's stable control of virus replication, while a lack of such coevolution eventually impaired r95071's ability to contain viremia, leading to its steadily increasing viremia.
CD8+ T cells may also exert control over immunodeficiency virus replication via noncytolytic mechanisms (56). Strong in vivo evidence for such activity emerged recently from two independent studies showing that transient CD8+ cell depletion had no effect on the decay of viremia in macaques initiating antiretroviral therapy (26, 58), suggesting that the mechanism by which CD8+ T cells control virus replication is not mainly via direct lysis of infected cells. Could the relative lack of diversification in Mamu-B*17-restricted epitopes in ECs' autologous virus indicate that ECs mount CD8+ T cell responses with particularly potent noncytolytic effector activities, irrespective of their epitope specificity? It seems likely that even noncytolytic effector functions in CD8+ T cells would have to be stimulated by T cell receptor interactions with MHC-I molecules presenting viral antigens. Efficacious CD8+ T cell responses associated with Mamu-B*17 expression would therefore be expected to exert selective pressure on replicating viruses irrespective of whether their main effector activities were cytolytic or noncytolytic. Indeed, in other studies, mutations in Mamu-B*17-restricted CD8+ T cell epitopes were commonly observed with the majority of progressors and even some ECs that express this MHC-I molecule (17, 34), but not in animals that lack Mamu-B*17, indicating that Mamu-B*17-restricted CD8+ T cells do indeed exert detectable selective pressure on the virus, whether through cytolytic or other means. Thus, our data cannot be used to distinguish the relative contributions of noncytolytic mechanisms and direct cytolysis to immune control.
It must be noted that our study has several important limitations. First, in our screens for cross-reactive CD8+ T cells, we used a single, relatively high peptide concentration (1 μM). It has previously been reported that the ability of variant peptides at such concentrations to stimulate cytokine secretion does not always correlate with antiviral activity (55), but in that study, there was a direct correlation between recognition of variant Nef IW9 peptides by T cells from a Mamu-B*17-positive EC in an ELISPOT assay and the ability of polyclonal CD8+ T cell lines from that animal to suppress mutant virus replication (55). Moreover, we have previously shown that Mamu-B*17-restricted CD8+ T cell responses from both controllers and progressors tend to be of low avidity, and even some progressor animals have CD8+ T cell populations capable of recognizing common epitope variants (34). Together, our data and those from previous studies suggest that Mamu-B*17-positive ECs/LTNPs make CD8+ T cell responses capable of recognizing some variant epitopes, but it seems likely that variant-specific CD8+ T cell responses cannot alone account for these LTNPs' resistance to superinfection with escape variant viruses.
Several studies have also shown that ECs/LTNPs tend to maintain “higher-quality,” polyfunctional HIV-specific CD8+ T cell responses longer than progressors do (4, 43) and that ECs' CD8+ T cells lack signs of functional exhaustion (11, 45, 53), but it is still not clear whether these attributes are the cause or the effect of nonprogressive infection. Unfortunately, investigations of CD8+ T cell response “quality” were beyond the scope of the present study, so we cannot draw conclusions about the impact of response “quality” on these animals' ability to control SIV replication and resist superinfection. Also, although the majority of epitope-specific CD8+ T cells are likely to secrete IFN-γ in response to peptide stimulation, here we did not specifically assay other effector functions, such as markers of cytolysis. CD8+ T cells capable of cytolysis, but not IFN-γ secretion, would therefore not have been detected in the present study.
Here we showed that 2 Mamu-B*17-positive LTNPs resisted superinfection with mutant viruses engineered to express escape mutations in Mamu-B*17-restricted CD8+ T cell epitopes. Since the sequence of the challenge viruses otherwise matched that of the original infecting strain, it is likely that a broad array of immune responses, including neutralizing antibodies, CD4+ T cells, and perhaps variant-specific CD8+ T cells, all played important roles in this resistance.
Although the number of animals in this study is small, we believe our results highlight an important concept: establishing effective control over AIDS virus replication and maintaining that control may in fact be distinct processes, with different immune responses playing differential roles in each process. Accordingly, in a previous study, Valentine et al. observed that SIVmac239 variants bearing escape mutations in 8 Mamu-B*08-restricted CD8+ T cell epitopes were poorly controlled by animals expressing this molecule, which is associated with highly effective control of wild-type SIVmac239 (54). Their study used previously uninfected animals and did not examine Mamu-B*08-positive ECs' ability to resist superinfection with the mutant viruses. We speculate that effective CD8+ T cell responses may be the main contributor to establishing elite control and that limiting virus replication to low levels soon after infection allows for the development of a full complement of immune responses that at later times can act in concert to stably contain virus replication and resist superinfection. If this hypothesis is correct, attempts to dissect the correlates of elite control may lead to different conclusions based on the stage of infection in which subjects (animal or human) are studied. Understanding both acute-phase correlates of establishing elite control or chronic-phase correlates of maintaining control and resisting superinfection will undoubtedly inform the search for an effective AIDS vaccine.
Acknowledgments
We gratefully acknowledge David I. Watkins and David O'Connor for helpful discussions and critical review of the manuscript. We also thank Eric Rogers, Eric Peterson, Kristin Crosno, Nancy Schultz-Darken, and members of the Wisconsin National Primate Research Center (WNPRC) Centralized Protocol Implementation and Veterinary Services Units for technical assistance.
This work was supported by National Institute for Allergy and Infectious Disease (NIAID) grants AI 068586 to T.C.F. and AI 055332 and AI 033292 to D.R.B. and in part by grant P51 RR000167 from the National Center for Research Resources (NCRR) to the WNPRC. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grants RR15459-01 and RR020141-01.
Footnotes
Published ahead of print on 20 October 2010.
REFERENCES
- 1.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434-439. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brumme, Z. L., C. J. Brumme, J. Carlson, H. Streeck, M. John, Q. Eichbaum, B. L. Block, B. Baker, C. Kadie, M. Markowitz, H. Jessen, A. D. Kelleher, E. Rosenberg, J. Kaldor, Y. Yuki, M. Carrington, T. M. Allen, S. Mallal, M. Altfeld, D. Heckerman, and B. D. Walker. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 82:9216-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 9.Cline, A. N., J. W. Bess, M. Piatak, Jr., and J. D. Lifson. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303-312. [DOI] [PubMed] [Google Scholar]
- 10.Crawford, H., W. Lumm, A. Leslie, M. Schaefer, D. Boeras, J. G. Prado, J. Tang, P. Farmer, T. Ndung'u, S. Lakhi, J. Gilmour, P. Goepfert, B. D. Walker, R. Kaslow, J. Mulenga, S. Allen, P. J. Goulder, and E. Hunter. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day, C. L., D. E. Kaufmann, P. Kiepiela, J. A. Brown, E. S. Moodley, S. Reddy, E. W. Mackey, J. D. Miller, A. J. Leslie, C. DePierres, Z. Mncube, J. Duraiswamy, B. Zhu, Q. Eichbaum, M. Altfeld, E. J. Wherry, H. M. Coovadia, P. J. Goulder, P. Klenerman, R. Ahmed, G. J. Freeman, and B. D. Walker. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350-354. [DOI] [PubMed] [Google Scholar]
- 12.Deeks, S. G., and B. D. Walker. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27:406-416. [DOI] [PubMed] [Google Scholar]
- 13.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frater, A. J., H. Brown, A. Oxenius, H. F. Gunthard, B. Hirschel, N. Robinson, A. J. Leslie, R. Payne, H. Crawford, A. Prendergast, C. Brander, P. Kiepiela, B. D. Walker, P. J. Goulder, A. McLean, and R. E. Phillips. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 81:6742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275-281. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich, T. C., A. B. McDermott, M. R. Reynolds, S. Piaskowski, S. Fuenger, I. P. De Souza, R. Rudersdorf, C. Cullen, L. J. Yant, L. Vojnov, J. Stephany, S. Martin, D. H. O'Connor, N. Wilson, and D. I. Watkins. 2004. Consequences of cytotoxic T-lymphocyte escape: common escape mutations in simian immunodeficiency virus are poorly recognized in naive hosts. J. Virol. 78:10064-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. R. Capuano, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, X., A. Bashirova, A. K. Iversen, J. Phair, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, M. Altfeld, S. J. O'Brien, and M. Carrington. 2005. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 11:1290-1292. [DOI] [PubMed] [Google Scholar]
- 19.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 21.Hessell, A. J., P. Poignard, M. Hunter, L. Hangartner, D. M. Tehrani, W. K. Bleeker, P. W. Parren, P. A. Marx, and D. R. Burton. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hessell, A. J., E. G. Rakasz, P. Poignard, L. Hangartner, G. Landucci, D. N. Forthal, W. C. Koff, D. I. Watkins, and D. R. Burton. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessell, A. J., E. G. Rakasz, D. M. Tehrani, M. Huber, K. L. Weisgrau, G. Landucci, D. N. Forthal, W. C. Koff, P. Poignard, D. I. Watkins, and D. R. Burton. 2010. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 84:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaizu, M., G. J. Borchardt, C. E. Glidden, D. L. Fisk, J. T. Loffredo, D. I. Watkins, and W. M. Rehrauer. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 59:693-703. [DOI] [PubMed] [Google Scholar]
- 25.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klatt, N. R., E. Shudo, A. M. Ortiz, J. C. Engram, M. Paiardini, B. Lawson, M. D. Miller, J. Else, I. Pandrea, J. D. Estes, C. Apetrei, J. E. Schmitz, R. M. Ribeiro, A. S. Perelson, and G. Silvestri. 2010. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 6:e1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 29.Loffredo, J. T., A. T. Bean, D. R. Beal, E. J. Leon, G. E. May, S. M. Piaskowski, J. R. Furlott, J. Reed, S. K. Musani, E. G. Rakasz, T. C. Friedrich, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 82:1723-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loffredo, J. T., J. Maxwell, Y. Qi, C. E. Glidden, G. J. Borchardt, T. Soma, A. T. Bean, D. R. Beal, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81:8827-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loffredo, J. T., J. Sidney, A. T. Bean, D. R. Beal, W. Bardet, A. Wahl, O. E. Hawkins, S. Piaskowski, N. A. Wilson, W. H. Hildebrand, D. I. Watkins, and A. Sette. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 182:7763-7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loh, L., J. Petravic, C. J. Batten, M. P. Davenport, and S. J. Kent. 2008. Vaccination and timing influence SIV immune escape viral dynamics in vivo. PLoS Pathog. 4:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maness, N. J., L. E. Valentine, G. E. May, J. Reed, S. M. Piaskowski, T. Soma, J. Furlott, E. G. Rakasz, T. C. Friedrich, D. A. Price, E. Gostick, A. L. Hughes, J. Sidney, A. Sette, N. A. Wilson, and D. I. Watkins. 2007. AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape. J. Exp. Med. 204:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maness, N. J., L. J. Yant, C. Chung, J. T. Loffredo, T. C. Friedrich, S. M. Piaskowski, J. Furlott, G. E. May, T. Soma, E. J. Leon, N. A. Wilson, H. Piontkivska, A. L. Hughes, J. Sidney, A. Sette, and D. I. Watkins. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques. J. Virol. 82:5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 37.McDermott, A. B., J. Mitchen, S. Piaskowski, I. De Souza, L. J. Yant, J. Stephany, J. Furlott, and D. I. Watkins. 2004. Repeated low-dose mucosal simian immunodeficiency virus SIVmac239 challenge results in the same viral and immunological kinetics as high-dose challenge: a model for the evaluation of vaccine efficacy in nonhuman primates. J. Virol. 78:3140-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 40.O'Connell, K. A., J. Xu, A. P. Durbin, L. G. Apuzzo, H. Imteyaz, T. M. Williams, S. C. Ray, J. B. Margolick, R. F. Siliciano, and J. N. Blankson. 2009. HIV-1 evolution following transmission to an HLA-B*5801-positive patient. J. Infect. Dis. 200:1820-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 77:9029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connor, S. L., J. J. Lhost, E. A. Becker, A. M. Detmer, R. C. Johnson, C. E. Macnair, R. W. Wiseman, J. A. Karl, J. M. Greene, B. J. Burwitz, B. N. Bimber, S. M. Lank, J. J. Tuscher, E. T. Mee, N. J. Rose, R. C. Desrosiers, A. L. Hughes, T. C. Friedrich, M. Carrington, and D. H. O'Connor. 2010. MHC heterozygote advantage in simian immunodeficiency virus-infected mauritian cynomolgus macaques. Sci. Transl. Med. 2:22ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen, R. E., J. W. Heitman, D. F. Hirschkorn, M. C. Lanteri, H. H. Biswas, J. N. Martin, M. R. Krone, S. G. Deeks, and P. J. Norris. 2010. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS 24:1095-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrovas, C., J. P. Casazza, J. M. Brenchley, D. A. Price, E. Gostick, W. C. Adams, M. L. Precopio, T. Schacker, M. Roederer, D. C. Douek, and R. A. Koup. 2006. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 203:2281-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 47.Piantadosi, A., B. Chohan, V. Chohan, R. S. McClelland, and J. Overbaugh. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piantadosi, A., M. O. Ngayo, B. Chohan, and J. Overbaugh. 2008. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res. Hum. Retroviruses 24:1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 50.Reynolds, M. R., A. M. Weiler, K. L. Weisgrau, S. M. Piaskowski, J. R. Furlott, J. T. Weinfurter, M. Kaizu, T. Soma, E. J. Leon, C. MacNair, D. P. Leaman, M. B. Zwick, E. Gostick, S. K. Musani, D. A. Price, T. C. Friedrich, E. G. Rakasz, N. A. Wilson, A. B. McDermott, R. Boyle, D. B. Allison, D. R. Burton, W. C. Koff, and D. I. Watkins. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 205:2537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 52.Streeck, H., B. Li, A. F. Poon, A. Schneidewind, A. D. Gladden, K. A. Power, D. Daskalakis, S. Bazner, R. Zuniga, C. Brander, E. S. Rosenberg, S. D. Frost, M. Altfeld, and T. M. Allen. 2008. Immune-driven recombination and loss of control after HIV superinfection. J. Exp. Med. 205:1789-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trautmann, L., L. Janbazian, N. Chomont, E. A. Said, S. Gimmig, B. Bessette, M. R. Boulassel, E. Delwart, H. Sepulveda, R. S. Balderas, J. P. Routy, E. K. Haddad, and R. P. Sekaly. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198-1202. [DOI] [PubMed] [Google Scholar]
- 54.Valentine, L. E., J. T. Loffredo, A. T. Bean, E. J. Leon, C. E. MacNair, D. R. Beal, S. M. Piaskowski, Y. C. Klimentidis, S. M. Lank, R. W. Wiseman, J. T. Weinfurter, G. E. May, E. G. Rakasz, N. A. Wilson, T. C. Friedrich, D. H. O'Connor, D. B. Allison, and D. I. Watkins. 2009. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J. Virol. 83:11514-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valentine, L. E., S. M. Piaskowski, E. G. Rakasz, N. L. Henry, N. A. Wilson, and D. I. Watkins. 2008. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 82:575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker, C. M., D. J. Moody, D. P. Stites, and J. A. Levy. 1986. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science 234:1563-1566. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, D. H. O'Connor, T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong, J. K., M. C. Strain, R. Porrata, E. Reay, S. Sankaran-Walters, C. C. Ignacio, T. Russell, S. K. Pillai, D. J. Looney, and S. Dandekar. 2010. In vivo CD8+ T-cell suppression of SIV viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog. 6:e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]