Abstract
RNA degradation, together with RNA synthesis, controls the steady-state level of viral RNAs in infected cells. The endoribonucleolytic cleavage of viral RNA is important not only for viral RNA degradation but for RNA recombination as well, due to the participation of some RNA degradation products in the RNA recombination process. To identify host endoribonucleases involved in degradation of Tomato bushy stunt virus (TBSV) in a Saccharomyces cerevisiae model host, we tested eight known endoribonucleases. Here we report that downregulation of SNM1, encoding a component of the RNase MRP, and a temperature-sensitive mutation in the NME1 gene, coding for the RNA component of RNase MRP, lead to reduced production of the endoribonucleolytically cleaved TBSV RNA in yeast. We also show that the highly purified yeast RNase MRP cleaves the TBSV RNA in vitro, resulting in TBSV RNA degradation products similar in size to those observed in yeast cells. Knocking down the NME1 homolog in Nicotiana benthamiana also led to decreased production of the cleaved TBSV RNA, suggesting that in plants, RNase MRP is involved in TBSV RNA degradation. Altogether, this work suggests a role for the host endoribonuclease RNase MRP in viral RNA degradation and recombination.
Many RNA viruses evolve rapidly due to a high frequency of mutations and genetic recombination as well as reassortment of genomic components (1, 52). Comparison of viral RNA genomes has revealed that recombination has shaped the evolution of many RNA viruses (52). RNA recombination is the process that joins two or more noncontiguous segments of the same RNA or two separate RNAs together (32). Recombination events can have small or dramatic effects on viral genomes by introducing insertions or duplications, combining new sequences, or leading to deletions or rearrangements. RNA recombination also functions to repair truncated or damaged viral RNA molecules (16, 17, 30, 37). The repair function of RNA recombination might compensate viruses for their high mutation rate, which could introduce detrimental mutations into the viral genomes and thus reduce the fitness of viral populations (38, 39). Therefore, depending on the nature of recombining RNAs, the locations of the recombinant junction sites, and the outcome of the recombination events, RNA recombination can guard the infectivity of the viral genome or can increase genome variability. Altogether, RNA recombination is a probabilistic event that can affect the population of viruses, contribute to virus variability, and function in genome repair that maintains the infectivity of RNA viruses (32, 39).
In addition to the roles of the viral replicase and the viral RNA (31, 32), host proteins are likely involved in viral RNA recombination as well. Accordingly, recent genome-wide approaches with Tomato bushy stunt virus (TBSV), a tombusvirus infecting plants, and with the model host Saccharomyces cerevisiae have led to the identification of more than 30 host genes affecting viral RNA recombination (20, 21, 27, 45, 46). Detailed analysis of a few host factors revealed that they could affect the properties/activities of the viral replicase; an example is the Pmr1 Ca2+/Mn2+ pump that controls the Mn2+ level in the cytosol (20). A high Mn2+ level in the absence of Pmr1 leads to high-frequency RNA recombination in yeast or plant cells as well as in a cell-free TBSV replication assay (20).
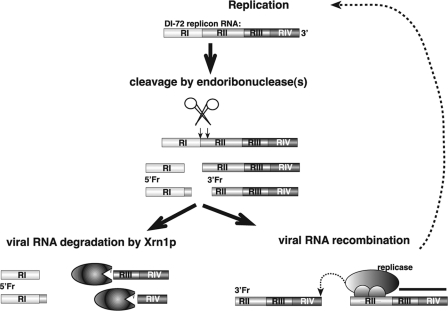
Studies on the role of host factors in TBSV RNA recombination also revealed the roles of the RNA degradation pathways. For example, a critical identified host factor was the cytosolic Xrn1p 5′-3′ exoribonuclease (called Xrn4 in plants), which suppresses TBSV recombination (5, 6, 19). Several partially cleaved TBSV replicon RNAs (repRNAs), termed degRNAs, were detected in xrn1Δ yeast or in plants silenced for XRN4, demonstrating that Xrn1p in yeast and Xrn4p in plants play an active role in TBSV RNA degradation (5, 6, 19). The profile of the degRNAs in xrn1Δ yeast or XRN4 knockdown plants suggested the existence of an endoribonuclease activity (or activities) (5, 6, 19, 23). Thus, this early work indicated that both exo- and endoribonucleases are involved in degradation of TBSV RNA, which is uncapped and lacks a 3′ poly(A) tail (51). Interestingly, several of the degRNAs actively participate in RNA recombination events, serving as recombination intermediates (6, 46). In the absence of Xrn1p, these recombination intermediates accumulate to high levels (Fig. 1), thus promoting recombination events. These findings invited attention to the role of RNA degradation in viral RNA accumulation as well as in RNA recombination (29).
FIG. 1.
Model of the roles of endo- and exoribonucleases in tombusvirus RNA degradation and recombination. During or after replication of TBSV DI-72 repRNA, a host endoribonuclease cleaves some of the viral RNAs (at places marked by arrows), producing 5′Fr and 3′Fr degRNAs. These cleaved repRNA products are then rapidly removed by the host Xrn1p 5′-3′ exoribonuclease. The rapid removal of the recombination substrates inhibits viral RNA recombination. The not-yet-degraded 3′Fr RNAs can participate in replicase-driven recombination events. Note that 5′Fr lacks important cis-acting replication sequences and does not seem to participate in RNA recombination.
There are several different RNA degradation pathways in yeast. The general RNA degradation pathway includes mostly exoribonuclease activities, starting with shortening of the poly(A) tail, followed by decapping/5′-3′ exoribonuclease activities in one pathway and by the exosome with 3′-5′ activities in the other pathway (7, 33). The specialized RNA degradation pathways include nonsense-mediated mRNA decay (NMD), nonstop mRNA decay (NSD), and no-go decay (NGD), which serve to degrade aberrant mRNAs (7, 33). The decay of some mRNAs also includes specific endoribonucleases recognizing sequence elements, such as ARE-mediated decay based on AU-rich instability sequences in the 3′-untranslated region (3′-UTR) (26, 33). Plants also have these conserved RNA degradation pathways, plus they have the RNA interference (RNAi)-based pathway based on dicer and AGO proteins (7). Several other endoribonucleases operate in mammalian cells, such as the viral-induced RNase L, the polysomal nuclease 1, and aldolase C (26). The components of the RNA silencing pathway have been shown to affect recombination of TBSV and a fungal RNA virus, suggesting that ribonucleases might contribute to the evolution of a range of RNA viruses (25, 48, 53).
Since generation of the internally cleaved TBSV RNA products, which are easily detectable in xrn1Δ yeast or in XRN4 knockdown plants, is likely due to cleavages by endoribonucleases present in yeast and plants, in this work we tested the effects of several endoribonucleases on the profile of TBSV RNA degradation products in yeast. We identified RNase MRP (RNase mitochondrial RNA processing) as one of the endoribonucleases cleaving the TBSV RNA both in vitro and in yeast, and possibly in plants. RNase MRP is a ribonucleoprotein endoribonuclease found in the nucleus, cytoplasm, and mitochondria (14, 28, 44). In yeast, RNase MRP is an essential enzyme involved in processing the rRNA precursor to the mature 5.8S rRNA (43). In the cytoplasm, RNase MRP has been shown to cleave the mRNA for Clb2 cyclin during mitosis (15); cleavage of this mRNA by RNase MRP is followed by rapid degradation of the mRNA by the Xrn1 exonuclease. Mutations in the gene for the RNA component of human RNase MRP cause short-limb dwarfism and immunodeficiency (18, 49). The yeast RNase MRP consists of a single RNA (encoded by the NME1 gene) and 10 proteins (Pop1p, Pop3-8p, Rpp1p, Snm1p, and Rmp1p). Eight of these proteins in RNase MRP are shared with the yeast RNase P, which is a closely related ribonucleoprotein endoribonuclease that processes tRNA precursors (28, 40). The RNA component of RNase MRP is also closely related to the RNA component of RNase P.
Here we show that downregulation of SNM1, encoding a component of the RNase MRP, and a temperature-sensitive mutation in the NME1 gene, coding for the RNA component, lead to reduced production of the cleaved TBSV RNA in yeast. We also show that the highly purified yeast RNase MRP cleaves the TBSV RNA in vitro, resulting in endonucleolytically processed TBSV RNAs similar in size to those observed in yeast cells. Virus-induced gene silencing (VIGS) of the NME1 homolog in Nicotiana benthamiana also led to decreased production of the cleaved TBSV RNA, suggesting that plant RNase MRP is involved in TBSV RNA degradation. Altogether, this work opens up the possibility that host endoribonuclease RNase MRP might be involved in viral RNA degradation and recombination.
MATERIALS AND METHODS
Yeast strains and expression plasmids.
S. cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), titratable yTHC yeast strains (TET::Snm1 and TET::Rmp1), and YKO deletion strains (HbsIΔ, Ngl2Δ, Ire1Δ, Ysh1Δ, Dom34Δ, Ngl1Δ, Ngl3Δ, and Rny1Δ strains) were obtained from Open Biosystems. Gene disruption of MET22 (met22::hphNT1) was done by homologous recombination in the indicated yeast strains. For this purpose, we amplified the hphNT1 marker of pFA6-hphNT1 (Euroscarf) (22) with primers 2581 (TGTATGGTGCAGATGGAGAGAGCTCGGACACATATGCGGCGTACGCTGCAGGTCGACGG) and 2590 (AGTAAAATATATGTTATTTAGGCGTTTCTTGACTGAATGACATCGATGAATTCGAGCTCG). The recombinant yeast strains were grown on yeast extract-peptone-dextrose (YPD) plates containing 200 μg/ml hygromycin B, and correct gene disruptions were confirmed by PCR, using primers 2591 (CTTGGATGGGAAGTCCAAGA) and 2501 (ATCCACGCCCTCCTACATC).
Yeast strains MES101 (MATa/MATα his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 trp1-Δ1/trp1-Δ1 ADE2/ade2-1 LYS2/lys2-801), MES111 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 nme1-Δ2::TRP1 pMES127 [CEN URA3 NME1]), MES111-140 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 nme1-Δ2::TRP1 pMES140 [CEN LEU2 NME1]), MES111-P6 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 nme1-Δ2::TRP1 pMES140-P6 [CEN LEU2 nme1-P6]), TLG105 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 nme1-Δ2::TRP1 xrn1-Δ1::KanMX4 pMES127 [CEN URA3 NME1]), TLG105-140 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 nme1-Δ2::TRP1 xrn1-Δ1::KanMX4 pMES140 [CEN LEU2 NME1]), TLG105-P6 (MATα his3-Δ200 leu2-3,112 ura3-52 trp1-Δ1 ade2 nme1-Δ2::TRP1 xrn1-Δ1::KanMX4 pMES140-P6 [CEN LEU2 nme1-P6]), and YSW1 (MATa POP4::TAPTAG::TRP1ks pep4::LEU2 nuc1::LEU2 sep1::URA3 trp1-1 his3-11,15 can-100 ura3-1 leu2-3,112) were described earlier (15, 42).
The expression plasmids pGAD-His92/CUP1 (containing the Cucumber necrosis virus [CNV] p92pol gene behind the CUP1 promoter), pGBK-His33/CUP1/DI-72/GAL1 (coexpressing 6×His-tagged p33 from the CUP1 promoter and DI-72 repRNA from the GAL1 promoter), pGBK-His33/DI72/CUP1 (coexpressing 6×His-tagged p33 from the ADH1 promoter and DI-72 repRNA from the CUP1 promoter), and pCM189-TET-His92 (expressing 6×His-tagged CNV p92 under the control of a doxycycline-regulatable promoter) have been described previously (20, 27, 35).
pTRV1 and pTRV2 VIGS vectors were obtained from S. P. Dinesh-Kumar (10). To generate pTRV2-NmeNb, primers 3232 (GGCGGAATTCCAGGAAAGTCCCCGGGCC) and 3235 (GGCGTCTAGAGTAAGCCCCGTTCAGTTA) were designed according to the sequence of tobacco 7-2/MRP RNA (24). The reverse transcription-PCR (RT-PCR) product amplified from N. benthamiana total RNA was digested with EcoRI/XbaI and ligated to EcoRI/XbaI-linearized pTRV2. The plasmids pTRV2-Xrn4Nb, pGD-CNV, and pGD-DI72 have been described previously (2, 19).
Yeast cultivation.
Yeast cells were cotransformed with the indicated plasmids or PCR products by using the lithium acetate-single-stranded DNA (ssDNA)-polyethylene glycol method (13), and transformants were selected by complementation of auxotrophic markers. The met22Δ/TET::snm1, met22Δ/TET::rmp1, and double deletion strains were cotransformed with pGBK-His33/CUP1/DI-72/GAL1 and pGAD-His92/CUP1. The transformed yeast strains were grown at 23°C in SC-UHL (synthetic complete medium without uracil, histidine, and leucine) medium containing 2% galactose with 50 μM CuSO4 to launch TBSV repRNA replication and containing 10 μg/ml doxycycline to downregulate gene expression in the yTHC (TET::) strains. After 7 h, the yeast cultures were resuspended in SC-UHL with glucose containing 50 μM CuSO4 and 10 μg/ml doxycycline for the yTHC strains. The yeast cultures were grown for an additional 18 h before being collected for RNA (Northern) analyses. Yeast strains MES111-140, MES111-P6, TLG105-140, and TLG105-P6 were cotransformed with pGBK-His33/DI72/CUP1 and pCM189-TET-His92. The transformed yeast strains were pregrown at 23°C for 12 h and then shifted to the indicated temperature in SC-UHL-glucose medium containing 50 μM CuSO4 for 48 h.
In vitro TBSV RNA cleavage assay with purified RNase MRP.
Purification of RNase MRP and the RNase MRP cleavage assay were performed as previously described (4, 15), with some modifications in order to make sure that all of the RNase P was removed from the preparation (28). The TBSV DI-72(+) RNA template and its derivatives were obtained by in vitro transcription with T7 RNA polymerase and, if indicated, labeled with [α-32P]UTP (36). The cleavage reaction was performed at 37°C for 60 min in buffer A (0.3 ml of 1 M HEPES-KOH, pH 7.4, 1 ml of 1 M potassium acetate, 0.02 ml of 1 M magnesium acetate, 0.1 M dithiothreitol [DTT], 10 U RNase inhibitor). After phenol-chloroform extraction and ammonium acetate-isopropanol precipitation, the [α-32P]UTP-labeled products were analyzed in a denaturing 5% PAGE-8 M urea gel, followed by phosphorimager analysis.
Primer extension.
The cleavage assay with purified RNase MRP was performed and RNA was purified as described above. Primer extension was carried out as previously described (40a), using oligonucleotides (labeled on the 5′ end with [γ-32P]ATP and polynucleotide kinase) that hybridize to three different positions in TBSV, namely, primers 313 (position 3820) (CCCAACAAGAGTAACCTGTATGCTATGCCA), 1160 (position 1510) (TTCTCTGCTTTTACGAAGGTAGT), and 4074 (position 708) (TAATACGACTCACTATAGGAGACAATCTGTCGCTTCTCT). After phenol-chloroform extraction and ammonium acetate-isopropanol precipitation, the [γ-32P]ATP-labeled cDNA products were analyzed in a denaturing 5% PAGE-8 M urea gel, followed by phosphorimager analysis.
Tombusvirus RNA analysis.
TBSV RNA was analyzed using total RNA preparations from yeast, plants, or in vitro reaction mixtures, and Northern blot analyses were performed as described previously (5, 6). For Northern blot detection of various regions of TBSV DI-72 repRNA, degRNAs, or recRNAs, we prepared α-32P-labeled probes by T7 transcription from PCR products obtained with the following primers: for TBSV region I, primers 20 (GGAAATTCTCCAGGATTTCTC) and 15 (GTAATACGACTCACTATAGGGCATGTCGCTTGTTTGTTGG); for TBSV region II, primers 1164 (AGAAACGGGAAGCTCGCTCGCACT) and 14 (GTAATACGACTCACTATAGGGTTCTCTGCTTTTACGAAG); for TBSV region III, primers 1165 (AGCGAGTAAGACAGACTCTTCA) and 23 (GTAATACGACTCACTATAGGGACCCAACAAGAGTAACCTG); for TBSV region IV, primers 1166 (ATTCCTGTTTACGAAAGTTAGGT) and 22 (GTAATACGACTCACTATAGGGCTGCATTTCTGCAATGTTCC); for CNV region IV, primers 312 (GCTGTCAGTCTAGTGGA) and 22 (GTAATACGACTCACTATAGGGCTGCATTTCTGCAATGTTCC). Northern blots were imaged with a Typhoon imager (GE Healthcare) and were analyzed by the ImageQuant program.
Virus-induced silencing of Nicotiana benthamiana plants.
The VIGS assay was described previously (19, 50). After 9 days of VIGS, the upper leaves were coagroinfiltrated with pGD-CNV and pGD-DI72. Leaf samples were collected from the coagroinfiltrated upper leaves 4 days after the second infiltration. Total RNA was extracted by grinding same-sized leaf samples in liquid nitrogen. After being grinded, the sample was resuspended in RNA extraction buffer (50 mM sodium acetate, pH 5.2, 10 mM EDTA, 1% SDS) and phenol and incubated for 5 min at 65°C in 96-deep-well plates. After another phenol-chloroform extraction and ethanol precipitation, the total RNA was dissolved in water, separated in a denaturing 5% PAGE-8 M urea gel, and electroblotted onto a nylon filter (Hybond N), and Northern blotting was performed with the indicated probes.
RESULTS
Downregulation of SNM1 expression decreases the accumulation of partially degraded TBSV repRNAs in yeast cells.
Previous work with TBSV revealed a connection between RNA degradation pathways and RNA recombination (6, 19, 46). Analysis of the recombination junctions in TBSV recombinant RNAs (recRNAs) and sequences at the ends of partially degraded TBSV RNA products suggested that the 3′ degradation products (3′Fr degRNAs) likely serve as recombination intermediates promoting template-switching events (Fig. 1). These partially degraded degRNAs are easily detectable when the Xrn1p 5′-to-3′ exoribonuclease is inactivated, as in xrn1Δ or met22Δ yeast strains (6, 21a, 46).
To identify host endoribonucleases involved in generation of TBSV degRNAs, we created a set of 6 double-knockout yeast strains missing known endoribonuclease genes, such as NGL1, NGL2, NGL3, IRE1, DOM34, and RNT1, and 2 strains with downregulatable essential endoribonuclease genes (YSH1 and SNM1) (26) in combination with a met22Δ mutation. Deletion of MET22 in these yeast strains helped to detect the generation of degRNAs, likely due to reduced Xrn1p activity (21a).
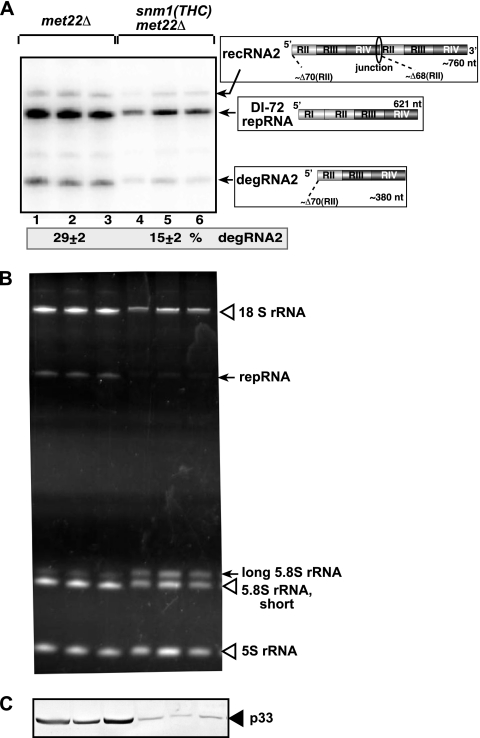
After launching TBSV repRNA replication from the GAL1 promoter by addition of galactose to the medium, we analyzed the accumulating TBSV repRNAs in the above double-knockout or downregulatable yeast strains. We found that none of the double-knockout yeast strains accumulated fewer degRNAs than the met22Δ strain used as a control (not shown). However, downregulation of expression of the SNM1 gene inhibited the accumulation of degRNAs ∼2-fold (Fig. 2A, lanes 4 to 6). These data suggest that Snm1p could be involved in TBSV repRNA degradation, while the other endoribonucleases tested play no role or their functions are redundant in repRNA degradation in yeast.
FIG. 2.
Downregulation of Snm1p level inhibits the accumulation of TBSV degRNA in yeast lacking Met22p bisphosphate-3′-nucleotidase. The met22Δ yeast strain shows decreased Xrn1p activity, facilitating the detection of endoribonucleolytically cleaved TBSV repRNA products. (A) Northern blot analysis for detection of plus strands of TBSV DI-72 repRNA and the newly formed degRNAs and recRNAs from met22Δ yeast strains. The accumulating repRNA, degRNA, and recRNA are shown with arrows. The numbers at the bottom of the panel show percentages of degRNA2 accumulation normalized to the level of DI-72 repRNA (100% in each sample). The various 5′-to-3′ sequences present in the repRNA and the generated recRNAs and degRNAs are shown on the right. The data were obtained from 24 independent yeast streaks. (B) Ethidium bromide-stained gel image showing partial processing of 5.8S rRNA in yeast with downregulated Snm1p. (C) Western blot image showing reduced expression level of p33 replication protein in yeast with downregulated Snm1p.
Reducing the Snm1p level in yeast reduced the accumulation level of the p33 replication protein (down to 33%) (Fig. 2C) and also decreased the level of the smaller 5.8S rRNA (by 40%) in this yeast strain (Fig. 2B, lanes 4 to 6) (3). The low p33 level is likely responsible for the 50% reduction in the TBSV repRNA accumulation level (21). Overall, Snm1p may affect TBSV repRNA degradation directly by leading to the cleavage of the TBSV repRNA, while inhibition of TBSV repRNA accumulation could be due to an indirect effect on the p33 level.
Overexpression of NME1, the RNA component of the RNase MRP, increases RNA recombination and the accumulation of partially degraded TBSV repRNAs in yeast cells.
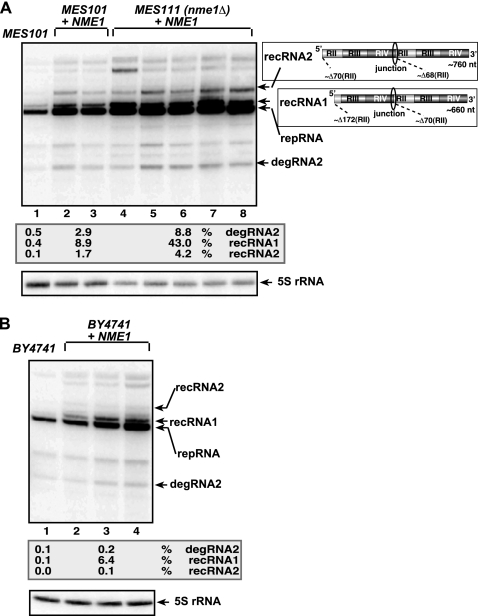
Since Snm1p is a component of the essential RNase MRP endoribonuclease complex, it is possible that RNase MRP is responsible for the endonucleolytic cleavage of the TBSV repRNA in yeast. To test this idea, we overexpressed NME1, which is the essential RNA component of the RNase MRP complex (47), from a plasmid in the wild-type (wt) MES101 and mutant MES111 (nme1Δ) yeast strains. The level of recRNAs increased ∼20- to 100-fold in yeast expressing the plasmid-borne wt NME1 (Fig. 3A, lanes 2 to 8) compared with the level in wt yeast expressing a normal level of wt NME1 (lane 1). As expected, the degRNA levels were also increased, up to ∼8-fold, in yeast overexpressing plasmid-borne wt NME1 (Fig. 3A). A similar trend of increased levels of recRNAs and degRNAs was also observed when we tested yeast strain BY4741, expressing plasmid-borne wt NME1 (Fig. 3B, lanes 2 to 4). Overall, these data suggest that the NME1 RNA is involved in endonucleolytic cleavage of TBSV repRNA and that overexpression of NME1 RNA stimulates TBSV RNA recombination in yeast.
FIG. 3.
Increased accumulation of TBSV recRNAs in yeast overexpressing plasmid-borne NME1 RNA. (A) Northern blot analysis of total RNA samples obtained from the shown yeast strains overexpressing wt NME1 RNA from a plasmid. The accumulating repRNA, degRNA, and recRNAs are shown with arrows. The numbers at the bottom of the panel show percentages of degRNA2, recRNA1, and recRNA2 accumulation normalized to the level of DI-72 repRNA (100% in each sample). The various sequences present in the (+)repRNA and the newly formed recRNAs are shown schematically on the right. (B) Northern blot analysis of total RNA samples obtained from yeast strain BY4741, with a normal level of NME1 RNA expression from the chromosome (lane 1) or overexpressing wt NME1 RNA from a plasmid and from the chromosome (lanes 2 to 4).
Expression of a temperature-sensitive mutant of NME1 decreases the accumulation of partially degraded TBSV repRNAs in yeast cells grown at a semipermissive temperature.
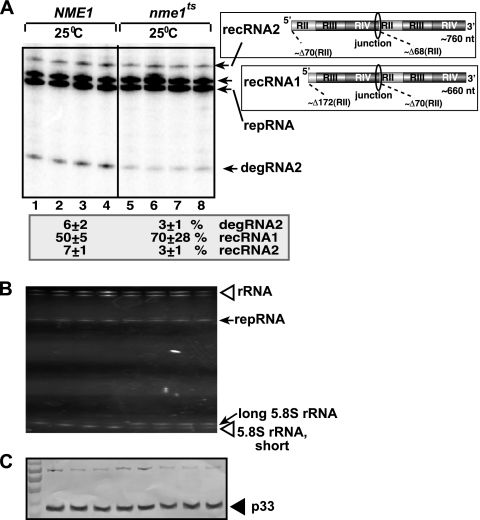
To further test the significance of NME1 RNA in endonucleolytic cleavage of TBSV repRNA, we expressed a temperature-sensitive mutant of NME1 (nme1ts) from a plasmid in an nme1Δ yeast strain. After launching TBSV repRNA replication, yeast cells were grown at various permissive and semipermissive temperatures, and TBSV repRNA accumulation was tested. Interestingly, TBSV degRNA accumulated at a 2-fold reduced level in the nme1ts yeast compared with that in yeast expressing the plasmid-borne wt NME1 RNA at the semipermissive temperature of 25°C (Fig. 4A). Indeed, nme1ts-containing RNase MRP was partially defective in processing the 5.8S rRNA at 25°C (Fig. 4B).
FIG. 4.
Reduced accumulation of TBSV degRNA in yeast expressing nme1ts RNA from a plasmid. (A) Northern blot analysis of total RNA samples obtained from nme1Δ yeast expressing plasmid-borne wt NME1 RNA or a temperature-sensitive nme1ts RNA at the semipermissive temperature of 25°C. The accumulating repRNA, degRNA, and recRNAs are shown with arrows. The numbers at the bottom of the panel show percentages of degRNA2, recRNA1, and recRNA2 accumulation normalized to the level of DI-72 repRNA (100% in each sample). The various sequences present in the (+)repRNA and the newly formed recRNAs are shown schematically on the right. (B) Ethidium bromide-stained gel image showing the accumulation of repRNA. (C) Western blot image showing the expression level of p33 replication protein in the above yeast strains.
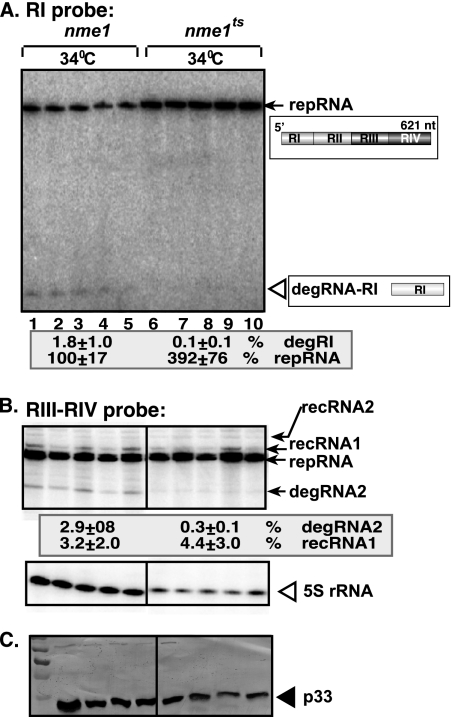
To test the effect of nme1ts on TBSV RNA degradation at a higher semipermissive temperature, we used 34°C during yeast growth, followed by testing of TBSV RNAs via Northern blotting (Fig. 5). We found that accumulation of degRNA-RI and degRNA2 decreased ∼20- and ∼10-fold, respectively, in yeast expressing nme1ts versus yeast with plasmid-borne wt NME1 RNA (Fig. 5A and B, lanes 6 to 10 versus lanes 1 to 5). Also, the accumulation of repRNA increased ∼4-fold in the yeast strain with plasmid-borne nme1ts versus that with plasmid-borne wt NME1 RNA (Fig. 5A, lanes 6 to 10 versus lanes 1 to 5). Overall, our results are consistent with the model that NME1 RNA is involved in TBSV repRNA degradation. Since both Snm1p and NME1 RNA are essential parts of the yeast RNase MRP, we suggest that the whole RNase MRP complex is involved in TBSV repRNA degradation as well as in viral RNA recombination in yeast.
FIG. 5.
Increased accumulation of TBSV repRNA and decreased level of degRNAs in yeast expressing NME1 or nme1ts RNA from a plasmid at the semipermissive temperature of 34°C. (A) Northern blot analysis of total RNA samples obtained from nme1Δ yeast cultured at 34°C. The yeast cells expressed plasmid-borne wt NME1 or nme1ts RNA. The accumulating repRNA and degRNA-RI were detected with an RI probe that does not detect recRNAs. The numbers at the bottom of the panel show percentages of repRNA and degRNA-RI accumulation (the latter was normalized to the level of DI-72 repRNA). (B) Northern blot analysis of the above total RNA samples, using an RIII/RIV probe that also detects recRNAs, in addition to repRNA and degRNAs. The bottom image shows a Northern blot of the above total RNA samples, using a 5S rRNA probe as a loading control. Note that the level of repRNA was normalized based on the loading control. (C) Western blot image showing expression level of the p33 replication protein in the above yeast strains.
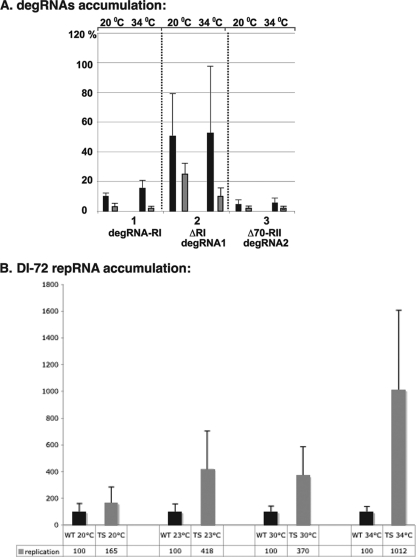
Since early work indicated that both exo- and endoribonucleases are involved in TBSV RNA degradation, we utilized a yeast strain lacking XRN1 and expressing nme1ts (nme1ts xrn1Δ). While yeast cells expressing wt Xrn1p and NME1 RNA from a chromosomal location do not accumulate TBSV degRNAs at a detectable level (46), double mutant yeast cells at the permissive temperature of 20°C (Fig. 5A) accumulated three degRNAs in easily detectable amounts, together reaching as much as ∼70% of the repRNA level (Fig. 6A). On the other hand, partial inactivation of nme1ts in yeast (nme1ts xrn1Δ) at 34°C reduced the total degRNA level to ∼20% of the repRNA level (Fig. 6A). The accumulation of repRNA was increased to an ∼10-fold higher level in nme1ts xrn1Δ yeast than in xrn1Δ yeast at the semipermissive temperature of 34°C (Fig. 6B). These data further strengthen the model that RNase MRP is involved in TBSV RNA degradation in yeast.
FIG. 6.
Partial inactivation of RNase MRP decreases formation of degRNAs and increases accumulation of repRNA in yeast. (A) Relative accumulation levels of three different degRNAs in nme1Δ xrn1Δ yeast (expressing the plasmid-borne nme1ts RNA) cultured at either the permissive temperature of 20°C or the semipermissive temperature of 34°C (gray bars). A comparable analysis was performed with degRNAs in nme1Δ xrn1Δ yeast expressing the plasmid-borne wt NME1 RNA, which is not temperature sensitive (black bars). The measurements are based on Northern blot analysis of total RNA samples obtained from the above nme1Δ yeast with either an RI probe or an RIV probe. Note that the above yeast strain lacks Xrn1p exoribonuclease to facilitate the detection of endoribonucleolytically cleaved degRNAs. (B) Relative accumulation of TBSV DI-72 repRNA in nme1Δ xrn1Δ yeast expressing plasmid-borne wt or nme1ts (TS) RNA. The yeast cells were cultured at either the permissive temperatures of 20°C and 23°C or the semipermissive temperatures of 30°C and 34°C. The measurements are based on Northern blot analysis of total RNA samples obtained from the above nme1Δ xrn1Δ yeast with an RIV probe. Note that the level of repRNA was normalized based on the 5S rRNA probe as a loading control.
Purified RNase MRP cleaves TBSV repRNA in vitro.
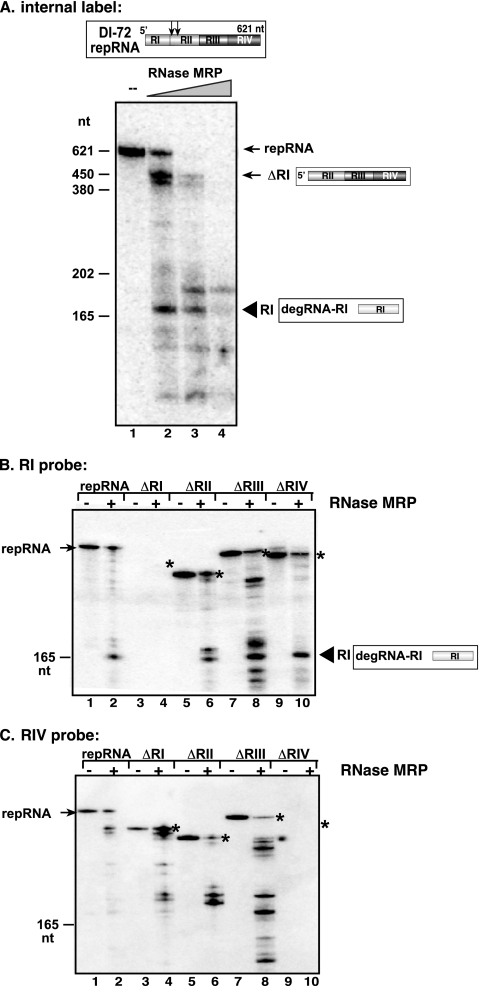
The above data suggested that the RNase MRP could be responsible for the endonucleolytic cleavage of TBSV repRNA in yeast cells. To test this directly, we used highly purified RNase MRP and 32P-labeled TBSV (+)repRNA in an in vitro cleavage assay. PAGE analysis of the products revealed that RNase MRP can cleave the TBSV repRNA in vitro and that the most abundant cleavage products have cleavage sites in the portion of the repRNA that is also cleaved in yeast cells, i.e., in the vicinity of the RI-RII border in DI-72 repRNA (Fig. 7A). Indeed, Northern blot analysis of the RNase MRP-treated repRNA and its deletion versions with RI and RIV probes confirmed that the one or two major cleavage sites are located in the vicinity of the RI-RII border (Fig. 7B and C). Thus, the in vitro cleavage data are consistent with the in vivo cleavage pattern (6), suggesting that RNase MRP could participate directly in partial degradation of TBSV repRNA.
FIG. 7.
In vitro endoribonucleolytic cleavage of TBSV DI-72 (+)repRNA by RNase MRP. (A) Increasing amounts of highly purified RNase MRP were added to internally labeled DI-72 RNA, followed by denaturing PAGE analysis. The bands likely representing DI-ΔRI and RI, which are due to endoribonucleolytic cleavage of the internally labeled DI-72 RNA, are marked. To test the origins of the different endoribonucleolytic cleavage products, we used RI (B) and RIV (C) probes and unlabeled repRNAs. Note that in addition to the full-length DI-72 RNA, we also used truncated derivatives lacking one of the four regions of DI-72, as indicated by the name of the given RNA. Asterisks indicate the input-sized RNAs (not visible are those input RNAs which had deletion of the region targeted by the probe), while the endoribonucleolytic cleavage product representing RI is marked with a filled arrowhead.
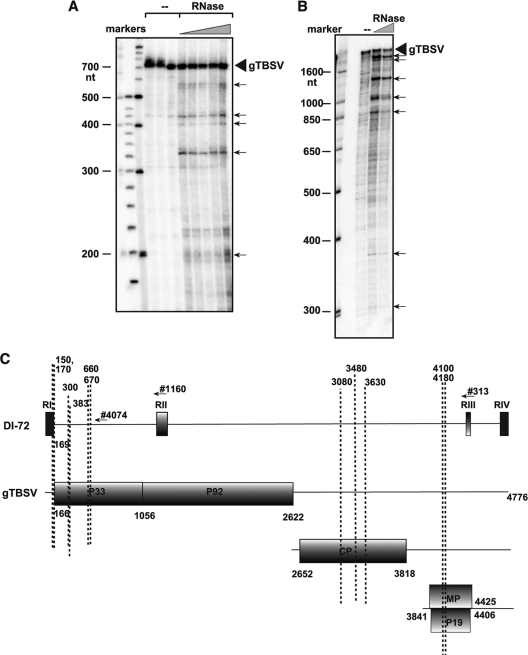
We also tested the cleavage sites in the 4,800-nucleotide TBSV genomic RNA (gRNA) in vitro (Fig. 8). RNase MRP was found to show endonuclease activity on TBSV gRNA as well, resulting in many discrete products in a reverse transcription-extension assay. Based on this in vitro result, we predict that the TBSV gRNA is also sensitive to RNase MRP-mediated endonucleolytic cleavage.
FIG. 8.
In vitro endoribonucleolytic cleavage of TBSV genomic (+)RNA by RNase MRP. (A) Increasing amounts of highly purified RNase MRP were added to unlabeled TBSV gRNA, followed by primer extension and denaturing PAGE analysis. The primer extension was performed with labeled 708R primer. Bands representing mapped endoribonucleolytically cleaved products are marked with arrows. We used a 5′-phosphorylated 100-bp ladder (NE Biolabs) and a 25-bp ladder (Gibco-BRL) as markers. (B) Similar cleavage-site mapping experiment to that in panel A, except that the primer extension was performed with labeled RIII (3820) primer. (C) Locations of mapped endoribonucleolytic cleavage sites in TBSV gRNA. Note that we show only the more frequent/repeatable endoribonucleolytic cleavage sites and their approximate locations.
Knockdown of expression of the plant homolog of NME1 also reduces the accumulation of TBSV degRNAs.
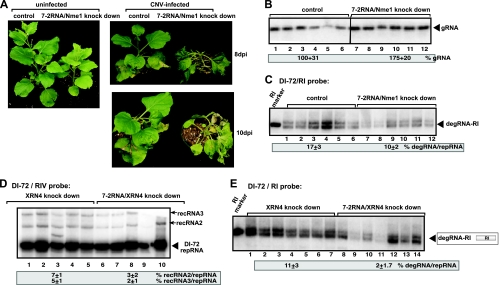
The RNase MRP is an essential and conserved endoribonuclease in plants and animals (24). To obtain evidence that the RNase MRP also plays a role in TBSV RNA degradation in plants, we cloned the homolog of the yeast NME1 gene, called 7-2 RNA (24), into a Tobacco rattle virus (TRV)-based silencing vector. Knockdown of the 7-2 RNA level in N. benthamiana plants resulted in a moderately stunting phenotype (Fig. 9A, left panel). Inoculation of 7-2 RNA knockdown plants with CNV helper tombusvirus resulted in more severe symptoms, followed by the death of plants 2 to 3 days earlier than the death of CNV-infected control plants (Fig. 9A, right panels). The severe phenotype correlated with the increased accumulation of CNV RNA (1.7-fold) in the 7-2 RNA knockdown plants (Fig. 9B, lanes 7 to 12 versus lanes 1 to 6). To test for TBSV replicon RNA degradation in the 7-2 RNA knockdown plants, we combined the TBSV DI-72 RNA with the CNV helper virus in the inoculum, followed by testing the accumulation of TBSV degRNAs in plants. This assay showed an almost 2-fold reduction in degRNA accumulation (specifically that of degRNA-RI), from 17% to 10%, in the control plants (Fig. 9C, lanes 7 to 12 versus lanes 1 to 6).
FIG. 9.
Role of RNase MRP in TBSV RNA degradation and RNA recombination in plants. (A) (Left) Phenotype of 7-2 RNA/NME1 knockdown N. benthamiana plants 14 days after agroinfiltration with TRV silencing vectors. (Right) Effect of knockdown of 7-2 RNA/NME1 on symptoms caused by CNV infection in N. benthamiana 8 and 10 days after the second agroinfiltration. VIGS was performed via agroinfiltration of vector (TRV) carrying the 7-2 RNA/NME1 sequence or the TRV empty vector (as a control). Coagroinfiltration to express TBSV DI-72 repRNA together with CNV gRNA was done 9 days after silencing of 7-2 RNA/NME1 expression by agroinfiltration. (B) Northern blot analysis of CNV gRNA accumulation in the agroinfiltrated leaves of 7-2 RNA/NME1 knockdown or control N. benthamiana plants 4 days after the second agroinfiltration. rRNA (not shown) was used as a loading control. The plant leaf samples were obtained from 5 separate experiments (total of 5 × 48 samples). (C) Decreased accumulation of TBSV degRNA-RI in 7-2 RNA/NME1 knockdown N. benthamiana plants compared to that in the control plants, which were agroinfiltrated with the TRV empty vector, 4 days after the second agroinfiltration. The accumulation level of degRNA-RI was normalized to the level of DI-72 repRNA. Note that the RI probe was specific for the TBSV repRNA and that the helper CNV gRNA or its degradation products were not detected. (D) Northern blot analysis of TBSV repRNA and newly formed recRNAs in agroinfiltrated leaves of 7-2 RNA/NME1 XRN4 double-knockdown or XRN4 single-knockdown control N. benthamiana plants. The numbers at the bottom of the panel show percentages of recRNA2 and recRNA3 accumulation normalized to the level of DI-72 repRNA (100% in each sample). rRNA (not shown) was used as a loading control. Note that the RIV probe was specific for the TBSV repRNA and that the helper CNV gRNA or its degradation products were not detected. (E) Decreased accumulation of TBSV degRNA-RI in 7-2 RNA/NME1 XRN4 double-knockdown plants compared to that in the control XRN4 single-knockdown N. benthamiana plants. See further details above as described for panel C.
Since the plant cytosolic Xrn4p exoribonuclease, similar to yeast Xrn1p, might quickly remove the partially degraded TBSV degRNAs (19) in 7-2 RNA knockdown plants, we also made Xrn4/7-2 RNA double-knockdown plants. We found that the level of TBSV degRNA-RI decreased ∼5-fold in the double-knockdown plants (Fig. 9E, lanes 8 to 14 versus lanes 1 to 7), suggesting that the RNase MRP is involved in TBSV RNA degradation in N. benthamiana. The effect of silencing of 7-2 RNA on viral RNA recombination was also measurable (decrease of ∼60%) in the double-knockdown plants (Fig. 9D, lanes 6 to 10 versus lanes 1 to 5). Overall, the obtained data suggest that RNase MRP is also involved in TBSV RNA degradation in plants, while its effect on TBSV RNA recombination seems to be moderate.
DISCUSSION
RNA degradation is an important process that, together with RNA synthesis, controls the steady-state level of host (7) and possibly viral RNAs in infected cells. It is thought that the general and specialized RNA degradation pathways of the cell also affect viral RNA levels in addition to RNAi, which is an induced antiviral RNA degradation pathway in plants (8, 9, 11). Since S. cerevisiae lacks components of the RNAi machinery, it is suitable to test the involvement of other RNA degradation pathways in TBSV RNA accumulation. Indeed, when the major 5′-3′ mRNA degradation pathway is debilitated by deletion of the Xrn1 exoribonuclease in yeast, many partial TBSV RNA degradation products become detectable (6, 34). This helped in the realization that yeast endoribonucleases are also involved in TBSV RNA degradation (6). Testing of 8 different yeast endoribonucleases in this work led to the identification of the role of RNase MRP in TBSV RNA degradation (Fig. 2). The 10 known protein components in RNase MRP, including Smn1p and the NME1 RNA, are essential for yeast viability (28, 40). Therefore, instead of knockout strains, we used yeast strains that either downregulated the Smn1p level or expressed a temperature-sensitive NME1 RNA. These studies showed that both Smn1p and NME1 RNA, likely as components of the RNase MRP, affect the amount of TBSV degRNAs (Fig. 2 and 3). Indeed, a highly purified RNase MRP showed endoribonuclease activity on TBSV repRNA in vitro (Fig. 7), leading to the generation of endonucleolytically cleaved products, including a cleavage close to the RI-RII border that is frequently observed in degRNAs from yeast as well (6, 20, 23). Since the genomic RNA of TBSV is also a substrate of the purified RNase MRP (Fig. 8), we propose that the TBSV RNA in general is subject to endonucleolytic attack in cells.
Based on the experimental data presented in this paper, it is not yet known if RNase MRP is the only endoribonuclease capable of cleaving TBSV repRNA in yeast or whether other endoribonucleases might also be involved or could play a partially complementary function in TBSV repRNA degradation. Accordingly, previous in vitro studies with purified Ngl2p endoribonuclease showed specific endoribonucleolytic cleavage of TBSV repRNA (6), but deletion of NGL2 did not decrease the amount of degRNAs in yeast (data not shown). Therefore, it is not known if Ngl2p is involved in TBSV degradation in yeast and its function is redundant due to the presence of other similar endoribonucleases, such as Ngl1p and Ngl3p. Moreover, Ngl2p may be localized to a location away from the TBSV RNA, since it has been shown to be involved in rRNA processing in the nucleolus. In contrast, RNase MRP and Xrn1 have clearly been shown to localize together in the cytoplasm (15). Since RNase MRP is essential for yeast viability (and for that of mammals and plants as well), we could not test the degradation of TBSV repRNA in the complete absence of functional RNase MRP in vivo. Downregulation of the Smn1p level and inhibition of nme1ts function by applying a semipermissive temperature likely caused only a partial reduction in RNase MRP activity, as suggested by the mixture of long and short 5.8S RNA species in the samples (Fig. 2B and 3B). The difference in the remaining RNase MRP activity in yeast when the Smn1p level was downregulated or the function of nme1ts was inhibited could also explain why the accumulation of TBSV repRNA was affected differently by these two approaches: downregulation of the Smn1p level reduced repRNA accumulation due to low levels of p33, while inactivation of the function of nme1ts RNA increased the TBSV repRNA level, likely due to a low level of viral RNA degradation. However, both approaches led to reductions of the relative levels of degRNAs (compared with the repRNA products), demonstrating that the RNase MRP is involved in endoribonucleolytic cleavage and degradation of the TBSV repRNA. Overall, the reduced level of TBSV degRNAs in the above yeast strains could be due to residual activity of the RNase MRP in yeast when the Smn1p level was downregulated or the function of nme1ts was inhibited. Alternatively, the presence of some TBSV degRNAs in these yeast strains could be due to additional endoribonucleases, whose number is still undetermined for yeast and other eukaryotic cells. Indeed, the number of known endoribonucleases has increased even since the beginning of this work, for example, by the recent discovery of an endoribonucleolytic activity of the exosome, including its Dis3p subunit (41). Similarly, the human PIN domain-containing SMG6 protein, an NMD component, shows endoribonuclease activity (12). Therefore, several as yet uncharacterized proteins/complexes could also be involved in endoribonucleolytic cleavage of TBSV repRNA in yeast, in addition to the characterized RNase MRP.
The endoribonucleolytic cleavage of TBSV RNAs seems to occur not only in yeast but also in plant protoplasts and in XRN4 knockdown whole plants as well (5, 6, 19). In addition, the profiles of the generated degRNAs are similar in yeast and plant cells, suggesting that comparable mechanisms of TBSV RNA degradation exist in these organisms. Accordingly, we showed previously that the cytosolic Xrn4p 5′-3′ exoribonuclease in N. benthamiana plays a comparable role to that of the yeast Xrn1p 5′-3′ exoribonuclease in TBSV RNA degradation (5, 19). Moreover, silencing of 7-2 RNA, a homolog of yeast NME1 RNA (24), in N. benthamiana led to accelerated development of virus-induced symptoms, increased tombusvirus gRNA accumulation, and decreased degRNA-RI accumulation (Fig. 9A and B), indicating that degradation of tombusvirus RNAs is inhibited in these plants with reduced RNase MRP activity. The reduced level of TBSV repRNA degradation was especially easily detectable in the XRN4/7-2 RNA double-knockdown plants, in which Xrn4p could not rapidly degrade the small TBSV degRNAs (Fig. 9E). Future experiments will address if the conserved POP factors or Rmp1 that are critical components of yeast and plant RNase MRP affect tombusvirus RNA degradation. Also, in addition to the RNase MRP, it is very likely that additional RNA degradation pathways are involved in TBSV RNA degradation, including the inducible RNA silencing pathway (8).
The endoribonucleolytic cleavage of TBSV RNA is important not only for viral RNA degradation but for RNA recombination as well, due to the participation of some RNA degradation products in the RNA recombination process (5, 19, 20, 23, 45, 46). In this work, we also obtained supporting evidence for the role of endoribonucleolytic cleavage of TBSV RNA, specifically by the RNase MRP, in TBSV RNA recombination (Fig. 3). Overexpression of the NME1 RNA component of RNase MRP from a plasmid resulted in increased levels of recRNA and degRNA accumulation (Fig. 3) compared with those in yeast showing a normal expression level of NME1 RNA from the regular chromosomal location. Also, XRN4/7-2 RNA double-knockdown plants supported 5-fold less degRNA and 2-fold less recRNA accumulation than did XRN4 knockdown plants (Fig. 9D), suggesting that recombination is less robust when the RNase MRP is less active in producing degRNAs. It is important that other RNA degradation pathways, such as 5′-3′ degradation via Xrn1p (Xrn4p) exoribonuclease, and the gene silencing pathways are also implicated in tombusvirus recombination (5, 6, 19, 25). These data firmly support the involvement of several RNA degradation pathways in tombusvirus RNA recombination.
Acknowledgments
We thank Judit Pogany and Daniel Barajas for their comments on the manuscript. We also thank Kunj Pathak for providing plasmids.
This work was supported by NIH/NIAID grants 5R21A1072170 and 5R01GM063798 to M.E.S.
Footnotes
Published ahead of print on 20 October 2010.
REFERENCES
- 1.Aaziz, R., and M. Tepfer. 1999. Recombination in RNA viruses and in virus-resistant transgenic plants. J. Gen. Virol. 80:1339-1346. [DOI] [PubMed] [Google Scholar]
- 2.Barajas, D., Y. Jiang, and P. D. Nagy. 2009. A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, T., T. R. Reilly, M. Cerio, and M. E. Schmitt. 1999. Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol. Cell. Biol. 19:7857-7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, T., and M. E. Schmitt. 2001. Characterization of ribonuclease MRP function. Methods Enzymol. 342:135-142. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, C. P., H. M. Jaag, M. Jonczyk, E. Serviene, and P. D. Nagy. 2007. Expression of the Arabidopsis Xrn4p 5′-3′ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology 368:238-248. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, C. P., E. Serviene, and P. D. Nagy. 2006. Suppression of viral RNA recombination by a host exoribonuclease. J. Virol. 80:2631-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiba, Y., and P. J. Green. 2009. mRNA degradation machinery in plants. J. Plant Biol. 52:114-124. [Google Scholar]
- 8.Csorba, T., V. Pantaleo, and J. Burgyan. 2009. RNA silencing: an antiviral mechanism. Adv. Virus Res. 75:35-71. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Pendon, J. A., and S. W. Ding. 2008. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46:303-326. [DOI] [PubMed] [Google Scholar]
- 10.Dinesh-Kumar, S. P., R. Anandalakshmi, R. Marathe, M. Schiff, and Y. Liu. 2003. Virus-induced gene silencing. Methods Mol. Biol. 236:287-294. [DOI] [PubMed] [Google Scholar]
- 11.Ding, S. W., and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130:413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberle, A. B., S. Lykke-Andersen, O. Muhlemann, and T. H. Jensen. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 16:49-55. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 14.Gill, T., J. Aulds, and M. E. Schmitt. 2006. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J. Cell Biol. 173:35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill, T., T. Cai, J. Aulds, S. Wierzbicki, and M. E. Schmitt. 2004. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 24:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, H., and A. E. Simon. 2000. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc. Natl. Acad. Sci. U. S. A. 97:12451-12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hema, M., K. Gopinath, and C. Kao. 2005. Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus. J. Virol. 79:1417-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermanns, P., A. A. Bertuch, T. K. Bertin, B. Dawson, M. E. Schmitt, C. Shaw, B. Zabel, and B. Lee. 2005. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum. Mol. Genet. 14:3723-3740. [DOI] [PubMed] [Google Scholar]
- 19.Jaag, H. M., and P. D. Nagy. 2009. Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386:344-352. [DOI] [PubMed] [Google Scholar]
- 20.Jaag, H. M., J. Pogany, and P. D. Nagy. 2010. A host Ca2+/Mn2+ ion pump is a factor in the emergence of viral RNA recombinants. Cell Host Microbe 7:74-81. [DOI] [PubMed] [Google Scholar]
- 21.Jaag, H. M., J. Stork, and P. D. Nagy. 2007. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368:388-404. [DOI] [PubMed] [Google Scholar]
- 21a.Jaag, H. M., and P. D. Nagy. 2010. The combined effect of environmental and host factors on the emergence of viral RNA recombinants. PLoS Pathog. 6:e1001156. [DOI] [PMC free article] [PubMed]
- 22.Janke, C., M. M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947-962. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, Y., C. P. Cheng, E. Serviene, N. Shapka, and P. D. Nagy. 2010. Repair of lost 5′ terminal sequences in tombusviruses: rapid recovery of promoter- and enhancer-like sequences in recombinant RNAs. Virology 404:96-105. [DOI] [PubMed] [Google Scholar]
- 24.Kiss, T., C. Marshallsay, and W. Filipowicz. 1992. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 11:3737-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, P., S. Uratsu, A. Dandekar, and B. W. Falk. 2009. Tomato bushy stunt virus recombination guided by introduced microRNA target sequences. J. Virol. 83:10472-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W. M., T. Barnes, and C. H. Lee. 2010. Endoribonucleases—enzymes gaining spotlight in mRNA metabolism. FEBS J. 277:627-641. [DOI] [PubMed] [Google Scholar]
- 27.Li, Z., D. Barajas, T. Panavas, D. A. Herbst, and P. D. Nagy. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82:6911-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Q., S. Wierzbicki, A. S. Krasilnikov, and M. E. Schmitt. 2010. Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components. RNA 16:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy, P. D. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46:217-242. [DOI] [PubMed] [Google Scholar]
- 30.Nagy, P. D., C. D. Carpenter, and A. E. Simon. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. U. S. A. 94:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy, P. D., and J. Pogany. 2010. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv. Virus Res. 76:123-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Newbury, S. F. 2006. Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 34:30-34. [DOI] [PubMed] [Google Scholar]
- 34.Panaviene, Z., T. Panavas, and P. D. Nagy. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panaviene, Z., T. Panavas, S. Serva, and P. D. Nagy. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao, A. L., and T. C. Hall. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67:969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 39.Roossinck, M. J. 2003. Plant RNA virus evolution. Curr. Opin. Microbiol. 6:406-409. [DOI] [PubMed] [Google Scholar]
- 40.Salinas, K., S. Wierzbicki, L. Zhou, and M. E. Schmitt. 2005. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 280:11352-11360. [DOI] [PubMed] [Google Scholar]
- 40a.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schaeffer, D., B. Tsanova, A. Barbas, F. P. Reis, E. G. Dastidar, M. Sanchez-Rotunno, C. M. Arraiano, and A. van Hoof. 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 16:56-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt, M. E., and D. A. Clayton. 1994. Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 8:2617-2628. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt, M. E., and D. A. Clayton. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7935-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt, M. E., and D. A. Clayton. 1992. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 6:1975-1985. [DOI] [PubMed] [Google Scholar]
- 45.Serviene, E., Y. Jiang, C. P. Cheng, J. Baker, and P. D. Nagy. 2006. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 80:1231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serviene, E., N. Shapka, C. P. Cheng, T. Panavas, B. Phuangrat, J. Baker, and P. D. Nagy. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 102:10545-10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shadel, G. S., G. A. Buckenmeyer, D. A. Clayton, and M. E. Schmitt. 2000. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene 245:175-184. [DOI] [PubMed] [Google Scholar]
- 48.Sun, Q., G. H. Choi, and D. L. Nuss. 2009. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 106:17927-17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel, C. T., D. Horn, B. Zabel, A. B. Ekici, K. Salinas, E. Gebhart, F. Ruschendorf, H. Sticht, J. Spranger, D. Muller, C. Zweier, M. E. Schmitt, A. Reis, and A. Rauch. 2005. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 77:795-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, R. Y., and P. D. Nagy. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178-187. [DOI] [PubMed] [Google Scholar]
- 51.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187-226. [DOI] [PubMed] [Google Scholar]
- 52.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, X., and D. L. Nuss. 2008. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U. S. A. 105:16749-16754. [DOI] [PMC free article] [PubMed] [Google Scholar]