Abstract
The downstream prion-like protein (doppel, or Dpl) is a paralog of the cellular prion protein, PrPC. The two proteins have ≈25% sequence identity, but seem to have distinct physiologic roles. Unlike PrPC, Dpl does not support prion replication; instead, overexpression of Dpl in the brain seems to cause a completely different neurodegenerative disease. We report the solution structure of a fragment of recombinant mouse Dpl (residues 26–157) containing a globular domain with three helices and a small amount of β-structure. Overall, the topology of Dpl is very similar to that of PrPC. Significant differences include a marked kink in one of the helices in Dpl, and a different orientation of the two short β-strands. Although the two proteins most likely arose through duplication of a single ancestral gene, the relationship is now so distant that only the structures retain similarity; the functions have diversified along with the sequence.
Keywords: doppel protein structure, NMR, protein structures, prion protein, prion diseases
The disease-causing isoform of the prion protein (PrP), designated PrPSc, causes neurodegeneration in humans and animals. These neurodegenerative diseases include scrapie in sheep, mad cow disease in cattle, and Creutzfeldt–Jakob disease in humans (1). PrPSc is derived from the benign, cellular isoform of the prion protein (PrPC) by a posttranslational process involving a profound conformational change (2). The three-dimensional structure of recombinant PrP from a number of species has been characterized by NMR (3–6). These structures show a folded domain at the C terminus that consists of a three-helix structure with a single disulfide bond and a pair of short β-strands. The N terminus of all of these proteins is unstructured under NMR conditions. The structure of PrPSc remains largely unknown, but molecular modeling and epitope mapping (7) have given important information about the formation of PrPSc from PrPC, which is presumed to occur through a process involving template-directed refolding (8). Conversion of the soluble form of PrP to an insoluble form with some of the characteristics of PrPSc has been demonstrated for mammalian PrPs and yeast prions (9, 10).
The function of PrPC is still unknown, although there is some evidence that the octarepeat sequence in the N-terminal unstructured region of the protein binds Cu(II) ions (11–13). Recent studies suggest that PrPC may function in signal transduction through a pathway involving Fyn kinase (14). It was hoped that deletion of the mouse Prnp gene might cause the appearance of a phenotype that would give a clue to the function of PrPC. Ablation of the PrP gene eliminates susceptibility to prion infection in Prnp0/0 knockout mice (15, 16). Some Prnp0/0 lines were viable and appeared phenotypically normal (17), whereas others were found to develop late-onset ataxia related to degeneration of Purkinje neurons (18). The recent discovery of a paralog of the Prnp gene, termed Prnd (19), with ≈25% identity to all known PrP genes, may provide new approaches to the investigation of prions. Although the encoded protein, doppel (Dpl), is normally not expressed in the central nervous system, it is up-regulated in the Prnp0/0 lines that develop late-onset ataxia (19), and the phenotype can be rescued by crossing the mice with those over-expressing wild-type mouse PrP (20). In addition, sequence alignment (Fig. 1) suggests that Dpl has the same folding topology as the structures of recombinant PrP (19). These observations suggest that Dpl may be capable of replacing PrPC in vivo.
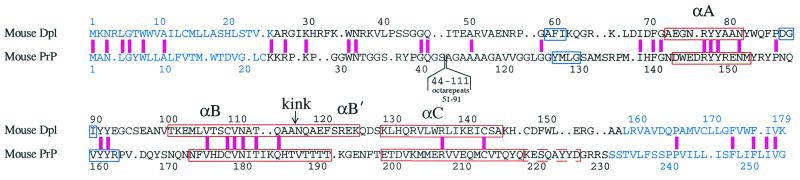
Figure 1.
Alignment of the amino acid sequences of mouse Dpl (19) and mouse PrPC (46). The sequences of the 26–157 construct of Dpl used in the present work, and of the 23–231 construct of mouse PrPC (47), are shown in black letters. Blue letters compose the signal sequence at the N terminus and the C-terminal hydrophobic region. The octarepeat sequence of PrPC (not present in Dpl) has been omitted for brevity. The pink bar indicates identical amino acids. Red and blue boxes indicate the location of α and β secondary structures in Dpl (this work) and mouse PrPC (3). Helices are labeled αA, αB, and αC; the location of the kink in the αB helix is shown. αB′ indicates the B helix C-terminal to the kink. The dashed red box at residues 220–227 of PrPC shows the location of the extended αC helix in Syrian hamster (4), bovine (5), and human (6) PrPC.
The expression of these two proteins in disease and nondisease states, together with their apparent structural similarity, may provide clues as to the physiological functions of PrP and Dpl, as well as to the neuropathology of prion disease. In this article, we report the expression and solution structure determination of recombinant Dpl protein. A comparison of the structures of recombinant Dpl with those of recombinant PrP reveals striking similarities in the secondary structure and overall similarities in the topology. Some significant local structural differences between PrP and Dpl may give insights into the underlying causes of lesions that occur when Dpl is overexpressed in the absence of PrPC. Our studies also suggest why Dpl is not converted into a PrPSc-like isoform.
Materials and Methods
Cloning and Expression of Dpl Residues 26–157 [Dpl(26–157)].
Two primers, 5′-GCGACACTGTCATATGGCAAGGGGCATAAAGCACAG-3′ and 5′-TCGTGACTGGATCCCTACGCAGCTCCCCTTTCCAGC-3′, were designed to amplify the gene encoding Dpl(26–157) by PCR from a template cDNA fragment containing the mouse doppel gene (pcDNAmodoppel1–179; ref. 19). The PCR product was then doubly digested with NdeI and BamHI at 37°C for 2 h and ligated with a NdeI/BamHI predigested pET-21a(+) vector. The new plasmid, with Dpl(26–157) inserted in the right reading frame, was sequenced and named pmodpl26–157. Expression of Dpl(26–157) followed standard protocol and is described here briefly. pmodpl26–157 was freshly transformed Escherichia coli strain BL21 (DE3) each time for expression. One colony from pmodpl26–157/BL21 (DE3) LB plate was inoculated in 50 ml of LB at 37°C until OD600 reached 0.6, and 10 ml of culture was transferred to 1 liter of LB or M9 minimal medium for isotopic labeling. M9 was supplemented with 1 g of 15NH4 and 6 g of glucose for 15N single labeling only, or 1 g of 15NH4 and 2.5 g of [U-13C6]glucose for double labeling. The density of the new culture was allowed to reach OD600 0.6 before 0.4 mM isopropyl β-d-thiogalactoside was added to induce the expression of Dpl(26–157). Cell paste was harvested after overnight induction and stored under −70°C as freezer stock. Target protein content was checked by SDS/PAGE before further work was carried out.
Purification of Dpl Protein.
The location of Dpl(26–157) in the inclusion bodies was determined by using a bacterial expression reagent (B-PER reagent; Pierce). Cell paste (10 g) was resuspended in 50 ml of 25 mM Tris⋅HCl buffer (pH 7.4) and sonicated five times for 1 min each. After centrifugation at 38,000 × g for 40 min, cell pellets were recovered and washed with Tris⋅HCl buffer and then resuspended in 6 M guanidinium chloride (Sigma) supplemented with 5 mM DTT. The resuspension mixture was stirred overnight in the cold, then poured into 1 liter of 50 mM Tris⋅HCl buffer (pH 8.5) containing 600 mM l-arginine, 0.2 mM PMSF protease inhibitor, 5 mM reduced and 0.5 mM oxidized glutathione. After overnight stirring, the solution was centrifuged at 10,000 × g for 30 min; the supernatant was concentrated to 50 ml before dialysis against two 4-liter batches of 50 mM Tris⋅HCl buffer (pH 7.4). The mixture was centrifuged at 8,000 × g for 40 min and was loaded onto an SP column (Amersham Pharmacia) at a flow rate of 2 ml/min. The column was washed with 25 mM Tris⋅HCl buffer (pH 7.4) until the absorbance at 215 nm ran to baseline or became flat. Dpl was eluted on a gradient of 0 M to 1 M NaCl in the same buffer applied in 50 min with a flow rate of 2 ml. The Dpl(26–157)-containing fraction came out at 0.5M NaCl as the dominant band in the elution. Fractions containing only pure Dpl, appearing as a 15-kDa band on Coomassie-stained SDS/PAGE, were pooled and used for analysis by matrix-assisted laser desorption ionization-time-of-flight and electrospray ionization mass spectroscopy, circular dichroism spectroscopy, free sulfhydryl analysis using the Ellmann reagent (Molecular Probes), and NMR spectroscopy. The presence of two disulfide bonds in Dpl was confirmed by mass spectrometry and by the reaction with Ellmann reagent.
NMR Spectroscopy.
NMR samples were prepared with 4 mM 15N or 2 mM 15N/13C uniformly labeled protein in 20 mM sodium acetate-d3 buffer (pH 5.2) in 90% H2O/10% D2O, with 0.005% NaN3 to prevent bacterial growth. All NMR data were acquired on Bruker Avance DRX 600 and DRX 800 spectrometers. Standard 15N and 13C spectra, heteronuclear single quantum correlation (HSQC), HNCO, HNCA (21), CBCA(CO)NH (22), HNCACB (23), C(CO)NH, H(CCO)NH (24) and HCCH-TOCSY (ref. 25; DIPSI-3 spin lock at 28.6 kHz for 15.2 ms) were collected at 26°C for sequential assignments. The 1H chemical shift was referenced internally to 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS) and 13C,15N chemical shifts were indirectly referenced (26). NMR data were acquired at 26°C, processed with nmrpipe (27), and analyzed with nmrview (28).
Structure Determination.
Because the N-terminal residues from 26 to 50 lack medium- or long-range nuclear Overhauser effects (NOEs), and are shown by {1H}-15N NOEs to be dynamically disordered, the structure calculation was performed only for residues 51 to 157. Disulfide bonds between Cys-109/Cys-143 and Cys-95/Cys-148 were enforced during all stages of calculation. Distance constraints were derived from a 15N-edited NOE spectroscopy (NOESY) spectrum (100-ms mixing time) and a 13C-edited NOESY spectrum (80-ms mixing time). Cross peak volumes were classified as strong, medium, or weak, and constraints were assigned to upper distance bounds of 2.7, 3.3, and 5.0 Å, respectively. Dihedral angle constraints were derived from consideration of the secondary structure indicated by chemical shift deviations from random coil values for Cα, together with the use of the secondary-structure prediction program talos (29). Dihedral angles for residues with backbone conformation in the α-region were restrained to −120° ≤ φ ≤ −20° and −80° ≤ ψ ≤ 20°, while the corresponding restraints for residues with backbone dihedral angles in the β-region were −170° ≤ φ ≤ −70° and 60° ≤ ψ ≤ 180°. All nonglycine residues were assigned to the negative φ region, by the application of a restraint −180° ≤ φ ≤ 0° to minimize the probability of spurious positive φ values in the calculated structures. Stereospecific assignments were made for eight residues, based on differences in intensities for intraresidue cross peaks in NOESY spectra. The χ1 angles were restrained for these residues, according to the rotamer indicated by the observed NOEs. Restraints were applied as −20° ≤ χ1 ≤ −100°, 140° ≤ χ1 ≤ 220°, and 20° ≤ χ1 ≤ 100° for the three possible rotamers. The two proline residues in the folded domain were restrained in the trans conformation, on the basis of the absence of strong dαα(i, i + 1) NOEs characteristic of cis-proline.
The initial 50 input structures were obtained by using dyana 1.5 (30) and were refined by using amber 6 (31) with a generalized Born solvent model (32). The refinement consisted of 5-ps molecular dynamics at 600 K, during which restraints (NOE, dihedral, and chiral) were gradually introduced, followed by annealing from 600 K to 0 K over a period of 20 ps. Force constants for the distance restraints were 20 kcal/mol⋅Å2, and for the dihedral angle restraints were 50 kcal/mol⋅rad2, except for the peptide bond ω, for which a force constant of 150 kcal/mol⋅rad2 was used. A second round of amber calculation was performed and the ensemble of 20 structures of lowest amber energy and restraint violation energy was used for further analysis.
Heteronuclear NOE Measurements.
{1H}-15N heteronuclear NOEs were measured at 600-MHz proton frequency by using standard methods (33). The recycle time was 1 s and protons were saturated by 120° pulses at an interval of 18 ms for a total time of 2.7 s.
Results
Design and Preparation of Recombinant Dpl.
Considerable effort was expended to obtain a homogeneous, well folded, monomeric sample of Dpl for NMR studies. A number of constructs, including the full-length sequence, were cloned from mouse cDNA containing the Dpl gene. Expression levels for the full-length sequence were low, but a construct containing residues 26–157 proved to express well in E. coli. It is likely that the low expression level for the full-length protein is related to the hydrophobic nature of the N- and C-terminal sequences (Fig. 1). Residues 1–26 were predicted to be a signal peptide, with a likely cleavage site between residues 26 and 27 (34). Both the N- and C-terminal sequences that were not included in the well expressed construct gave high probabilities for transmembrane helices (35). The gene for residues 26–157 was over-expressed in inclusion bodies in E. coli; it was found that allowing the protein to refold in the crude mixture before purification gave the best yield of the desired product. Refolding conditions included the presence of a mixture of reduced and oxidized glutathione, which allowed the formation of the correct disulfide bonds in the final product. Application of this refolding technique to the protein after purification by HPLC gave less satisfactory results.
After NMR analysis (see below), it became clear that the residues 26–50 of the Dpl(26–157) construct were not structured. A second construct including residues 51–157 was prepared, but was found to be less soluble than the longer construct. Full NMR assignments and secondary structure analysis, as well as solution structure determination were, therefore, performed on the longer construct Dpl(26–157).
Disulfide Bonding.
Full-length Dpl 1–179 contains five cysteines; sequence alignment with PrPC suggests that Cys-109 and Cys-143 should be linked by a disulfide bond. If the topology of Dpl is similar to that of PrPC, then a second disulfide bond, between cysteines 95 and 148, is possible. The remaining cysteine in the full-length sequence, Cys-169, is part of the C-terminal hydrophobic sequence. The presence of two disulfide bonds in the recombinant Dpl construct 26–157 is confirmed by several results. The reaction of the correctly refolded protein with Ellmann reagent [5,5′-dithiobis(2-nitrobenzoic acid)] gave a negative result, consistent with the absence of free thiols. The mass spectrum of the folded protein shows a single peak of the correct molecular weight for a monomer, thus eliminating the possibility that intermolecular disulfide bonds had been formed. The presence of two disulfides in Dpl is also validated by the values of the Cβ chemical shifts of the four Cys residues, which are all close to 38 ppm, compared with the random coil Cβ chemical shift of reduced (cysteine) of 28.0 ppm and oxidized (cystine) of 41.1 ppm. The values are closer to those of oxidized Cys, slightly lowered by the presence of the disulfides in helical or near-helical regions. The disulfide pattern was confirmed by extensive networks of NOEs between Cys-95 and Cys-148, and between Cys-109 and Cys-143. Reaction of Dpl with DTT leads to precipitation of the protein, suggesting that the disulfides are required for its structural integrity.
Resonance Assignments.
Virtually complete backbone and side-chain resonance assignments have been made for Dpl(26–157). The assignments have been deposited in the BioMagResBank (accession no. 4938). The 15N-1H HSQC spectrum of Dpl is shown in Fig. 2. The spectrum indicates that most of the protein is folded into a globular domain. However, the chemical shifts are close to those characteristic of a random coil for residues 26–57, suggesting that this region is unstructured. This suggestion is confirmed by the absence of significant medium- or long-range NOEs associated with this sequence. The chemical shifts show the presence of three helices in the folded domain (residues 58–153), as would be expected if the topology of Dpl resembles that of PrPC; the locations of these helices are confirmed by the presence of medium-range NOEs. The presence and location of the two short β-strands that are present in PrPC are not obvious from the chemical shifts, but can be inferred from long-range NOE connectivities.
Figure 2.
1H-15N HSQC spectrum of Dpl(26–157) showing resonance assignments. Insets show assignments for the crowded central region (Left) and for the Gln and Asn side-chain amides (Right). Cross peaks for Gly-89 and Glu-93 are folded to the opposite side of the spectrum.
Three-Dimensional Structure of Dpl.
The three-dimensional structure was calculated from a total of 1,262 NOE restraints and 155 dihedral-angle restraints, by using the programs dyana (30) and amber (31, 32). A family of 20 structures is shown in Fig. 3a, and parameters characterizing the solution structure of Dpl are shown in Table 1. The secondary structure elements (three α-helices and two 3-residue β-strands) are well defined, with an rms deviation for the backbone heavy atoms in regular secondary structure of 0.55 Å from the mean.
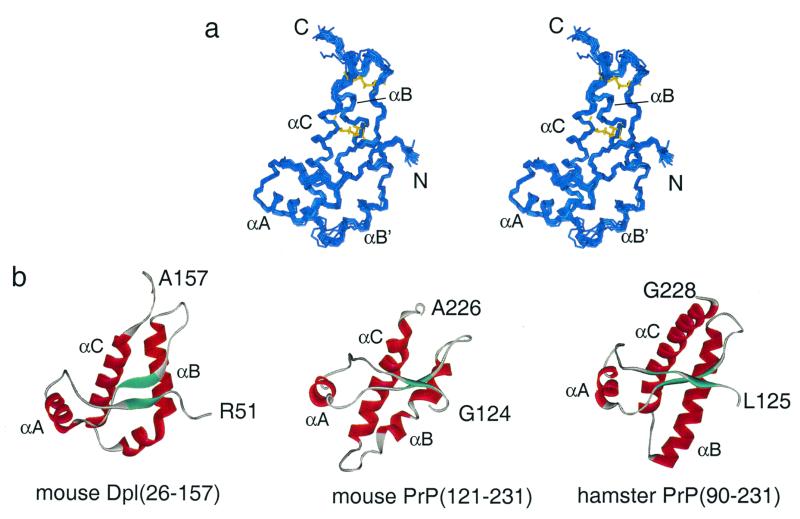
Figure 3.
Solution structure of mouse Dpl. (a) Stereo representation of the backbone of a family of 20-solution structures of Dpl(26–157) showing residues 51–157. The N and C termini are labeled, and the positions of the helices αA, αB, and αC are indicated, with the portion of the α-B helix C-terminal to the kink labeled αB′. The positions of the two disulfide bonds in one of the structures are shown in yellow. (b) Comparison of the backbone topology of mouse Dpl(26–157), mouse PrP(121–231) (3), and Syrian hamster PrP(90–231) (4).
Table 1.
Structural statistics for the globular domain of mouse Dpl(26–157)
| Restraints | |
| NOE restraints (total) | 1,262 |
| Short range: (i, i) or (i, i + 1) | 256 |
| Medium range: (i, j) [(i + 1) < j < (i + 4)] | 408 |
| Long range: | 456 |
| Dihedral angle restraints (total) | 155 |
| φ | 88 |
| ψ | 59 |
| χ1 | 8 |
| rms deviation (from mean), Å | |
| Backbone (residues 55–151) | 0.72 ± 0.14 |
| Regular secondary structure—backbone | 0.55 ± 0.18 |
| Average total amber energy, kcal⋅mol−1 | |
| (including solvation energy of the system) | −5158 |
| Average total violation energy, kcal⋅mol−1 | 5.0 |
| Maximum violations | |
| Distance, Å | 0.20 |
| Angle, ° | 6.3 |
| Average number of distance violations per structure | |
| 0.1–0.2 Å | 4.3 |
| >0.2 Å | 0 |
| Average number of dihedral angle violations | |
| ≤5° | 0.3 |
| 5–10° | 0.15 |
| procheck (45) statistics | |
| Most favored regions, % | 86.8 |
| Additionally allowed regions, % | 13.1 |
| Generously allowed regions, % | 0.1 |
| Disallowed regions, % | 0.0 |
Heteronuclear NOEs.
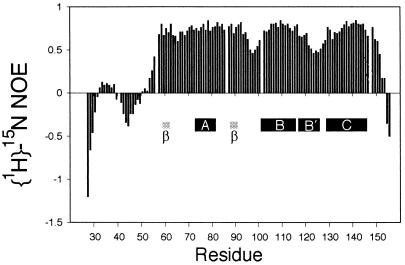
The flexibility of folded Dpl(26–157) was estimated by measuring the {1H}-15N NOE. The NOE value determined for each residue is plotted in Fig. 4. As for most proteins, the residues at the N and C termini are disordered, with negative values for the NOE. The well folded part of the molecule has NOE values between 0.5 and 0.8, and thus is localized between residues 56 and 150. Within the folded region, there are areas, at residues 96–98 and 123–125, of significantly greater flexibility, corresponding to loops between areas of secondary structure. In addition, the N terminus seems not to be uniformly flexible; an area between residues 32 and 38 has small positive values of the NOE, a possible indication that it is sampling a structure where it is associated more closely with the folded part of the molecule.
Figure 4.
Plot of the heteronuclear {1H}-15N NOE values observed for each residue in Dpl(26–157). The positions of secondary structure elements are denoted by labeled black bars for helices and gray bars for the two β-strands.
Discussion
Comparison of the Structures of Dpl and PrPC.
The structured region of Dpl is remarkably similar to that of PrPC. A comparison of the structures of the folded domains of mouse Dpl(51–157), mouse PrP(121–231) (3), and Syrian hamster PrP(90–231) (4) is shown in Fig. 3b. It is clear that the overall topology of the Dpl and PrP proteins is very similar. The three helices, αA, αB, and αC, and the two β-strands are mapped onto the amino acid sequences of Dpl and PrPC in Fig. 1. In the main, the secondary structure elements fall in the same positions in the two proteins, with some variation in the length of the helices. The β-strands are shorter in Dpl, and are displaced by one or two residues from those of PrP. The orientation of the two-strand β-sheet differs between Dpl and PrP; in Dpl, the plane of the β-sheet is parallel to the axis of helices αB and αC, whereas in PrP, it is perpendicular. A significant difference between the two structures occurs in helix αB, which in Dpl contains a marked kink. This feature is well validated by a number of NMR parameters, including the chemical shift index for the two residues Ala-116 and Asn-117, which have Cα and CO chemical shifts that are not typical of helix.
The third helix (αC) is significantly shorter in Dpl than in Syrian hamster (4), bovine (5), or human (6) PrP, but, interestingly, the mouse PrP structure (3) has a shorter helix αC, more comparable with the Dpl structure. The variation in the length of the αC helix in different PrPC structures appears to be a real difference, reflected in the chemical shifts and NOEs observed for this region in the three proteins (6) and consistent with the small but significant sequence differences between these species. On the other hand, because the Dpl sequences from species as diverse as human, mouse, and rat are virtually identical (19), the structures of the Dpl proteins are likely to be highly conserved.
Flexibility of Dpl and PrPC.
Fig. 4 shows that the N-terminal 30 residues or so of Dpl(26–157) have low or negative values of the heteronuclear {1H}-15N NOE, and are, therefore, flexible. Interestingly, there seems to be a degree of variation in the flexibility in this region, with residues 33–39 having generally higher NOEs (positive values) than the surrounding residues. The reason for this difference is not clear from the heteronuclear NOE measurements alone. One possibility is that this region of the polypeptide chain makes transient contact with the folded C-terminal portion of the protein, but confirmation of this hypothesis will likely require a more comprehensive analysis of the relaxation behavior of the construct.
There is a significant variation in the heteronuclear NOE values within the folded domain. Interestingly, the region encompassing the two β-strands and the αA helix (residues 59–91) is rather uniformly of low flexibility, despite the length of the loops between the three short secondary structures in this sequence. By contrast, the loop between the second β-strand and helix αB is relatively flexible, with lowered NOE values. The portion of helix αB C-terminal to the kink (termed α-B′) is quite flexible, despite the presence of a well defined secondary structure. The heteronuclear NOE results for hamster PrP(90–231) and PrP(29–231) also show flexibility at the C terminus of helix αB (36).
Biological Significance of the Structural Similarity of Dpl and PrPC.
The structural similarity between Dpl and PrPC suggests that Dpl might be capable of functional substitution for PrPC in Prnp0/0 mice. Support for this suggestion comes from the observation that overexpression of Dpl produces Purkinje cell degeneration that can be prevented by coexpression of PrPC (20). There are two major regions of the folded structure where groups of residues are identical in Dpl and PrPC (Fig. 1). These segments are associated with helix A and its preceding loop, and with helix B. In the first of these regions, the conserved residues are located largely on the surface, projecting out from the molecule. In the second, the residues are clearly part of the hydrophobic core, and include one of the cysteines of the disulfide bond common to both proteins. This observation suggests that the sequence identity in the helix B region is associated with the structural integrity of the protein, and that whatever functional similarity is preserved between Dpl and PrPC may reside in the helix A region. However, the absence of the octarepeat sequences in the Dpl protein raises doubts as to whether Dpl can actually assume the function of PrPC. The octarepeat sequences are highly conserved in PrPC from various mammalian species, and similar repeating sequences are conserved in birds and reptiles (37). This finding argues for the functional significance of these sequences that is borne out by their apparent specificity in the binding of Cu(II) (13). If, indeed, PrPC has a function in copper metabolism, then Dpl would clearly be an imperfect if not dysfunctional substitute in Prnp0/0 mice.
No relationship has so far been established between Dpl and the replication of prions. Our structures suggest a possible reason for this observation. Several studies implicate residues 90–145 as the conformationally flexible component in the conversion of PrPC to PrPSc (8, 38). Experiments on PrP fragments have suggested that the two β-strands could be extended, and that a palindromic portion of the polypeptide (Ala-Gly-Ala-Ala-Ala-Gly-Ala) encompassing residues 112–119 (before the first β-strand) is likely to be recruited into the β-structure in PrPSc. Dpl also contains two short β-strands, but lacks this palindrome. Presumably, these sequence differences, coupled with the second disulfide bridge in the C-terminal portion of the molecule, preclude the conversion of Dpl into a scrapie-like conformation. To answer the question whether PrP gained the convertible sequence or Dpl lost it will require the identification of other Dpl-like and PrP-like genes.
Whether Dpl is capable of modifying dominant-negative inhibition of prion replication remains to be established. Epidemiological studies of Japanese individuals with the Q219K PrP polymorphism (39) and Suffolk sheep with the Q171R substitution (40, 41) suggested that these sequences cannot be converted into PrPSc. Because Japanese heterozygotes are protected from prion disease, it seemed that PrP(Q219K) exerts a dominant-negative inhibition on prion formation. Studies of transfected neuroblastoma cells (42, 43) and transgenic mice (V. Perrier, F.E.C., and S.B.P., unpublished data) show that residues 167, 171, 215, and 219 exhibit a dominant-negative phenotype when positively charged amino acids are substituted at these positions. Mouse Dpl has a K instead of Q at the position analogous to position 171 (Fig. 1). If variants of PrP exert dominant-negative inhibition via protein binding, it is possible that Dpl could play a similar role. However, it is unlikely that Dpl will take part in a truly homologous interaction with this binding partner, because the second disulfide bridge involves a position analogous to 219 in PrP.
It is unclear why PrPC and Dpl produce central nervous system disease by such different mechanisms yet have quite similar structures. On the one hand, PrPC must be converted into PrPSc before causing disease, except when PrPC overexpression is extreme (44). On the other hand, simple overexpression of Dpl at modest levels is sufficient to cause Purkinje-cell loss and concomitant ataxia; moreover, no scrapie-like form of Dpl has so far been detected. Constructing chimeric Dpl-PrP proteins should produce some particularly informative results; the sequences are quite different but the structures are very similar, as described here. Such chimeric proteins not only may give important insights into the mechanism by which Dpl causes cerebellar dysfunction, but they may also help decipher the process by which PrPSc is formed.
Acknowledgments
We thank Maria Martinez-Yamout and Linda Tennant for valuable assistance in cloning and expression, Sam Xie for assistance with refolding Dpl(26–157), John Chung and Brian Lee for assistance with NMR data acquisition, and Darlene Groth for helpful discussions. This work was supported by Grant NS14069 from the National Institutes of Health (to S.B.P.).
Abbreviations
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- HSQC
heteronuclear sequential quantum correlation
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1I17).
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 4938).
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan K M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner SB. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 4.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez G F, Zahn R, Riek R, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahn R, Liu A, Luhrs T, Riek R, Von Schroetter C, Lopez G F, Billeter M, Calzolai L, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peretz D, Williamson R A, Matsunaga Y, Serban H, Pinilla C, Bastidas R B, Rozenshteyn R, James T L, Houghten R A, Cohen F E, et al. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 8.Cohen F E, Prusiner S B. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 9.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 10.Sparrer H E, Santoso A, Szoka F C, Jr, Weissman J S. Science. 2000;289:595–599. doi: 10.1126/science.289.5479.595. [DOI] [PubMed] [Google Scholar]
- 11.Hornshaw M P, McDermott J R, Candy J M, Lakey J H. Biochem Biophys Res Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 12.Brown D R, Qin K F, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, Von Bohlen A, Schulz-Schaeffer W, et al. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 13.Viles J H, Cohen F E, Prusiner S B, Goodin D B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche J L, Lehmann S, Launay J M, Kellermann O. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 15.Bueler H, Aguzzi A, Sailer A, Greiner R A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang S L, DeArmond S J. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp H P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Nature (London) 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 18.Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, Sugimoto T, Nakatane A, Kataoka Y, Houtani T, Shirabe S, et al. Nature (London) 1996;380:528–531. doi: 10.1038/380528a0. [DOI] [PubMed] [Google Scholar]
- 19.Moore R C, Lee I Y, Silverman G L, Harrison P M, Strome R, Heinrich C, Karunaratne A, Pasternak S H, Chishti M A, Liang Y, et al. J Mol Biol. 1999;292:797–817. doi: 10.1006/jmbi.1999.3108. [DOI] [PubMed] [Google Scholar]
- 20.Nishida N, Tremblay P, Sugimoto T, Shigematsu K, Shirabe S, Petromilli C, Erpel S P, Nakaoke R, Atarashi R, Houtani T, et al. Lab Invest. 1999;79:689–697. [PubMed] [Google Scholar]
- 21.Bax A, Grzesiek S. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 22.Grzesiek S, Bax A. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 23.Grzesiek S, Bax A. J Magn Reson. 1992;99:201–207. [Google Scholar]
- 24.Grzesiek S, Anglister J, Bax A. J Magn Reson Ser B. 1993;101:114–119. [Google Scholar]
- 25.Bax A, Clore G M, Gronenborn A M. J Magn Reson. 1990;88:425–431. [Google Scholar]
- 26.Wishart D S, Bigam C G, Yao J, Abildgaard F, Dyson H J, Oldfield E, Markley J L, Sykes B D. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F, Grzesiek S, Vuister G W, Guang Z, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Johnson B A, Blevins R A. J Chem Phys. 1994;29:1012–1014. [Google Scholar]
- 29.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 30.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 31.Case D A, Pearlman D A, Caldwell J W, Cheatham T E, III, Ross W S, Simmerling C L, Darden T A, Merz K M, Stanton R V, Cheng A L, et al. amber. San Francisco: University of California; 1999. Version 6. [Google Scholar]
- 32.Tsui V, Case D A. J Am Chem Soc. 2000;122:2489–2498. [Google Scholar]
- 33.Farrow N A, Muhandiram R, Singer A U, Pascal S M, Kay C M, Gish G, Shoelson S E, Pawson T, Forman-Kay J D, Kay L E. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 36.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonic T, Duga S, Strumbo B, Asselta R, Ceciliani F, Ronchi S. FEBS Lett. 2000;469:33–38. doi: 10.1016/s0014-5793(00)01232-1. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko K, Ball H L, Wille H, Zhang H, Groth D, Torchia M, Tremblay P, Safar J, Prusiner S B, DeArmond S J, et al. J Mol Biol. 2000;295:997–1007. doi: 10.1006/jmbi.1999.3386. [DOI] [PubMed] [Google Scholar]
- 39.Shibuya S, Higuchi J, Shin R W, Tateishi J, Kitamoto T. Ann Neurol. 1998;43:826–828. doi: 10.1002/ana.410430618. [DOI] [PubMed] [Google Scholar]
- 40.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 41.Hunter N, Moore L, Hosie B D, Dingwall W S, Greig A. Vet Rec. 1997;140:59–63. doi: 10.1136/vr.140.3.59. [DOI] [PubMed] [Google Scholar]
- 42.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zulianello L, Kaneko K, Scott M, Erpel S, Han D, Cohen F E, Prusiner S B. J Virol. 2000;74:4351–4360. doi: 10.1128/jvi.74.9.4351-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westaway D, DeArmond S J, Cayetano-Canlas J, Groth D, Foster D, Yang S L, Torchia M, Carlson G A, Prusiner S B. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski R A, Rullmann J A C, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 46.Westaway D, Goodman P A, Mirenda C A, McKinley M P, Carlson G A, Prusiner S B. Cell. 1987;51:651–662. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 47.Hornemann S, Korth C, Oesch B, Riek R, Wider G, Wüthrich K, Glockshuber R. FEBS Lett. 1997;413:277–281. doi: 10.1016/s0014-5793(97)00921-6. [DOI] [PubMed] [Google Scholar]