Abstract
Irrespective of their effects on ongoing host protein synthesis, productive replication of the representative alphaherpesvirus herpes simplex virus type 1, the representative gammaherpesvirus Kaposi's sarcoma herpesvirus, and the representative betaherpesvirus human cytomegalovirus [HCMV] stimulates the assembly of the multisubunit, cap-binding translation factor eIF4F. However, only HCMV replication is associated with an increased abundance of eIF4F core components (eIF4E, eIF4G, eIF4A) and the eIF4F-associated factor poly(A) binding protein (PABP). Here, we demonstrate that the increase in translation factor concentration was readily detected in an asynchronous population of HCMV-infected primary human fibroblasts, abolished by prior UV inactivation of virus, and genetically dependent upon viral immediate-early genes. Strikingly, while increased mRNA steady-state levels accompanied the rise in eIF4E and eIF4G protein levels, the overall abundance of PABP mRNA, together with the half-life of the polypeptide it encodes, remained relatively unchanged by HCMV infection. Instead, HCMV-induced PABP accumulation resulted from new protein synthesis and was sensitive to the mTORC1-selective inhibitor rapamycin, which interferes with phosphorylation of the mTORC1 substrate p70 S6K and the translational repressor 4E-BP1. While virus-induced PABP accumulation did not require p70 S6K, it was inhibited by the expression of a dominant-acting 4E-BP1 variant unable to be inactivated by mTORC1. Finally, unlike the situation in alpha- or gammaherpesvirus-infected cells, where PABP is redistributed to nuclei, PABP accumulated in the cytoplasm of HCMV-infected cells. Thus, cytoplasmic PABP accumulation is translationally controlled in HCMV-infected cells via a mechanism requiring mTORC1-mediated inhibition of the cellular 4E-BP1 translational repressor.
Herpesvirus mRNAs contain methyl-7-GTP caps and 3′ polyadenylate tails like their host cell counterparts and are primarily translated by a cap-dependent mechanism. Assembly of the cap-binding protein eIF4E, eIF4G, and the RNA helicase eIF4A into the active, cap-binding, multisubunit translation initiation factor eIF4F represents a key step regulating translation (reviewed in reference 33). In addition to controlling small ribosome subunit recruitment to the mRNA 5′ end, whereupon a scanning mechanism commences to locate the initiator AUG codon, eIF4F assembly is responsive to a diverse assortment of cell stress and signaling inputs, including viral infection (24). Typically, eIF4E is bound to the translational repressor 4E-BP1. Hyperphosphorylation of 4E-BP1 by activated mTORC1 relieves this repression, releasing eIF4E and exposing the binding site for eIF4G, a large assembly platform bound to eIF4A. eIF4G also binds eIF3, which directly associates with the 40S ribosome subunit. The cellular poly(A) binding protein (PABP) and the eIF4E kinase Mnk are eIF4F-associated proteins that physically associate with eIF4G and act to stimulate translation. Bound to both the 3′ poly(A) tail and eIF4G, PABP mediates an interaction between the mRNA 3′ and 5′ ends (reviewed in reference 33). To ensure that their mRNAs are effectively translated and the proteins required for their productive replication are synthesized, herpesviruses go to great lengths to successfully commandeer eIF4F. Despite the fundamental nature of this task, notable similarities and differences have emerged in how eIF4F is regulated in cells infected with different herpesvirus subfamily members, most notably, human cytomegalovirus (HCMV).
Productive HCMV replication, like the replicative growth of the representative alphaherpesvirus herpes simplex virus type 1 (HSV-1) and the representative gammaherpesvirus Kaposi's sarcoma herpesvirus (KSHV), promotes the binding of eIF4E to eIF4G and thereby stimulates eIF4F assembly (2, 14, 38, 40). In all cases, this involves inactivation of the 4E-BP1 translational repressor by virus-encoded functions that activate mTOR signaling and promote 4E-BP1 hyperphosphorylation. While each virus uses a distinct mechanism to activate mTOR, differences in the sensitivity of 4E-BP1 hyperphosphorylation to the mTORC1-selective inhibitor rapamycin have been observed (14, 15, 25, 32, 38, 40). An additional step controlling eIF4F assembly has been defined in HSV-1-infected cells, where a direct interaction between eIF4G and the virus-encoded protein ICP6 stimulates binding of eIF4G to eIF4E (39). Finally, eIF4F assembly in representative alpha-, beta-, and gammaherpesvirus-infected cells is accompanied by Mnk-mediated eIF4E phosphorylation. Moreover, interfering with eIF4E phosphorylation inhibits productive replication of the representative herpesvirus family members examined (2, 38-40).
Irrespective of these similarities, significant differences regarding how eIF4F core and associated components are regulated distinguish cells infected with HCMV from cells infected with alpha- or gammaherpesviruses. Ultimately, these features may have an impact upon how ongoing cellular mRNA translation is managed in herpesvirus-infected cells.
In HSV-1- and KSHV-infected cells, host mRNA translation is impaired and steady-state eIF4F subunit and PABP levels remain unchanged (2, 38). However, PABP accumulates in the nucleus and is excluded from eIF4F complexes (2, 5, 16, 29, 40). In contrast, host protein synthesis proceeds and overall steady-state levels of certain translation initiation factors, including eIF4E, eIF4G, eIF4A, and PABP, rise dramatically in growth-arrested, HCMV-infected cells (11, 34, 40). Here, we explore the basis for this unprecedented elevation in host eIF4F concentration and establish that alteration of cellular translation initiation factor homeostasis in HCMV-infected cells requires viral gene expression. While augmented eIF4E and eIF4G protein levels were accompanied by elevated mRNA abundance, increased PABP levels were regulated translationally. This translation-driven increase in PABP was rapamycin sensitive and independent of the mTORC1 substrate p70 S6K but was inhibited by the expression of a dominant-acting 4E-BP1 allele that could not be phosphorylated by mTORC1. Finally, PABP accumulated in the cytoplasm of HCMV-infected cells and did not detectably accumulate in nuclei, as reported for HSV-1- and KSHV-infected cells. Together, these findings demonstrate that HCMV increases host PABP cytoplasmic levels via a mechanism requiring inactivation of the cellular translational repressor 4E-BP1.
MATERIALS AND METHODS
Cell culture, viruses, and chemicals.
Primary normal human dermal fibroblasts (NHDFs; Clonetics, Walkersville, MD) were propagated in 5% CO2 incubators with Dulbecco's modified Eagle's medium (DMEM) plus 5% fetal bovine serum (FBS) or serum deprived as described previously (38). Retroviruses expressing wild-type (WT) and AA mutant (a double-mutant derivative with alanine substituted for each of the critical T37 and T46 phosphoacceptor residues) FLAG-hemagglutinin (HA)-4E-BP1 were produced using a pBABE retroviral vector (gifts from Robert Schneider; see reference 3) and used to infect early-passage NHDFs. HCMV strain AD169 and the murine cytomegalovirus (MCMV) Smith strain were obtained from ATCC. HCMV was propagated or UV inactivated as previously described (40). S6K1(−/−)/S6K2 (−/−) doubly deficient primary mouse embryo fibroblasts (MEFs) and WT parental MEFs (28) were provided by S. Kozma and G. Thomas (University of Cincinnati). Antibodies and chemical inhibitors were described previously (37, 38, 40), except for anti-MCMV IE1 mouse monoclonal CROMA 101 (gift of S. Jonic, University of Rijeka) and the eEF2 (no. 2332), S6 (no. 2217), and S6K1/2 (no. 9430) antibodies from Cell Signaling Technology (Beverly, MA).
Metabolic labeling and protein half-life determinations.
NHDFs were seeded into 12-well plates in DMEM plus 5% FBS, infected at a high multiplicity of infection (MOI) with HCMV, metabolically labeled, and analyzed for total protein synthesis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (40). For pulse-chase experiments, NHDFs were infected with HCMV (MOI = 5), incubated for 48 h, pulse-labeled for 2 h in methionine- and cysteine-deficient DMEM supplemented with 70 μCi/ml [35S]methionine-cysteine (Express; Perkin-Elmer), washed with phosphate-buffered saline (PBS), and chased with fresh complete DMEM for the indicated times. Cells were washed with PBS and harvested in 250 μl of cold NP-40 lysis buffer (NLB), and extracts were prepared as described previously (40). After preadsorbing extracts with normal rabbit serum and protein A-Sepharose (PAS), 2 μl anti-PABP antibody was added to the precleared supernatants and the samples were incubated for 1 h at 4°C. PAS (0.1 ml, 10% slurry in NLB) was added, and the incubation was continued for an additional 1 h with rocking. Beads were collected by centrifugation and washed three times with 0.4 ml of NLB. Radiolabeled proteins were separated by SDS-PAGE, visualized by autoradiography, and quantified by densitometry.
RNA interference (RNAi).
To interfere with IE2-86 or UL38 expression, small interfering RNA (siRNA) transfections were performed as previously reported (41) and cultures were subsequently infected with HCMV (MOI = 5). IE2 (sense target, 5′-AAACGCAUCUCCGAGUUGGAC-3′; efficacy demonstrated in reference 41), and control, nonsilencing siRNAs were composed of synthetic 21-nucleotide complementary RNAs with 2-nucleotide overhangs (Dharmacon).
Immunofluorescence microscopy.
NHDFs seeded onto 22-mm coverslips were allowed to grow for 24 h, subsequently serum starved for 72 h in 0.2% FBS DMEM, and then infected with HCMV (MOI = 5). At specified times, cells were washed once with PBS, fixed with 3.7% formaldehyde-PBS at room temperature for 20 min, and permeabilized with 0.1% Triton X-100-PBS for 20 min, and nonspecific binding was blocked with PBS plus FBS plus saponin at 37°C for 30 min. Cells were incubated with the diluted primary antibody (PABP at 1:600 or pp28 at 1:600) for 60 min at 37°C, washed with PBS (three times, 5 min), and next incubated with the secondary antibody (fluorescein isothiocyanate-conjugated anti-mouse [Vector Laboratories] or anti-rabbit Alexa Fluor 633 [Molecular Probes] antibody) for 60 min at 37°C. After washing with PBS (three times, 5 min), fluorescence images were collected with a Zeiss LSM510 Meta confocal laser scanning microscope.
Real-time PCR.
Total RNA was isolated using Tri-reagent (MRC, Inc.). RNA quality and quantity were determined by agarose gel electrophoresis and NanoDrop spectrophotometry. Following reverse transcription (RT) to obtain first-strand cDNA using specific primers for eIF4GI, eIF4E, PABP1, and β-actin, cDNAs were amplified by real-time PCR. All real-time PCR quantification was performed using the Bio-Rad iCycler iQ system, and double-stranded DNA was measured by fluorescence assay using SYBR green I fluorogenic dye (Sigma). A fluorescence cycle threshold (CT) value was calculated for each sample and normalized against the CT value obtained for β-actin. The relative expression ratio [ratio = 2−(ΔCT sample − ΔCT control)] was used to determine the n-fold induction for each mRNA.
RESULTS
HCMV-triggered increase in host translation initiation factor concentration requires viral gene expression.
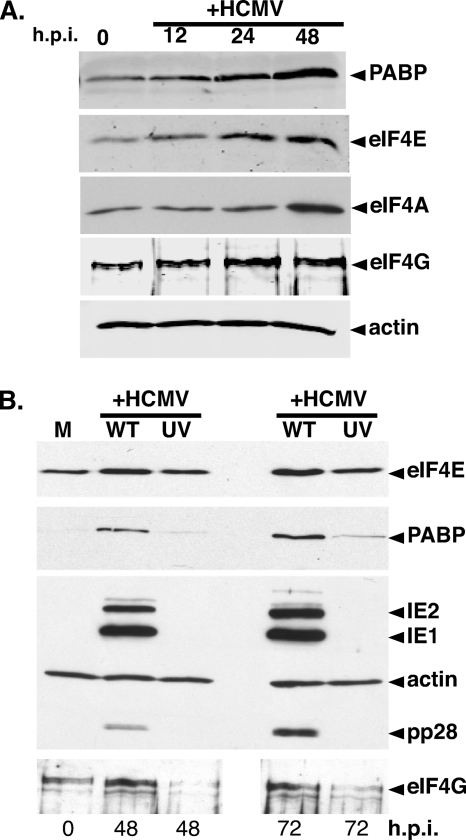
Increased abundance of cellular translation initiation factors eIF4E, eIF4G, eIF4A, and PABP was initially observed upon HCMV infection of growth-arrested primary human cells, raising the possibility that this resulted from simply infecting quiescent cells (40). To discern if increased translation initiation factor levels were a coincidental effect of CMV generally upregulating S-phase proliferation and housekeeping genes, an asynchronous population of subconfluent, actively dividing NHDFs was mock infected or infected with HCMV. Total protein isolated at different times postinfection was analyzed by immunoblotting using antibodies specific for eIF4E, eIF4G, eIF4A, or PABP (Fig. 1A). Significantly, steady-state PABP, eIF4E, eIF4A, and eIF4G levels all increased in HCMV-infected, asynchronously growing, subconfluent cells continuously maintained in full serum (Fig. 1A). Thus, exit of growth-arrested primary cells from quiescence upon HCMV infection was therefore unlikely to account for increased translation factor abundance. Instead, the possibility that viral gene expression specifically induced the accumulation of certain translation initiation factors was considered.
FIG. 1.
Elevation of the translation initiation factor concentration in HCMV-infected cells requires viral gene expression. (A) Asynchronous, subconfluent NHDFs were mock infected (0 h.p.i.) or infected with HCMV. At the indicated times postinfection, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera. (B) UV inactivation of HCMV abrogates the increase in translation initiation factor levels. NHDFs were either mock infected (M) or infected (MOI = 5) with WT or UV-inactivated (UV) HCMV. At the indicated times postinfection, total protein was analyzed by immunoblotting as described for panel A.
Besides introducing the viral genome into the nucleus, herpesviruses deliver a complex polypeptide mixture into the host cytoplasm that acts prior to viral gene expression (23). While UV inactivation precludes expression from incoming viral genomes, it does not prevent virus entry and virion protein deposition into the cytoplasm. To determine if increased translation factor abundance requires viral gene expression, NHDFs were mock infected, infected with active HCMV, or infected with UV-inactivated HCMV. Total protein was harvested at various times postinfection, and overall viral (IE1, IE2, pp28) and cellular antigen levels were evaluated by immunoblotting (Fig. 1B). The increase in PABP, eIF4E, and eIF4G levels easily seen by 48 h postinfection (hpi) with active virus was substantially impaired, even after 72 hpi with UV-inactivated virus. This supports the notion that viral gene expression was required to increase the host translation factor concentration.
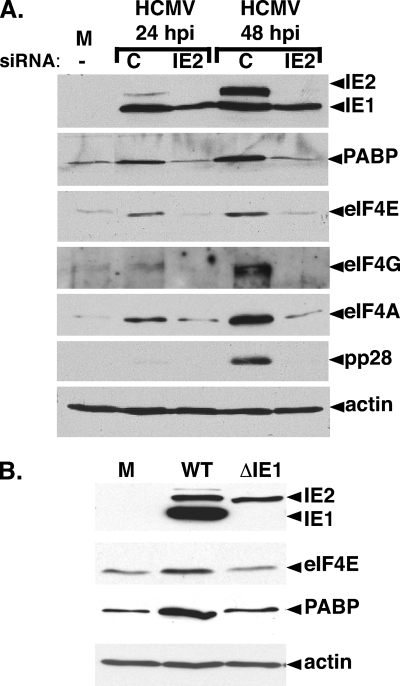
To rule out nonspecific effects of UV inactivation, viral gene expression was interrupted by an independent means using RNAi (41). IE2 was targeted, as it is essential for viral replication and IE2-deficient viruses do not express early or late genes (36). Encoded by the HCMV major immediate-early (IE) gene region, IE2 is one of two master regulators (IE1, IE2) whose synthesis begins at IE times and continues throughout infection (reviewed in reference 23). After transfection of nonsilencing control siRNA or IE2 siRNA, cells were infected with HCMV, total protein was harvested at the indicated times, and the abundance of certain antigens was measured by immunoblotting. In control siRNA-transfected cells, eIF4E, eIF4A, eIF4G, and PABP levels all increased over time (Fig. 2A). Similarly, IE2 accumulated and viral gene expression progressed into the late phase, as evidenced by accumulation of the late protein pp28 (Fig. 2A). IE2 siRNA, however, precluded IE2 protein accumulation and blocked progression of the viral life cycle into the late phase, as evidenced by the inability to detect pp28 at 48 hpi. Significantly, interfering with IE2 expression prevented eIF4E, eIF4A, eIF4G, and PABP accumulation (Fig. 2A).
FIG. 2.
HCMV-mediated translation factor accumulation is dependent upon viral IE gene expression. (A) Inhibition of translation factor accumulation by RNAi-mediated IE2 silencing. NHDFs transiently transfected with control noninterfering siRNA (C), IE2 siRNA (IE2), or no siRNA (−) were either mock infected (M) or infected with HCMV (MOI = 5). Total protein was harvested at the indicated times postinfection and analyzed by immunoblotting with the indicated antisera. (B) Quiescent NHDFs were either mock infected (M) or infected with WT HCMV (Towne strain) or an IE1-deficient mutant (ΔIE1). Total protein was harvested at 60 hpi and analyzed by immunoblotting as described for panel A.
Additional genetic evidence supporting a role for viral gene expression triggering increased host translation factor abundance was obtained using a well-characterized IE1-deficient virus (8). While IE1 is not essential for productive replication, IE1-deficient viruses are replication impaired at lower MOIs, where it acts together with IE2 to stimulate viral gene expression. Indeed, eIF4E and PABP abundance in NHDFs infected with an IE1-deficient virus remained comparable to levels in mock-infected cells (Fig. 2B). This establishes that the observed increase in the host translation initiation factor concentration in HCMV-infected cells is IE1 dependent and therefore under the genetic control of the virus, as it requires viral gene expression. Furthermore, since the IE1 mutant is in a Towne strain background, it also demonstrates that the HCMV-mediated increase in translation factor abundance occurs in different viral strains and was not limited to the commonly used AD169 laboratory strain. Thus, by multiple independent criteria, HCMV gene expression was required to increase host translation initiation factor levels in infected cells, whereas virion protein deposition, which occurs normally in cells infected with ΔIE1- or siRNA-treated cultures, was insufficient. Finally, IE1 and IE2 individually were insufficient to trigger translation initiation factor accumulation, as IE1 levels in IE2-silenced cultures and IE2 levels in ΔIE1-infected cells were near WT levels (Fig. 2A and B).
Transcriptional and posttranscriptional mechanisms account for the HCMV-induced increase in eIF4F core and associated proteins.
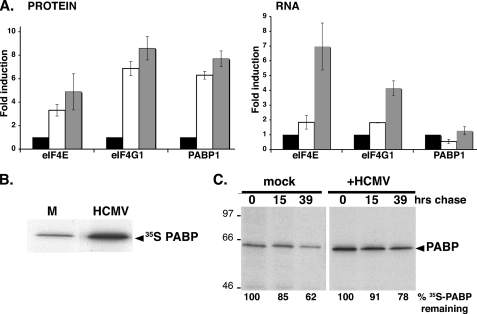
To determine if elevated mRNA abundance contributes to increased translation factor levels, total RNA was isolated from mock-infected or HCMV-infected NHDFs and the overall amount of eIF4E, eIF4G, or PABP mRNA relative to that of a control (actin) mRNA was evaluated by real-time PCR. Increased total eIF4E and eIF4G protein levels were both accompanied by 4- to 7-fold increases in steady-state mRNA levels (Fig. 3A). Remarkably, while PABP polypeptide levels increased 7-fold by 96 hpi, PABP mRNA abundance did not significantly change under conditions where 2-fold differences were readily detected (Fig. 3A). This suggested that increased PABP levels in HCMV-infected cells did not require mRNA abundance changes but instead involved altering either protein stability or mRNA translation.
FIG. 3.
Translational control of PABP levels in HCMV-infected cells. (A) Analysis of eIF4E, eIF4G, and PABP mRNA abundance in HCMV-infected cells. Serum-starved NHDFs were mock infected (0 h) or infected with HCMV (MOI = 5). At 0 hpi (black bars), 70 hpi (white bars), and 96 hpi (gray bars), total protein (left) was fractionated by SDS-PAGE and analyzed by immunoblotting with the indicated antisera. Chemiluminescence images were captured and quantified with a Bio-Rad ChemiDoc XR5 system. Total RNA (right) was isolated, and eIF4G1, eIF4E, and PABP mRNA levels were determined by real-time RT-PCR (normalized to actin). (B) Stimulation of new PABP synthesis in HCMV-infected cells. NHDFs were infected as described for panel A. At 24 hpi, cultures were pulse-labeled for 2 h with [35S]Met-Cys, and PABP was immunoprecipitated. Immune complexes were fractionated by SDS-PAGE and visualized by autoradiography. (C) PABP stability is similar in both mock- and HCMV-infected cells. NHDFs infected as described for panel A were pulse-labeled at 50 hpi as described for panel B and then returned to unlabeled medium for the indicated chase times. Total protein was subsequently isolated, and PABP was immunoprecipitated and analyzed as described for panel B. The mobility of molecular mass standards (in kilodaltons) is shown to the left. The percentage of [35S]PABP remaining over time was quantified by densitometry. In each case (mock versus HCMV infection), the amount of radiolabeled PABP after the pulse (0 h chase) was set at 100%.
To examine if increased PABP abundance was accompanied by new PABP synthesis, mock-infected and HCMV-infected cultures were metabolically labeled with 35S-labeled amino acids and PABP was immunoprecipitated from cell-free lysates and analyzed by SDS-PAGE. Indeed, HCMV infection stimulated new PABP synthesis, as evidenced by 35S-labeled amino acid incorporation (Fig. 3B). To determine if PABP stability was affected, pulse-labeled cultures were chased by the addition of excess unlabeled amino acids and the incubation was continued for 39 h. PABP half-life was approximately analogous, irrespective of HCMV infection (Fig. 3C). The minor stabilization of PABP after the 39-h chase was marginal compared to the observed increase in overall abundance. Together, these data suggest that new PABP synthesis in CMV-infected cells is controlled posttranscriptionally and is primarily responsible for the increasing PABP abundance observed.
Stimulation of PABP mRNA translation in HCMV-infected cells is rapamycin sensitive and requires site-specific 4E-BP1 phosphorylation.
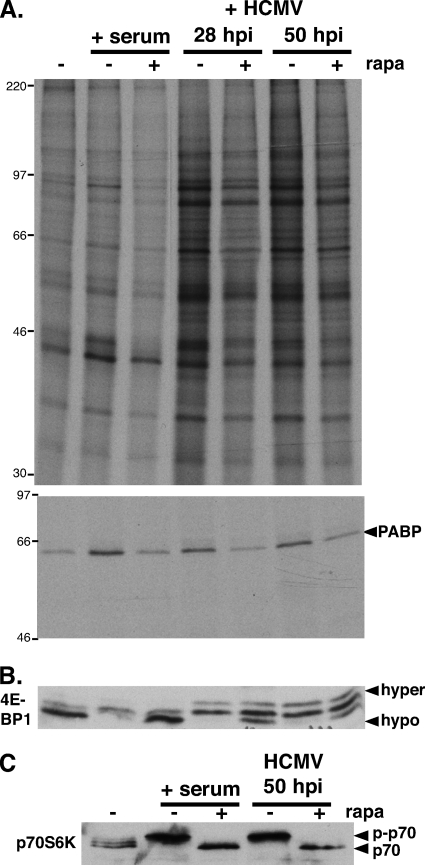
mTOR is a critical cellular component that integrates a wide variety of physiological inputs, including nutrient availability and stress, to properly regulate translation initiation. The catalytic mTOR subunit is a component of at least two discrete kinase complexes, mTORC1 and mTORC2 (43). Substrates of both complexes have the potential to influence protein synthesis, as mTORC2 phosphorylates Akt while mTORC1 targets p70 S6K and 4E-BP1. To determine if mTORC1 signaling contributes to the HCMV-induced PABP increase, the ability of the mTORC1-selective inhibitor rapamycin to interfere with this process was examined. At various times, proteins synthesized in HCMV-infected NHDFs were metabolically labeled with 35S-labeled amino acids with or without rapamycin. Cell extracts were prepared, and PABP immunoprecipitates were analyzed by SDS-PAGE followed by autoradiography (Fig. 4A). As a control, PABP was immunoprecipitated from parallel cultures of serum-stimulated NHDFs. Whereas both serum and HCMV infection stimulated global 35S incorporation into protein, rapamycin was more effective in reducing bulk protein synthesis in uninfected, serum-stimulated cells (Fig. 4A). This correlated with the effectiveness of rapamycin in preventing hyperphosphorylation and inactivation of the translational repressor 4E-BP1. Consistent with other reports, 4E-BP1 phosphorylation was only partially rapamycin sensitive in HCMV-infected cells (Fig. 4B), implicating other mTORC complexes in the phosphorylation of 4E-BP1 (14, 15, 40). Rapamycin also prevented p70 S6 kinase activation (S6K) and, most significantly, inhibited new PABP synthesis in response to both serum and HCMV infection (Fig. 4A and C). Thus, HCMV-induced PABP synthesis and S6K activation were similarly sensitive to the mTORC1 inhibitor rapamycin.
FIG. 4.
Rapamycin sensitivity of HCMV-induced PABP mRNA translation and p70RSK activation. (A, top) Growth-arrested NHDFs were either serum stimulated (+ serum) or infected with HCMV (MOI = 5). At 26 or 46 hpi, HCMV-infected cells were treated with either DMSO or rapamycin (rapa) for 30 min and subsequently pulse-labeled with [35S]Met-Cys with (+) or without (−) rapa for 1.5 h. Uninfected cells (with or without rapa) were similarly metabolically labeled following stimulation with 20% FBS for 20 min. Total protein (top) and PABP immunoprecipitates (bottom) were fractionated by SDS-PAGE and visualized by autoradiography. The migration of molecular mass standards (in kilodaltons) is shown on the left. (B, C) Lysates from panel A were fractionated by SDS-PAGE and analyzed by immunoblotting using the indicated antibodies. The hyper- and hypophosphorylated 4E-BP1 forms are designated. p-p70, phosphorylated p70 S6K.
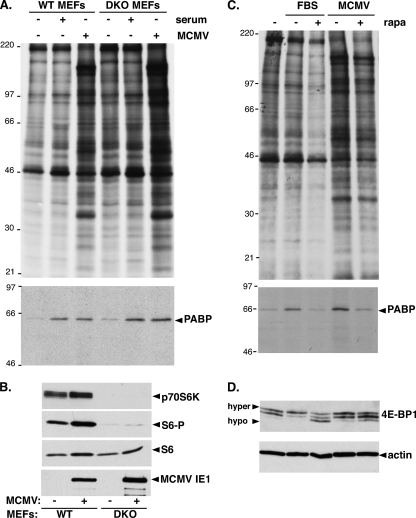
Since S6K activation and new PABP synthesis in HCMV-infected cells were both rapamycin sensitive, the requirement for ribosomal protein S6 kinases was evaluated by infecting murine cells deficient in the two S6 kinases (S6K1, S6K2) with MCMV. Growth-arrested parental (WT) or S6K1/S6K2 doubly deficient (DKO) cells were mock infected, exposed to serum for 20 min, or infected with MCMV. Cell extracts were prepared from metabolically labeled cells, and PABP immunoprecipitates were analyzed by SDS-PAGE followed by autoradiography. Serum stimulation and MCMV infection induced new PABP synthesis to similar extents, irrespective of the DKO deficiency (Fig. 5A). As expected, neither p70 S6K1/2 nor phospo-S6 accumulation was detected in DKO cells, and viral IE1 protein accumulation proved the DKO cells were indeed infected (Fig. 5B). Finally, serum- or MCMV-induced new PABP synthesis remained rapamycin sensitive in DKO cells (Fig. 5C). In addition, whereas 4E-BP1 phosphorylation in serum-stimulated cells was rapamycin sensitive, it was partially rapamycin sensitive in MCMV-infected cells (Fig. 5D), similar to findings in HCMV-infected cells (14, 40). Thus, the rapamycin-sensitive target controlling new PABP synthesis did not require the mTORC1 substrate p70 S6K1/2.
FIG. 5.
Induction of PABP mRNA translation by MCMV is preserved in p70 S6K-deficient cells. (A) Growth-arrested WT or p70 S6K1/K2 doubly deficient primary MEFs (DKO) were mock infected (−), infected (+) with MCMV (MOI = 5), or serum stimulated. At 24 hpi, cultures were metabolically labeled with [35S]Met-Cys for 2 h. Cell-free lysates (top) and PABP immunoprecipitates (bottom) were fractionated by SDS-PAGE and analyzed by autoradiography. The mobility of molecular weight standards (in kilodaltons) is shown on the left. (B) Total protein isolated from MEFs infected as described for panel A was fractionated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. S6-P, phospho-specific S6. (C) At 22 hpi, MEFs (infected as described for panel A) were pulse-labeled with [35S]Met-Cys for 2 h with or without rapamycin. Cell-free lysates (top) and PABP immunoprecipitates (bottom) were analyzed as described for panel A. FBS, fetal bovine serum. (D) Samples described in panel C were analyzed by immunoblotting with the indicated antisera. The hyper- and hypophosphorylated forms of 4E-BP1 are indicated.
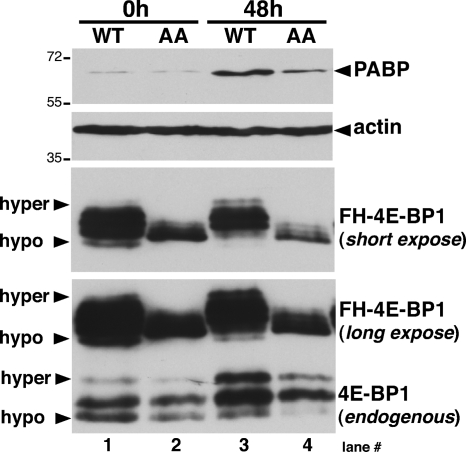
Given that the HCMV-mediated PABP increase did not require S6K but was rapamycin sensitive, a role for the mTORC1 substrate 4E-BP1 was considered. To determine if phosphorylation of the 4E-BP1 translational repressor contributed to increasing PABP levels in HCMV-infected cells, NHDFs were isolated that stably expressed epitope-tagged WT 4E-BP1 or a double-mutant derivative with alanine substituted for each of the critical T37 and T46 phosphoacceptor residues (AA mutant). Unable to be inactivated by mTORC1, the AA mutant behaves as a constitutively activated translational repressor (6). Total protein was isolated from WT or AA mutant 4E-BP1-expressing NHDFs which were either mock infected or HCMV infected, and 4E-BP1 phosphorylation and PABP accumulation were analyzed by immunoblotting. Using anti-4E-BP1 antibody, both endogenous and ectopically expressed 4E-BP1 species were readily detected but the ectopically expressed protein was more abundant. While hyperphosphorylated endogenous 4E-BP1 accumulated in all WT and AA mutant 4E-BP1-expressing cultures (Fig. 6, endogenous panel; compare lanes 1 and 2 with 3 and 4), only epitope-tagged WT 4E-BP1 was hyperphosphorylated upon infection (Fig. 6, FH-4E-BP1 panel; compare lane 1 with lane 3 and lane 2 with lane 4). Despite substantial overexpression of WT 4E-BP1, PABP accumulation increased upon HCMV infection (Fig. 6). However, significantly lower levels of AA-4E-BP1 suppressed PABP accumulation in infected cells (Fig. 6). This demonstrates that the HCMV-induced rise in PABP levels requires phosphorylation and inactivation of the host translational repressor 4E-BP1. Moreover, it implies that 4E-BP1 is the target that mediates the effects of mTORC1 on PABP synthesis.
FIG. 6.
Regulation of PABP accumulation by 4E-BP1 phosphorylation in HCMV-infected cells. NHDFs stably expressing epitope-tagged (FH) WT or AA mutant 4E-BP1 were growth arrested by serum deprivation and either mock infected (0 h) or infected with HCMV (MOI = 5). After 48 h, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with anti-4E-BP1 antibody which recognizes both endogenous and ectopically expressed (FH-4E-BP1) forms. Long and short exposures of the ectopically expressed 4E-BP1 protein are shown.
PABP accumulates in the cytoplasm of HCMV-infected cells.
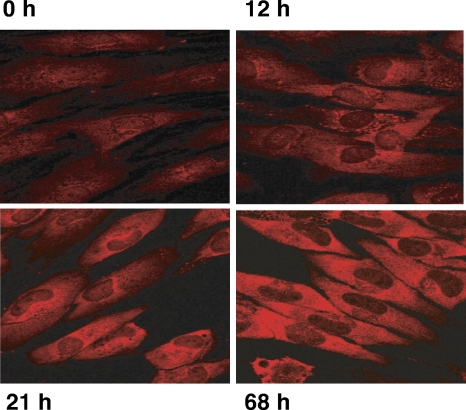
The remarkable overall increase in steady-state PABP levels raises questions about its subcellular distribution, as PABP is an RNA-binding protein capable of shuttling between the nucleus and the cytoplasm (1). To determine how PABP subcellular distribution responds to HCMV infection, NHDFs were infected at a high MOI and processed for indirect immunofluorescence assay using anti-PABP antibody at different times postinfection. In uninfected cells (0 h), PABP exhibited the expected distribution of a shuttling RNA-binding protein and was readily detected throughout the cell in both the nucleus and the cytoplasm (Fig. 7). Over time, the overall intensity of PABP staining in the cytoplasm paralleled the overall accumulation observed by immunoblotting. While increased staining was observed throughout the cytoplasm, between 12 and 21 hpi, the protein accumulated the most in perinuclear areas (Fig. 7). By 68 hpi, the cytoplasmic distribution appeared more uniform. It is noteworthy that while the overall intensity of cytoplasmic staining increased, detectable levels within nuclei remained relatively constant (Fig. 7). This is particularly striking as PABP is redistributed and accumulates in nuclei of HSV-1- and KSHV-infected cells (2, 5, 16, 29). Thus, compared to alpha- and gammaherpesvirus-infected cells, PABP homeostasis is remarkably different in HCMV-infected cells, where its overall abundance in the cytoplasm increases.
FIG. 7.
Cytoplasmic accumulation of PABP in HCMV-infected cells. NHDFs were either mock infected (0 h) or infected with HCMV (MOI = 5). At the indicated times postinfection (12 h, 21 h, 68 h), cells were fixed and processed for indirect immunofluorescence assay using anti-PABP.
DISCUSSION
Unlike cells infected with alpha- or gammaherpesviruses, where host protein synthesis is impaired in part due to accelerated global mRNA turnover (7, 26), host protein synthesis proceeds uninterrupted in cells infected with the betaherpesvirus HCMV (35). Curiously, overall steady-state levels of critical, limiting translation initiation factors eIF4E, eIF4G, and PABP increase in HCMV-infected cells (40). Here, we establish that the increase in certain translation initiation factor levels was not dependent upon prior growth arrest of the cells and that viral gene expression was required. Furthermore, although eIF4E and eIF4G mRNA abundance was considerably elevated in infected cells, PABP mRNA levels and protein stability remained similar. The virus-induced PABP increase was achieved by a translational control mechanism that was rapamycin sensitive, independent of p70 S6K, and inhibited by the expression of a 4E-BP1 derivative incapable of being phosphorylated by mTORC1. Finally, PABP accumulated in the cytoplasm of infected cells, unlike the nuclear retention described in HSV- and KSHV-infected cells. Thus, HCMV manipulates PABP homeostasis in a manner distinct from that of other herpesvirus subfamily members, as it increases host PABP cytoplasmic levels via a mechanism requiring inactivation of the cellular translational repressor 4E-BP1.
Among herpesviruses, cytomegalovirus has a distinct impact on host translational regulatory circuits, as it does not impair host protein synthesis during its acute-phase replication cycle (35). Increasing steady-state eIF4E, eIF4G, and PABP levels likely contribute to HCMV replication by allowing viral mRNA translation to proceed concurrently with ongoing cellular mRNA translation. Indeed, raising the initiation factor concentration is thought to enable discrete classes of mRNAs to better compete for limiting initiation factors (12, 13, 18-20, 22). Perhaps a similar strategy is operative in HCMV-infected cells to provide viral mRNAs better access to limiting host initiation factors.
Earlier microarray studies found increased eIF4E (4-fold) and eIF4A (36-fold) mRNA abundance by 24 hpi, but changes in eIF4G mRNA levels were not reported (44). Even though increased eIF4E and eIF4G mRNA abundance accompanied the virus-induced rise in protein levels, a role for translational control in regulating their protein levels cannot be completely discounted. Nevertheless, mechanisms dependent upon the viral IE1/2 transactivators are probably responsible for increasing eIF4E and eIF4G mRNAs. Although never validated, a 5-fold decrease in PABP mRNA at 24 hpi was also reported using microarrays (44). Our study clearly shows that HCMV infection increased PABP levels without detectably altering PABP mRNA abundance by using real-time PCR. Thus, translational control appears to be responsible for increasing the PABP concentration, as the protein's half-lives remains similar in infected and uninfected cells.
An HCMV-encoded component(s) is likely required to trigger the process that results in increased eIF4F core and associated components, followed by eIF4F assembly, as viral gene expression is required. Taken together, our experiments suggest that one or more viral genes responsive to the master regulatory gene products IE1/2 are potentially involved. Given that IE2 accumulates in cells infected with an IE1-deficient mutant and that IE1 is not essential for replication at high MOIs (8), IE1/2 are unlikely to be directly involved. Instead, downstream viral and/or cellular effectors responsive to IE1/2 are likely to cause the rise in translation factor levels. Indeed, in HSV-1-infected cells, multiple viral gene products act to regulate eIF4F assembly. While eIF4F assembly is dependent upon the HSV-1 ICP0 IE master regulatory protein, it is not absolutely required at high MOIs, similar to IE1 in HCMV-infected cells (38). In particular, the HSV-1 ICP6 gene, which is responsive to ICP0, encodes a protein product that directly associates with eIF4G, promotes binding to eIF4E, and facilitates active eIF4F complex assembly (39). Significantly, the overall levels of eIF4F core and associated components remain constant in HSV-1-infected cells (38). Our efforts to detect specific HCMV-encoded eIF4G-associated proteins have been unsuccessful to date (unpublished data), raising the possibility that different strategies are utilized in HCMV-infected cells to drive eIF4F assembly. In this regard, a coordinate rise in eIF4F core and associated factor subunit concentrations could conceivably contribute to eIF4F assembly in HCMV-infected cells and be a critical determinant controlling viral mRNA translation. Thus, HCMV-mediated increases in the translation initiation factor concentration may promote viral replication, in contrast to strategies used by other viruses that reduce the active translation factor concentration (17).
Multiple distinct translational control pathways act to maintain PABP homeostasis in cells. Besides an adenine-rich autoregulatory sequence involved in repressing translation (42), the PABP 5′ untranslated region contains a terminal oligopyrimidine (TOP) element found in mRNAs whose translation is stimulated in response to mitogenic, growth, and nutritional stimuli (9, 10). While mechanistic details of both strategies are under investigation, TOP mRNA translation is thought to involve mTOR signaling, albeit in a raptor- or rictor-independent manner in uninfected, insulin-treated cells (27). Conceivably, the recently identified HCMV-encoded UL38 mTOR activator might play a role in increasing the PABP concentration in infected cells (25). This could, however, be complicated by the altered substrate specificity of raptor-containing mTORC1 versus rictor-containing mTORC2 complexes in HCMV-infected cells (4, 15).
Unlike PABP abundance in alpha- and gammaherpesvirus-infected cells, where it is constant and the protein is redistributed to the nuclei, PABP abundance increases in HCMV-infected cells and the protein accumulates in the cytoplasm. Furthermore, enhanced PABP association with eIF4F complexes is only evident in HCMV-infected cells (2, 5, 40). The distinct subcellular distribution and eIF4F association of PABP in HCMV-infected cells, compared with those in alpha- and gammaherpesvirus-infected cells, could be important for the persistence of cellular mRNA translation in HCMV-infected cells. Indeed, a strong correlation can now be made involving PABP subcellular distribution, association of PABP with eIF4F, and impairment of host protein synthesis in herpesvirus-infected cells. Representative alpha- and gammaherpesviruses that impair host protein synthesis do not enhance PABP binding to eIF4F and restrict PABP to nuclei, whereas betaherpesviruses recruit PABP to eIF4F and accumulate cytoplasmic PABP. Finally, as the poly(A) tail contributes to mRNA stability, PABP levels and subcellular distribution may also influence mRNA decay (reviewed in references 21 and 31). While possibilities for future investigations abound, it is likely that the unique features of PABP homeostasis in HCMV-infected cells play a critical role in how the virus controls the host translational machinery to benefit its productive replication.
Acknowledgments
We are grateful to Simon Morley, Bob Schneider, Sara Kozma, Stipan Jonić, Edward Mocarski, and Wolfram Brune for generously providing antisera, cell lines, and siRNAs. In addition, we thank Carolina Arias, Angus Wilson, Derek Walsh, and Bob Schneider for many spirited, helpful discussions and Derek Walsh, Angus Wilson, and Rafa Cuesta-Sanchez for their critical reviews of the manuscript.
This work was supported by grants from the NIH to I.M. C.P. was the recipient of an ASM Watkins graduate fellowship and was subsequently supported in part by an NIH training grant (T32 AI007647). U.C. was also supported in part by an NIH training grant (T32 AI007647). I.M. is a scholar of the Irma T. Hirshl Trust. Purchase of the confocal microscope was funded by a shared instrumentation grant from the NIH (S10 RR017970).
Footnotes
Published ahead of print on 27 October 2010.
REFERENCES
- 1.Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273:13015-13021. [DOI] [PubMed] [Google Scholar]
- 2.Arias, C., D. Walsh, J. Harbell, A. C. Wilson, and I. Mohr. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 5:e1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunstein, S., K. Karpisheva, C. Pola, J. Goldberg, T. Hochman, H. Yee, J. Cangiarella, R. Arju, S. C. Formenti, and R. J. Schneider. 2007. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell 28:501-512. [DOI] [PubMed] [Google Scholar]
- 4.Buchkovich, N. J., Y. Yu, C. A. Zampieri, and J. C. Alwine. 2008. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signaling pathway. Nat. Rev. Microbiol. 6:266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrikova, E., M. Shveygert, R. Walters, and M. Gromeier. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its sub-cellular distribution. J. Virol. 84:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaunsinger, B. A., and D. E. Ganem. 2006. Messenger RNA turnover and its regulation in herpesviral infection. Adv. Virus Res. 66:337-394. [DOI] [PubMed] [Google Scholar]
- 8.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton, T. L., M. Stoneley, K. A. Spriggs, and M. Bushell. 2006. TOPs and their regulation. Biochem. Soc. Trans. 34:12-16. [DOI] [PubMed] [Google Scholar]
- 10.Hornstein, E., A. Git, I. Braunstein, D. Avni, and O. Meyuhas. 1999. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J. Biol. Chem. 274:1708-1714. [DOI] [PubMed] [Google Scholar]
- 11.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabat, D., and M. R. Chappell. 1977. Competition between globin messenger ribonucleic acids for a discriminating initiation factor. J. Biol. Chem. 252:2684-2690. [PubMed] [Google Scholar]
- 13.Koromilas, A. E., A. Lazaris-Karatzas, and N. Sonenberg. 1992. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 11:4153-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc. Natl. Acad. Sci. U. S. A. 103:14182-14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, Y. J., and B. A. Glaunsinger. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 7:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd, R. E. 2006. Translational control by viral proteinases. Virus Res. 119:76-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodish, H. F. 1974. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251:385-388. [DOI] [PubMed] [Google Scholar]
- 19.Ma, S., R. B. Bhattacharjee, and J. Bag. 2009. Expression of poly(A)-binding protein is upregulated during recovery from heat shock in HeLa cells. FEBS J. 276:552-570. [DOI] [PubMed] [Google Scholar]
- 20.Mamane, Y., E. Petroulakis, Y. Martineau, T. A. Sato, O. Larsson, V. K. Rajasekhar, and N. Sonenberg. 2007. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One 2:e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeehan, W. L. 1974. Regulation of hemoglobin synthesis. Effect of concentration of messenger ribonucleic acid, ribosome subunits, initiation factors, and salts on ratio of alpha and beta chains synthesized in vitro. J. Biol. Chem. 249:6517-6526. [PubMed] [Google Scholar]
- 23.Mocarski, E., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 24.Mohr, I. J., T. Pe'ery, and M. B. Mathews. 2007. Protein synthesis and translational control during viral infection, p. 545-595. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Moorman, N. J., I. M. Cristea, S. S. Terhune, M. P. Rout, B. T. Chait, and T. Shenk. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patursky-Polischuk, I., M. Stolovich-Rain, M. Hausner-Hanochi, J. Kasir, N. Cybulski, J. Avruch, M. A. Rüegg, M. N. Hall, and O. Meyuhas. 2009. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol. Cell. Biol. 29:640-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pende, M., S. H. Um, V. Mieulet, M. Sticker, V. L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S. C. Kozma, and G. Thomas. 2004. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salaun, C., A. I. MacDonald, O. Larralde, L. Howard, K. Lochtie, H. M. Burgess, M. Brook, P. Malik, N. K. Gray, and S. V. Graham. 2010. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J. Virol. 84:8539-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandri-Goldin, R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241-5256. [DOI] [PubMed] [Google Scholar]
- 31.Smith, R. W., and N. K. Gray. 2010. Poly(A)-binding protein (PABP): a common viral target. Biochem. J. 426:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Sodhi, A., R. Chaisuparat, J. Hu, A. K. Ramsdell, B. D. Manning, E. A. Sausville, E. T. Sawai, A. Molinolo, J. S. Gutkind, and S. Montaner. 2006. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell 10:133-143. [DOI] [PubMed] [Google Scholar]
- 33.Sonenberg, N., and A. G. Hinnebusch. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanton, R. J., B. P. McSharry, C. R. Rickards, E. C. Wang, P. Tomasec, and G. W. Wilkinson. 2007. Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J. Virol. 81:7860-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stinski, M. F. 1977. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J. Virol. 23:751-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stinski, M. F., and D. T. Petrik. 2008. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 325:133-152. [DOI] [PubMed] [Google Scholar]
- 37.Walsh, D., C. Arias, C. Perez, D. Halladin, M. Escandon, T. Ueda, R. Watanabe-Fukunaga, R. Fukunaga, and I. Mohr. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 28:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, D., and I. Mohr. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18:660-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh, D., and I. Mohr. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20:461-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh, D., C. Perez, J. Notary, and I. Mohr. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79:8057-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiebusch, L., M. Truss, and C. Hagemeier. 2004. Inhibition of human cytomegalovirus replication by small interfering RNAs. J. Gen. Virol. 85:179-184. [DOI] [PubMed] [Google Scholar]
- 42.Wu, J., and J. Bag. 1998. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J. Biol. Chem. 273:34535-34542. [DOI] [PubMed] [Google Scholar]
- 43.Wullschleger, S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell 124:471-484. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]