Abstract
We used expression cloning to isolate cDNAs encoding a microsomal 3β-hydroxy-Δ5-C27-steroid oxidoreductase (C27 3β-HSD) that is expressed predominantly in the liver. The predicted product shares 34% sequence identity with the C19 and C21 3β-HSD enzymes, which participate in steroid hormone metabolism. When transfected into cultured cells, the cloned C27 3β-HSD cDNA encodes an enzyme that is active against four 7α-hydroxylated sterols, indicating that a single C27 3β-HSD enzyme can participate in all known pathways of bile acid synthesis. The expressed enzyme did not metabolize several different C19/21 steroids as substrates. The levels of hepatic C27 3β-HSD mRNA in the mouse are not sexually dimorphic and do not change in response to dietary cholesterol or to changes in bile acid pool size. The corresponding human gene on chromosome 16p11.2-12 contains six exons and spans 3 kb of DNA, and we identified a 2-bp deletion in the C27 3β-HSD gene of a patient with neonatal progressive intrahepatic cholestasis. This mutation eliminates the activity of the enzyme in transfected cells. These findings establish the central role of C27 3β-HSD in the biosynthesis of bile acids and provide molecular tools for the diagnosis of a third type of neonatal progressive intrahepatic cholestasis associated with impaired bile acid synthesis.
Introduction
Bile acids are important components of normal physiology with essential functions in the liver and small intestine. Their synthesis in the liver provides a metabolic pathway for the catabolism of cholesterol and their detergent properties promote the solubilization of essential nutrients and vitamins in the small intestine. Inherited conditions that prevent the synthesis of bile acids can cause the accumulation of cholesterol and liver dysfunction (cholestasis), underscoring the essential role of bile acids in metabolism.
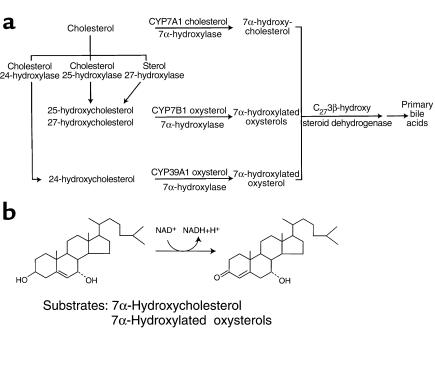
The synthesis of bile acids was initially thought to involve a single major pathway progressing from cholesterol to the primary bile acids cholate and chenodeoxycholate (reviewed in ref. 1). In fact, this synthesis is more complex and may involve as many as three separate pathways (Figure 1a). Each pathway differs in the initial steps and in the involvement of distinct sterol 7α-hydroxylase enzymes that add an essential hydroxyl group to carbon seven of the sterol ring. After the addition of this group, the next step in each pathway is catalyzed by a 3β-hydroxy-Δ5-C27-steroid oxidoreductase, which isomerizes the Δ5 bond to the Δ4 position and oxidizes the 3β-hydroxyl group to a 3-oxo moiety on intermediates with 27 carbon atoms (Figure 1b). For historical reasons this enzyme is often referred to in the literature as a C27 3β-hydroxysteroid dehydrogenase (C27 3β-HSD). The branching of the early steps in the bile acid synthesis pathways and the structural diversity of the resulting 7α-hydroxylated sterol intermediates raises the following questions. First, is there one C27 3β-HSD enzyme that participates in all three pathways, or are there multiple enzymes specific for each pathway? Second, how does the C27 3β-HSD enzyme of bile acid synthesis relate to the previously characterized family of C19/21 3β-HSD enzymes involved in steroid hormone metabolism?
Figure 1.
(a) Schematic showing branching in the early steps of the bile acid synthesis pathways. Individual enzymes and their catalyzed reactions are indicated. (b) The reaction catalyzed by the C27 3β-hydroxysteroid dehydrogenase isolated in this report is shown for the bile acid synthesis pathway involving cholesterol 7α-hydroxylase.
Progressive neonatal intrahepatic cholestasis is marked by jaundice, fat-soluble vitamin deficiency, and lipid malabsorption and is a rare condition of diverse etiologies. Among the causes of this disorder are inborn errors of metabolism that affect the production and secretion of bile (2). In the absence of a normal bile flow and bile acid pool size, the end products of heme metabolism are not secreted, and their accumulation causes the characteristic jaundice. Dietary fat-soluble vitamins are not effectively taken up in the intestine, leading to deficiencies in hemostasis, and hydrophobic lipids such as long-chain fatty acids and cholesterol are poorly absorbed causing fatty stools. Mutations in one of several genes encoding enzymes that synthesize bile acids (3–5) or that actively transport these steroids across the canalicular membranes of the liver into the bile (6) often underlie familial cases of intrahepatic cholestasis manifested in childhood. Knowledge of the exact molecular basis of the disorder is important because it can determine the therapeutic course (7). For example, defects in bile acid synthesis can often be treated by oral bile acid therapy, whereas the absence of a transporter requires orthotopic liver transplantation.
Deficiencies in three enzymes involved in bile acid synthesis lead to neonatal cholestasis. These include the CYP7B1 oxysterol 7α-hydroxylase (5), the C27 3β-HSD (3, 8), and a 3-oxo-Δ4-steroid 5β-reductase (4). These enzymes normally catalyze sequential steps in bile acid synthesis and, when missing, a characteristic spectrum of sterol intermediates accumulates. The diagnosis of which enzyme is defective can thus be made by gas chromatography-mass spectrometry analyses of serum or urine. In cases of 3-oxo-Δ4-steroid 5β-reductase deficiency (9, 10) or CYP7B1 oxysterol 7α-hydroxylase deficiency (5), the encoding genes have been isolated, allowing a molecular diagnosis to be made. The gene specifying C27 3β-HSD has not yet been cloned, and, consequently, the molecular genetics of cholestasis arising from a deficiency of this enzyme remain unknown.
To characterize the C27 3β-HSD enzyme(s) that participates in bile acid synthesis and to gain insight into the molecular genetics of progressive intrahepatic cholestasis caused by C27 3β-HSD deficiency, we used expression cloning to isolate cDNAs and genes encoding this enzyme activity in the mouse and human. Analysis of these DNAs reveals that a single C27 3β-HSD enzyme acts on intermediates arising in the three known pathways of bile acid synthesis. The C27 3β-HSD enzyme shares approximately 34% sequence identity with the known C19/21 3β-HSD enzymes. In addition, we have characterized a mutation in the C27 3β-HSD gene from a patient with neonatal cholestasis, which confirms the central role of this enzyme in bile acid synthesis.
Methods
Expression cloning.
A cDNA library in the expression vector pCMV6 was constructed as described previously (11). Poly (A)+ RNA was prepared from a pooled liver sample derived from three male mice (mixed strain C57BL/6J/129SvEv) and converted into cDNA by treatment with reverse transcriptase using the reagents of a Superscript Plasmid Kit (Life Technologies, Bethesda, Maryland, USA). Size-fractionated cDNA (>1.5 kb) was ligated into NotI/SalI-digested pCMV6 expression vector. Plasmid DNA from the ligation reaction was purified, and an aliquot was used for transformation of Electromax Escherichia coli DH10B cells (Life Technologies). The resulting library, which contained approximately 1.2 × 106 independent recombinants harboring cDNAs of average insert size 1.8 kb, was divided into pools of 500 colonies each. Plasmid DNAs were purified from each pool and used to transfect HEK 293 cells, which were plated on day 0 at a density of 7 × 105 cells per 60-mm dish in medium A (DMEM containing 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate). On day 1, cells were transfected using FuGene reagent (Boehringer Mannheim Biochemicals Inc., Indianapolis, Indiana, USA) with a mixture of plasmid DNAs that included 2.25 μg of a cDNA pool, 2.25 μg of pCYP7B1, a vector expressing the mouse CYP7B1 oxysterol 7α-hydroxylase (12), and 0.5 μg of pVA-1, a vector expressing the adenovirus type 5 VA1 gene (13). On day 2, medium A containing the transfection reagent was replaced with medium A supplemented with 1.2 μM 25-[3H]hydroxycholesterol (81.5 Ci/mmol; NEN Life Science Products, Boston, Massachusetts, USA). On day 3, lipids were extracted from the medium using Folch reagent (chloroform/methanol; 2:1 vol/vol) and separated by thin-layer chromatography in a solvent system containing toluene/ethylacetate (2:3 vol/vol) on prescored LK5DF silica gel thin-layer chromatography plates (Whatman, Hillsboro, Oregon, USA). Plates were subsequently analyzed using phosphorimaging on a BAS1000 apparatus (Fuji Medical Systems, Stamford, Connecticut, USA).
Pools of cDNAs expressing C27 3β-HSD enzyme activity were transformed into E. coli, and the resulting colonies were subdivided into subpools of approximately 80 as described (11). Plasmid DNAs were purified from the subpools and transfected into HEK 293 cells, which were then assayed for C27 3β-HSD enzyme activity. Positive pools from this secondary screen were transformed into E. coli, and individual colonies were grown and arranged in a matrix. Pools of plasmid DNAs from the rows and columns of the matrix were screened by transfection for clones expressing C27 3β-HSD enzyme activity. Plasmids at the intersections of positive rows and columns were isolated, screened again by transfection, and characterized as described in the text.
Analysis of substrate specificity.
HEK 293 cells were transfected with mixtures of expression plasmids using the FuGene reagent. A plasmid encoding the rat C19/21 3β-HSD type I enzyme was a kind gift of Stefan Andersson, University of Texas Southwestern Medical Center. The plasmid (pCYP7A1) encoding the rat CYP7A1 cholesterol 7α-hydroxylase was isolated as described previously (14). Plasmid DNAs encoding C27 3β-HSD enzymes were isolated as described above.
The 3β-HSD activity was assayed in transfected cells as detailed above, except that substrate was included in medium A at a concentration of 3 μM. The tested substrates were: [14C]cholesterol (51 mCi/mmol; NEN); 24-[3H]hydroxycholesterol (50 Ci/mmol; American Radiolabeled Chemicals, St. Louis, Missouri, USA); 25-[3H]hydroxycholesterol (81.5 Ci/mmol; NEN); 27-[3H]hydroxycholesterol (80 Ci/mmol; NEN); [14C]dehydroepiandrosterone (55.5 mCi/mmol; NEN;, [3H]pregnenolone (22.5 Ci/mmol; NEN); and [3H]androst-5-ene-3β,17β-diol (42 Ci/mmol; NEN). When cholesterol was used as a substrate, the transfected cells were incubated with 20 mg/ml 2-hydroxypropyl-β-cyclodextrin (Sigma Chemical Co., St. Louis, Missouri, USA) in medium A for 1 hour before the addition of radiolabeled cholesterol to the medium. This treatment reduces the intracellular concentration of cholesterol and consequently increases the sensitivity of the assay (11). Thin-layer chromatography was carried out as described above for the C27 substrates or in a solvent system containing chloroform/ethylacetate (3:1 vol/vol) when dehydroepiandrosterone, pregnenolone, or androst-5-ene-3β, 17β-diol were used as substrates.
Isolation of human C27 3β-HSD cDNA.
A 186-kb human genomic DNA clone (GenBank/EBI Data Bank accession no. AC021142) containing several segments with high sequence identity to the mouse C27 3β-HSD cDNA was identified by basic local alignment search tool (BLAST) search of the Human Genome database (http://www.ncbi.nlm.nih.gov/BLAST/). Based on this sequence, two oligonucleotides of the following sequences were designed that were predicted to correspond to the 5′- and 3′-untranslated regions of the C27 3β-HSD mRNA: 5′-GCTAGTCGACTCTCTTCCCCAGCCAGGC-3′ (forward primer) and 5′-ATCGCGGCCGCGCTGTATCTGGGCCTCCA-3′ (reverse primer). These oligonucleotides also contained SalI-restriction sites (forward primer) and NotI-restriction sites (reverse primer) at their 5′ ends. This primer pair was used in a PCR to amplify C27 3β-HSD cDNAs from an aliquot (105 CFU) of a human liver cDNA library obtained from Life Technologies (catalog no. 10422-012). The thermocycler (GeneAmp 9600; Perkin-Elmer Applied Biosystems, Foster City, California, USA) was programmed to perform one cycle of 94°C for 60 seconds, followed by 30 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. The amplified cDNA fragment was purified by gel-filtration chromatography and then digested for 2 hours with the restriction enzymes SalI and NotI (New England Biolabs, Beverly, Massachusetts, USA). The resulting major cDNA fragment of approximately 1.1 kb was purified by electrophoresis through a 0.8% agarose gel recovered from the matrix using the reagents of a QIAquick gel extraction kit (QIAGEN GmbH, Hilden, Germany) and ligated into a SalI/NotI-digested pCMV6 vector. Plasmid DNA was prepared, and the identity of the human C27 3β-HSD cDNA confirmed by sequencing. The resulting expression plasmid was designated pC27-3β-HSD-Human.
DNA sequencing and RNA blotting.
DNA sequencing was performed on an ABI Prism 377 sequencer (Perkin-Elmer) using thermocycler sequencing protocols and fluorescent dye terminators. Assembly of contiguous DNA sequences and sequence alignments were performed using a Lasergene software package (DNASTAR Inc., Madison, Wisconsin, USA).
For RNA blotting, mouse and rat multiple-tissue RNA blots (catalog no. 7762-1 and no. 7764-1; CLONTECH, Palo Alto, California, USA) were hybridized overnight in a buffer containing 50% (vol/vol) formamide at 42°C with a [32P]-radiolabeled probe derived from a near full-length mouse C27 3β-HSD cDNA. Human multiple-tissue RNA blots (catalog no. 7760-1 and no. 7759-1; CLONTECH) were hybridized to a radiolabeled, near full-length, human C27 3β-HSD cDNA probe using similar conditions. In both experiments, the cDNAs were radiolabeled with [α32P]dCTP by random nonamer priming (Megaprime Labeling Kit; Amersham Life Sciences, Arlington Heights, Illinois, USA). Blots were washed stringently at 65°C in 0.1× SSC containing 0.1% (wt/vol) SDS and were exposed to Kodak X-OMAT AR film using an intensifying screen for the times indicated in the legend to Figure 6.
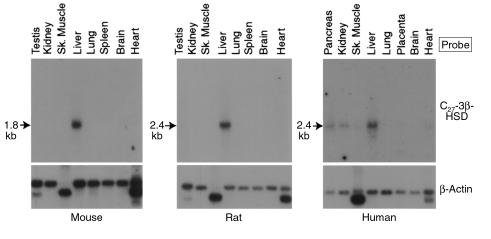
Figure 6.
Tissue distribution of C27 3β-HSD mRNAs in the mouse, rat, and human. Aliquots (2 μg) of poly(A)+-enriched mRNA from the indicated tissues and species were size fractionated by agarose-gel electrophoresis, transferred to nylon membranes, and subjected to blot hybridization using radiolabeled probes derived from the mouse (mouse and rat membranes) or human (human membrane) C27 3β-HSD cDNAs. After washing, the mouse, rat, and human filters were exposed to x-ray film for 72, 16, and 168 hours, respectively. The size of the C27 3β-HSD mRNA detected in each experiment is shown to the left of the autoradiograms. To ensure that mRNA was present in each lane of the individual membranes, they were stripped of radioactivity after the initial hybridization and rehybridized with a [32P]-labeled probe derived from a human β-actin cDNA. The filters were then washed and exposed to x-ray film. The resulting autoradiograms, with signals in each lane, are shown below those derived with the C27 3β-HSD cDNA probes.
Diet studies.
Mixed-strain (C57BL/6J/129SvEv) littermate mice that were either wild-type at the cholesterol 7α-hydroxylase locus (Cyp7a1+/+) or carried an induced null allele (Cyp7a1–/–) (15) were generated at the University of Texas Southwestern Medical Center. Mice were fed a cereal-based diet (7001; Harlan Teklad, Madison, Wisconsin, USA), which contained ≥ 4% (wt/wt) fat, ≥ 24% (wt/wt) protein, and ≤ 5% (wt/wt) fiber. Where indicated, the diet was supplemented with 1% (wt/vol) cholesterol (ICN Radiochemicals, Irvine, California, USA). Mice of the indicated sex (n = 6 per group, age 3 months) were fed either the normal or supplemented diets for two weeks. On the last day of the experiment, animals were sacrificed and the livers dissected. RNA was isolated by guanidinium isothiocyanate acid phenol extraction. Equal amounts of RNA from individual tissues from animals of each group were pooled, poly(A)+ RNA was isolated by oligo(dT) cellulose chromatography, and the samples were analyzed by RNA blotting using standard methods (16).
Clinical history of patient MU2.
This Saudi Arabian boy was the fifth child of parents who were first cousins. An older sister and brother died of complications of progressive liver disease. He weighed 4 kg at birth and exhibited jaundice 2 days later. He was brought to clinical attention in Great Britain as a 3-month-old infant (3). He had moderate jaundice, hepatomegaly, and an accumulation of 3β,7α-dihydroxy- and 3β,7α,12α-trihydroxy-5-cholenoic acids in his urine. The family returned to Saudi Arabia, and the patient was lost to follow up until age 2 years, 9 months, when he was seen at the King Faisal Specialist Hospital. He was underdeveloped (25th percentile for weight, less than fifth percentile for height), had persistent jaundice, pruritus, rickets, and hepatomegaly (8). Treatment with chenodeoxycholic acid led to an increased well-being, a decrease in pruritus, and a normalization of urinary steroids (17). He is now 17 years old and doing well on cholic acid therapy.
Genetic analysis.
The C27 3β-HSD gene was isolated from genomic DNA extracted from the white blood cells of a normal subject and from the fibroblasts of patient MU2 (3, 8). PCRs were performed with an Advantage-GC Genomic PCR kit (catalog no. K1908-1; CLONTECH) using the human C27 3β-HSD oligonucleotide primers described above. The thermocycler program consisted of one cycle of 94°C for 15 seconds, followed by 30 cycles of 94°C for 5 seconds, 70°C for 180 seconds, and a final cycle of 70°C for 60 seconds. The amplified 2.7-kb DNA fragments from the control and patient genomes were purified, digested with SalI and NotI as described above, and ligated into pCMV6 to yield expression plasmids pC27-3β-HSD-Normal and pC27-3β-HSD-MU2, respectively. The DNA sequences of the cloned inserts were determined, and the resulting plasmids were used in transfection experiments to express C27 3β-HSD activity.
The exon-intron structure of the human C27 3β-HSD gene was deduced from the DNA sequence deposited in GenBank/EBI Data Bank (accession no. AC021142) by comparison with the cloned cDNA sequence. This structure was confirmed by DNA sequence analysis of the gene amplified from the genomic DNA of a normal control. A 2-bp deletion in patient MU2 was identified by amplification of individual exons from genomic DNA, then by sequence analysis.
Gene mapping.
The chromosomal location of the human C27 3β-HSD gene was determined by radiation hybrid mapping using the Stanford G3 panel (Research Genetics, Huntsville, Alabama, USA). The primer pair used for amplification of the panel DNAs by the PCR was 5′-AAGGGCACTCAGGGGTGTGT-3′ (forward primer) and 5′-CGGGACCTGGGTGGATGTGC-3′ (reverse primer). This primer pair corresponds to nucleotides 62,583–62,602 and 62,093–62,112, respectively, of the human gene sequence (GenBank/EBI Data Bank accession no. AC021142). The thermocycler program consisted of one cycle of 94°C for 15 seconds, followed by 30 cycles of 94°C for 5 seconds, 70°C for 180 seconds, and a final cycle of 70°C for 60 seconds. The radiation hybrid data were analyzed through the Stanford Genome Center server (http://shgc-www.stanford.edu/RH/index.html/), which indicated linkage of the human C27 3β-HSD gene to the SHGC-57014 marker (lod score = 15.1, cR_1000 = 5) on the short arm of chromosome 16 in the vicinity of Giemsa band p11.2–12. Despite many attempts, we were unable to determine the chromosomal location of the mouse gene by analysis of the T31 radiation hybrid panel available from the Jackson Laboratory (Bar Harbor, Maine, USA).
Results
In the course of transfection experiments with cDNAs encoding enzymes that catalyze early steps in the bile acid synthesis pathways (Figure 1a), we noted that 7α-hydroxylated oxysterols were good substrates for an endogenous C27 3β-HSD enzyme present in several different cell lines. For example, as shown in Figure 2 (lane 1), the addition of cholest-5-ene-3β,25-diol (25-hydroxycholesterol) to the medium of cultured human embryonic HEK 293 cells led to the formation of a 7α-hydroxylated oxysterol (cholest-5-ene-3β,7α,25-triol) and the conversion of this compound to 7α,25-dihydroxy-cholest-4-ene-3-one by the C27 3β-HSD activity. Time-course experiments indicated that cholest-5-ene-3β,7α,25-triol was the first product formed, followed by its conversion to 7α,25-dihydroxy-cholest-4-ene-3-one (data not shown). Transfection of a cDNA encoding the CYP7B1 oxysterol 7α-hydroxylase increased the synthesis of the 7α-hydroxylated intermediate but had little effect on the formation of the 3-oxo-Δ4 sterol (Figure 2, lane 2). These results suggested that cDNAs encoding C27 3β-HSD enzymes could be identified by adding 25-hydroxycholesterol to cells transfected with the CYP7B1 oxysterol 7α-hydroxylase and screening a cDNA library for clones that would increase the formation of the 3-oxo-Δ4 sterol intermediate.
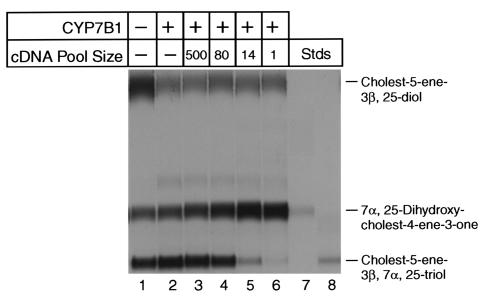
Figure 2.
Expression cloning of mouse cDNAs encoding C27 3β-HSD enzyme activity. Cultured HEK 293 cells were cotransfected with a cDNA encoding the CYP7B1 oxysterol 7α-hydroxylase and pools of cDNAs derived from a mouse liver library. After 24 hours, the transfected cells were assayed for enzyme activity by thin-layer chromatography using [3H]cholest-5-ene-3β,25-diol (25-hydroxycholesterol, 1.2 μM) as substrate (see Methods). In lane 1, cells transfected with a control plasmid lacking a cDNA insert exhibited basal oxysterol 7α-hydroxylase activity, which converts 25-hydroxycholesterol into cholest-5-ene-3β,7α,25-triol, and low levels of C27 3β-HSD enzyme activity, which converts the product of the CYP7B1 enzyme into 7α,25-dihydroxy-cholest-4-ene-3-one. In lane 2, transfection with the CYP7B1 oxysterol 7α-hydroxylase cDNA alone increased the synthesis of cholest-5-ene-3β,7α,25-triol. In lane 3, transfection of a pool of 500 cDNAs containing a cDNA encoding C27 3β-HSD enzyme activity increased the synthesis of 7α,25-dihydroxy-cholest-4-ene-3-one. In lanes 4–6, transfection of progressively smaller (more enriched) pools containing the cDNA encoding the C27 3β-HSD activity resulted in a gradual increase in the synthesis of 7α,25-dihydroxy-cholest-4-ene-3-one and a decrease in the 7α-hydroxylated intermediate. In lanes 7 and 8 are radiolabeled sterol standards whose identities were confirmed previously by gas chromatography-mass spectrometry (12).
Biochemical studies indicate that C27 3β-HSD enzyme activity is abundant in the livers of several species, including the mouse (18–20). For this reason, we prepared a cDNA library in an expression vector using mouse hepatic mRNA as a template. The library was divided into pools containing approximately 500 independent cDNAs each for screening purposes. Individual pools were then introduced into HEK 293 cells together with the CYP7B1 oxysterol 7α-hydroxylase cDNA, and the transfected cells were incubated with radiolabeled 25-hydroxycholesterol. In an initial screen of 60 pools, several were found that increased the formation of the 3-oxo-Δ4 compound (e.g., Figure 2, lane 3). One such active pool was progressively subdivided and screened by transfection to identify a single cDNA encoding C27 3β-HSD activity (Figure 2, lanes 4–6).
The purified plasmid contained a cDNA insert of 1.75 kb that encoded a 5′-untranslated sequence of 36 bp, a translated sequence of 1107 bp, and a 3′-untranslated region of 603 bp ending in a poly(A) sequence. The translated sequence specified a protein of 369 amino acids (Figure 3a). Database searches revealed a highly homologous rat cDNA sequence (GenBank/EBI Data Bank accession no. AB000199), which was identified previously in a screen for mRNAs that accumulated in growth-arrested cells and named cca2 (21). The rat cDNA encoded a sequence of 338 amino acids that was contiguous with that of the mouse, except for a sequence of 31 amino acids unique to the mouse protein (Figure 3a).
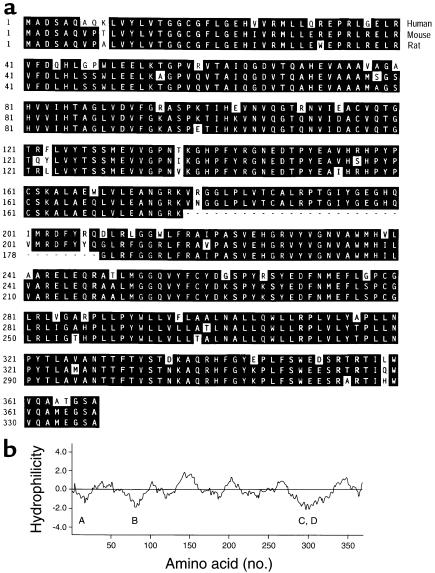
Figure 3.
(a) Alignment of C27 3β-HSD enzyme sequences deduced from the human, mouse, and rat cDNAs. Identities between enzymes are shaded in black. Amino acids are numbered on the left. A sequence of 31 amino acids present in the human and mouse enzymes but absent in that of the rat is indicated by dashes. The GenBank/EBI Data Bank accession numbers for the human, mouse, and rat sequences are AF277719, AF277718, and BAA22931, respectively. (b) A hydropathy plot was generated for the amino acid sequence of the human C27 3β-HSD using the Kyte-Doolittle algorithm. The window size was 17 amino acids. Hydrophobic sequences fall below the central dividing line, and hydrophilic sequences rise above this line. Letters below the plot indicate four putative transmembrane domains.
A search of the human genome sequence database revealed an extended sequence (GenBank/EBI Data Bank accession no. AC021142) that in certain sections shared approximately 84% sequence identity with the mouse cDNA. By comparing these two sequences, we deduced the predicted 5′- and 3′-untranslated regions of the human C27 3β-HSD mRNA, and based on these data, synthesized two oligonucleotide primers that were oppositely oriented and complementary to these regions. This primer pair was used in RT-PCRs to amplify a cDNA from human liver mRNA that encoded the C27 3β-HSD enzyme. The human cDNA specified a protein of 369 amino acids that was contiguous along its length with the mouse protein (Figure 3a).
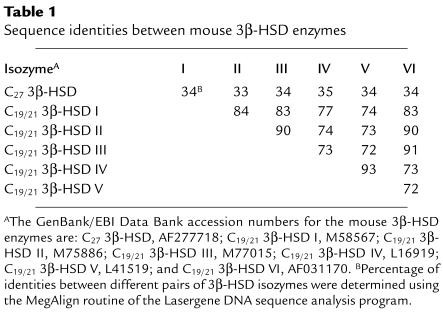
Several features of these predicted protein sequences suggested that they were C27 3β-HSD enzymes. First, the human, mouse, and rat sequences shared 86–94% sequence identity in pairwise comparisons, which suggested that they were orthologs of the same enzyme. Second, their amino-terminal sequences were similar to a sequence of 18 amino acids that was established from a highly purified preparation of pig liver C27 3β-HSD (20). Third, hydropathy analyses of the sequences showed four regions of extended hydrophobicity, each approximately 20 amino acids, which may represent membrane-spanning domains (Figure 3b). The presence of these putative transmembrane sequences was consistent with the enrichment of C27 3β-HSD enzyme activity in hepatic liver membranes (18–20). Finally, these protein sequences shared approximately 34% sequence identity with six 3β-HSD enzymes that act on steroid substrates containing 19 carbon atoms (C19) or 21 carbon atoms (C21) (Table 1).
Table 1.
Sequence identities between mouse 3β-HSD enzymes
To determine the substrate specificities of the mouse and human C27 3β-HSD enzymes, we transfected HEK 293 cells with expression vectors containing the respective cDNAs and then measured the ability of the transfected cells to metabolize different radiolabeled C27 sterols that arise in the bile acid synthesis pathways (Figure 1a). The C27 3β-HSD enzymes are only active against 7α-hydroxylated sterol substrates, which are not readily available. This problem was overcome by generating 7α-hydroxylated sterols in situ through cotransfection of the appropriate sterol 7α-hydroxylases. To these ends, 7α-hydroxylated oxysterols were generated by adding oxysterols to cells transfected with a CYP7B1 oxysterol 7α-hydroxylase, and 7α-hydroxycholesterol was produced by addition of cholesterol to cells transfected with a CYP7A1 cholesterol 7α-hydroxylase cDNA. Subsequent incubations were done under conditions that minimized the contribution of the endogenous C27 3β-HSD enzyme activity present in the HEK 293 cells. Finally, the substrate specificities of the C27 3β-HSD enzymes were compared and contrasted to those of a C19/21 3β-HSD type I enzyme.
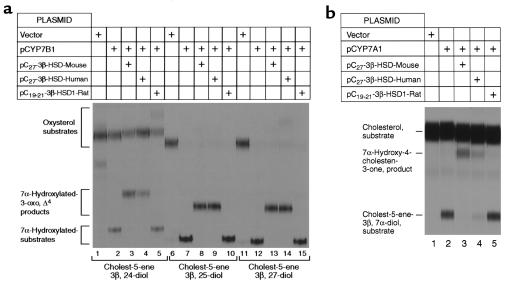
As shown in Figure 4a, the expressed mouse and human C27 3β-HSD enzymes were active against the 7α-hydroxylated forms of 24-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol. The percentage of starting substrate converted into the Δ4, 3-oxo product in each case was proportional to the amount of 7α-hydroxylated oxysterol formed in the cell, which suggested that the C27 3β-HSD enzymes did not show a preference for one 7α-hydroxylated oxysterol substrate over another. Both enzymes also were active against 7α-hydroxycholesterol (Figure 4b). In experiments not shown, the metabolism of radiolabeled 7α-hydroxylated oxysterols by the expressed C27 3β-HSD enzymes was inhibited by the addition of unlabeled 7α-hydroxycholesterol to the culture medium, and conversely, the metabolism of 7α-hydroxycholesterol was inhibited by the addition of 7α-hydroxylated oxysterols. None of these 7α-hydroxylated C27 substrates were metabolized to their Δ4, 3-oxo forms by cells transfected with a cDNA encoding a C19/21 3β-HSD type I enzyme (Figure 4, a and b).
Figure 4.
Substrate specificities of the human and mouse C27 3β-HSD enzymes. Cultured HEK 293 cells were transfected with the indicated cDNA-expression plasmids and assayed for 3β-HSD enzyme activity using different radiolabeled sterol intermediates of the bile acid synthesis pathways. In (a), intermediates arising in the oxysterol 7α-hydroxylase pathways were added to cells at a final concentration of 3 μM, and the incubations continued for 24 hours: lanes 1–5, results obtained with [3H]cholest-5-ene-3β,24-diol (24-hydroxycholesterol); lanes 6–10, [3H]cholest-5-ene-3β,25-diol (25-hydroxycholesterol); and lanes 11–15, [3H]cholest-5-ene-3β,27-diol (27-hydroxycholesterol). In (b), [14C]cholesterol was added to cyclodextrin-treated cells at a final concentration of 3 μM, and the incubations continued for 24 hours. The positions to which sterols of known structure migrated on the thin-layer chromatograms are shown on the left.
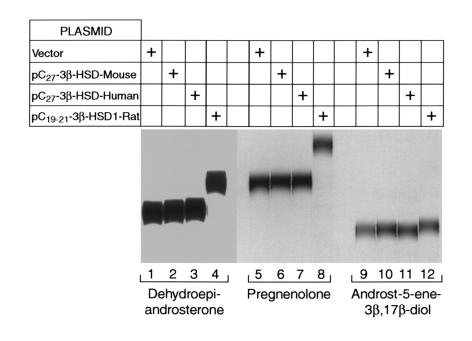
In contrast to these results, cells transfected with the C19/21 3β-HSD type I cDNA avidly metabolized dehydroepiandrosterone (C19), pregnenolone (C21), and androst-5-ene-3β, 17β-diol (C19), whereas the patterns of metabolites generated from these substrates in cells transfected with the mouse and human C27 3β-HSD cDNAs were no different from those of mock-transfected cells (Figure 5). Cotransfection of a CYP7B1 cDNA, which is known to produce the 7α-hydroxylated derivatives of dehydroepiandrosterone and pregnenolone (22), did not alter this outcome (data not shown). We concluded from this series of experiments that the C27 3β-HSD enzymes encoded by the isolated cDNAs metabolize C27 intermediates that arise in the three known pathways of bile acid synthesis and that these enzymes probably do not act on intermediates of steroid hormone biosynthesis and catabolism.
Figure 5.
Activities of different 3β-HSD enzymes against C19/21 steroid substrates. Cultured HEK 293 cells were transfected with the indicated cDNA expression plasmids and assayed for 3β-HSD enzyme activity using different radiolabeled intermediates of steroid hormone metabolism. In lanes 1–4, [14C]dehydroepiandrosterone (C19) was added to cells at a concentration of 3 μM and the incubation continued for 24 hours. In lanes 5–8, [3H]pregnenolone (C21) was added, and in lanes 9–12, [3H]androst-5-ene-3β, 17β-diol (C19) was added.
The tissue distributions of the mouse, rat, and human C27 3β-HSD mRNAs were examined next by RNA blotting (Figure 6). Analysis of eight mouse tissues revealed a single hybridizing species of approximately 1.8 kb that was present only in the liver (Figure 6, left). A similar pattern of liver-specific expression was found among eight rat tissues (Figure 6, middle). In contrast, the distribution of the human C27 3β-HSD mRNA was more widespread (Figure 6, right). In this species, a mRNA of approximately 2.4 kb was detected at high levels in the liver, pancreas, and kidney, and at lower levels in the heart, skeletal muscle, and placenta. The abundance of these C27 3β-HSD mRNAs in the liver was consistent with a role for the encoded enzyme in bile acid synthesis.
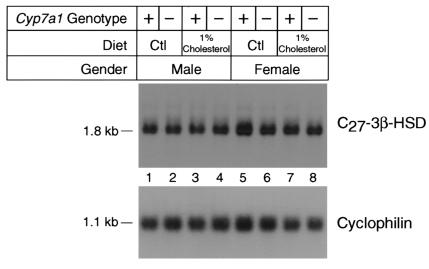
To determine if the hepatic levels of C27 3β-HSD mRNA were regulated in response to changes in the enteroheptic flux of cholesterol or bile acids, groups of male and female mice were fed a normal chow diet or a diet supplemented with 1% cholesterol for a period of 21 days. Thereafter, the animals were sacrificed, mRNA was purified from the liver, and aliquots of the mRNA were analyzed by blot hybridization. The data of Figure 7 show that the levels of C27 3β-HSD mRNA did not change between the control and experimental animals. In addition, there was no detectable sexual dimorphism in the levels of this mRNA. Similar results were obtained when mice were fed diets supplemented with 0.5% cholate, 2% cholesterol plus 0.5% cholate, or 2% colestipol (data not shown). Furthermore, when cholesterol feeding was carried out in mice lacking the cholesterol 7α-hydroxylase gene (Cyp7a1), which have much reduced bile acid–pool sizes (23), the levels of C27 3β-HSD mRNA did not change relative to those in wild-type animals (Figure 7). We concluded from this series of experiments that the steady-state level of mouse C27 3β-HSD mRNA is immune to dietary supplements that affect the expression of other genes encoding bile acid–synthesis enzymes and to large alterations in the size of the circulating bile acid pool.
Figure 7.
Expression of C27 3β-HSD mRNA in wild-type and cholesterol 7α-hydroxylase (Cyp7a1) knockout mice fed low- and high-cholesterol diets. Poly(A)+-enriched mRNA was isolated from the livers (n = 5) of male and female wild-type mice (+) or cholesterol 7α-hydroxylase (Cyp7a1) knockout mice (–) fed normal chow containing 0.02% (wt/wt) cholesterol (Ctl) or chow supplemented with 1% (wt/wt) cholesterol for 21 days. Aliquots (5 μg) of this RNA were size fractionated with agarose-gel electrophoresis, transferred to nylon membranes, and subjected to blot hybridization using a radiolabeled probe derived from the mouse C27 3β-HSD cDNA. After washing, the filter was exposed to x-ray film for 72 hours. The location to which the C27 3β-HSD mRNA migrated in the experiment and its interpolated size (1.8 kb) are shown on the left of the autoradiogram. To ensure that equal amounts of mRNA were present in each lane of the membrane, it was stripped of radioactivity after the initial hybridization and rehybridized with a [32P]-labeled probe derived from a rat cyclophilin cDNA. The filter was washed and exposed to x-ray film. The resulting autoradiogram, with a signal of equal intensity in each lane, is shown below that derived with the C27 3β-HSD cDNA probe. The calculated size of the cyclophilin mRNA (1.1 kb) is shown to the left of the autoradiogram.
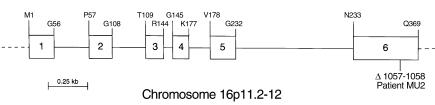
To analyze the molecular genetics of C27 3β-HSD deficiency, the structure of the human gene was determined. This proved to be a straightforward task as an extended sequence containing the gene was deposited in the database during the course of these studies (GenBank/EBI Data Bank accession no. AC021142). By comparing the sequences of the genomic DNA to those of the mouse and human cDNAs isolated as described above, we deduced that the human C27 3β-HSD gene contained six exons and spanned approximately 3 kb of DNA (Figure 8). This predicted structure was confirmed by DNA-sequence analysis of the gene after its amplification from genomic DNA by PCR. Analysis of a radiation hybrid panel mapped the human gene to chromosome 16p11.2-12.
Figure 8.
Structure of the human C27 3β-HSD gene. A schematic of the gene is shown drawn to scale with six exons indicated by numbered boxes and five introns indicated by connecting lines. Individual amino acids occurring at the exon-intron junctions are shown in single letter code above the gene. The location of a 2-bp deletion (Δ1057-1058) found in homozygous form in the DNA of a patient (MU2) with progressive intrahepatic cholestasis is shown in exon 6 below the gene. Radiation hybrid panel–mapping experiments indicated that the human C27 3β-HSD gene is located on chromosome 16p11.2-12 (see Methods).
Several patients presenting with progressive neonatal cholestasis due to C27 3β-HSD enzyme deficiency have been reported in the literature. We were successful in obtaining a genomic DNA sample from only one such affected individual. The C27 3β-HSD gene was amplified from this sample and subjected to DNA sequencing. Several differences were detected between the patient’s gene and a normal control, most of which were likely to be random polymorphisms. However, the patient’s gene also contained a 2-bp deletion in exon 6 in homozygous form (Figure 8), which was not present in 25 healthy controls and was predicted to shift the translational reading frame causing premature truncation and removal of 23 amino acids from the carboxyl-terminus of the protein.
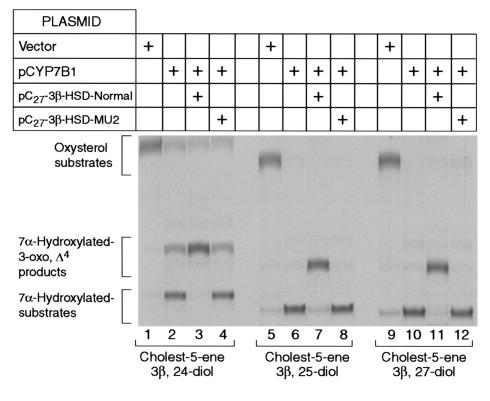
To determine if this deletion affected the expression of active C27 3β-HSD enzyme, a control gene and the patient’s gene were amplified by PCR and then ligated individually into an eukaryotic expression vector. The two cloned genomic DNA inserts were sequenced, and the plasmids were introduced into HEK 293 cells. After a 16-hour period to allow expression, the transfected cells were assayed for C27 3β-HSD activity. The data of Figure 9 show that the 2-bp deletion prevented the production of enzyme capable of isomerizing and oxidizing the 7α-hydroxylated forms of 24-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol. A similar loss of active enzyme was realized when 7α-hydroxycholesterol was used as a substrate (data not shown). Whether loss of activity in these experiments was due to a failure of the protein to accumulate in cells or whether it was due to the production of an inactive protein was not determined.
Figure 9.
Expression of normal and mutant C27 3β-HSD genes in cultured HEK 293 cells. The C27 3β-HSD gene was amplified using PCR from the genomic DNA of either a normal subject (Normal) or an individual with intrahepatic cholestasis (MU2) and inserted into the plasmid pCMV6 as described in Methods. The resulting expression vectors were introduced together with a plasmid expressing the mouse CYP7B1 oxysterol 7β-hydroxylase cDNA (pCYP7B1) into HEK 293 cells by cotransfection. After 24 hours, the ability of the transfected cells to convert [3H]cholest-5-ene-3β,24-diol (24-hydroxycholesterol, lanes 1–4), [3H]cholest-5-ene-3β,25-diol (25-hydroxycholesterol, lanes 5–8), and [3H]cholest-5-ene-3β,27-diol (27-hydroxycholesterol, lanes 9–12) was determined by thin-layer chromatography. The chromatogram was exposed to x-ray film for 6 days.
Discussion
In this paper we report the isolation of cDNAs encoding mouse and human enzymes with C27 3β-HSD activity. Each enzyme contains 369 amino acids and is predicted to be associated with the membrane. They share 86% sequence identity and are both active against several 7α-hydroxylated sterols that contain 27 carbons. These enzymes do not appear to use steroid substrates that contain 19 or 21 carbons. C27 3β-HSD mRNA is present in the liver of rats and mice and is more widely distributed in humans. The levels of this mRNA do not change in the mouse when diets containing different amounts of cholesterol are consumed or when bile acid synthesis via the classical pathway is impaired. The human C27 3β-HSD gene spans approximately 3 kb of DNA on chromosome 16p11.2-12 and contains six exons separated by five short introns. A 2-bp deletion was present in this gene in a patient with progressive intrahepatic cholestasis due to C27 3β-HSD enzyme deficiency. We conclude from these studies that a single C27 3β-HSD enzyme is active in the three defined pathways of bile acid synthesis and that a deficiency in this enzyme leads to one form of neonatal liver failure.
The 3β-HSD enzyme family consists of a large number of proteins that are present in both prokaryotes and eukaryotes. They are proposed to play a wide variety of anabolic and catabolic roles in intermediary metabolism, and, consistent with this broad function, they are expressed in abundance in organisms ranging from viruses to humans. For example, at least six 3β-HSD enzymes that act on Δ5, 3β-hydroxysteroids have been described in the mouse (24). Despite the widespread distribution of these proteins and their large number, in only a few cases has an essential function been assigned to a single 3β-HSD enzyme. These exceptions include the human C19/21 3β-HSD type II enzyme, which is required for the synthesis of all steroid hormone classes (25); the product of the Nsdh1 gene in the mouse, which catalyzes reactions in the terminal steps of cholesterol biosynthesis (26); the 3β-HSD enzyme encoded by the Sax1 gene of the plant Arabidopsis, which is required for brassinosteroid biosynthesis (27); and the presently described C27 3β-HSD enzyme of bile acid biosynthesis.
It is not yet clear why there are so many 3β-HSD enzymes and whether, as is the case with the four enzymes noted above, each member plays a unique metabolic role. The C27 3β-HSD protein and the C19/21 3β-HSD type I protein share approximately 34% sequence identity and are clearly related (Table 1), yet the substrate specificities of these two enzymes are nonoverlapping: the C27 3β-HSD enzyme is specific for 27-carbon substrates whereas the C19/21 3β-HSD type I enzyme acts on 19- or 21-carbon substrates (Figure 4 and Figure 5). These findings in transfected cells agree with biochemical studies in which purified preparations of C27 3β-HSD enzyme activity from rabbit liver (19) and pig liver (20) were shown to prefer C27 sterol substrates. A difference in the substrate specificities of the C27 3β-HSD versus the type II C19/21 3β-HSD is also apparent from the distinct phenotypes of individuals lacking these enzymes. C27 3β-HSD enzyme deficiency is marked by accumulation of C27 sterol intermediates of bile acid synthesis, progressive intrahepatic cholestasis, and no endocrine abnormalities (28). In contrast, type II C19/21 3β-HSD deficiency causes accumulation of C19 and C21 intermediates of steroid hormone biosynthesis, congenital adrenal hyperplasia, and no cholestasis (29). Taken together, these results suggest that there are few functional redundancies between these two types of 3β-HSD enzymes and thus that individual 3β-HSD enzymes play distinct physiological roles.
Alternate forms of each 3β-HSD enzyme may also exist based on sequence comparisons between the human, rat, and mouse C27 3β-HSD proteins (Figure 3a). Compared with the other two, the deduced rat sequence (21) is missing 31 amino acids from the middle of the enzyme. The deleted sequence does not correspond to an exon in the human gene (Figure 8), and thus the mechanism by which the encoding transcript arises is not evident. When this cDNA was introduced into cells, it did not express an enzyme with 3β-HSD activity against C19/21 steroids (21), however, C27 substrates were not tested. It is conceivable that this form of the C27 3β-HSD, which is expressed by cultured rat cells grown to confluence (21), may play a different role from the full-length enzyme expressed in the liver.
Clayton and colleagues first reported progressive familial intrahepatic cholestasis due to C27 3β-HSD enzyme deficiency in 1987 (3). The typical subject with this disorder is a young child with jaundice, fat-soluble vitamin deficiency, and steatorrhea, who responds favorably to bile acid supplementation (17, 30). Analysis of urinary and plasma steroids reveals an accumulation of 3β,7α-dihydroxy-5-cholenoic acid and 3β,7α,12α-trihydroxy-5-cholenoic acid, as well as the sulfated and conjugated forms of these hepatotoxic steroids (3). The C27 3β-HSD enzyme activity is expressed in normal fibroblasts and is absent from these cells in affected individuals (8). Occasional individuals do not develop clinical liver disease as neonates but do present with rickets and failure to thrive in childhood (31). The disease is extremely rare, and only a few patients (approximately six) have been reported in the clinical literature.
We describe the molecular basis of C27 3β-HSD deficiency in one of these affected individuals in whom the course of the disease and the attendant symptoms have been described in detail (3, 8, 17, 30). Patient MU2 is the fifth child of Saudi Arabian parents who were first cousins. As expected from the parental consanguinity, MU2 is homozygous for a 2-bp deletion in exon 6 of the C27 3β-HSD gene. This mutation disrupts the normal translational reading frame and causes a premature truncation that inactivates the enzyme (Figure 8 and Figure 9). When these findings are considered with the substrate specificity of the enzyme (Figure 4) and the phenotype of the patient (3), it is clear that loss of this C27 3β-HSD enzyme prevents the synthesis of all primary bile acids. Inasmuch as this patient is representative of others with this genetic disease, the mode of inheritance of C27 3β-HSD deficiency would appear to be autosomal recessive.
The prognosis of patients who inherit mutations in the C27 3β-HSD gene is good, provided that the disorder is diagnosed early and bile acids are prescribed (17, 30). The isolation of the gene and the elucidation of its structure provides a simple and rapid method of screening for this genetic disease. A molecular genetic analysis can be coupled with more specialized gas chromatography-mass spectrometry assays and with chemical colorimetric tests (17) to obtain an unambiguous diagnosis of this inborn error.
Mice lacking various enzymes involved in bile acid biosynthesis have been produced. The analysis of these animals has in every instance revealed one or more redundant pathways that compensate for the missing enzyme (32–35). If we extrapolate from the results obtained in humans and those reported here, it would appear that there is only a single C27 3β-HSD involved in bile acid synthesis and that a deficiency of this enzyme can be treated by oral administration of bile acids. Thus, a deletion of this gene in mice may provide a useful animal model in which the levels and synthesis of bile acids can be regulated.
Acknowledgments
We thank Jill Abadia for assistance with tissue culture; Mala Mahendroo and Kristi Cala for a liver cDNA expression library; and Mike Brown, Joe Goldstein, Helen Hobbs, Jay Horton and Jean Wilson for critical reading of the manuscript. This research was supported by grants from the NIH (HL-20948 to D.W. Russell and DK-07745 to M. Schwarz) and the Robert A. Welch Foundation (I-0971 to D.W. Russell).
References
- 1.Russell DW, Setchell KDR. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 2.Gourley, G.R. 1994. Bilirubin metabolism and neonatal jaundice. In Liver disease in children. F.J. Suchy, editor. Mosby. St. Louis, Missouri, USA. 105–125.
- 3.Clayton PT, et al. Familial giant cell hepatitis associated with synthesis of 3β,7α-dihydroxy- and 3β,7α,12α-trihydroxy-5-cholenoic acids. J Clin Invest. 1987;79:1031–1038. doi: 10.1172/JCI112915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Setchell KD, et al. Δ4-3-oxosteroid 5β-reductase deficiency described in identical twins with neontal hepatitis. A new inborn error in bile acid synthesis. J Clin Invest. 1988;82:2148–2157. doi: 10.1172/JCI113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Setchell KDR, et al. Identification of a new inborn error in bile acid synthesis: mutation of the oxysterol 7α-hydroxylase gene causes severe neonatal liver disease. J Clin Invest. 1998;102:1690–1703. doi: 10.1172/JCI2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trauner M, Meier PJM, Boyer JL. Molecular pathogenesis of cholestasis. N Engl J Med. 1998;339:1217–1227. doi: 10.1056/NEJM199810223391707. [DOI] [PubMed] [Google Scholar]
- 7.Riely, C.A. 1994. Familial intrahepatic cholestasis syndromes. In Liver disease in children. F.J. Suchy, editor. Mosby. St. Louis, Missouri, USA. 443–459.
- 8.Buchmann MS, et al. Lack of 3β-hydroxy-Δ5-C27-steroid dehydrogenase/isomerase in fibroblasts from a child with urinary excretion of 3β-hydroxy-Δ5-bile acids. J Clin Invest. 1990;86:2034–2037. doi: 10.1172/JCI114939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo KH, et al. Cloning and expression of cDNA of human Δ4-3-oxosteroid 5β-reductase and substrate specificity of the expressed enzyme. Eur J Biochem. 1994;219:357–363. doi: 10.1111/j.1432-1033.1994.tb19947.x. [DOI] [PubMed] [Google Scholar]
- 10.Charbonneau A, Luu-The V. Assignment of steroid 5β-reductase (SRD5B1) and its pseudogene (SRD5BP1) to human chromosome bands 7q32-q33 and 1q23-25, respectively, by in situ hybridization. Cytogenet Cell Genet. 1999;84:105–106. doi: 10.1159/000015230. [DOI] [PubMed] [Google Scholar]
- 11.Lund EG, Kerr TA, Sakai J, Li W-P, Russell DW. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem. 1998;273:34316–34348. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz M, Lund EG, Lathe R, Bjorkhem I, Russell DW. Identification and characterization of a mouse oxysterol 7α-hydroxylase cDNA. J Biol Chem. 1997;272:23995–24001. doi: 10.1074/jbc.272.38.23995. [DOI] [PubMed] [Google Scholar]
- 13.Schneider RJ, Shenk T. Impact of virus infection on host cell protein synthesis. Annu Rev Biochem. 1987;56:317–332. doi: 10.1146/annurev.bi.56.070187.001533. [DOI] [PubMed] [Google Scholar]
- 14.Jelinek DF, Andersson S, Slaughter CA, Russell DW. Cloning and regulation of cholesterol 7α-hydroxylase, the rate limiting enzyme in bile acid biosynthesis. J Biol Chem. 1990;265:8190–8197. [PMC free article] [PubMed] [Google Scholar]
- 15.Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW. Disruption of cholesterol 7α-hydroxylase gene in mice. J Biol Chem. 1996;271:18017–18023. doi: 10.1074/jbc.271.30.18017. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Plainview, New York, USA.
- 17.Ichimiya H, et al. Bile acids and bile alcohols in a child with hepatic 3β-hydroxy-Δ5-C27-steroid dehydrogenase deficiency: effects of chenodeoxycholic acid treatment. J Lipid Res. 1991;32:829–841. [PubMed] [Google Scholar]
- 18.Kandutsch AA. 3β-Hydroxy-C27-steroid dehydrogenase in mouse liver microsomes. Steroids. 1967;10:31–48. doi: 10.1016/0039-128x(67)90017-7. [DOI] [PubMed] [Google Scholar]
- 19.Wikvall K. Purification and properties of a 3β-hydroxy-Δ5-C27-steroid oxidoreductase from rabbit liver microsomes. J Biol Chem. 1981;256:3376–3380. [PubMed] [Google Scholar]
- 20.Furster C, Zhang J, Toll A. Purification of a 3β-hydroxy-Δ5-C27-steroid dehydrogenase from pig liver microsomes active in major and alternative pathways of bile acid synthesis. J Biol Chem. 1996;271:20903–20907. doi: 10.1074/jbc.271.34.20903. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi Y, Kiyono T, Fujita M, Ishibashi M. Isolation of a novel cDNA whose corresponding mRNA is accumulated in growth-arrested confluent but not in growing sub-confluent rat 3Y1 cells. Biochim Biophys Acta. 1997;1352:145–150. doi: 10.1016/s0167-4781(97)00051-1. [DOI] [PubMed] [Google Scholar]
- 22.Rose KA, et al. Cyp7b, a novel brain cytochrome P450, catalyzes the synthesis of neurosteroids 7α-hyroxy DHEA and 7α-hydroxypregnenolone. Proc Natl Acad Sci USA. 1997;94:4925–4930. doi: 10.1073/pnas.94.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res. 1998;39:1833–1843. [PubMed] [Google Scholar]
- 24.Payne AH, Abbaszade IG, Clarke TR, Bain PA, Park C-HJ. The multiple murine 3β-hydroxysteroid dehydrogenase isoforms: structure, function,and tissue- and developmentally specific expression. Steroids. 1997;62:169–175. doi: 10.1016/s0039-128x(96)00177-8. [DOI] [PubMed] [Google Scholar]
- 25.Morel Y, et al. Structure-function relationships of 3β-hydroxysteroid dehydrogenase: contribution made by the molecular genetics of 3β-hydroxysteroid dehydrogenase deficiency. Steroids. 1997;62:176–184. doi: 10.1016/s0039-128x(96)00178-x. [DOI] [PubMed] [Google Scholar]
- 26.Liu XY, et al. The gene mutated in bare patches and striated mice encodes a novel 3β-hydroxysteroid dehydrogenase. Nat Genet. 1999;22:182–187. doi: 10.1038/9700. [DOI] [PubMed] [Google Scholar]
- 27.Ephritikhine G, et al. The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 1999;18:315–320. doi: 10.1046/j.1365-313x.1999.00455.x. [DOI] [PubMed] [Google Scholar]
- 28.Bjorkhem, I., and Boberg, K.M. 1995. Inborn errors in bile acid biosynthesis and storage of sterols other than cholesterol. In The metabolic and molecular bases of inherited disease. 7th edition. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill. New York, New York, USA. 2073–2075.
- 29.Orth, D.N., and Kovacs, W.J. 1998. The adrenal cortex. In Williams textbook of endocrinology. J.D. Wilson, D.F. Foster, H.M. Kronenberg, and P.R. Larsen, editors. W.B. Saunders Co. Philadelphia, Pennsylvania, USA. 517–664.
- 30.Horslen SP, Lawson AM, Malone M, Clayton PT. 3β-Hydroxy-Δ5-C27-steroid dehydrogenase deficiency: effect of chenodeoxycholic acid therapy on liver histology. J Inherit Metab Dis. 1992;15:38–46. doi: 10.1007/BF01800342. [DOI] [PubMed] [Google Scholar]
- 31.Akobeng AK, Clayton PT, Miller V, Super M, Thomas AG. An inborn error of bile acid synthesis (3β-hydroxy-Δ5-C27-steroid dehydrogenase deficiency) presenting as malabsorption leading to rickets. Arch Dis Child. 1999;80:463–465. doi: 10.1136/adc.80.5.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz M, et al. Disruption of cholesterol 7α-hydroxylase gene in mice. J Biol Chem. 1996;271:18024–18031. doi: 10.1074/jbc.271.30.18024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen H, et al. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J Biol Chem. 1998;273:14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
- 34.Li-Hawkins J, Lund EG, Bronson AD, Russell DW. Expression cloning of an oxysterol 7α-hydroxylase selective for 24-hydroxycholesterol. J Biol Chem. 2000;275:16543–16549. doi: 10.1074/jbc.M001810200. [DOI] [PubMed] [Google Scholar]
- 35.Baes M, et al. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J Biol Chem. 2000;275:16329–16336. doi: 10.1074/jbc.M001994200. [DOI] [PubMed] [Google Scholar]