Abstract
The importance of the 2′-5′ oligoadenylate synthetase (OAS)/RNase L and double-stranded RNA (dsRNA)-dependent protein kinase (PKR) pathways in host interferon induction resulting from virus infection in response to dsRNA has been well documented. In poxvirus infections, the interactions between the vaccinia virus (VV) genes E3L and K3L, which target RNase L and PKR, respectively, serve to prevent the induction of the dsRNA-dependent induced interferon response in cell culture. To determine the importance of these host genes in controlling VV infections, mouse single-gene knockouts of RNase L and PKR and double-knockout mice were studied following intratracheal infection with VV, VVΔK3L, or VVΔE3L. VV caused lethal disease in all mouse strains. The single-knockout animals were more susceptible than wild-type animals, while the RNase L−/− PKR−/− mice were the most susceptible. VVΔE3L infections of wild-type mice were asymptomatic, demonstrating that E3L plays a critical role in controlling the host immune response. RNase L−/− mice showed no disease, whereas 20% of the PKR−/− mice succumbed at a dose of 108 PFU. Lethal disease was routinely observed in RNase L−/− PKR−/− mice inoculated with 108 PFU of VVΔE3L, with a distinct pathology. VVΔK3L infections exhibited no differences in virulence among any of the mouse constructs, suggesting that PKR is not the exclusive target of K3L. Surprisingly, VVΔK3L did not disseminate to other tissues from the lung. Hence, the cause of death in this model is respiratory disease. These results also suggest that an unanticipated role of the K3L gene is to facilitate virus dissemination.
Small-animal (mouse) models to study poxvirus pathogenesis have been employed for decades and have provided invaluable insight into host-virus interactions and mechanistic insight as to how individual viral genes influence the host response. Infection of mice by various routes continues to be productive for study of a number of orthopoxviruses, including vaccinia virus (VV), ectromelia virus, cowpox virus, and rabbitpox virus (23, 24, 44, 45, 48-50, 67, 75, 77). The general purpose of these infected mouse studies is to provide surrogate models for human smallpox virus infection and host response to evaluate antiviral drug efficacy and the effectiveness of various smallpox vaccines. Advantages of mouse models include their relatively low cost, small size of the animals, and the fact that there are many mouse mutant strains available for genes that control responses to infection.
A number of routes for the initiation of poxvirus infections in mice have been studied, including intranasal (i.n.), intratracheal (i.t.), and intradermal (i.d.) (ear pinnae, footpad) routes and scarification. Other routes, such as the intracranial (i.c.) or intraperitoneal (i.p.) route, are rarely used today because they do not effectively reproduce “natural” infection conditions. Arguably, the most relevant routes of infection in terms of human disease are the i.n. and i.t. routes, because smallpox is transmitted via the respiratory route. The majority of the experimental data on respiratory infections in mice were generated using the i.n. route of infection (35, 51, 60), although the i.t. method has several notable advantages (45).
The i.n. route exhibits some animal-to-animal variation due in part to the tendency of the animal to expel a portion of the virus inoculum through sneezing or, alternatively, from swallowing a portion of the inoculum or from unintended access to the brain. The i.n. route is also problematic in that a significant portion of the retained virus inoculum remains in the nasal passages, giving a somewhat variable lung infection. It is clear, however, that the initial i.n. infection is then followed by an efficient spread to most organs within the animal, a characteristic feature of many orthopoxvirus diseases.
We have chosen instead to utilize the recently described i.t. route (45) of vaccinia virus infection, which we find gives a much more reproducible lung infection and requires lower doses of the virus to cause disease and fatal symptoms than i.n. inoculation. In our earlier study (45), we also chose to use C57BL/6 mice to evaluate both clinical and pathological disease progression, rather than the more susceptible BALB/c strain, because of the large number of available mouse mutant strains constructed on the C57BL/6 background. The C57BL/6 mouse has also been evaluated as a strain in which to study ectromelia virus infections (50).
We studied i.t. infections of different C57BL/6 mouse strains in order to evaluate the roles of the host genes encoding double-stranded RNA-dependent protein kinase (PKR) and RNase L in controlling poxvirus infections. The PKR and RNase L genes were two of the first host genes implicated as targets for viral gene products to serve as a means of controlling the interferon (IFN) response to poxvirus infection (2).
The PKR and RNase L genes are both key components of the host response to virus infection linked to the interferon response (31, 58, 68). Activation of PKR from an inactive monomeric molecule involves dimerization mediated by double-stranded RNA (dsRNA), followed by autophosphorylation (46, 47), to produce the active molecule. One key activity of PKR is to phosphorylate and inactivate the translation factor eIF2α (57, 79). RNase L is an endoribonuclease that is activated by 2′-5′ adenylic acid, which is the product of the enzyme 2′-5′ oligoadenylate synthetase (OAS) (33, 85). OAS is activated by double-stranded RNA initiation or highly structured RNA, in a process that results in the activation of RNase L and ultimately in RNA degradation. Both PKR and RNase L/2′-5′ oligoadenylate system proteins and functions have been reviewed recently (26, 34, 52, 55, 56, 64, 65, 66, 72). It is now known that at least two virus-carried genes, the E3L and K3L genes, protect the virus from the effects of interferon and that their encoded proteins interact with host RNase L and PKR proteins (10, 11, 19, 64). Much of what is known about the presumed mechanism of action of the K3L and E3L genes on the host response has been inferred from studies of infected cells in culture.
The E3L protein is a double-stranded DNA binding protein comprised of at least two distinct functional domains (6). The C terminus of E3L contains a dsRNA binding domain (12, 46, 83), whereas the N-terminal region of the protein contains sequences associated with binding to Z-DNA and PKR (30, 38, 39). Both regions are required for virulence in mice (6). The binding of E3L to Z-DNA allows the protein to be considered a member of the larger Z-DNA binding family (39). Furthermore, E3L has some sequence similarity to DAI (DNA-dependent activator of IFN regulatory factors), a protein involved in DNA pattern recognition that can trigger the innate response (20, 39, 70). It is also known that deletion of the E3L gene from the virus leads to a restricted host range of the virus (1, 2, 13, 63) in cell culture and to profound attenuation in mice, leading to consideration of VVΔE3L mutants as vaccine candidates (6, 78).
The K3L protein is considered a substrate mimic for eIF2α, a natural substrate for phosphorylation by PKR. K3L interacts with active PKR to prevent PKR from phosphorylating and inactivating eIF2α (10, 17, 18, 19, 21, 37, 62). eIF2α phosphorylation is associated with interferon induction (32, 53, 68, 73). Like the E3L gene, the K3L gene has been reported to control host range (41). A dynamic relationship between K3L and PKR was suggested by recent experiments in which K3L and PKR appeared to have coevolved as an example of ongoing competition between host and virus genes (22) and by the observation that mutants of PKR can overcome inhibition by K3L (54, 61). There is little available information on the effects of virus deleted for K3L in animal models.
C57BL/6 mouse strains are available which are singly deleted for either the PKR (PKR−/−) or RNase L (RNase L−/−) gene. In addition, there is a mouse deleted for both the PKR and RNase L genes (PKR−/− RNase L−/−) (59, 86). The availability of these mouse strains, together with virus mutants in the controlling genes E3L and K3L, allows the roles of both virus and host genes to be evaluated within the context of each other in animal models of orthopoxvirus disease. An earlier study showed that a vaccinia virus (VV) deleted for the E3L gene (VVΔE3L) produced no plaques in cells from wild-type mice and a reduced number of plaques in cells from RNase L−/− or PKR−/− mice and was attenuated in PKR−/− RNase L−/− mice (81). We show here that i.t. infection of animals with VV leads to rather precise, well-defined focal lung lesions restricted to the bronchi, involving bronchial epithelial cells, rather than to a generalized, indiscriminate infection of the lung. In wild-type VV infections, viremia ensues rapidly, with dissemination throughout the animal. Despite a localized infection within the lung, we also show that overall gene expression patterns of the entire infected lung are altered dramatically between 3 and 4 days after infection and that the response of the lung is global and not limited to the bronchi and bronchioles. We also report on the roles of both host and virus genes within the context of disease, lung pathology, virus growth and spread, tissue histopathology, and immunohistochemistry (IHC).
MATERIALS AND METHODS
Cells and viruses. (i) Cell culture.
CV-1, BHK-21, and PK15 cells were maintained in minimum essential medium (MEM) with Earle's salts (Gibco, Grand Island, NY) supplemented with 2 mM glutamine (Media Tech, Herndon, VA), 50 U/ml penicillin G (Media Tech), 50 μg/ml streptomycin (Media Tech), 1 mM sodium pyruvate (Media Tech), 0.1 mM nonessential amino acids (Media Tech), and 10% (vol/vol) fetal bovine serum (FBS) (Gibco).
(ii) Construction of VVgfp (VVΔATI::gfp).
Wild-type vaccinia virus strain Western Reserve (a gift from R. Condit, University of Florida) was used as a control virus. Vaccinia virus expressing enhanced green fluorescent protein (EGFP) under the control of the poxvirus synthetic early/late promoter was used as the screening fluorophore. The PE/L EGFP coding sequence was engineered into the ATI locus in VV, using ligation-mediated PCR. In this reaction, 3 fragments are generated: a left virus-flanking sequence, a right virus-flanking sequence, and a middle reporter gene under the control of its own promoter. The 3 fragments then undergo restriction enzyme digestion (with enzymes from NEB) and ligation, as in the field standard; however, rather than transforming the fragments into a vector, PCR of the ligation mixture is performed immediately. PCR using primers located in the left and right flanks allows for specific amplification of the desired product for future use. The left flank was amplified using primers IDT536 (CGGAGCTCGTCTACAGTCGTTCAACTGTC) and IDT571 (CATGGTACCAAGGAGGATCTTGATAAGGCC). The right flank was amplified using primers IDT572 (TCTAGGCCTCGAGTCTGGACAGTTCCCATTCG) and IDT539 (CCGGTACCGAATGCGGAAATAGTGGAGAG). The gfp gene and the PE/L promoter fragment were amplified from plasmid pSC65gfpPCT by use of primers IDT30 (CCAGACATTGTTGAATTAGATCG) and IDT130b (GCTGGTACCGGTGGGTTTGGAATTAGTG). Each fragment underwent digestion with the corresponding restriction enzyme following the manufacturer's directions (New England Biolabs). The pieces were then ligated with T4 DNA ligase, using traditional protocols. The ligation mixture was used as a PCR template with the outermost primers, IDT536 and IDT539. The corresponding band was then used for transfection and subsequent screening for recombinant viruses following standard methods. The resulting virus, VVgfp, was grown and plaque purified on CV-1 cells.
(iii) Construction of VVΔE3L (VVΔE3L::gfp).
The E3L::gfp fragment (gfp fragment flanked by E3L sequences) was generated in a PCR-mediated ligation reaction as previously described. In this reaction, 3 fragments are generated: a left virus-flanking sequence, a right virus-flanking sequence, and a middle reporter gene under the control of its own promoter. The 3 fragments then undergo restriction enzyme digestion (with enzymes from NEB) and ligation, like the field standard; however, rather than transforming the fragments into a vector, PCR of the ligation mixture is performed immediately. PCR using primers located in the left and right flanks allows for specific amplification of the desired product for future use. The E3L::gfp PCR fragment was amplified using primers IDT345 (GAGAATTCCACAAACATCAATGGCGGTAAC) and IDT594 (AGCTCCTTCGATTCC). The right flank was amplified using primers IDT344 (GAGAATTCCTTGGTTCATACATGAAATGATC) and IDT595 (CTCGTTTAGATTTTCC).
The gfp gene and the PE/L promoter fragment used were the same as those mentioned previously. Primers IDT599 (GAGGTTCGTCAGCGGC) and IDT598 (GTGATAATTTATGTGTGAGGC) were then used for template amplification.
The deletion of E3L was confirmed by a PCR using the following primers: IDT529 (ATAGCCTTGTCCTCGTGCAG), IDT530 (CGCAATCGATACATGAAAACA), IDT344 (GAGAATTCCTTGGTTCATACATGAAATGATC), and IDT272 (GAGTTATAGTTGTATTCCAGCT).
VVΔE3L::gfp was generated by infection/transfection and subsequent plaque purification on BHK-21 cells, using standard methods. BHK-21 cells were the only cell line tested that would allow for the growth of VVΔE3L::gfp, and therefore they were the cells chosen for screening.
VVΔE3L::gfp was rescued by infection/transfection with the wild-type E3L gene fragment amplified with primers IDT344 and IDT345. The resulting viruses were plaque purified on PK15 cells. Due to the host range exhibited by VVΔE3L::gfp, only viruses that have been rescued successfully will form plaques on PK15 cells. The resulting virus, RVVE3L+, was tested in cell culture and mice and was found to exhibit the wild-type phenotype.
(iv) Construction of VVΔK3L (VVΔK3L::gfp).
Cloning of the K3L::gfp fragment was performed using ligation-mediated PCR as previously described. The K3L::gfp left flank was amplified using primers IDT639 (CTTTTGTATAATCAACTCTAA) and IDT531 (ATGCAGGCAATAGCGACATA), and the right flank was amplified using primers IDT638 (CCTTCTCGTATACTCTGCCC) and IDT532 (AGATGCTCCACATGTAT). The previously described gfp fragment was the fluorophore used. Primers IDT531 and IDT532 were used for PCR generation of the K3L::gfp PCR fragment. The K3L locus from bp 45 to 199 was deleted, with gfp replacing this region.
The deletion of K3L was confirmed by PCR, using primers IDT531, IDT532, IDT419 (AGGCATCCCATTAGACATACCGGATCTACG), and IDT272 (AGCTGGAATACAACTATAACTC).
VVΔK3L::gfp was rescued by infection/transfection with the wild-type K3L gene fragment amplified with IDT531 and IDT532. The resulting viruses were plaque purified on CV-1 cells. The resulting virus, RVVK3L+, was tested in cell culture and mice and found to exhibit the wild-type phenotype.
(v) Virus growth.
VV, VVgfp, VVΔK3L::gfp, RVVE3L+, and RVVK3L+ were grown and titrated on CV-1 cells by standard methods. VVΔE3::gfp was grown and titrated on BHK-21 cells. All viruses for animal infections were pad purified over 36% sucrose by standard methods and resuspended in phosphate-buffered saline (PBS) (15, 16).
Animals and animal methodology. (i) Mouse breeding.
C57BL/6, PKR−/−, RNase L−/−, and PKR−/− RNase L−/− double-knockout mice were provided by Robert Silverman of the Cleveland Clinic and were maintained by sibling mating and genotyped using methods published by Williams et al. and Silverman et al. (82, 87).
(ii) Intratracheal infection of mice.
Infections were performed as previously described (45). Briefly, mice were subjected to general anesthesia (isoflurane) prior to inoculation. All mice maintained a surgical plane of anesthesia during the procedure. A 3-mm ventromedial incision was made adjacent to the trachea for subcutaneous insertion of a microchip to monitor body temperature and animal identification number (BioMedic Data Systems, Seaford, DE). Thirty microliters of virus diluted in PBS was injected into the trachea. The incision was closed using surgical glue (Nexaband; Abbott Animal Health, Chicago, IL).
(iii) Monitoring of animals.
Each mouse was microchipped at the time of infection to transmit its body temperature and identification number to a DAS-5007 reader (BioMedic Data Systems, Seaford, DE). Weight, temperature, and physical observations of the mice (grooming habits, facial swelling, secretions, removal of hair, and respiratory distress) were recorded daily. Criteria for euthanizing the mice included open-mouth breathing, severity of dyspnea, hypothermia (body temperature of ≤30°C), and weight loss of >30% of initial body weight (69). All animal procedures were carried out according to the University of Florida IACUC guidelines.
(iv) Tissue processing.
Sections of the lung, liver, spleen, gonad, and brain were flash frozen until processed. The tissues were homogenized in 2-ml screw-top tubes (Sarstedt) with ∼1.0 ml sterile 24-grit silicon carbide (Electro Abrasives, Buffalo, NY) and 1 ml growth medium without serum (MEM; Invitrogen), using a Mini-Beadbeater 8 instrument (Biospec, Bartlesville, OK) for 1.5 min on the homogenization setting. Virus titers in the supernatant were evaluated by plaque purification on CV-1 cells, using standard procedures (15, 16).
Tissue sections for histopathology were fixed in 10% buffered formalin (Fisher Scientific) for 12 to 18 h at room temperature and then paraffin embedded.
Four-micrometer-thick sections of tissue were stained with hematoxylin and eosin (H&E) by standard protocols at the Molecular Pathology Core at the University of Florida. Immunohistochemistry was performed on 4-μm paraffin-embedded sections for detection of GFP (Abcam) (25), STAT-1 (Abcam) (4), and Ki67 (Abcam) (80). STAT-1 and Ki67 IHC required antigen retrieval with citra solution (10 mM citric acid, 0.05% Tween 20, pH 6.0), using standard protocols. Mach 2 goat anti-rabbit-horseradish peroxidase (HRP) polymer (Biocare Medical, Concord, CA) was the secondary antibody used for all immunohistochemical staining. Cardassian DAB chromogen (Biocare Medical) was then used according to the manufacturer's protocol to generate the final brown color observed in positive samples, and the nuclei were counterstained with hematoxylin (Vector) following the manufacturer's recommendations.
Microarrays.
Entire lungs without the bronchi were flash frozen in liquid nitrogen and maintained at −80°C until used for RNA isolation. Lung tissue was homogenized with Trizol (Invitrogen) and 24-grit silicon carbide (Electro Abrasives, Buffalo, NY) in a Mini-Beadbeater 8 apparatus. RNA isolation was performed with Trizol (Invitrogen) following the manufacturer's protocol. The resulting RNA pellets were then placed through a Micro-to-Midi total RNA purification system (Invitrogen) for the removal of any residual cellular DNA according to the manufacturer's protocol.
Two micrograms of total purified RNA was used for the generation of cRNAs for use as probes for the Affymetrix microarrays. First-strand cDNA synthesis was performed using a T7-(dT)24 primer and Superscript II reverse transcriptase (RT) (Invitrogen). Second-strand synthesis was performed using second-strand reaction buffer (Invitrogen), Escherichia coli DNA ligase (NEB), E. coli DNA polymerase I (NEB), E. coli RNase H (NEB), and T4 DNA polymerase (NEB). cDNAs were then purified using a GeneChip sample cleanup module (Affymetrix). cRNAs were then made using an Affymetrix 3′-labeling IVT kit (Affymetrix) and cleaned up using a GeneChip sample cleanup module (Affymetrix). The samples were then fragmented, hybridized, and detected on Affymetrix mouse 430.2A arrays (Affymetrix).
Analysis of the microarray data was performed using BRB Array Tools (NIH). The median probe intensity was used to normalize the arrays, with a median intensity of 84 and a percent call of ∼50% across all arrays. The arrays were subjected to an F test with random variance, with a significance level of 0.001, in which all arrays were assigned to classes based upon the day postinfection that the lungs were harvested. The resulting 1,811 probe sets were then represented pictorially in a heat map generated by BRB Array Tools.
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE23738 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23738).
RESULTS
Patterns of disease in mice.
The initial baseline clinical parameters of VV infection via the i.t. route were standardized by inoculating different strains of mice with various dilutions of VV and evaluating the animals for 10 days, with daily measurements of temperature, body weight, overall clinical appearance, and survival rates. No animals were allowed to die of natural causes; therefore, the time of death indicated on the survival curves is the time at which an animal was euthanized due to severe disease. Criteria for euthanasia included weight loss of >30%, hypothermia (body temperature of 30°C or less), or dyspnea.
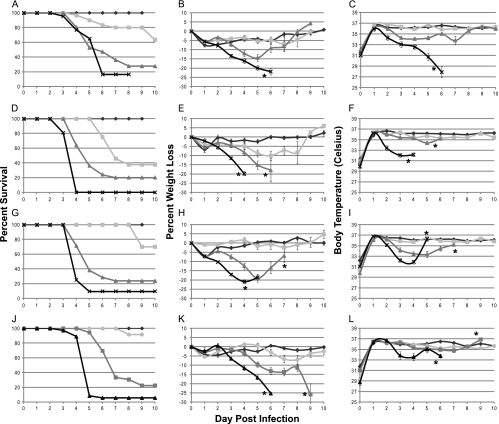
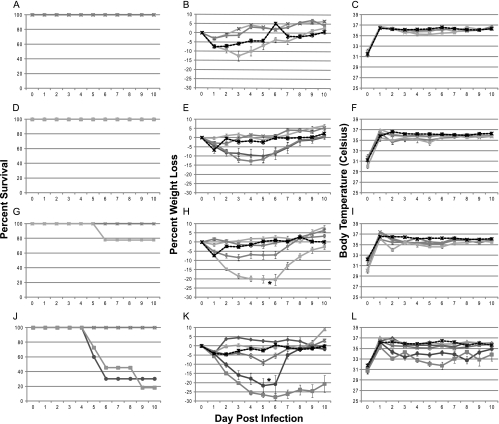
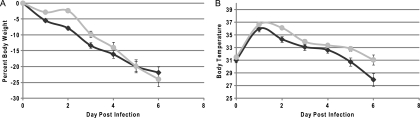
The clinical responses to VV for wild-type mouse strains are summarized in Fig. 1A to C, and those for mutant mouse strains are shown in Fig. 1D to L. Survival curves for each mouse strain are shown in Fig. 1A, D, G, and J. Mice that received lethal doses of virus exhibited rapid and continued weight loss of up to 30% (Fig. 1B, E, H, and K) compared to the initial body weight of the animal. Weight loss serves as an excellent indication of severity of disease, as previously described (74). This degree of weight loss and the timing at which it occurred were dependent upon the dose of virus the animal received. Although it is difficult to ascertain the exact cause of an animal's dramatic weight loss over a period of 4 days (days 3 to 6 postinfection), it is most likely due to combined anorexia and dehydration. As the animals' body weights decreased, their body temperatures also decreased, as shown in Fig. 1C, F, I, and L. Body temperature was measured by a microchip placed subcutaneously into the mouse at the time of infection. While this method of temperature assessment may not represent a true measure of core body temperature, it does serve as a relative temperature indicator and an adequate predictor of disease severity and likelihood of survival.
FIG. 1.
Survival, temperatures, and weights of mice following VV infection. Mice were infected intratracheally with VV, during which a microchip was inserted for measurement of temperature. At least 10 mice were evaluated for each time point. The overall significance of the curves (P < 0.001) was determined using Kaplan-Meier survival analysis. (A to C) C57BL/6 mice; (D to F) RNase L−/− mice; (G to I) PKR−/− mice; (J to L) RNase L−/− PKR−/− mice. Panels A, D, G, and J indicate survival curves for the mice. Panels B, E, H, and K indicate average percent weight loss based upon weight at the time of infection. Panels C, F, I, and L indicate average body temperatures. Stars denote the points at which all lethally infected mice were euthanized, and survivors were no long considered beyond those points. Diamonds, PBS; circles, 1 × 103 PFU; squares, 1 × 104 PFU; triangles, 1 × 105 PFU; ×, 1 × 106 PFU.
There was a dose-dependent response to infection. Mice infected with higher doses of virus showed a more rapid and increased disease severity. Clinical symptoms were more severe and appeared sooner with higher doses of virus. The experimental dose that indicated the necessity for euthanasia in 80% of animals (LD80) was determined to be 1 × 106 PFU for C57BL/6 animals. The LD80s for VV-infected mice were 10-fold less for the PKR−/− and RNaseL−/− mice and 100-fold less for the PKR−/− RNaseL−/− mice, as determined from the data in Fig. 1, and were the doses used for all subsequent experiments. The LD80 value for each mouse construct was calculated using regression analysis, and the values are summarized in Table 1. These values demonstrate that PKR−/−, RNase L−/−, and PKR−/− RNase L−/− mice are 4.8, 1.9, and 27 times more susceptible to VV than C57BL/6 mice. These experimentally determined LD80s are about 10-fold lower than the values published for VV infections of wild-type C57BL/6 mice via the i.n. route (5, 6, 76). The average day to euthanasia for C57BL/6 mice infected with VV was determined to be 4.95 days at the LD80, with all mice being euthanized by day 6. These results are also consistent with previously published data (5).
TABLE 1.
Calculated LD80 values for all mouse strains infected with VV, VVΔE3L, and VVΔK3La
| Mouse construct | LD80 value (PFU) |
||
|---|---|---|---|
| VV | VVΔE3L | VVΔK3L | |
| C57BL/6 | 4.1 × 105 | >1 × 109 | 2.1 × 106 |
| RNase L−/− | 8.5 × 104 | >1 × 109 | 5.9 × 105 |
| PKR−/− | 2.1 × 105 | 3.2 × 108 | 5.4 × 105 |
| RNase L−/− PKR−/− | 1.5 × 104 | 1.3 × 107 | 3.6 × 105 |
Values were calculated using regression analysis.
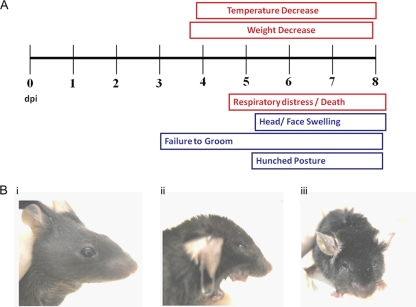
Mice that succumbed to infection had a distinct, easily recognizable, and reproducible overt disease presentation, which is summarized in Fig. 2A. Disease onset began at 4 days postinfection and increased in severity until the time of euthanasia. Symptoms included hunched posture, a failure to groom (Fig. 2B, panel iii), infraorbital sinusitis (Fig. 2B, panel ii [compare to uninfected mouse in Fig. 2B, panel i]), dyspnea, weight loss, and hypothermia. The hunched posture and failure to groom are typical of a lethal infection. Occasional lacrimal secretions were observed, as were gas-filled intestines at postmortem.
FIG. 2.
Disease progression in intratracheally infected mice. (A) Measurable clinical symptoms (weight and body temperature) are shown in boxes above the time line, while observational symptoms are depicted in boxes below the time line. (B) Photographs of infected mice at 5 days postinfection. (i) Uninfected mouse; (ii) head and face swelling with open mouth breathing; (iii) lack of grooming, typical of lethally infected mice.
Virus dissemination from the site of inoculation.
To evaluate dissemination from the site of infection, animals were infected with the mouse construct-specific LD80 dose of VV i.t. and sacrificed on days 2 to 6 postinfection. Ten mice were harvested per day. The tissues harvested for analysis included lung, liver, spleen, gonad, and brain tissues for all constructs of mice. Table 2 shows the levels of virus found in each of these organs in each mouse strain over time.
TABLE 2.
Tissue titers for all mouse constructs infected with VV, VVΔE3L, and VVΔK3L
| Mouse construct | Dose (log10) | Organ | Titer (PFU/mg) at day postinfectiona |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VV |
VVΔE3L |
VVΔK3L |
|||||||||
| 2 | 4 | 6 | 2 | 4 | 6 | 2 | 4 | 6 | |||
| Wild type | 6 | Lung | 6.1 | 6.5 | 5.9 | 3.7 | 2.7 | 2.6 | 5.1 | 6.0 | 5.1 |
| RNase L−/− | 5 | Lung | 4.9 | 5.6 | 4.8 | 2.4 | 2.3 | ND | 4.3 | 5.4 | 5.1 |
| PKR−/− | 5 | Lung | 4.8 | 5.7 | 5.6 | 3.9 | 3.6 | 1.8 | 4.1 | 5.4 | 5.6 |
| RNase L−/− PKR−/− | 4 | Lung | 5.4 | 5.7 | 6.1 | 3.5 | 3.7 | 2.6 | 2.2 | 4.2 | 4.2 |
| Wild type | 6 | Liver | 3.6 | 6.4 | 7.6 | ND | ND | ND | 0.9 | 0.9 | 0.9 |
| RNase L−/− | 5 | Liver | 2.9 | 5.4 | 6.3 | ND | ND | ND | 1.3 | 0.4 | 0.7 |
| PKR−/− | 5 | Liver | 2.8 | 5.5 | 6.9 | ND | ND | ND | 1.2 | 1.1 | 0.9 |
| RNase L−/− PKR−/− | 4 | Liver | 3.0 | 3.9 | 3.7 | ND | ND | ND | 1.3 | 1.1 | 2.3 |
| Wild type | 6 | Spleen | 4.5 | 5.9 | 6.0 | ND | ND | ND | 0.8 | 1.7 | 0.8 |
| RNase L−/− | 5 | Spleen | 4.7 | 5.8 | 5.9 | ND | ND | ND | 1.2 | 0.9 | 0.7 |
| PKR−/− | 5 | Spleen | 4.1 | 6.2 | 6.6 | ND | ND | ND | 1.2 | 1.6 | 1.8 |
| RNase L−/− PKR−/− | 4 | Spleen | 2.6 | 4.8 | 4.2 | ND | ND | ND | 1.3 | 1.2 | 2.4 |
| Wild type | 6 | Gonad | 3.7 | 5.1 | 7.3 | ND | ND | ND | 1.0 | 0.8 | 3.1 |
| RNase L−/− | 5 | Gonad | 3.9 | 5.0 | 7.4 | ND | ND | ND | 1.3 | 1.0 | 0.7 |
| PKR−/− | 5 | Gonad | 3.7 | 5.3 | 6.2 | ND | ND | ND | 1.3 | 1.9 | 2.6 |
| RNase L−/− PKR−/− | 4 | Gonad | 4.4 | 5.0 | 6.1 | ND | ND | ND | 1.3 | 1.1 | 1.1 |
| Wild type | 6 | Brain | 5.8 | 6.4 | 7.3 | ND | ND | ND | 1.3 | 3.1 | 3.7 |
| RNase L−/− | 5 | Brain | 3.6 | 6.8 | 6.7 | ND | ND | ND | 1.1 | 2.4 | 2.1 |
| PKR−/− | 5 | Brain | 4.2 | 6.6 | 7.2 | ND | ND | ND | 1.1 | 2.4 | 3.6 |
| RNase L−/− PKR−/− | 4 | Brain | 6.5 | 5.9 | 6.1 | ND | ND | ND | 1.3 | 1.4 | 2.7 |
Values are averages for ≥10 mice per condition. ND, no virus detected. The limit of detection for this assay is 0.2 PFU/mg.
The data in Table 2 indicate that virus dissemination from the lungs to all organs occurred within the first 2 days following infection in all cases, reaching 106 to 107 PFU/mg in some tissues by 6 days postinfection in the case of VV infections. Also noteworthy for VV infections is the fact that lung titers reached a level of 106 PFU/mg by 2 days after infection in C57BL/6 animals but changed very little thereafter. This general pattern of spread was repeated with PKR−/− and RNase L−/− mice. However, in the case of the PKR−/− RNase L−/− construct, there appeared to be normal spread to the liver and spleen; however, virus growth did appear to be impaired, with comparatively less accumulation of virus in the liver and spleen. There was no difference found between males and females for virus spread or titers of virus in the organs harvested (data not shown), and the levels of virus in the male and female gonads were comparable. This observation is different from that observed with ectromelia virus-infected animals, which do exhibit a sex-dependent susceptibility (7, 8). The error estimations for the viral titers at early times were large, which is reflective of significant variation in the onset of disease. However, later in the infection, these values were relatively small, as the infections became more synchronous with time.
Histopathology.
We analyzed the histopathology of infection and the presence of virus in various tissues. Cells infected with VVgfp were detected by the expression of gfp. This virus, which has the gfp gene inserted into the defective VV ATI gene, has been evaluated in animals (data not shown) and exhibits a wild-type VV phenotype for survival and clinical symptoms. While spread of VV was observed at high levels throughout the mice, as determined by tissue titers (Table 2), our initial interest centered about the response and analysis of the lung throughout the period of infection and, at later times, those of the liver and spleen. Both H&E staining and IHC were performed on 2 or more sections from all 3 tissues and from at least 2 different mice at days 2 to 6 postinfection for C57BL/6 mice and days 3 and 5 postinfection for all other mouse constructs. IHC for GFP allowed for localization of virus within the lung.
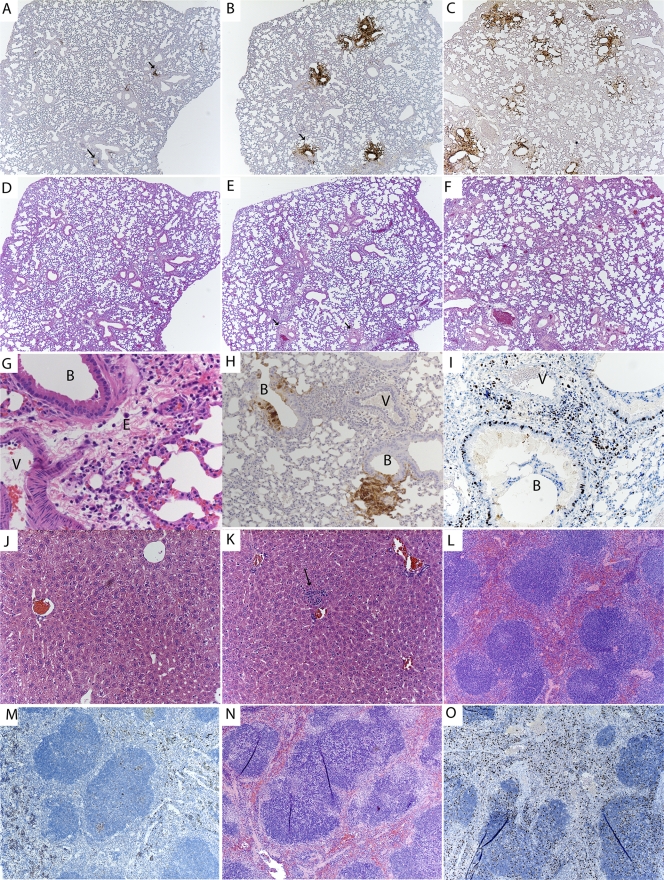
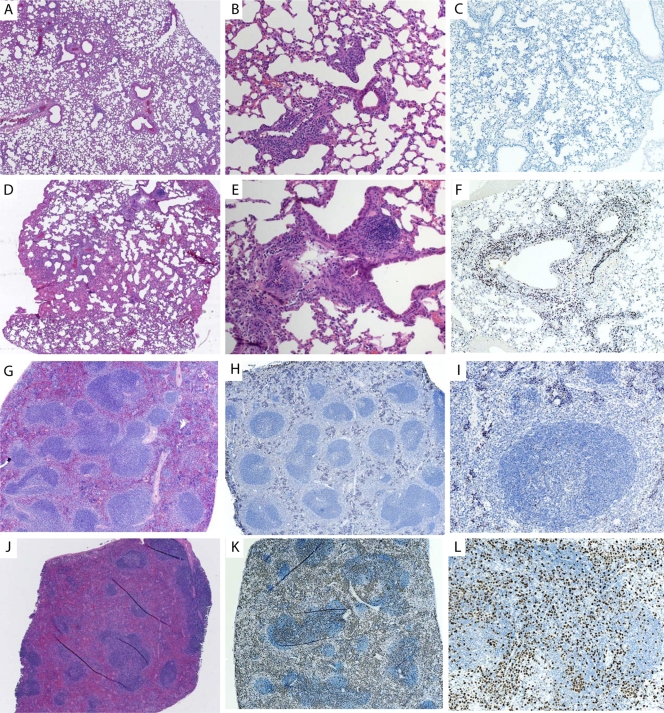
The lung sections from day 2 postinfection showed small focal areas of viral infection surrounding the large bronchi, with the majority of the infected cells being bronchiolar epithelial cells, based on IHC. These localized areas of infection enlarged throughout the time course, with the bronchiolar epithelial cells being the cell type exhibiting the highest rate of infection (Fig. 3A to C and H). Histological staining of GFP (virus) was undetectable in alveolar type I or type II pneumocytes observed at early time points (Fig. 3A). However, late in the infection, as the infected foci became larger, the infection did spread to both type I and II pneumocytes in the alveolar tissue (Fig. 3B, C, and H). Macrophage infiltrates containing VV antigen were also observed late in infection (data not shown). When H&E-stained samples were examined, there was also mild edema around infected bronchial epithelial cells, with infiltrating neutrophils, early in infection that increased over the time course (Fig. 3D to F). Late in infection, the areas surrounding the infected bronchial epithelial cells exhibited mild to moderate edema and cell infiltrates containing neutrophils and macrophages (Fig. 3G). Regardless of the virus dose administered, the histological findings for all 4 mouse constructs were similar, indicating that the properties of the infection were not altered in the PKR−/−, RNase L−/−, or PKR−/− RNase L−/− mice (data not shown).
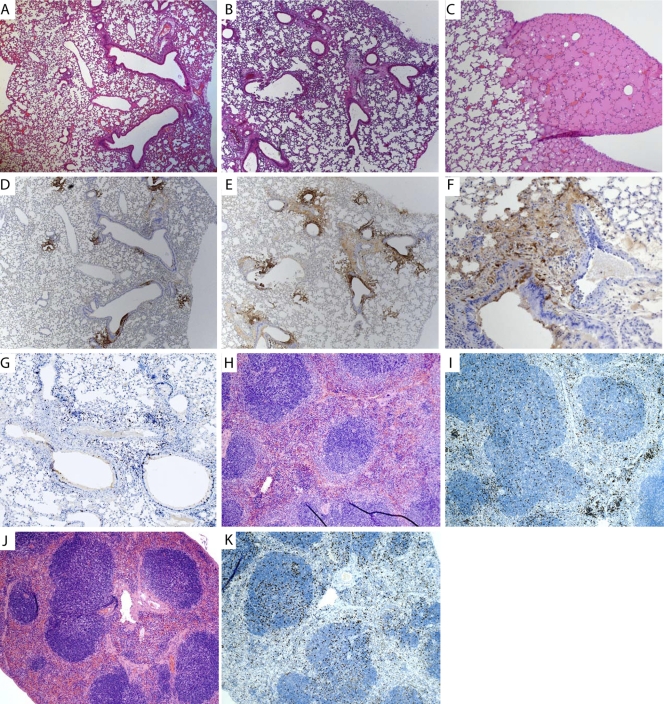
FIG. 3.
(A to I) Analysis of virus-infected cells in lung tissue over time, with arrows identifying infected bronchi. Sections are shown for day 2 (A), day 4 (B), and day 6 (C) postinfection. Magnification, ×5. H&E staining of lung tissue over time is shown for day 2 (D), day 4 (E), and day 6 (F) postinfection. Magnification, ×5. (G) H&E-stained infected bronchus and nearby blood vessel, with cells and fluid entering the surrounding tissues. Magnification, ×40. B, infected bronchus; V, position of a blood vessel; E, area of edema. (H) Infected bronchus and surrounding tissue at day 5 postinfection, showing localized infection upon IHC staining for virally infected cells. Magnification, ×10. (I) IHC staining for Ki67, showing activation of immune cells within the infected lung at day 5. (J and K) H&E staining of liver section from PBS-treated mouse (J), showing normal pathology, and from mouse at day 5 postinfection (K), showing neutrophilic areas within the liver (arrow). Magnification, ×20. (L and M) H&E staining of normal spleen (L) and Ki67 staining of normal spleen (M). (N and O) H&E staining of spleen at day 5 postinfection, showing neutrophilic and histiocytic aggregates within lymphoid follicles (N), and Ki67 staining for activation in spleen (O).
IHC staining for Ki67, a cellular marker of proliferation, was performed on lung sections to determine the level of cellular proliferation and activation of the immune response. Lung sections demonstrated staining for Ki67 in both the alveolar and bronchiolar cells, with cell infiltrates immediately concentrated in areas surrounding the site of infection (Fig. 3I) late in infection. Samples from early time points stained poorly for Ki67 (data not shown).
The liver sections of infected animals showed no pathology at early time points (data not shown), but after 5 days, there were areas of random foci of necrosis, which are typical of viral infections (Fig. 3K), compared to sections from PBS-treated animals (Fig. 3J). Virus replication was not detected in the liver at early time points, based on a lack of immunostaining for virus-expressed GFP, but a few random epithelial cells were immunopositive for VV late in infection (data not shown). We were surprised that there was no virus replication detected in the liver while virus levels were high. One explanation is that the virus detected by plaque assay was not from infected liver cells but rather was attached to the cells or traveling through the liver in the blood.
Spleen sections were normal early in infection, with no immunopositivity for virus replication by IHC. However, tissue sections isolated late in infection showed evidence of an immune response, with neutrophilic and histiocytic aggregates within the follicles (Fig. 3N), compared to uninfected spleen sections (Fig. 3L). Ki67 staining of the spleen at day 5 postinfection showed an increase of staining (Fig. 3O) compared to sections from uninfected animals (Fig. 3M), indicating an immune response to the infection. The overall conclusion from these studies is that the virus controls the host response very well.
Microarray analysis of vaccinia virus-infected lung tissue.
Our studies of the infected lung (Fig. 3) indicated that the infection was focal and that the histological response to infection and damage to the lung were minimal for a period of at least 6 days and localized to areas around the infected bronchi, with minimal cellular influx. This seemed to conflict with the fact that there was an acute clinical lung response leading to mortality within 6 days (Fig. 2). Therefore, we decided to determine whether there were significant changes in patterns of gene expression in the lung over the course of infection by using microarrays. Microarray technology has been utilized in the context of poxvirus-infected cells in culture to identify key genes that undergo an expression change after infection in cell culture. These studies defined changes in gene profiles which occur during the infectious process (9, 27-29, 40, 43). One key feature of infections is the overall shutdown of host transcript levels observed within 4 h of infection at high multiplicities of infection (9). This is a major consideration in the analysis of the data in terms of choosing the appropriate normalization standards (9). This information is useful, although it does not necessarily allow for extrapolation to the responses observed in animals. Infections in animals involve cell populations that can constantly change with the recruitment of lymphocytes and other cells to the site of infection. It is also impossible to have all the cells in a given tissue of an animal infected simultaneously, so there are subpopulations of cells at various stages of infection that give different profiles, as well as neighboring uninfected cells that may or may not be responding to signals sent from the infected cells. Nevertheless, we asked the question of whether there are detectable changes in gene expression in the overall lung tissue during the course of infection and, if so, at what time these changes are observed.
C57BL/6 mice infected with 1 × 106 PFU of wild-type VV were sacrificed at days 0 to 5 postinfection, and the whole lungs with the major bronchial tissue removed were harvested for microarray analysis. Three female mice per time point were used for this study. While both males and females exhibited identical susceptibilities to VV infection, only females were used to eliminate the possibility of sex-related transcriptional differences. The mice exhibited the expected clinical symptoms later in the time course before being sacrificed.
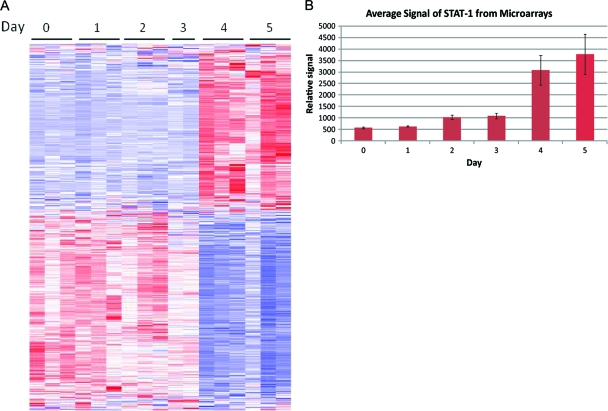
Affymetrix mouse 430.2A microarrays were used for analysis of host gene transcriptional differences over the course of infection (see Materials and Methods). The arrays exhibited no decrease in overall transcript levels over time, as previously observed in studies of tissue culture, due to the low percentage of infected cells in the lung compared to data in the in vitro studies, in which high-multiplicity infections were used. The arrays were subjected to an F test in BRB Array Tools, which gave 1,811 probe sets that were differentially expressed over time, with significance set at a P value of 0.001. A heat map showing the relative up- or downregulation compared to the average values for the probe sets identified in the F test as being expressed differentially over time is shown in Fig. 4A. This heat map shows a clear change in the gene expression pattern between days 3 and 4 postinfection. This change in expression profile occurred at approximately the same time that the clinical symptoms of disease began in the animals.
FIG. 4.
Gene profiling of virus-infected lungs. (A) Microarray analysis heat map of probes differentially expressed over time in lung tissues of infected mice, as determined by an F test performed on the data. Three samples per time point were analyzed. Each horizontal line represents a single probe expression profile. Vertical collections of lines represent the overall expression profile for a sample. Blue represents downregulation of the probe compared to the average expression value, white is the average expression value, and red is upregulation of the probe compared to the average expression value. (B) Average relative signal for 4 probe sets for STAT-1 for each time point. There was an increase of 3.5 times from day 3 to day 4 postinfection, demonstrating the significant change in the lung environment that occurred between days 3 and 4 postinfection.
The probe sets identified were from a wide variety of pathways, with no consensus for up- or downregulation within a pathway. Software from Ingenuity and the BioCarta classifications in BRB Array Tools were used for the pathway classifications to examine the relative regulation changes within pathways. Specific classes of probe sets were analyzed to determine whether the changes in expression patterns could be attributed to any pathway or cell type. The pathway and cell type expression analysis, however, showed that the changes observed were not easily categorized into a single pathway. These data do suggest that the response is highly focused within the immune response pathways, but it is still not possible to discern from these data the most important genes or pathways.
STAT-1 staining of lung tissue.
Infection of the lungs was highly focal, yet differences in gene expression could be detected readily via microarray analysis. This finding raised the question of whether the changes in gene expression were due primarily to changes within the focal areas of infection or, alternatively, to a global response of the overall lung to virus infection. There was also a question of whether the changes in gene transcripts correlated with changes in protein levels. To answer these questions, immunohistochemical detection of STAT-1 was performed. The STAT-1 gene was one of the genes identified through the microarray data analysis as upregulated (3.5-fold) between days 3 and 5 (Fig. 4B), and STAT-1 is a known member of the host immune response involving interferon activity (36, 42, 71).
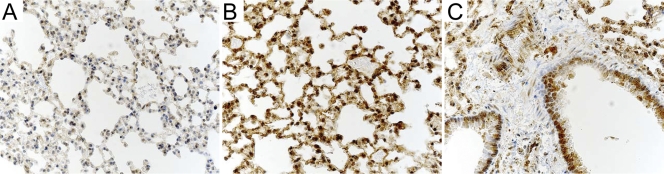
Lung tissues isolated from mice 3 and 5 days after infection with VV were stained for the presence of STAT-1. The day 3 postinfection lung sections clearly showed some low initial levels of STAT-1, and most cells exhibited a light, mostly cytoplasmic staining pattern similar to that of the uninfected sections (data not shown). However, the staining of the day 5 postinfection sections (Fig. 5B and C) was far more intense than that observed in both the uninfected lung sections (Fig. 5A) and day 3 postinfection sections (not shown), and the staining pattern was distributed throughout the cell. The increased STAT-1 staining observed in the lung was not limited to the sites of infection (the bronchial epithelial cells) shown in Fig. 3 but rather was distributed throughout the lung. By day 5, the infected epithelial cells again showed a pattern of staining of both the nucleus and cytoplasm, while the infiltrating cells showed no staining (Fig. 5C). Thus, while it was shown that the virus develops at rather restricted sites within the lung, the STAT-1 staining patterns indicate that the response of the lung is global.
FIG. 5.
Immunohistochemistry staining of STAT-1 protein in lung tissue at 5 days postinfection. Nuclei were stained with hematoxylin and appear blue; STAT-1 staining appears brown. Uninfected lung tissue (A), alveolar tissue distal from infected bronchi from an infected mouse (B), and infected bronchial regions (C) are shown. Magnification, ×40.
VVΔE3L infection of mice.
We next examined the behavior of VVΔE3L in C57BL/6, PKR−/−, RNase L−/−, and PKR−/− RNase L−/− mice. In the absence of the E3L gene, levels of free double-stranded RNA should be elevated markedly, triggering a vigorous host interferon response and virus attenuation. VVΔE3L has been reported to be highly attenuated in mice (6, 82). In agreement with these previously reported results, we likewise found VVΔE3L to be very attenuated compared to VV in C57BL/6 mice infected via the i.t. route, as expected (compare Fig. 6 and Fig. 1). We were unable to observe any serious illness in C57BL/6 mice at even the highest concentration (1 × 108 PFU) of VVΔE3L used. Essentially the same results were noted in PKR−/− and RNase L−/− mice, with the only serious illness being noted in PKR−/− mice infected with 1 × 108 PFU. The lack of serious disease in these mice was reflected by only a slight weight loss and little effect on temperature even at the highest dose of virus used (1 × 108 PFU). However, when PKR−/− RNase L−/− mice were inoculated with very high doses of VVΔE3L, serious illness was observed, occasionally at a dose of 1 × 107 PFU and consistently at a dose of 1 × 108 PFU (Fig. 6J to L). The disease observed, based on weight loss and temperature decrease, strongly resembled the wild-type disease (Fig. 1K and L). The calculated LD80 values for all mouse strains infected with VVΔE3L are shown in Table 1. Due to the mild nature of the disease and to the fact that no mice required euthanasia, LD80 values for C57BL/6 and RNase L−/− mice were unable to be determined and most likely cannot be ascertained experimentally due to the lack of disease caused by infections with VVΔE3L in these mice. The LD80 values for the RNase L−/− PKR−/− and PKR−/− mouse constructs were determined to be 1.3 × 107 and 3.2 × 108, respectively. Serious illness has never been reported following VVΔE3L infection of mice (6). The revertant virus of VVΔE3L, RVVE3L+, was analyzed in mice and demonstrated characteristics identical to those of wild-type VV.
FIG. 6.
Survival, temperature, and weight variations following VVΔE3L infection. Mice were infected intratracheally, and at least 10 mice were evaluated for each time point. The overall significance of the curves (P < 0.001) was determined using Kaplan-Meier survival analysis. (A to C) C57BL/6 mice; (D to F) RNase L−/− mice; (G to I) PKR−/− mice; (J to L) RNase L−/− PKR−/− mice. Panels A, D, H, and J indicate survival curves for the mice. Panels B, E, H, and K indicate the average percent weight loss from the weight at the time of infection. Panels C, F, I, and L indicate average body temperatures. Stars, PBS; diamonds, 1 × 104 PFU; triangles, 1 × 105 PFU; ×, 1 × 106 PFU; circles, 1 × 107 PFU; squares, 1 × 108 PFU.
We examined infections caused by VVΔE3L at the wild-type virus doses that are lethal to the respective mouse constructs (i.e., 1 × 106 PFU for C57BL/6 mice, 1 × 105 PFU for RNase L−/− and PKR−/− mice, and 1 × 104 PFU for RNase L−/− PKR−/− mice) for virus spread over time. By day 2 postinfection, the levels of virus in the lungs were 2 log lower than those observed in VV infections in all mouse constructs (Table 2). The virus titers continued to decrease in the lung over time. The ability of VVΔE3L to spread from the lung to distal sites in the animal was also examined. There was no detectible virus found in the liver, spleen, gonad, or brain in any of the mouse constructs, suggesting that VVΔE3L is unable to spread successfully in non-lethally infected mice. Upon increasing the virus inoculum in RNase L−/− PKR−/− mice to 1 × 108 PFU, virus dissemination was observed, consistent with the lethal infection observed, which resembled VV infections (data not shown). However, lethal doses of VVΔE3L in these animals were extraordinarily high compared to those of VV (compare Fig. 1J and 6J).
We compared the histological characteristics of lung, liver, and spleen sections from C57BL/6 and RNase L−/− PKR−/− animals infected with low (1 × 104 PFU) and high (1 × 108 PFU) VVΔE3L doses at day 3 and day 5 postinfection. At the lower dose of VVΔE3L in the mice (1 × 104 PFU), there was little evidence of virus growth in the lung, whereas in VV-infected lungs, focal sites of replication were readily visible (data not shown). Indeed, the only evidence of VVΔE3L infection at the lower dose, regardless of the mouse construct used, was in the increased influx of immune cells into the infected lung (data not shown). This is in sharp contrast with what was seen when VVΔE3L infections were initiated at 1 × 108 PFU (Fig. 7) for C57BL/6 mice (panels A to C and G to I) and RNase L−/− PKR−/− mice (panels D to F and J to L). The results for day 5 only are shown in Fig. 7 for C57BL/6 mice (resistant to VVΔE3L infection) and RNase L−/− PKR−/− mice (susceptible to VVΔE3L infection) infected with a dose of 1 × 108 PFU. At this higher dose in RNase L−/− PKR−/− mice, there were visible foci of virus growth similar to those seen in VV infections, whereas there were no detectible levels of virus replication in C57BL/6 mice (data not shown). However, massive cellular influx into the infected C57BL/6 and RNase L−/− PKR−/− mouse lungs was unique to the VVΔE3L infection (Fig. 7A, B, D, and E).
FIG. 7.
Comparative lung histology of VVΔE3L-infected mice. All mice were infected with 1 × 108 PFU of virus. (A) C57BL/6 mouse lung at day 5. Magnification, ×5. (B) H&E-stained mouse lung at day 5. Magnification, ×20. (C) Day 5 lung stained for Ki67. Magnification, ×10. (D) H&E-stained PKR−/− RNase L−/− mouse lung at day 5. Magnification, ×5. (E) Same lung as in panel D. Magnification, ×20. (F) Ki67 staining of day 5 mouse lung. Magnification, ×10. (G) H&E-stained C57BL/6 mouse spleen at day 5. Magnification, ×5. (H) Ki67 staining of same spleen. Magnification, ×5. (I) Ki67 staining of same spleen. Magnification, ×20. (J) H&E-stained PKR−/− RNase L−/− mouse spleen at day 5. Magnification, ×5. (K) Ki67 staining of same spleen. Magnification, ×5. (L) Ki67 staining of same spleen. Magnification, ×20.
C57BL/6 mice infected with VVΔE3L were asymptomatic and survived the virus infection. Unlike animals infected with VV, VVΔE3L-infected C57BL/6 mice exhibited immune infiltrates and mild edema around the infected bronchi of the lung, but very little damage was noted to the lung as a whole (Fig. 7A and B). Upon staining of the lung for Ki67, it was noted that there was no elevation in staining at day 5 postinfection in the lungs of C57BL/6 mice, suggesting little or no virus-induced cellular proliferation (Fig. 7C). Liver sections at all time points were normal on H&E staining (data not shown). Spleen sections stained with H&E showed follicular hyperplasia in VVΔE3L-infected C57BL/6 mice at both days 3 and 5 postinfection (Fig. 7G). Staining for Ki67 showed some positive cells, confirming limited splenic cellular proliferation and a limited immune response (Fig. 7H and I).
RNase L−/− PKR−/− mice infected with the high virus inoculum (1 × 108 PFU), unlike VVΔE3L-infected C57BL/6 mice, exhibited large areas of immune infiltrate around infected bronchioles and into the alveolar tissue of the lung (Fig. 7D). These lung sections also exhibited lymphoid tissue proliferation, mostly contained in small areas, causing obstruction of entire sections of the lung tissue (Fig. 7E). Lung samples from days 3 and 5 postinfection showed staining for virus replication around epithelial cells of bronchi, areas of edema, and infiltrates, as observed with IHC for VV infection (data not shown). Staining for Ki67 showed large numbers of positive cells, suggesting an active immune response to the infection within the lung, with most of the positive cells located around infected bronchioles and bronchi (Fig. 7F). Liver sections at all time points were normal on H&E staining (data not shown). Spleen sections were normal on H&E staining at day 3 postinfection and exhibited severe lymphoid depletion at day 5 postinfection (Fig. 7J), as evidenced by the absence of large numbers of well-defined follicles. Staining for Ki67 showed very large numbers of positive cells throughout the spleen, confirming proliferation and an immune response (Fig. 7K and L).
Clinical symptoms also appeared at approximately the same time and at the level of severity observed for mice infected with a lethal dose of VV. It appears that if the initial host barriers to infection are overcome, the infection can proceed and lead to development of clinical symptoms and a lethal infection. It is clear that the E3L gene is a major contributor to controlling the host responses to infection and that loss of the E3L gene can be compensated only partially by deleting both PKR and RNase L from the mouse host, and very little by deleting PKR alone. This is in contrast to what is found in cell culture, where deleting the PKR gene alone from HeLa cells compensates for deletion of the E3L gene (84).
VVΔK3L infection of mice.
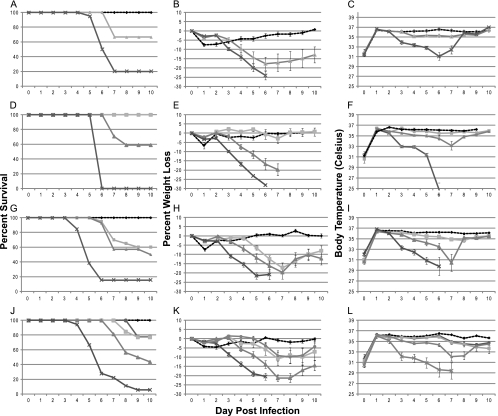
The vaccinia virus K3L gene, when deleted, also confers interferon sensitivity on VV-infected cells (2, 3). The use of PKR−/−, RNase L−/−, and PKR−/− RNase L−/− mice and VV strains deleted for the K3L gene allowed the role of K3L interactions with PKR as well as RNase L to be evaluated. The results of these experiments are shown in Fig. 8. When VVΔK3L was examined in any of the four mouse constructs, there was very little difference in inherent susceptibility of the mice to VVΔK3L (Fig. 8A, D, G, and J). This similarity was also recapitulated when either weight loss (Fig. 8B, E, H, and K) or decreases in temperature (Fig. 8C, F, I, and L) were examined. There was little, if any, decrease in susceptibility of C57BL/6 mice infected by the i.t. route when the K3L gene was deleted from the virus genome, nor was there an increased susceptibility if the mice lacked either PKR, RNase L, or both. The lack of attenuation of VVΔK3L is mimicked if one compares clinical symptoms exhibited by VV and VVΔK3L infections of C57BL/6 mice. A comparison of weight loss and body temperature is shown in Fig. 9, demonstrating that both indicators of disease are comparable for the two viruses.
FIG. 8.
Survival, temperature, and weight variations following VVΔK3L infection. Mice were infected intratracheally, and at least 10 mice were evaluated for each time point. The overall significance of the curves (P < 0.001) was determined using Kaplan-Meier survival analysis. (A to C) C57BL/6 mice; (D to F) RNase L−/− mice; (G to I) PKR−/− mice; (J to L) PKR−/− RNase L−/− mice. Panels A, D, H, and J indicate survival curves for the mice. Panels B, E, H, and K indicate percent weight loss over time. Panels C, F, I, and L indicate body temperatures. Surviving C57BL/6 animals infected with 1 × 106 PFU of virus were terminated at day 6. RNase L−/− or PKR−/− mice infected at 1 × 105 PFU of virus were terminated at day 5, and PKR−/− RNase L−/− animals infected with 1 × 105 PFU, which were all less ill than the other animals, were terminated at day 6. Stars, PBS; circles, 1 × 103 PFU; squares, 1 × 104 PFU; triangles, 1 × 105 PFU; ×, 1 × 106 PFU.
FIG. 9.
Comparative temperatures and weight losses for VV- and VVΔK3L-infected C57BL/6 mice. (A) Average weight losses. (B) Average body temperatures. Error bars represent standard errors of the means. Diamonds, VV-infected animals; circles, VVΔK3L-infected animals.
Despite this similarity in disease and overt symptoms, when we compared the histopathology of the lungs and spread of the virus to distal tissues and organs, significant differences were noted between the two viruses. Both histopathology of the lung and growth and dissemination of the virus to distal tissues revealed some unique properties of the K3L gene that have not been seen before. At early time points (day 3 postinfection), the pattern of virus growth was similar to that observed with VV by IHC (compare Fig. 10D and Fig. 3A) and histologically, as revealed by H&E staining (compare Fig. 10A and Fig. 3D). By 5 days postinfection, VVΔK3L had primarily targeted and infected the bronchial epithelial regions of the lung (Fig. 10E and F). Perivascular edema and necrosis with a neutrophilic infiltrate and cellular debris were observed in the lung by 5 days postinfection, indicating activation of the immune response and the presence of lymphocytes attempting to clear the infection (Fig. 10B). Moderate perivascular lymphocytic aggregates and fibrinoid necrosis were also observed, indicating damage to the blood vessels and subsequent leaking of fluid from the blood vessels into the lung (Fig. 10C). This was not observed in VV infections. The areas of virus infection detected by IHC appeared larger and more widespread in the VVΔK3L-infected mice than in VV-infected mice, with bronchiolar and peribronchiolar epithelial cells staining positive for the presence of virus (compare Fig. 10E with Fig. 3C). Ki67 staining of lung sections also showed a moderate level of staining, with the majority of the proliferative cells located around the areas of infection (Fig. 10G).
FIG. 10.
Analysis of virus spread and lung histology of mice infected with VVΔK3L at 5 days postinfection. Images are for C57BL/6 mice infected with 1 × 106 PFU of virus. (A) H&E staining of lung at 3 days postinfection. Magnification, ×5. (B) H&E staining of lung at 5 days postinfection. Magnification, ×5. (C) H&E staining of lung at 5 days postinfection, showing severe edema with no virus present. Magnification, ×10. (D) Immunohistochemistry staining for virus replication (GFP) in the lung at 3 days postinfection. Magnification, ×5. (E) Immunohistochemistry staining for virus replication in the lung at 5 days postinfection. Magnification, ×5. (F) Immunohistochemistry staining for virus replication in the lung at 5 days postinfection. Magnification, ×40. (G) Ki67 staining of lung tissue at day 5 postinfection, showing active immune response at the site of infection. Magnification, ×10. (H) H&E staining of the spleen at day 3 postinfection. Magnification, ×10. (I) Ki67 staining at day 3 postinfection. Magnification, ×10. (J) H&E staining of day 5 spleen. Magnification, ×10. (K) Ki67 staining of day 5 spleen. Magnification, ×10.
Liver sections from infected animals demonstrated random neutrophilic foci of necrosis for all time points taken. No virus replication was detected in the liver by IHC (data not shown). Spleen sections showed mild to moderate lymphoid depletion early in infection (Fig. 10H) that progressed to severe lymphoid depletion late in infection (Fig. 10J), indicating a strong immune response to the virus infection. There was a small subset of spleen sections in which the follicles were completely depleted (data not shown). No virus replication was detected by IHC in the spleen, as was observed with VV-infected animals. Ki67 staining of the spleen showed high levels of staining throughout the time course, again indicating proliferation and a strong immune response (Fig. 10I and K).
What was most surprising was a comparison of dissemination of newly replicated viruses to distal tissues, which was markedly impaired in VVΔK3L-infected mice compared to that in VV-infected mice. While the titers of virus in VVΔK3L-infected mice remained unchanged from those observed for VV-infected lungs, there was a clear difference in virus dissemination (Table 2). These results indicate that replication per se of VVΔK3L in the lung is not impaired but rather that the virus is markedly inhibited in the ability to disseminate to other target sites. A detailed comparative analysis of the pattern of dissemination of the lung resulting from VVΔK3L infection is shown in Table 2. Comparing the data for VV infections and VVΔK3L infections, unlike what was observed with VV-infected animals, there was very little spread of VVΔK3L from the lung. Later in the infection, there were low levels of dissemination of VVΔK3L, first into the brain and then into the gonads. This pattern was strikingly different from what was seen with wild-type virus-infected animals. These data allow us to speculate on the cause of death of the animals. The courses of disease in VVΔK3L- and VV-infected animals were clinically indistinguishable in terms of time of onset and severity of symptoms. The fact that VVΔK3L was restricted mostly to the lungs suggests that the cause of death for mice infected lethally via the intratracheal route was the result of respiratory illness. Finally, to ensure that the failure of virus to spread from the lung was due solely to deletion of the K3L gene, we reconstructed wild-type VV from VVΔK3L (RVVK3L+). When analyzed in mice, RVVK3L+ behaved identically to VV and was rapidly disseminated from the lung to distal tissue.
DISCUSSION
In this study, we examined vaccinia virus-infected wild-type mice and mice defective in RNase L, PKR, or both by using wild-type VV and VV mutants in the E3L and K3L genes, genes known to control these host factors, in an attempt to ascertain the relative contribution of each to the disease process. VV infection of wild-type C57BL/6 mice was shown to produce severe disease in which 80% of mice had to be euthanized when animals were infected with 106 PFU of virus via the i.t. route. The clinical signs of disease in mice were characterized by rapid weight loss, dyspnea, and hypothermia. A decrease in grooming and development of nose and facial swelling late in disease were also characteristic of VV infection by the i.t. route.
The primary site of infection was the lung, where virus growth was observed to be quite focal, being restricted to the bronchiolar epithelial cells, keeping the infection localized to and around the bronchi. The localized, focal infection within the lung was unanticipated and surprising, given that the animals were infected with 106 PFU of VV directly but with no other targeting to the lung. The subsequent spread of virus from the lungs was rapid, showing dissemination to the brain, liver, spleen, and gonads within 2 days of infection. The levels of virus found in the lung remained constant over the course of infection, suggesting a steady-state situation where newly formed virus in the lung was rapidly disseminated to the other organs. This rapid virus dissemination from the lung is consistent with the continued localized areas of infection observed within the lung at later times. Virus dissemination and replication in many other distal tissues are well-documented and accepted features of vaccinia virus infections. Clearly, the virus is capable of growing in a variety of tissues and cells, but within the lung it does not cause the massive amount of tissue damage that would be expected from a direct but relatively nonspecific inoculation into the lung. Within the animal, it is likely that some of the clinical symptoms (weight loss and lack of grooming behaviors) were a consequence of systemic infection of the animal. On the other hand, the respiratory distress and ultimate respiratory failure which necessitated euthanasia were most likely due to the lung damage caused by the virus, directly or indirectly, suggesting that the small amount of visible damage observed within the lung generated sufficient collateral pathology to cause death. Long-distance collateral effects occurred, as indicated by the global induction of STAT-1 in the infected lung (Fig. 4 and 5). Indeed, the fact that we observed identical clinical diseases with VV- and VVΔK3L-infected animals, with no observed attenuation of VVΔK3L infections—where dissemination from the lung was inhibited, resulting in increased titers of virus in the lung—adds further credence that the primary cause of death was the result of respiratory disease.
The host proteins PKR and RNase L play a role in controlling the host innate immune response to viral infections. Both are activated in part by the synthesis of dsRNA, which serves to induce an interferon response. Poxviruses produce dsRNA late in infection due to the bidirectionality of the open reading frames and polydispersity in lengths of late transcripts. Double-stranded RNA serves to activate both PKR and RNase L (26, 66). Vaccinia virus carries two genes, the E3L and K3L genes, which are hypothesized to control the host immune response to dsRNA by interacting with RNase L and PKR, respectively. Therefore, examining virus mutants in either of these viral genes with mouse strains lacking either RNase L, PKR, or both allowed us to assess the relative contributions of the host and viral genes to the infection.
The viral E3L gene is thought to act preferentially to control RNase L activity by binding to dsRNA, which is an activator of RNase L. RNase L is a component within the host antiviral response pathway which binds dsRNA and thereby serves to activate the OAS pathway, leading to the destruction of RNA within the cell and to activation of the antiviral interferon pathway. RNase L−/− mice exhibited a somewhat increased sensitivity (10-fold) to VV infection compared to C567BL/6 mice, but this increase in sensitivity was not as profound as might be expected, perhaps because a functional viral E3L gene remained present. However, in the absence of the E3L gene, VVΔE3 infections of C57BL/6 were profoundly attenuated, consistent with the proposed role of the E3L gene in controlling dsRNA responses. Yet the only mild increase in sensitivity to VVΔE3 infections that we observed in RNase L−/− or PKR−/− mice suggests that sequestering RNA is important for critical host response pathways other than the RNase L and PKR pathways or, alternatively, that the E3L protein has functions totally unrelated to the binding of dsRNA, as previously suggested by Xiang et al. (81). The E3L protein is also a double-stranded DNA binding protein. There are two distinct functional domains that have been identified within E3L (6). The C terminus of E3L contains a dsRNA binding domain, whereas the N-terminal region of the protein contains sequences associated with binding to Z-DNA and PKR (38). Both regions are required for virulence in mice (6). E3L is now considered a member of the larger Z-DNA binding family (39). Furthermore, E3L has some sequence similarity to DAI, a protein involved in DNA pattern recognition that can trigger the innate immune response (20, 39, 70). Therefore, E3L may control the pathogen-associated pattern recognition process mediated by dsDNA. Consistent with additional roles of the E3L protein is the fact that the immune response to VVΔE3 infections is quite robust in the lung and spleen compared to that of VV-infected animals. It is interesting that when RNase L−/− PKR−/− mice were infected with very high doses of VVΔE3, virus dissemination was observed (albeit it at lower levels), together with a large immune response which was absent in VV-infected animals. It would therefore appear that after the innate immune barrier has been breached, the virus is relatively unrestricted in growth and dissemination.
The K3L protein is believed to regulate the activity of PKR. PKR, like RNase L, is activated by the presence of dsRNA. However, PKR is inhibited by interacting with the K3L protein, which serves to prevent the activation of PKR by phosphorylation. PKR is then unable to phosphorylate eIF2α, which would otherwise lead to an inhibition of protein translation in the cell (14, 21, 73). PKR has a role in the host control of VV infection, as indicated by the increase in sensitivity of PKR−/− mice to VV infection, but again, this increase in sensitivity was relatively modest. Interestingly, PKR−/− mice infected with VVΔK3L exhibited no difference in susceptibility to infection from C57BL/6 mice or any of the other mutant mice with VV infection. These data suggest that the interaction between PKR and K3L in the intratracheal model is not important in modulating disease and that K3L, like E3L, may have alternative functions in compromising the host.
Indeed, one such role is the completely unanticipated requirement of K3L for dissemination of virus from the lung to the distal organs. Generally, dissemination of virus from initial sites of infection to secondary target tissue, although well known, is a very poorly understood aspect of poxvirus infections. In the case of VV, the mechanism by which K3L promotes dissemination of the virus is completely unknown. The K3L-mediated control of dissemination is independent of either the PKR or RNase L gene and defines an entirely new, dynamically regulated process of virus infection which clearly merits further study.
Acknowledgments
We acknowledge M. Cecilia Lopez, M. Delano, and Cynthia L. Tannahill for their contributions to the microarray data and analysis. We thank R. H. Silverman and B. R. Williams for the gifts of founder breeding mice for all knockout strains utilized in this study. We thank Andrew J. Smith, Dorothy Smith, and JoAnne Anderson for their technical assistance during the mouse studies and for management of the breeding colony.
We acknowledge support from NIH grant AI015722.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Beattie, E., E. B. Kauffman, H. Martinez, M. E. Perkus, B. L. Jacobs, E. Paoletti, and J. Tartaglia. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes 12:89-94. [DOI] [PubMed] [Google Scholar]
- 2.Beattie, E., E. Paoletti, and J. Tartaglia. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L− and E3L− mutant viruses. Virology 210:254-263. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, E., J. Tartaglia, and E. Paoletti. 1991. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183:419-422. [DOI] [PubMed] [Google Scholar]
- 4.Blance, R. N., A. H. Sims, E. Anderson, A. Howell, and R. B. Clarke. 2009. Normal breast tissue implanted into athymic nude mice identifies biomarkers of the effects of human pregnancy levels of estrogen. Cancer Prev. Res. (Philadelphia) 2:257-264. doi: 10.1158/1940-6207.CAPR-08-0161. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, T., M. C. Heck, S. Vijaysri, G. M. Jentarra, J. M. Cameron, and B. L. Jacobs. 2005. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology 333:263-270. [DOI] [PubMed] [Google Scholar]
- 6.Brandt, T. A., and B. L. Jacobs. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownstein, D., P. N. Bhatt, and R. O. Jacoby. 1989. Mousepox in inbred mice innately resistant or susceptible to lethal infection with ectromelia virus. V. Genetics of resistance to the Moscow strain. Arch. Virol. 107:35-41. [DOI] [PubMed] [Google Scholar]
- 8.Brownstein, D. G., and L. Gras. 1995. Chromosome mapping of Rmp-4, a gonad-dependent gene encoding host resistance to mousepox. J. Virol. 69:6958-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brum, L. M., M. C. Lopez, J. C. Varela, H. V. Baker, and R. W. Moyer. 2003. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 315:322-334. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, K., O. Elroy Stein, B. Moss, and R. Jagus. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 268:12837-12842. [PubMed] [Google Scholar]
- 11.Chang, H.-W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, H. W., and B. L. Jacobs. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537-547. [DOI] [PubMed] [Google Scholar]
- 13.Chang, H. W., L. H. Uribe, and B. L. Jacobs. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J. J. 2007. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood 109:2693-2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condit, R. C., and A. Motyczka. 1981. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology 113:224-241. [DOI] [PubMed] [Google Scholar]
- 16.Condit, R. C., A. Motyczka, and G. Spizz. 1983. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology 128:429-443. [DOI] [PubMed] [Google Scholar]
- 17.Craig, A. W., G. P. Cosentino, O. Donze, and N. Sonenberg. 1996. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J. Biol. Chem. 271:24526-24533. [DOI] [PubMed] [Google Scholar]
- 18.Dar, A. C., and F. Sicheri. 2002. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol. Cell 10:295-305. [DOI] [PubMed] [Google Scholar]
- 19.Davies, M. V., O. Elroy Stein, R. Jagus, B. Moss, and R. J. Kaufman. 1992. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 66:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng, L., P. Dai, T. Parikh, H. Cao, V. Bhoj, Q. Sun, Z. Chen, T. Merghoub, A. Houghton, and S. Shuman. 2008. Vaccinia virus subverts a mitochondrial antiviral signaling protein-dependent innate immune response in keratinocytes through its double-stranded RNA binding protein, E3. J. Virol. 82:10735-10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. doi:S0092867402006426. [DOI] [PubMed] [Google Scholar]
- 22.Elde, N. C., S. J. Child, A. P. Geballe, and H. S. Malik. 2008. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457:485-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteban, D. J., and R. M. Buller. 2005. Ectromelia virus: the causative agent of mousepox. J. Gen. Virol. 86:2645-2659. [DOI] [PubMed] [Google Scholar]
- 24.Fahy, A. S., R. H. Clark, E. F. Glyde, and G. L. Smith. 2008. Vaccinia virus protein C16 acts intracellularly to modulate the host response and promote virulence. J. Gen. Virol. 89:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foust, K. D., A. Poirier, C. A. Pacak, R. J. Mandel, and T. R. Flotte. 2008. Neonatal intraperitoneal or intravenous injections of recombinant adeno-associated virus type 8 transduce dorsal root ganglia and lower motor neurons. Hum. Gene Ther. 19:61-70. doi: 10.1089/hum.2007.093. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, M. A., E. F. Meurs, and M. Esteban. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799-811. [DOI] [PubMed] [Google Scholar]
- 27.Grinde, B., M. Gayorfar, and G. Hoddevik. 2007. Modulation of gene expression in a human cell line caused by poliovirus, vaccinia virus and interferon. Virol. J. 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerra, S., L. A. Lopez-Fernandez, R. Conde, A. Pascual-Montano, K. Harshman, and M. Esteban. 2004. Microarray analysis reveals characteristic changes of host cell gene expression in response to attenuated modified vaccinia virus Ankara infection of human HeLa cells. J. Virol. 78:5820-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerra, S., J. L. Najera, J. M. Gonzalez, L. Lopez, N. Climent, J. M. Gatell, T. Gallart, and M. Esteban. 2007. Distinct gene expression profiling after infection of immature human monocyte-derived dendritic cells by the attenuated poxvirus vectors MVA and NYVAC. J. Virol. 81:8707-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha, S. C., N. K. Lokanath, Q. D. Van, C. A. Wu, K. Lowenhaupt, A. Rich, Y. G. Kim, and K. K. Kim. 2004. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a yatapoxvirus Zalpha bound to DNA. Proc. Natl. Acad. Sci. U. S. A. 101:14367-14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. doi:S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 33.Hovanessian, A. G. 1991. Interferon-induced and double-stranded RNA-activated enzymes: a specific protein kinase and 2′,5′-oligoadenylate synthetases. J. Interferon Res. 11:199-205. [DOI] [PubMed] [Google Scholar]
- 34.Hovanessian, A. G. 2007. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2′-5′oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev. 18:351-361. [DOI] [PubMed] [Google Scholar]
- 35.Hutchens, M. A., K. E. Luker, J. Sonstein, G. Nunez, J. L. Curtis, and G. D. Luker. 2008. Protective effect of Toll-like receptor 4 in pulmonary vaccinia infection. PLoS Pathog. 4:e1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katze, M. G., Y. He, and M. Gale. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 37.Kawagishi-Kobayashi, M., J. B. Silverman, T. L. Ung, and T. E. Dever. 1997. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2alpha. Mol. Cell. Biol. 17:4146-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, Y. G., K. Lowenhaupt, D. B. Oh, K. K. Kim, and A. Rich. 2004. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: implications for development of a therapy for poxvirus infection. Proc. Natl. Acad. Sci. U. S. A. 101:1514-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, Y. G., M. Muralinath, T. Brandt, M. Pearcy, K. Hauns, K. Lowenhaupt, B. L. Jacobs, and A. Rich. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:6974-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laassri, M., C. A. Meseda, O. Williams, M. Merchlinsky, J. P. Weir, and K. Chumakov. 2007. Microarray assay for evaluation of the genetic stability of modified vaccinia virus Ankara B5R gene. J. Med. Virol. 79:791-802. [DOI] [PubMed] [Google Scholar]
- 41.Langland, J. O., and B. L. Jacobs. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133-141. [DOI] [PubMed] [Google Scholar]
- 42.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. doi:S1359610100000277. [DOI] [PubMed] [Google Scholar]
- 43.Ludwig, H., J. Mages, C. Staib, M. H. Lehmann, R. Lang, and G. Sutter. 2005. Role of viral factor E3L in modified vaccinia virus Ankara infection of human HeLa cells: regulation of the virus life cycle and identification of differentially expressed host genes. J. Virol. 79:2584-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luker, K. E., M. Hutchens, T. Schultz, A. Pekosz, and G. D. Luker. 2005. Bioluminescence imaging of vaccinia virus: effects of interferon on viral replication and spread. Virology 341:284-300. [DOI] [PubMed] [Google Scholar]
- 45.MacNeill, A. L., L. L. Moldawer, and R. W. Moyer. 2009. The role of the cowpox virus crmA gene during intratracheal and intradermal infection of C57BL/6 mice. Virology 384:151-160. [DOI] [PubMed] [Google Scholar]
- 46.McCormack, S. J., D. C. Thomis, and C. E. Samuel. 1992. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology 188:47-56. [DOI] [PubMed] [Google Scholar]
- 47.McKenna, S. A., D. A. Lindhout, I. Kim, C. W. Liu, V. M. Gelev, G. Wagner, and J. D. Puglisi. 2007. Molecular framework for the activation of RNA-dependent protein kinase. J. Biol. Chem. 282:11474-11486. [DOI] [PubMed] [Google Scholar]
- 48.Moulton, E. A., J. P. Atkinson, and R. M. Buller. 2008. Surviving mousepox infection requires the complement system. PLoS Pathog. 4:e1000249. doi: 10.1371/journal.ppat.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker, A. K., S. Parker, W. M. Yokoyama, J. A. Corbett, and R. M. Buller. 2007. Induction of natural killer cell responses by ectromelia virus controls infection. J. Virol. 81:4070-4079. doi: 10.1128/JVI.02061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker, S., A. M. Siddiqui, C. Oberle, E. Hembrador, R. Lanier, G. Painter, A. Robertson, and R. M. Buller. 2009. Mousepox in the C57BL/6 strain provides an improved model for evaluating anti-poxvirus therapies. Virology 385:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quenelle, D. C., R. M. Buller, S. Parker, K. A. Keith, D. E. Hruby, R. Jordan, and E. R. Kern. 2007. Efficacy of delayed treatment with ST-246 given orally against systemic orthopoxvirus infections in mice. Antimicrob. Agents Chemother. 51:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raven, J. F., and A. E. Koromilas. 2008. PERK and PKR: old kinases learn new tricks. Cell Cycle 7:1146-1150. [DOI] [PubMed] [Google Scholar]
- 53.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Invest. 110:1383-1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rothenburg, S., E. J. Seo, J. S. Gibbs, T. E. Dever, and K. Dittmar. 2009. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 16:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadler, A. J., and B. R. Williams. 2007. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316:253-292. [DOI] [PubMed] [Google Scholar]
- 56.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samuel, C. E. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603-7606. [PubMed] [Google Scholar]
- 58.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samuel, M. A., K. Whitby, B. C. Keller, A. Marri, W. Barchet, B. R. Williams, R. H. Silverman, M. Gale, Jr., and M. S. Diamond. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sbrana, E., R. Jordan, D. E. Hruby, R. I. Mateo, S. Y. Xiao, M. Siirin, P. C. Newman, A. P. Da Rosa, and R. B. Tesh. 2007. Efficacy of the antipoxvirus compound ST-246 for treatment of severe orthopoxvirus infection. Am. J. Trop. Med. Hyg. 76:768-773. [PubMed] [Google Scholar]
- 61.Seo, E. J., F. Liu, M. Kawagishi-Kobayashi, T. L. Ung, C. Cao, A. C. Dar, F. Sicheri, and T. E. Dever. 2008. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc. Natl. Acad. Sci. U. S. A. 105:16894-16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharp, T. V., J. E. Witzel, and R. Jagus. 1997. Homologous regions of the alpha subunit of eukaryotic translational initiation factor 2 (eIF2 alpha) and the vaccinia virus K3L gene product interact with the same domain within the dsRNA-activated protein kinase (PKR). Eur. J. Biochem. 250:85-91. [DOI] [PubMed] [Google Scholar]
- 63.Shors, T., K. V. Kibler, K. B. Perkins, R. Seidlerwulff, M. P. Banaszak, and B. L. Jacobs. 1997. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology 239:269-276. [DOI] [PubMed] [Google Scholar]
- 64.Silverman, R. H. 1994. Fascination with 2-5A-dependent RNase: a unique enzyme that functions in interferon action. J. Interferon Res. 14:101-104. [DOI] [PubMed] [Google Scholar]
- 65.Silverman, R. H. 2007. A scientific journey through the 2-5A/RNase L system. Cytokine Growth Factor Rev. 18:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silverman, R. H. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith, G. L., J. A. Symons, A. Khanna, A. Vanderplasschen, and A. Alcami. 1997. Vaccinia virus immune evasion. Immunol. Rev. 159:137-154. [DOI] [PubMed] [Google Scholar]
- 68.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 69.Stern, R. J., J. P. Thompson, and R. W. Moyer. 1997. Attenuation of B5R mutants of rabbitpox virus in vivo is related to impaired growth and not an enhanced host inflammatory response. Virology 233:118-129. [DOI] [PubMed] [Google Scholar]
- 70.Takaoka, A., Z. Wang, M. K. Choi, H. Yanai, H. Negishi, T. Ban, Y. Lu, M. Miyagishi, T. Kodama, K. Honda, Y. Ohba, and T. Taniguchi. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501-505. [DOI] [PubMed] [Google Scholar]
- 71.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8:907-922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 72.Takeuchi, O., and S. Akira. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka, H., and C. E. Samuel. 1994. Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 91:7995-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson, J. P., P. C. Turner, A. N. Ali, B. C. Crenshaw, and R. W. Moyer. 1993. The effects of serpin gene mutations on the distinctive pathobiology of cowpox and rabbitpox virus following intranasal inoculation of Balb/c mice. Virology 197:328-338. [DOI] [PubMed] [Google Scholar]
- 75.Tscharke, D. C., P. C. Reading, and G. L. Smith. 2002. Dermal infection with vaccinia virus reveals roles for virus proteins not seen using other inoculation routes. J. Gen. Virol. 83:1977-1986. [DOI] [PubMed] [Google Scholar]
- 76.Turner, G. S. 1967. Respiratory infection of mice with vaccinia virus. J. Gen. Virol. 1:399-402. [DOI] [PubMed] [Google Scholar]
- 77.Turner, P. C., and R. W. Moyer. 2002. Poxvirus immune modulators: functional insights from animal models. Virus Res. 88:35-53. [DOI] [PubMed] [Google Scholar]
- 78.Vijaysri, S., G. Jentarra, M. C. Heck, A. A. Mercer, C. J. McInnes, and B. L. Jacobs. 2008. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: intra-nasal vaccination. Vaccine 26:664-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams, B. R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112-6120. [DOI] [PubMed] [Google Scholar]
- 80.Xia, H., D. Diebold, R. Nho, D. Perlman, J. Kleidon, J. Kahm, S. Avdulov, M. Peterson, J. Nerva, P. Bitterman, and C. Henke. 2008. Pathological integrin signaling enhances proliferation of primary lung fibroblasts from patients with idiopathic pulmonary fibrosis. J. Exp. Med. 205:1659-1672. doi: 10.1084/jem.20080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang, Y. L., L. F. Reis, J. Pavlovic, A. Aguzzi, R. Schafer, A. Kumar, B. R. Williams, M. Aguet, and C. Weissmann. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu, L., and S. Shuman. 1996. Mutational analysis of the RNA triphosphatase component of vaccinia virus mRNA capping enzyme. J. Virol. 70:6162-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang, P., B. L. Jacobs, and C. E. Samuel. 2008. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J. Virol. 82:840-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 86.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]
- 87.Zhou, A. M., J. Paranjape, T. L. Brown, H. Q. Nie, S. Naik, B. H. Dong, A. S. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]