Abstract
Elevated expression of steroid receptor coactivator-3 (SRC-3), a member of the p160 family of nuclear receptor coactivators has been implicated in tamoxifen resistance of breast tumors while the involvement of the two other members of this family, SRC-1 and SRC-2 is less well characterized. In this study employing siRNA-based silencing, the role of each SRC coactivator in the growth of the LCC2 estrogen-independent and tamoxifen-resistant breast cancer cell line was evaluated. Loss of SRC-1, SRC-2 or SRC-3 did not significantly alter LCC2 proliferation or cell cycle distribution of 4-hydroxytamoxifen- versus vehicle-treated cells. However, depletion of SRC-2 and SRC-3, but not SRC-1, decreased basal cell proliferation and increased apoptosis. Cell cycle analyses further illustrated the divergent contributions of SRC-2 and SRC-3 with depletion of the former increasing the percentage of cells in the G0G1 and sub-G0G1 phases of cell cycle yet maintaining sensitivity to estradiol and ICI 182,780 antiestrogen, while SRC-3 depletion increased cells in the sub-G0G1 phase and ablated response to ERα ligands. Surprisingly, the effects of SRC coactivator depletion on ERα transcriptional activity, as measured by luciferase reporter gene did not correspond to the observed effects on proliferation (e.g. SRC-1 knock-down increases ERα activity). Collectively these data indicate that SRC control of basal and hormone-regulated proliferation is not solely mediated by ERα, and suggest that targeting growth inhibition by disrupting SRC-2 and SRC-3 function may be an effective approach to inhibit the growth of tamoxifen resistant breast cancer.
Keywords: coactivators, tamoxifen resistance, estrogen receptor, proliferation, breast
INTRODUCTION
The female sex steroid 17β-estradiol (E2) is important in normal growth and development of mammary gland, although an elevated exposure to this hormone plays a crucial role in progression of carcinogenesis (Nilsson et al. 2001). Because of the connection between estrogens, cell proliferation and breast cancer, there has been great interest in the use of estrogen receptor (ER) antagonists to combat this disease. Over the past few decades, tamoxifen has been the most widely used endocrine therapy employed to treat ER-positive breast cancer and its use as an adjuvant significantly improves the survival of early stage breast cancer patients (Early Breast Cancer Trialists' Collaborative Group 1998). Tamoxifen resistance is, however a major problem in the treatment of breast cancer. A significant number of patients, despite the presence of ERα in their breast tumors exhibit de novo tamoxifen resistance, while others who initially responded to tamoxifen therapy will develop resistance to this therapy. Accordingly, one of the major challenges in adjuvant treatment of breast cancer is to better understand the molecular mechanisms of such resistance in anticipation that this will facilitate the development of approaches to avoid resistance to endocrine therapy.
The transcriptional activity of ERα is dependent on the nature of the ligand that occupies its ligand binding pocket (e.g agonist or antagonist) and as a consequence the receptor’s interaction with various coregulators that positively (coactivators) or negatively (corepressors) influence gene expression. The three members of the p160 family of steroid receptor coactivators (SRCs) are strong regulators of ERα transcriptional activity and are amongst the best characterized coregulators for nuclear receptors. The family members include SRC-1 (NCoA1), SRC-2 (TIF2/GRIP-1/NCoA2) and SRC-3 (AIB1/ACTR/pCIP/RAC3/TRAM-1/NCoA3). These coactivators interact with agonist-bound ERα and in so doing recruit other coactivators such as CBP/p300 or CARM1/PRMT1 that possess chromatin remodeling enzymatic activities such as histone acetyltransferase and methyltransferase. These in turn relax chromatin structure and increase the accessibility of basal components of the transcriptional machinery to ERα target genes.
The p160 family of coactivators share significant structural and functional similarity, and all of them can stimulate ERα activity. Functionally, exogenous expression of all of the p160 coactivators stimulates E2-dependent ERα activity in trans-activation assays conducted in HeLa cells (Xu & Li 2003) while small interfering RNA (siRNA) mediated depletion of each SRC coactivator decreases ERα activity in MCF-7 breast cancer cells (Karmakar et al. 2009). Likewise, all three p160 coactivators positively regulate E2-dependent expression of the endogenous ERα target gene, pS2 (Labhart et al. 2005). However, the distinct phenotypes of SRC coactivator knockout mice suggest that these molecules are not redundant (Karmakar et al. 2009). Consistent with this, siRNA depletion of individual SRC coactivators reveals SRC-specific effects in the regulation of endogenous ER target genes other than pS2 such as progesterone receptor and Bcl-2 in MCF-7 cells (Karmakar et al. 2009). The SRCs also exhibit differential regulation of nuclear receptor transcriptional activity in a MMTV model system where SRC-1 preferentially activated progesterone receptor (PR) and SRC-2 showed preference for glucocorticoid receptor (GR) (Li et al. 2003). There are also instances where SRC-2 but not SRC-1 or SRC-3 is required for the repressive effects of GR and ER on ligand down-regulated genes (Rogatsky et al. 2002; Cvoro et al. 2006). Thus, there is increasing evidence that the p160 coactivators can play distinct as well as overlapping roles depending on the biological context.
The p160 family of coactivators also has been implicated as mediators of endocrine resistance in breast cancer. Initial studies on SRC-3 overexpression in transient transfection assays suggested that elevated expression of this coactivator increases the agonistic activity of ER and reduces the growth inhibitory activity of tamoxifen (Smith et al. 1997). Subsequently, two members of the p160 family of coactivators (SRC-1 and SRC-3) have been demonstrated to be involved in tamoxifen resistance in breast cancer patients. High expression of SRC-3 in conjunction with erbB2 overexpression is correlated with poor disease-free and overall survival (Osborne et al. 2003). There is also evidence indicating an association between SRC-3 protein expression and breast tumor recurrence in erbB2 positive breast tumors (Fleming et al. 2004). In cell culture studies, depletion of SRC-3 from a tamoxifen resistant, erbB2-positive breast cancer cell line BT474 increased tamoxifen sensitivity (Su et al. 2008). Collectively, these studies indicate that SRC-3, perhaps via crosstalk with the HER2 signaling pathway, blocks the antagonistic activity of tamoxifen. Although the involvement of SRC-1 in tamoxifen resistance is less well documented than for SRC-3, examination of SRC-1 expression in ER/PR-positive breast tumors of patients that have undergone chemotherapy and tamoxifen adjuvant treatment revealed that SRC-1 expression was associated with disease recurrence in a subgroup patients whose tumors were also HER2-positive (Fleming et al. 2004). SRC-1 has been found to be an independent predictor of disease recurrence, irrespective of endocrine treatment (Redmond et al. 2009), and there is cell-based evidence that increased expression of SRC-1 results in tamoxifen agonism (Shang & Brown 2002; Smith et al. 1997). Thus, the collective, available data suggest that expression of SRC-1 and SRC-3 are associated with resistance to tamoxifen.
In a previous study, we demonstrated that p160 coactivators play unique roles in the growth of ligand-responsive MCF-7 breast cancer cells (Karmakar et al. 2009). However, the role of this coactivator family in proliferation of ERα positive but ligand-insensitive (i.e. E2 or 4HT) breast cancer cells is unknown. This is important because multiple lines of evidence supports an association between SRC-1 and SRC-3 expression and tamoxifen resistance of ERα-positive breast tumors, but to date no studies directly compare the relative contribution of SRC-3 and the two other p160 coactivators to growth of tamoxifen-resistant breast cancer cells. In this study experiments were initiated to examine the contribution of each individual member of the p160 coactivator family in proliferation and survival of an estrogen-independent and tamoxifen-resistant breast cancer cell line, LCC2. This line was generated by a multi-step process beginning with a selection for MCF-7 cell growth in ovariectomized nude mice; the estrogen-independent cell line derived thereof was called LCC1 (Brunner et al. 1993a). These cells were then subjected to stepwise selection in increasing concentrations of 4HT to produce the LCC2 cell line that grows independently of estrogen and is resistant to growth inhibition by 4HT in vitro and in vivo (Brunner et al. 1993b). Importantly, LCC2 cells express levels of ERα comparable to MCF-7 cells and growth of this cell line is sensitive to ICI compounds indicating that they still require functional ER responses (Brunner et al. 1993b). Experiments presented herein suggest that the SRC-2 and SRC-3 coactivators exert a blend of ligand-dependent and ligand-independent effects in the regulation of cell growth and apoptosis of LCC2 cells. However, sensitivity of cell proliferation to tamoxifen is not re-acquired by siRNA-mediated depletion of any of the p160 coactivators in these cells. Our data demonstrates that each of the coactivators differentially contributes to ERα activity and plays distinctive roles in regulating the proliferation and survival of estrogen-independent and tamoxifen-resistant LCC2 breast cancer cells.
MATERIALS AND METHODS
Chemicals, siRNAs and Plasmids
17β-estradiol (E2) and the partial anti estrogen, 4-hydroxytamoxifen (4HT) were obtained from Sigma Chemical Company (St. Louis, MO). The pure antiestrogen ICI 182,780 (ICI) was obtained from Tocris (Ellisville, MO). All siRNAs were chemically synthesized by Ambion (Austin, TX) as oligonucleotide duplexes, described earlier (Karmakar et al. 2009). A sequence targeting luciferase was used as the non-specific siRNA control in all experiments except for those employing luciferase reporter genes which used Ambion’s Silencer #2 as the negative control. The expression plasmids pCR3.1-ERα, pCR3.1-PRB and corresponding reporter genes ERE-E1b-Luc and PRE-E1b-Luc, respectively, have been described previously (Nawaz et al. 1999). The pEGFP-SRC3-AAA mutant expression vector was made by substituting a ClaI to XbaI cassette from pCMV-Tag2B-SRC3-AAA in which all three LXXLL motifs have been mutated to LXXAA (Zheng et al. 2005) into the wild type pEGFP-SRC-3 plasmid (Amazit et al. 2007). The control pEGFP plasmid is from Clontech (Clontech Laboratories Inc, Mountain View, CA).
Cell Culture and Growth assays
The LCC2 cell line, a kind gift of Dr. Robert Clarke, Georgetown University, was maintained in phenol red-free Improved Modified Eagle’s medium (IMEM) with 5% charcoal-stripped FBS (sFBS). MCF-7 human breast cancer cells were obtained from American Type Cell Culture (ATCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). For six day growth assays, 25,000 LCC2 cells and 100,000 MCF-7 cells were plated in each well of six-well plate and grown overnight in IMEM containing 5% sFBS and DMEM containing 10% FBS, respectively. The next day, MCF-7 cells were washed with Hanks’ Balanced Salt Solution (HBSS) and fed with DMEM containing 10% sFBS (day 0). Thereafter, cells were treated with VEH (0.1 % ethanol), 1 nM E2 or 100 nM 4HT and allowed to grow for another six days with media plus hormone replacement at day 4. Cells were harvested on day 0, 4 and 6 from parallel set of cultures with 0.25% Trypsin-EDTA (Invitrogen), and cell number was determined with a Beckman Z1 Dual Coulter Counter (Beckman Coulter, Fullerton, CA). To determine the effect of depletion of SRCs on cell growth, one day prior to transfection, 2 × 105 LCC2 cells were plated into each well of a six-well multiplate and grown overnight in IMEM containing 5% sFBS. The next day, cells were transfected with 10 pmol siRNA targeting luciferase (siControl), SRC-1 (5 pmol siSRC-1a and 5 pmol siSRC-1b; siSRC-1), SRC-2 (siSRC-2) or SRC-3 (siSRC-3) using Oligofectamine reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). After four hours, cells were treated with VEH (0.1 % ethanol), 1 nM E2, 100 nM 4HT, or 100 nM ICI. Two days later, media was replaced and fresh hormone was added. On day 5 post-transfection, cells were harvested and counted. In addition, cells were also harvested in parallel and whole cell lysates were prepared and assessed by Western blot to verify the reduction of coactivator protein levels by siRNA treatments. To determine the effect of overexpression of SRC-3 (wild-type or mutated) on cell growth, one day prior to transfection, 2 × 105 LCC2 cells were plated into each well of a six-well multiplate and grown overnight in IMEM containing 5% sFBS. The next day, cells were transfected with 1 µg control vector (pEGFP) or vector harboring wild-type (wt, pEGFP-SRC3) or mutated (mt, pEGFP-SRC3-AAA) SRC-3 using Fugene 6 reagent according to the manufacturer’s protocol (Roche, Indianapolis, IN). Four to five hours post transfection, the cells were fed with phenol-red free IMEM supplemented with 5% sFBS and on day 5 post-transfection, cells were harvested and counted.
Cell Cycle Distribution and Apoptosis Assays
LCC2 cells were plated at a density of 2 × 105 cells/well of 6-well plate in IMEM without phenol red supplemented with 5% stripped serum. The next day (day 0), the cells were transfected with 10 pmol siRNA/100 mm dish (siControl, siSRC-1, siSRC-2 or siSRC-3) using Oligofectamine. Four to five hours post transfection, the cells were fed with phenol-red free IMEM supplemented with 5% sFBS and incubated for an additional 48 h followed by treatment with VEH (0.1% ethanol), 1 nM E2, 100 nM 4HT or 100 nM ICI for 24 h. Thereafter, adherent cells were collected for flow cytometric analysis and both adherent cells and cell supernatant were collected for apoptosis assay (see below).
Analysis of Cell Cycle Distribution by Flow Cytometry
Adherent cells were collected, washed once with PBS, resuspended in 0.9% NaCl and fixed with 100% ethanol. The cells were then washed again with PBS, resuspended in PBS containing 50 µg/ml propidium iodide (Sigma) and incubated with RNase A (Worthington Biochemical Corporation, Lakewood, NJ) for 30 min at 37 °C. Samples were analyzed on a Beckman-Coulter EPICS XL-MCLs and the resulting data was examined using WinMDI free flow cytometric analysis program (Scripps Research Institute, La Jolla, CA).
Analysis of Apoptosis by Cell Death ELISA
Apoptosis was detected by following the protocol for the Cell Death ELISA kit from Roche Applied Sciences (Indianapolis, IN). Briefly, cells were collected, appropriately diluted and incubated on ice for 20–30 min for cell lysis. Following centrifugation, supernatants were stored at −20 °C until assay. Samples were then assessed for levels of histone-associated DNA fragments by ELISA assay. Absorbance was measured at 405 nm with an Original MultiSkan PLUS plate reader (Thermo Electron Corporation, Milford, MA).
Trans-activation assays for ER and PR activity
Three × 105 LCC2 cells were plated into each well of a six-well multiplate and grown overnight in IMEM containing 5% sFBS. The following day, siSRC-1, siSRC-2 or siSRC-3 were transfected as described above and Ambion’s Silencer #2 was used as a negative control (siControl). Twenty-four hours thereafter, cells were transfected with 1 µg ERE-E1b-LUC reporter construct (ER trans-activation assay) or 50 ng PRB plasmid and 1 µg PRE-E1b-LUC reporter construct (PR trans-activation assay) using TransIT®-LT1 following the manufacturer’s instructions (Mirus, Madison, WI), and one day later, cells were treated with VEH (0.1% ethanol) or hormone (1 nM E2 or 100 nM P4, as appropriate) for an additional 24 h in phenol red-free IMEM containing 5% sFBS. Duplicate wells were harvested, and cell extracts were prepared for luciferase activity using the Luciferase Assay System Kit (Promega, Madison, WI) and a Luminoskan Ascent Thermo Labsystems (Thermo Electron Corporation). Relative luciferase units (RLU) were normalized to total cellular protein measured by Bio-Rad protein assay (Bio-Rad, Hercules, CA). A third well was harvested in parallel from which whole cell lysates were extracted to verify coactivator down-regulation by western blot analysis.
Reverse transcription and quantitative real-time PCR
For monitoring endogenous ER-responsive genes, LCC2 cells were plated in phenol red-free IMEM with 5% sFBS. Cells were transfected with the specified siRNAs and placed in phenol red-free IMEM with 5% sFBS. Forty-eight hours later they were treated with VEH (0.1% ethanol) or 1 nM E2 for 24 h and harvested for RNA isolation using Trizol (Invitrogen). RNA was reverse transcribed with random primers and SuperScript II reverse transcriptase according to Invitrogen’s protocol. Reactions for quantitative PCRs (qPCRs) were conducted using TaqMan (PR and 18S) or Power SYBR Green (pS2) PCR master mixes (Applied Biosystems, Foster City, CA), and samples were amplified with the ABI Prism 7500 sequence detector (Applied Biosystems). Primers for pS2 (Labhart et al. 2005) and primer and probe sequences for PR have been published (Peterson et al. 2007). All mRNA quantities were normalized against 18S RNA using Eukaryotic 18S rRNA endogenous control reagent (Applied Biosystems).
Western Blot Analyses
Western blot analyses were performed following the protocol as described earlier (Karmakar et al. 2009). Briefly, equal amounts of protein from cell lysates were resolved by SDS-PAGE (using Invitrogen’s precast 3–8% Tris-acetate gels), electro-transferred to nitrocellulose membrane and probed with primary antibodies (see below) and the appropriate HRP-conjugated secondary antibody. Signals were detected by chemiluminescence using ECL Plus (GE healthcare) with XO-1 blue film (Kodak; Branchburg, NJ). The primary antibodies are as follows: anti-ERα (HC-20, Santa Cruz Biotechnology), anti-ERβ (PAI-311, Pierce Biotechnology, Rockford, IL) anti-SRC-1 (612378, BD Transduction Laboratories), anti- SRC-2 (610985, BD Transduction Laboratories), anti-SRC-3 (611095, BD Transduction Laboratories), anti-PR (1294; a kind gift from Dr. Dean Edwards, Baylor College of Medicine) and anti-actin (MAB1501R; Chemicon, Temecula, CA). Recombinant ERβ (human long form) was purchased from Invitrogen.
RESULTS
The work of several investigators have implicated elevated expression of SRC-3 in conjunction with erbB2 as molecular events that enable breast cancer cells to grow in the presence of tamoxifen (Osborne et al. 2003; Lahusen et al. 2007). The overall objective of this study was to determine the role of each of the p160 family coactivators in the growth of the LCC2 tamoxifen resistant breast cancer cell line. This cell line was derived from MCF-7 cells via a multistep process such that their growth was estrogen independent and tamoxifen insensitive. However, like MCF-7 cells, LCC2 cells express ERα and retain sensitivity to ICI 182,780 and ICI 164,384 (Brunner et al. 1993b; Coopman et al. 1994) indicating that they require functional ERα responses. The proliferative response of LCC2 and MCF-7 cells to ER ligands was compared in our laboratory by a growth assay. As shown in Figure 1, the proliferation profile of LCC2 cells differ from that of tamoxifen-sensitive MCF-7 cells. Under basal conditions, MCF-7 cell number increased ~3-fold from day 0 to day 6. As expected, in comparison to vehicle-treated controls, E2 significantly increased proliferation, both after 4 and 6 days of treatment indicating that MCF-7 cells are strongly growth stimulated by E2 treatment, while tamoxifen showed partial agonistic activity (Fig. 1A). In contrast, cell number for basal LCC2 growth conditions increased by ~17-fold from day 0 to day 6 and only mild estrogenic growth stimulation was observed with respect to the vehicle controls on days 4 and 6 (Fig. 1B). Importantly, proliferation of these cells was unaffected by tamoxifen on days 4 or 6 compared to the respective vehicle-treated control cells indicating that the robust ligand-independent growth of LCC2 cells was insensitive to tamoxifen treatment. Thus, the proliferation of these LCC2 cells is weakly estrogen sensitive and tamoxifen resistant. To test the possibility that the disparity in growth pattern between these to cell lines could be due to differential expression of p160 coactivators or estrogen receptors (α or β), their levels were examined in MCF-7 and LCC2 cells by Western blotting. Comparable levels of each of the SRC coactivators (Fig. 1C) and the 66 kDA band of ERα (Fig. 1D) were found for each cell type while ERβ expression was undetectable. This indicates that the hormonal insensitivity of the proliferation of LCC2 cells is not due to variation in expression of these factors.
Figure 1. Comparison of cell proliferation and expression of SRC coactivators and estrogen receptors in MCF-7 and LCC2 breast cancer cells.
MCF-7 (A) and LCC2 (B) cells cultured in phenol red-free DMEM containing 10% sFBS and phenol red-free IMEM containing 5% sFBS, respectively, were treated with Veh (0.1% ethanol), E2 (1 nM) or 4HT (100 nM) and allowed to grow for six days. Cells were harvested from parallel sets of cultures and counted on day 0 (on the day of hormone addition), day 4 and day 6. The experiment was repeated three times in triplicate and data were plotted as relative change of cell number with time, setting the number corresponding to E2 treated cells on day 6 as 100. Data are reported as the average ± SEM. Statistical analyses by student’s t-test indicate *p<0.05, **p<0.001 and ap=0.052 versus each group’s vehicle control. Expression of (C) SRC coactivators and (D) ERα and ERβ were assessed for each cell type by Western blot. Actin was used as a loading control and recombinant ERβ (rERβ) was employed in panel D as a positive control. Expression analyses were conducted three times and representative blots are shown.
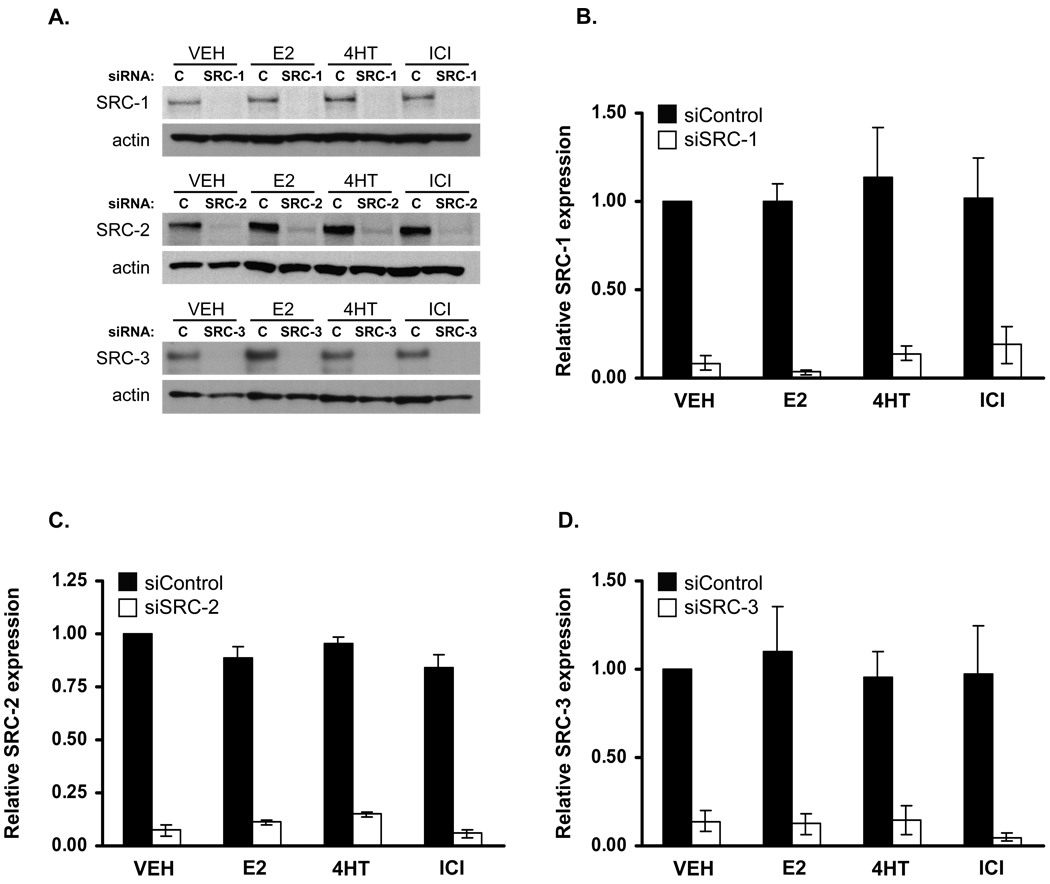
In order to test the importance of the SRC-3 coactivator for growth of the LCC2 breast cancer cells in comparison to the other two members of the p160 coactivator family, SRC-1 and SRC-2, the expression of each coactivator was depleted by siRNA. Our previous work in MCF-7 cells demonstrated that these siRNAs were specific for their respective targets and reduced coactivator expression within 24h of transfection (Karmakar et al. 2009). To test the effectiveness of the siRNAs in LCC2 cells, they were transfected with either control siRNA or siRNA directed against SRC-1, SRC-2 or SRC-3 followed by treatment with either vehicle, 1 nM E2, 100 nM 4HT or 100 nM ICI for 5 days, and harvested for Western blot analyses. Each of the p160 coactivators siRNAs effectively down regulated their respective targets, and this down regulation was maintained for at least 5 days after transfection indicating that SRC depletion could be maintained for a sufficient period of time to conduct cell proliferation assays (Fig. 2). The expression of the SRC coactivators was not significantly impacted by treatment with either E2 or antiestrogens.
Figure 2. Efficient silencing of p160 family coactivators five days after siRNA transfection.
LCC2 cells were transfected with 10 pmol of control siRNA or siRNA directed against SRC-1, SRC-2 or SRC-3 in each well of six well plates. Four hours after transfection, cells were treated with VEH (0.1% ethanol), E2 (1 nM), 4HT (100 nM) or ICI (100 nM) and allowed to grow for five days. Thereafter, cells were harvested, lysed and total proteins were resolved by SDS-PAGE followed by Western blotting for SRC coactivators and actin. (B – D) Blots from two independent experiments were quantitated by densitometry and normalized to actin. Bars represent the average ± the range of individual values.
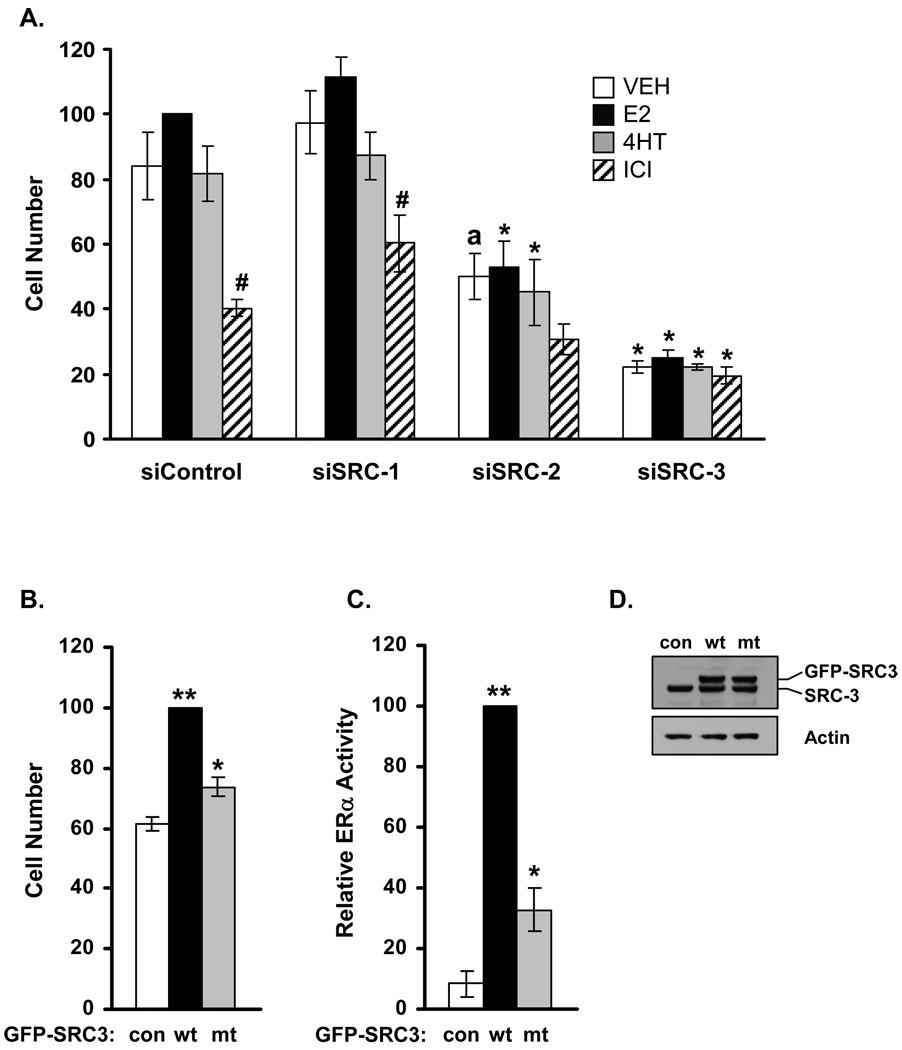
Having established the ability of siRNAs to suppress SRC coactivator expression for at least five days, the effect of SRC depletion on LCC2 cell proliferation was assessed. As expected, 4HT did not affect the growth of the tamoxifen-resistant LCC2 cell line transfected with the control siRNA while E2 treatment modestly increased and ICI significantly reduced cell number compared to that observed for vehicle treated controls (Fig. 3A). Depletion of SRC-1 slightly increased LCC2 cell proliferation both in the presence or absence of estrogen; this effect was not significant. Inhibition of the proliferation of SRC-1 depleted LCC2 cells by ICI indicated that these cells retained ERα-dependent, but ligand-independent growth. In contrast, depletion of either SRC-2 or SRC-3 inhibited the growth of LCC2 cells. The growth of E2 or 4HT treated cultures of SRC-2 depleted LCC2 cells were significantly reduced in comparison to siControl cultures. The difference between control and SRC-2 depleted vehicle-treated cells approached significance (p=0.053) suggesting that loss of SRC-2 expression reduced the ligand-independent growth of vehicle-treated cells. It seems likely that growth of SRC-2 depleted cells retains some requirement for ERα as there was a trend (p=0.090) towards reduced cell number in ICI-treated cells. Depletion of SRC-3 produced the strongest inhibitory effect on LCC2 growth, with comparable cell numbers for vehicle and each of the ligand-treated cultures. Growth inhibition by ICI was not apparent for SRC-3 depleted cultures indicating that the ligand-independent and ERα-dependent growth of LCC2 cells is largely reliant on SRC-3 expression. Moreover, the cell number for these cultures was lower than for ICI-treated, control siRNA cells suggestive that growth inhibition associated with SRC-3 depletion reflects more than loss of ERα function.
Figure 3. Regulation of LCC2 cell proliferation by SRC family coactivators.
(A) LCC2 cells were transfected with siRNAs as indicated and 24 h post-transfection, cells were treated with VEH (0.1% ethanol), E2 (1 nM), 4HT (100 nM) or ICI (100 nM) for five days and counted. Data represent the average ± SEM (n=3) and are expressed as relative change in cell number, setting the number corresponding to the E2-treated siControl group as 100. Statistical analyses were done by students’ t-test where ‘*’ indicates p<0.05 and ‘a’ indicates p=0.053 versus the respective treatment (VEH, E2, 4HT or ICI) group in the siControl cells; ‘#’ indicates p<0.05 and ‘b’ indicates p=0.090 versus the vehicle control within the group. (B) LCC2 cells were transfected with plasmids for GFP control (con), wild-type GFP-SRC-3 (wt) or a GFP-SRC-3 mutant (mt) in which each of the 3 receptor interacting motifs of SRC3 were mutated (LXXLL→LXXAA), allowed to grow for five days and counted. Results are plotted as relative cell number, setting the value for wild type SRC-3 transfected cells as 100 (n=4). *, p < 0.05; or **, p < 0.001 vs. vector control group. (C) Transcriptional activity of wild-type and mutated GFP-SRC-3 in comparison to control GFP plasmid was determined in a trans-activation assay in HeLa cells co-transfected with ERα and ERE-e1b-Luc. Data represent the average ± SEM of three experiments. *, P < 0.05; or **, P < 0.001 vs. vector control group. (D) LCC2 cells transfected as in panel B were assessed for expression of wt SRC-3 and GFP-SRC3 by Western blot using antibody specific for SRC-3. Actin was employed as a loading control.
To further test the role of SRC-3 in mediating growth, LCC2 cells were transfected with expression vectors for either wild type SRC-3 or a SRC-3 mutant in which all three LXXLL motifs utilized for binding to ERα were mutated. Growth assays revealed a strong increase in LCC2 cell number induced by wild type SRC-3 while the SRC-3-AAA mutant induced a significant but much lower increase in LCC2 cell proliferation (Fig. 3B). These results confirm the ability of SRC-3 to promote ligand-independent growth of LCC2 cells and indicate that a portion of this simulation is independent of LXXLL-mediated SRC-3 binding to ERα. Coactivators were GFP-tagged in this experiment in order to assess the relative levels of exogenous wild type versus mutant SRC-3 which were found to be similar by Western blot analyses (Fig. 3D). Trans-activation assays confirmed the ability of SRC-3 to stimulate ERα transcriptional activity was largely dependent on intact LXXLL motifs, however the weaker activity of the SRC-3 mutant indicated a modest ability to enhance ERα function independent of these interaction sites (Fig. 3C) potentially via the receptor’s AF-1 domain (Webb et al. 1998; Dutertre & Smith 2003). Thus the ability of the mutant SRC-3 to promote LCC2 proliferation reflects an LXXLL-motif independent effect mediated via ERα and/or other transcription factors.
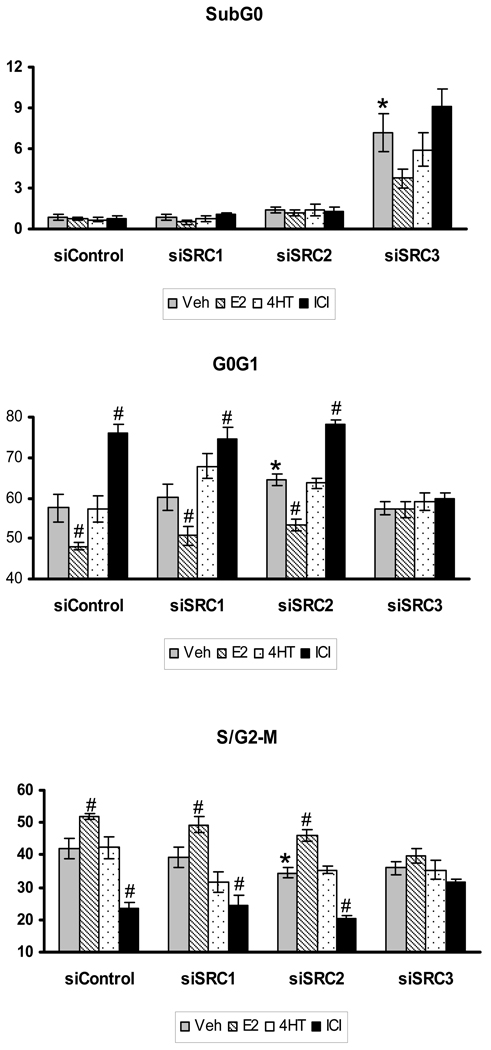
To further investigate the impact of SRC coactivator depletion on LCC2 cell proliferation, the distribution of p160-depleted cells throughout the cell cycle was evaluated by flow-cytometric analyses. In vehicle-treated cultures, 3 d after transfection with control siRNA, cells are found primarily in the G0G1 (57.48 ± 3.31%) and S/G2-M (42.06 ± 3.20%) phases of cell cycle, with a small population of cells (0.88 ± 0.19%) in the sub-G0G1 phase (Fig. 4). Treatment with E2 for 24 h produced an increase in the percentage of cells in S/G2-M at the expense of a corresponding decrease of cells in the G0G1 phase. In contrast, 24 h of ICI treatment shifted the distribution of cells from the S/G2-M to the G0G1 phase of cell cycle, consistent with this antiestrogen’s strong growth inhibitory effect. It was noted that the ICI and in particular E2 effects on cell cycle distribution were more prominent than those observed for the 5d growth assay described above, and this likely reflects that these ligand treatments were applied to cells grown under conditions that would synchronize MCF-7 cells (e.g. reduced serum concentrations) for only 24 h. Treatment with 4HT did not alter cell cycle distribution in comparison to vehicle-treated cells and this demonstrates the absence of a growth regulatory effect of this antiestrogen in LCC2 cells, even under assay conditions that emphasize the ability of ligands to regulate cell proliferation.
Figure 4. Down-regulation of SRCs differentially affects the cell cycle distribution of LCC2 cells.
LCC2 cells were subjected to siRNA treatment and 48 h after transfection were treated with VEH (0.1% ethanol), 1 nM estradiol (E2), 100 nM tamoxifen (4HT) or 100 nM ICI for an additional 24 h. Thereafter, cells were collected, fixed and stained with propidium iodide followed by flow-cytometric analysis. Data represents average of four independent experiments ± SEM where ‘#’ indicates p<0.05 versus the corresponding Veh-treated control and ‘*’ indicates significance at p<0.05 versus the siControl vehicle group. Statistical analyses were done by student’s t-test.
Depletion of SRC-1 did not alter the distribution pattern of cells throughout the cycle, consistent with the results obtained for the cell proliferation experiments. Likewise, depletion of SRC-2 did not substantially alter the ability of E2 or ICI to influence the distribution of cells throughout the cycle. Loss of SRC-2 did not enable 4HT to alter cell distribution relative to vehicle treated cells. However, a hormone-independent effect was observed with an ~7% increase in the percentage of cells in G0G1 (to 64.46 ± 1.56%) in vehicle-treated cells relative to the vehicle-treated siControl group and a near doubling of the cells present in the sub-G0G1 phase (1.41 ± 0.26%) that was compensated for by an ~8.5% decrease in the percentage of cells in S/G2-M. Thus, depletion of SRC-2 appears to decrease the basal growth of LCC2 cells with little effect on the ability of the cells to respond to ERα ligands.
Consistent with the cell growth data, depletion of SRC-3 blocked the ability of ER ligands to alter the cell cycle distribution of LCC2. In absence of ligand, the percentage of cells in G0G1 or S/G2-M phases was unaltered by SRC-3 depletion, as compared to that in control siRNA cells. Moreover, none of the tested ER ligands was able to induce a shift in those two phases in SRC-3 depleted cells relative to the vehicle treated control. Depletion of SRC-3 did, however, provoke a >8-fold increase in the percentage of cells in the sub-G0G1 population. Estrogen treatment tended to decrease cells in the sub-G0G1 population (7.13 ± 1.39 to 3.76 ± 0.72%) at the expense of a corresponding increase in the cells in the S/G2-M phase (35.93 ± 2.14 to 39.59 ± 2.20%), but this was not significant (p=0.098 and p=0.299, respectively). Collectively, these results indicate that depletion of SRC-2 and SRC-3 reduces cell proliferation via different mechanisms with loss of SRC-2 working primarily through a hormone-independent shift in the baseline distribution of cells from S/G2-M to the G0G1 phase with retention of ERα activity, while SRC-3 depletion results in a shift of cells into the sub-G0G1 phase and a significant loss of ER ligand control of cell cycle distribution. In no case did SRC coactivator depletion enable 4HT to alter cell cycle distribution in comparison to vehicle-treated cells.
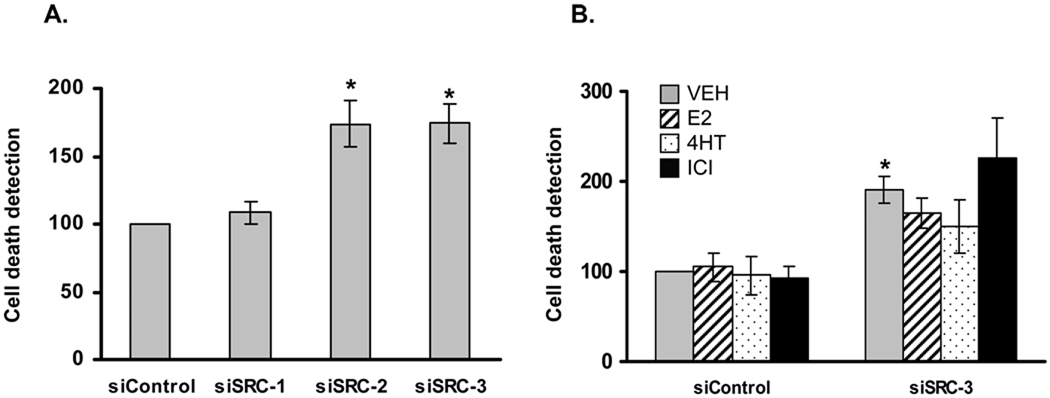
The increase in SRC-2 and SRC-3 depleted cells in the sub-G0G1 population suggested that loss of these coactivators may increase apoptosis. To test independently for this, LCC2 cells were transfected with control and p160 coactivator siRNAs, exposed to vehicle and 24 h thereafter were assessed by a Cell Death ELISA which measures levels of nucleosome-DNA complexes. A significant increase in apoptosis was observed for cultures depleted of SRC-2 or SRC-3 (Fig. 5A). The effect of ligand treatment on SRC-3-depleted cells was also tested by Cell Death ELISA because of the variation in the percentage of subG0 cells detected by the cell cycle distribution assays (Fig. 4). Consistent with the results above, there was no significant difference in the extent of apoptotic cells among the hormone-treated groups (Fig. 5B). These results in conjunction with the cell cycle distribution data indicate that SRC-2 and SRC-3 provide a hormone-independent survival function to LCC2 cells.
Figure 5. Depletion of SRC-2 and SRC-3 but not SRC-1 induces apoptosis in LCC2 cells.
(A) LCC2 cells were subjected to siRNA transfection and then allowed to grow 48 h in phenol-red free IMEM containing 5% stripped serum followed by exposure to VEH (0.1% ethanol) for an additional 24 h. Thereafter cells (both adherent and non-adherent) were collected and assayed by Cell Death ELISA. The experiment was repeated six times and data represent the average ± SEM, setting the number corresponding to the control group as 100. Statistical analyses was done by t-test and indicates *p<0.05 versus siControl cells. (B) LCC2 cells were transfected with control or SRC-3 siRNA as above and 48 h thereafter were treated with VEH (0.1% ethanol), E2 (1 nM), 4HT (100 nM) or ICI (100 nM) for 24 h followed by assessment by Cell Death ELISA. Data represent average ± SEM of three experiments. ‘*’ indicates p<0.05 versus VEH treated siControl cells.
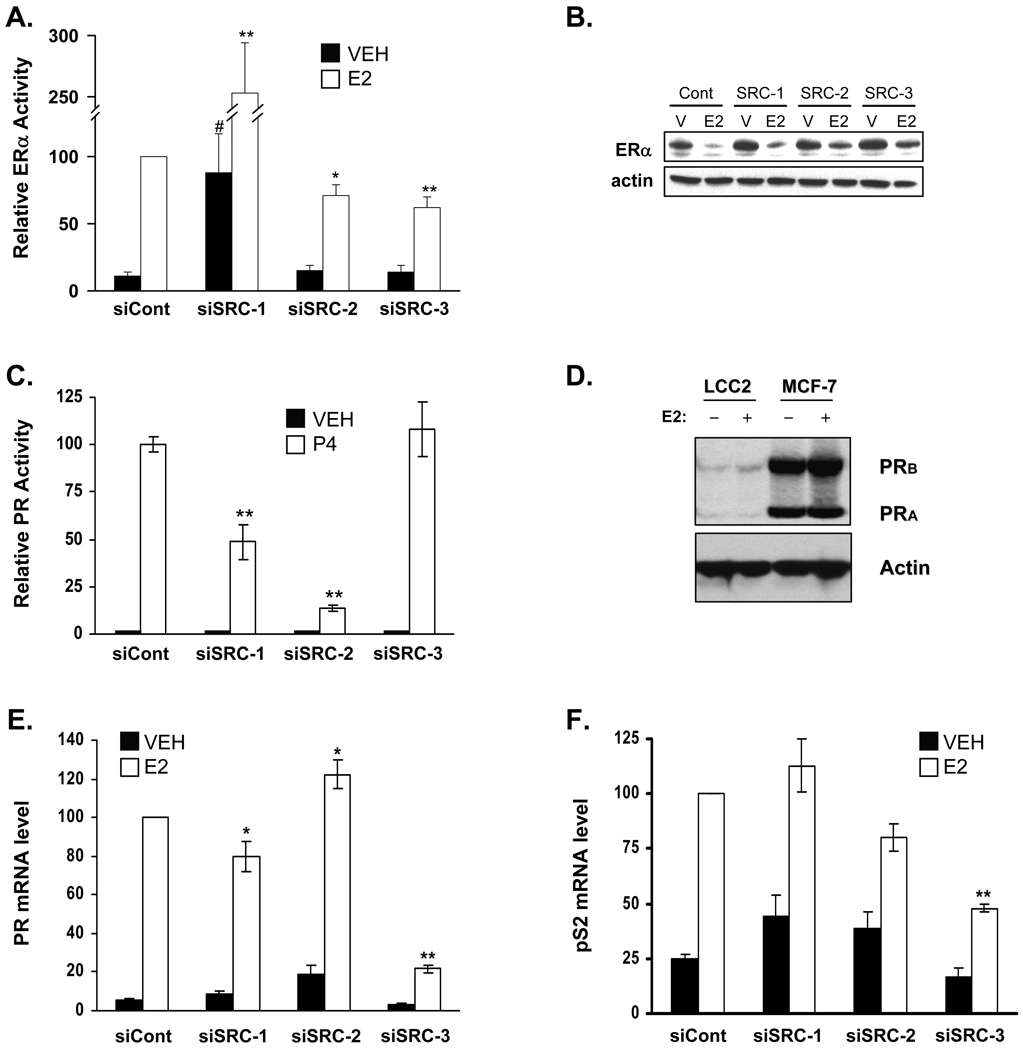
To determine if the impact of SRC coactivator depletion on hormonal regulation of cell proliferation and cell cycle distribution were reflected in changes in ERα transcriptional activity, trans-activation assays were conducted in LCC2 cells with or without p160 coactivator depletion. This enables ERα activity to be assessed largely independently of the other transcription factors that would be present on endogenous ERα target genes. In control cells, E2 treatment significantly increased ER transcriptional activity (Fig. 6A), suggesting that E2-ERα activity measured with an ERE-luciferase reporter gene is operational in LCC2 cells even though the receptor’s transcriptional activity is not reflected in cell growth. The transcriptional activity of ERα was greatly increased in SRC-1 depleted versus siControl cells either in absence of hormone or with estrogen treatment indicating a repressive effect of this p160 coactivator on the expression of the luciferase reporter gene. As for the control cells, there is no correlation between SRC-1 regulation of ERα activity measured by luciferase activity, and the lack of impact of SRC-1 depletion on LCC2 cell growth. In contrast, inhibition of SRC-2 or SRC-3 expression reduced E2-induced ERα activity without altering the receptor’s basal activity which differs from the significantly decreased proliferation of vehicle-treated SRC-2 and SRC-3 depleted cells. Overall, there is little to no correlation on the impact of p160 coactivator depletion between ERα transcriptional activity and regulation of LCC2 cell growth. To ensure that SRC family coactivator depletion did not impact expression of ERα, Western blot analyses of parallel cultures were performed. Depletion of the coactivators did not alter levels of ERα in vehicle-treated cells, and E2 treatment induced down-regulation of ERα expression (Fig. 6B).
Figure 6. ERα and PR transcriptional activity and endogenous ERα target gene expression is differentially altered by down regulation of SRCs in LCC2 cells.
Twenty-four hours after the indicated siRNA transfection, LCC2 cells were further transfected with ERE-e1b-Luc (A) or PRB and PRE-e1b-Luc (C) reporter genes and 24 h thereafter were subjected to treatment with vehicle (VEH; 0.1% ethanol), 1 nM E2 or 100 nM progesterone (P4) for an additional 24 h. Cells were then harvested, lysed and measured for luciferase activity followed by normalization against total protein. The experiment was conducted three times in duplicate and data are expressed as the average ± SEM. Statistical analyses were carried out using student’s t-test, where *p<0.05 and **p<0.001 versus the siControl E2 group and ‘#’ indicates p<0.05 versus siControl vehicle groups. (B) Parallel sets of samples from experiment (A) were assessed by 765 Western blot to determine the impact of E2 treatment on ERα expression. (D) Expression of PRA and PRB in LCC2 and MCF-7 cells treated with VEH or 1 nM E2 for 24 h was determined by Western blot. A representative blot from three experiments is shown. The expression of the ERα target genes PR (panel E) and pS2 (panel F) was assessed by RT-qPCR for LCC2 cells transfected with the indicated siRNA and treated 48 h later with VEH or 1 nM E2 for 24 h. Data are normalized to 18S and E2-treated control siRNA samples were set to 100. Values represent the mean ± SEM of four experiments. *, p < 0.05; or **, p < 0.001 vs. E2 treated control siRNA-transfected cells.
The ability of SRC-1 to repress ERα transcriptional activity was surprising, and therefore to determine if this extended to other steroid receptors, the impact of p160 coactivator depletion was tested for progesterone receptor (PR). The much lower levels of PR in LCC2 than MCF-7 cells (Fig. 6D) prompted us to transfect a PRB expression plasmid along with the luciferase reporter gene for this trans-activation assay. This also avoided the potentially confounding influence of SRC depletion impacting expression of PR, itself an ER target gene. In contrast to ERα, PR activity requires both SRC-1 and SRC-2 and is independent of SRC-3 expression (Fig. 6C). Thus the ability of SRC coactivators to regulate transcriptional activity is dependent on the identity of the steroid receptor. To determine whether the contributions of the individual p160 coactivators to ERα transcriptional activity measured by reporter assay was reflected in the regulation of endogenous genes, the impact of SRC coactivator depletion on the expression of two well-characterized ERα target genes (PR and pS2) was examined (Figs. 6E & F). For both genes, E2-induced expression is significantly decreased by SRC-3 depletion. However, the effect of SRC-1 and SRC-2 siRNA treatment is gene-dependent; pS2 is unaffected by silencing of either coactivator, while PR mRNA expression is down- and up-regulated, respectively.
DISCUSSION
Several independent studies have reported an association of SRC-3 expression with tamoxifen resistance, both in the clinical setting as well as in breast cancer cell lines (Osborne et al. 2003; Fleming et al. 2004; Su et al. 2008). However, the contribution of the two other p160 family members to the growth and survival of tamoxifen resistant breast cancer cells is less well understood. In this study, we used the E2-independent, 4HT-resistant LCC2 cell line which is derived from MCF-7 cells to address the role of each of the p160 coactivators in this tamoxifen-resistant phenotype. Our results demonstrate that loss of SRC-1, SRC-2 or SRC-3 did not significantly alter LCC2 proliferation or cell cycle distribution in 4HT- versus vehicle-treated cells. However, a differential requirement of the various p160 coactivators for growth and survival of LCC2 cells was observed as depletion of SRC-2 and SRC-3 but not SRC-1 decreased cell proliferation. Cell cycle analyses further illustrated the different contributions of SRC-2 and SRC-3 in control of LCC2 proliferation with SRC-2 depletion increasing the percentage of cells in the G0G1 and sub-G0G1 phases of cell cycle yet maintaining sensitivity to E2 and ICI, while SRC-3 depletion resulted in a large increase in sub-G0G1 phase cells and loss of response to estrogen and ICI antiestrogen. Consistent with prior reports (Brunner et al. 1993b), LCC2 cells express ERα and are growth inhibited by ICI antiestrogen indicating a requirement of ERα for growth and survival. It was therefore surprising that the effects of p160 coactivator depletion on ERα transcriptional activity measured by luciferase assay did not correspond to the observed effects on proliferation, thus indicating that mechanisms other than, or in addition to, regulating ERα activity are determinants of SRC coactivator control of basal and hormone/antihormone regulated proliferation.
This study clearly demonstrates that SRC-1 has little to no role in regulating the overall growth of LCC2 cells, and this is consistent with prior in vitro experiments in which SRC-1 had little impact on proliferation of MCF-7 cells (Karmakar et al. 2009). The absence of a role for SRC-1 in breast tumor growth also is observed in a mouse mammary tumor virus-polyoma middle T (PyMT) breast cancer mouse model where tumor formation and growth were similar in wild type versus SRC-1−/− mice, although SRC-1 was found to play an intrinsic role in tumor cell metastasis (Wang et al. 2009), and subsequently shown to regulate the expression of the metastasis regulator, Twist (Qin et al. 2009). The lack of effect of SRC-1 depletion on the growth of tamoxifen-treated cultures was somewhat surprising as SRC-1 expression has been shown to be important for cell cycle progression of tamoxifen-treated Ishikawa endometrial carcinoma cells as well as tamoxifen induction of the c-myc cell cycle regulator (Shang & Brown 2002). However, there were a small but not significant increase and decrease in cells in G0G1 and S-G2/M phases, respectively, in tamoxifen- versus vehicle-treated, SRC-1-depleted cells that were not observed in control cells; these trends suggest that the antagonistic properties of tamoxifen may be slightly enhanced in SRC-1 depleted cells. Some, but not all studies have found a correlation between increased SRC-1 expression and tamoxifen resistance in breast cancer (Myers et al. 2004; Morrison et al. 2003; Sarvilinna et al. 2006), and our results argue that further studies are warranted to evaluate the contribution of SRC-1 to endocrine resistance in breast cancer.
There also was no impact of SRC-1 depletion on apoptosis. In contrast, depletion of SRC-3 provoked a strong increase in the percent of LCC2 cells in the sub-G0G1 population while increases induced by SRC-2 depletion were subtle. The percentage of cells in sub-G0G1 phase was not, however, reflected in the apoptosis assay suggesting that for SRC-3 depleted cultures the majority of cells in the sub-G0G1 phase are not reflective of apoptosis, but rather point toward non-apoptotic mechanisms such as autophagy or necrosis. Nonetheless, our data clearly implicate SRC-2 and SRC-3 as hormone-independent survival factors in these breast cancer cells. We had found previously that depletion of SRC-1 and SRC-3 reduced expression of the anti-apoptotic protein, Bcl-2 in LCC2 cells (Karmakar et al. 2009), and the lack of correlation between reduced Bcl-2 expression and apoptosis for the SRC-1 coactivator depletion indicates that other anti-apoptotic genes or pathways compensate for the loss of Bcl-2.
Depletion of either SRC-2 or SRC-3 reduced cell proliferation. For SRC-2, this was reflected in an increased proportion of cells in G0G1 and an accompanying decrease in the S-G2/M population of cells, independent of ligand. There was little effect on ICI- or E2-dependent shifts in cell cycle distribution suggesting that SRC-2 exerts effects on cell proliferation and prevention of apoptosis that are largely independent of ERα action; estrogen and antiestrogen effects are relatively subtle in the 5 d growth assay because of the strong effect of SRC-2 depletion on basal cell growth. Little is known regarding the mechanisms by which SRC-2 impacts cell proliferation and apoptosis, but one study employing overexpression of this coactivator found increased proliferation and induction of CXCL12 expression, a chemokine that contributes to the survival and proliferation of cancer cells (Kishimoto et al. 2005). In view of the differences in the ability of SRC-2 and SRC-3 depletion to negatively impact breast cancer cell proliferation, it would be interesting to compare the effect of their individual depletion in RNA profiling experiments to determine the extent to which these coactivators overlap and differ in their regulation of gene expression.
Prior studies have shown that knock-down of SRC-2 and SRC-3 reduces ERα activity to a similar extent in luciferase reporter gene assays conducted in MCF-7 cells, while the impact of these depletions on endogenous ERα targets was gene specific (Karmakar et al. 2009). For instance, while some genes are unaffected (e.g. c-Myc) or down-regulated (e.g. pS2) by knock-down of SRC-2 and SRC-3, some genes (e.g. PR, cyclin D1 or Bcl2) are affected differentially by their depletion. Consistent with this, the decrease in E2-induced luciferase activity for SRC-2 and SRC-3 depleted cells was not uniformly mirrored in changes in PR or pS2 mRNA expression in LCC2 cells. Interestingly, the impact of p160 coactivator depletion on E2-induced PR and pS2 gene expression varied between MCF-7 and LCC2 cells, indicating that gene-specific coactivator usage is altered in tamoxifen-sensitive versus resistant cells. For instance, pS2 mRNA expression in MCF-7 cells is sensitive to depletion of each of the p160 coactivators, while in LCC2 cells only SRC-3 depletion reduces this gene’s expression. While this is consistent with the concept that gene expression in hormone-independent breast cancer cells is less dependent on p160 coactivators (Naughton et al. 2007), PR expression is impacted by all three SRC coactivators in LCC2 cells, while only SRC-3 depletion reduces its expression in MCF-7 cells (Karmakar et al. 2009). Moreover, SRC-2 exerts a repressive role in control of PR mRNA levels. This argues against global p160 coactivator-independent gene expression as a feature of hormone-independent breast cancer, and suggests that an understanding of the importance of SRC coactivators to gene expression in hormone-sensitive versus insensitive breast cancer cells requires a genome-wide profiling approach.
In comparison to other SRC family members, the role of SRC-3 in control of cell proliferation has been relatively well characterized. Previous studies by laboratories including ours demonstrated that depletion of SRC-3 reduced MCF-7 cell growth (List et al. 2001; Karmakar et al. 2009) and it has been shown that SRC-3 coactivates both ERα and E2F1 transcriptional activities (Louie et al. 2004). Failure to activate E2F1 transcriptional activity effectively blocks fibroblasts from entering S phase of the cell cycle (Louie et al. 2006) and our prior demonstration that SRC-3 depletion inhibited E2-induction of cyclin D1 expression and progression of MCF-7 cells into S-phase is in accord with a role for SRC-3 in this phase of the cell cycle (Karmakar et al. 2009). Consistent with these prior reports, our experiments demonstrate a role for SRC-3 in G1 to S phase transition in LCC2 cells as only SRC-3 depletion blocked the E2-dependent increase in cells in the S-G2/M phase. LCC2 cells treated with SRC-3 siRNA also lost their responsiveness to ICI treatment, indicating that ERα was unable to regulate basal cell proliferation in SRC-3 depleted cells and thereby highlighting the importance of this coactivator for ligand-independent ERα activity. Moreover, the SRC-3-AAA mutant confirms the ability of SRC-3 to promote the ligand-independent activity of ERα. It also raises the possibility that SRC-3 promotes proliferation via transcription factors other than ERα, and this is consistent with our finding that growth of SRC-3 depleted cells was lower than for ICI-treated control cells.
These studies revealed little to no correlation between the impact of SRC depletion on E2-stimulated ERα transcriptional activity and cell proliferation. Depletion of SRC-2 or SRC-3 had significant effects on cell growth, but no effect on basal and only a modest effect on E2-dependent luciferase reporter gene activity. This would suggest that the primary, p160 coactivator-dependent control of LCC2 proliferation is not via ERα, but rather by other transcription factor(s) whose activity is regulated by SRC-2 and SRC-3. There are a number of other transcription factors that could fulfill this role such as NF-κB, AP-1, STATs, ETS, p53 and E2F1 (Xu et al. 2009), and SRC-3 regulation of proliferation via E2F1 has been described (Louie et al. 2004; Louie et al. 2006). Overexpression of SRC-3 in the mammary gland of transgenic mice is associated with elevated expression of IGF-1 and activation of the PI3K/Akt signaling pathway (Torres-Arzayus et al. 2004), and this raises the possibility of a reduction in the activity of a SRC-3-dependent signaling pathway that is important for basal proliferation of LCC2 cells. Taken together, our data suggests that the ability of SRC-2 and SRC-3 to contribute to LCC2 cell growth reflects the loss of ERα transcriptional activity (e.g. ligand effect on cell cycle) for SRC-3 depleted cells as well as loss of the activity of other transcription factors and/or signal transduction pathways that control the ligand-independent growth of these cells.
Prior experiments with estrogen- and tamoxifen-sensitive MCF-7 cells revealed that depletion of SRC-1 had little effect on ERα transcriptional activity while loss of SRC-2 and SRC-3 reduced ERα transcriptional activity by at least 55% as measured by luciferase reporter gene assay (Karmakar et al. 2009). In contrast, the impact of depleting SRC-2 or SRC-3 on ERα transcriptional activity in LCC2 cells, measured herein by the same methodology, is relatively modest thereby indicating that the receptor’s transcriptional activity has become less dependent on these coactivators. A more dramatic difference is found in comparing the impact of SRC-1 depletion between the two cells types; SRC-1 has little influence on ERα-dependent ERE reporter gene expression in MCF-7 cells but is repressive in LCC2 cells. In the latter cell type, the unchanged pS2 expression and mild reduction in E2-induced PR mRNA levels in SRC-1 depleted LCC2 cells indicates that SRC-1 control of transcription also is target-specific. A contrast for reporter versus target gene expression also is observed for SRC-2 which stimulates luciferase levels but inhibits E2-induction of PR mRNA. Taken together, these data indicate that the acquisition of estrogen-insensitivity and/or tamoxifen resistance in LCC2 cells is accompanied by alterations in the ability of SRC family coactivators to regulate ERα transcriptional activity. It is unclear how this activity becomes less dependent on SRC coactivators in these endocrine resistant cells or if other coactivators become more important for regulation of ERα. There are differences in cellular responses to the cAMP-inducing agents, forskolin and IBMX in MCF-7 versus LCC2 cells (Al-Dhaheri & Rowan 2006) as well as TGFβ expression (Arteaga et al. 1999). The activity of both ERα (Murphy et al. 2009) as well as SRC family coactivators (Xu et al. 2009) can be altered by cell signaling pathways (e.g. cAMP, MAPKs, etc.) and cell signaling can modulate physical interactions between ERα and p160 coactivators, particularly under ligand-independent conditions (Dutertre & Smith 2003). This raises the possibility that differences in signaling between MCF-7 and LCC2 cells may reduce functional relationships between ERα and p160 coactivators while promoting ERα’s utilization of non-p160 coactivators.
In conclusion, we have demonstrated that SRC-2 and SRC-3 but not SRC-1 controls the growth of the estrogen-independent and tamoxifen-resistant LCC2 breast cancer cell line, largely through control of basal cell growth, and that there is a poor correlation between the ability of individual p160 coactivators to control ERα transcriptional activity and regulate LCC2 proliferation. The shift in coactivator regulation of ERα activity relative to proliferation may represent a key step in acquisition of an estrogen-independent and/or tamoxifen-resistant breast cancer cell growth phenotype, and suggests that targeting growth inhibition via SRC-2 and SRC-3 may be a more effective approach to inhibit the growth of tamoxifen resistant breast cancer.
Acknowledgements
Funding: This work was supported by the National Institutes of Health (DK53002 and DK64038; CLS), the Department of Defense (W81XWH-04-1-0424; CLS) and Susan G. Komen for the Cure Foundation (PDF0707868; SK).
We thank Patricia Dillard, Javier Pacheco and Judy Roscoe for technical assistance.
Footnotes
Publisher's Disclaimer: Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in Endocrine-related Cancer, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the Society for Endocrinology accepts no responsibility for any errors or omissions it may contain. The definitive version is now freely available at http://dx.doi.org/10.1677/ERC-09-0285.
Declaration of interest: The authors have no conflict of interest.
REFERENCES
- Al-Dhaheri MH, Rowan BG. Protein kinase A exhibits selective modulation of estradiol dependent transcription in breast cancer cells that is associated with decreased ligand binding, altered estrogen receptor alpha promoter interaction and changes in receptor phosphorylation. Mol Endocrinol. 2006;21:439–456. doi: 10.1210/me.2006-0059. [DOI] [PubMed] [Google Scholar]
- Amazit L, Pasini L, Szafran AT, Berno V, Wu RC, Mielke M, Jones ED, Mancini MG, Hinojos CA, O'Malley BW, Mancini MA. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol Cell Biol. 2007;27:6913–6932. doi: 10.1128/MCB.01695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga CL, Koli KM, Dugger TC, Clarke R. Reversal of tamoxifen resistance of human breast carcinomas in vivo by neuralizing antibodies to transforming growth factor-β. J Natl Cancer Inst. 1999;91:46–53. doi: 10.1093/jnci/91.1.46. [DOI] [PubMed] [Google Scholar]
- Brunner N, Boulay V, Fojo A, Freter CE, Lippman ME, Clarke R. Acquisition of hormone-independent growth in MCF-7 cells is accompanied by increased expression of estrogen-regulated genes but without detectable DNA amplifications. Cancer Research. 1993a;53:283–290. [PubMed] [Google Scholar]
- Brunner N, Frandsen TL, Holst-Hansen C, Bei M, Thompson EW, Wakeling AE, Lippman ME, Clarke R. MCF7/LCC2: a 4-hydroxytamoxifen resistant human breast cancer variant that retains sensitivity to the steroidal antiestrogen ICI 182,780. Cancer Research. 1993b;53:3229–3232. [PubMed] [Google Scholar]
- Coopman P, Garcia M, Brunner N, Derocq D, Clarke R, Rochefort H. Anti-proliferative and anti-estrogenic effects of ICI 164,384 and ICI 182,780 in 4-OH-tamoxifen-resistant human breast-cancer cells. Int J Cancer. 1994;56:295–300. doi: 10.1002/ijc.2910560225. [DOI] [PubMed] [Google Scholar]
- Cvoro A, Tzagarakis-Foster C, Tatomer D, Paruthiyli S, Fox MS, Leitman DC. Distinct roles of unliganded estrogen receptors in transcriptional repression. Mol Cell. 2006;21:555–564. doi: 10.1016/j.molcel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Dutertre M, Smith CL. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-α: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol Endocrinol. 2003;17:1296–1314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- Fleming FJ, Myers E, Kelly G, Crotty TB, O'Higgins NJ, Hill AD, Young LS. Expression of SRC-1, AIB1 and PEA3 in HER2 mediated endocrine resistant breast cancer: a predictive role for SRC-1. J Clin Pathol. 2004;57:1069–1074. doi: 10.1136/jcp.2004.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar S, Foster EA, Smith CL. Unique roles of p160 coactivators for regulation of breast cancer cell proliferation and estrogen receptor-α transcriptional activity. Endocrinology. 2009;150:1588–1596. doi: 10.1210/en.2008-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto H, Wang Z, Bhat-Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1alpha/CXCL12. Carcinogenesis. 2005;26:1706–1715. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- Labhart P, Karmakar S, Salicru EM, Egan BS, Alexiadis V, O'Malley BW, Smith CL. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci USA. 2005;102:1339–1344. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahusen T, Fereshteh M, Oh A, Wellstein A, Riegel AT. Epidermal growth factor receptor tyrosine phosphorylation and signaling controlled by a nuclear receptor coactivator, amplified in breast cancer 1. Cancer Res. 2007;67:7256–7265. doi: 10.1158/0008-5472.CAN-07-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai M-J, O'Malley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List H-J, Lauritsen KJ, Reiter R, Powers C, Wellstein A, Riegel AT. Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells. Journal of Biological Chemistry. 2001;276:23763–23768. doi: 10.1074/jbc.M102397200. [DOI] [PubMed] [Google Scholar]
- Louie MC, Revenko AS, Zou JX, Yao J, Chen H-W. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol Cell Biol. 2006;26:3810–3823. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen H-W. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Herrick RE, Heinsohn EC, Schiff R, Osborne CK. Dominant-negative nuclear receptor corepressor relieves transcriptional inhibition of retinoic acid receptor but does not alter the agonist/antagonist activities of the tamoxifen-bound estrogen receptor. Mol Endocrinol. 2003;17:1543–1554. doi: 10.1210/me.2001-0144. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Skliris GP, Rowan BG, Al-Dhaheri M, Williams C, Penner C, Troup S, Begic S, Parisien M, Watson PH. The relevance of phosphorylated forms of estrogen receptor in human breast cancer in vivo. J Steroid Biochem Mol Biol. 2009;114:90–95. doi: 10.1016/j.jsbmb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Myers E, Fleming FJ, Crotty TB, Kelly G, McDermott EW, O'Higgins NJ, Hill AD, Young LS. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br J Cancer. 2004;91:1687–1693. doi: 10.1038/sj.bjc.6602156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Macleod K, Kuske B, Clarke R, Cameron DA, Langdon SP. Progressive loss of estrogen receptor α (ERα) cofactor recruitment in endocrine resistance. Molecular Endocrinology. 2007;21:2615–2626. doi: 10.1210/me.2007-0110. [DOI] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai M-J, O'Malley BW. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S, Mäkelä S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson J-Å. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SAW, Wong J, Allred DC, Clark GM, Schiff R. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL. The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor-α transcriptional activity. Mol Cell Biol. 2007;27:5933–5948. doi: 10.1128/MCB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Research. 2009;69:3819–3827. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond AM, Bane FT, Stafford AT, McIlroy M, Dillon MF, Crotty TB, Hill AD, Young LS. Coassociation of estrogen receptor and p160 proteins predicts resistance to endocrine treatment: SRC-1 is an independent predictor of breast cancer recurrence. Clin Cancer Res. 2009;15:2098–2106. doi: 10.1158/1078-0432.CCR-08-1649. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvilinna N, Eronen H, Miettinen S, Vienonen A, Ylikomi T. Steroid hormone receptors and coregulators in endocrine-resistant and estrogen-independent breast cancer cells. Int J Cancer. 2006;118:832–840. doi: 10.1002/ijc.21431. [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Smith CL, Nawaz Z, O'Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- Su Q, Hu S, Gao H, Ma R, Yang Q, Pan Z, Wang T, Li F. Role of AIB1 for tamoxifen resistance in estrogen receptor-positive breast cancer cells. Oncology. 2008;75:159–168. doi: 10.1159/000159267. [DOI] [PubMed] [Google Scholar]
- Torres-Arzayus MI, de Mora JF, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Wang S, Yuan Y, Liao L, Kuang S-Q, Tien JC-Y, O'Malley BW, Xu J. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci USA. 2009;106:151–156. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen MP, Chen D, Huang S-M, Subramanian S, McKinerney E, Katzenellenbogen BS, Stallcup MR, Kushner PJ. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng FF, Wu R-C, Smith CL, O'Malley BW. Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Mol Cell Biol. 2005;25:8273–8284. doi: 10.1128/MCB.25.18.8273-8284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]