Abstract
Exemestane is a potent and irreversible steroidal aromatase inhibitor drug used for the treatment of estrogen receptor-positive breast cancer. Our aim was to identify and assess the contribution of the specific cytochromes P450 (P450s) responsible for exemestane primary in vitro metabolism. With the use of high-performance liquid chromatography and liquid chromatography-tandem mass spectrometry analytical techniques, 17-hydroexemestane (MI) formation and 6-hydroxymethylexemestane (MII) formation were found to be the predominant exemestane metabolic pathways. In a bank of 15 well characterized human liver microsomes with known P450 isoform-specific activities, the MI formation rate correlated significantly with CYP1A2 (Spearman r = 0.60, p = 0.02) and CYP4A11 (Spearman r = 0.67, p = 0.01) isoform-specific activities, whereas the MII production rate significantly correlated with CYP2B6 (Spearman r = 0.57, p = 0.03) and CYP3A (Spearman r = 0.76, p = 0.005) isoform-specific activities. Recombinant CYP1A1 metabolized exemestane to MI with a catalytic efficiency (Clint) of 150 nl/pmol P450 × min that was at least 3.5-fold higher than those of other P450s investigated. Recombinant CYP3A4 catalyzed MII formation from exemestane with a catalytic efficiency of 840 nl/pmol P450 × min that was at least 4-fold higher than those of other P450s investigated. Among a panel of 10 chemical inhibitors tested, only ketoconazole and troleandomycin (CYP3A-specific chemical inhibitors) significantly inhibited the formation of MII by 45 and 95%, respectively. None of them markedly inhibited the formation of MI. In summary, exemestane seems to be metabolized to MI by multiple P450s that include CYP4A11 and CYP1A1/2, whereas its oxidation to MII is primarily mediated by CYP3A.
Introduction
Exemestane (6-methylenandrosta-1,4-diene-3,17-dione), which belongs to the steroidal group of aromatase inhibitors, is U.S. Food Drug Administration-approved and indicated for sequential adjuvant treatment of postmenopausal women with estrogen receptor-positive breast cancer who have received 2 to 3 years of initial adjuvant tamoxifen therapy (Clemett and Lamb, 2000). The landmark International Exemestane Study demonstrated that patients who switched to exemestane after 2 to 3 years of tamoxifen endocrine treatment had significantly more clinical benefits than those who continued tamoxifen therapy for 5 years. The survival advantage was based on a relative reduction in risk of breast cancer recurrence or death of 24% with exemestane compared with tamoxifen, which corresponded to a 3.3% absolute reduction in recurrence or death at the end of 5 years (Coombes et al., 2004, 2007). However, emerging data suggest high interindividual variability in the beneficial and adverse effects of exemestane that include skeletal events (musculoskeletal disorders, joint pains, and bone fractures) and cardiovascular events (Robinson, 2009). Exemestane is primarily cleared by metabolism, and there is evidence that its pharmacokinetics vary widely among patients (Mauras et al., 2003; Jannuzzo et al., 2004; Rivera et al., 2004; Corona et al., 2009).
The phase I cytochrome P450 superfamily of drug-metabolizing enzymes is responsible for the metabolism of most commercially available drugs (Wojnowski and Kamdem, 2006). The expression and activity of each P450 enzyme have been shown to vary from one individual to another because of environmental and genetic factors (Meyer, 1994). Allelic variants are important sources of interindividual variation in P450 activity. Amino acid substitution can increase or decrease P450 enzyme activity. Some of the genetic factors that influence P450 activity identified thus far were summarized (Nagata and Yamazoe, 2002). The environmental factors that are known to affect P450 expression include medications, food, social habits, and disease status. Therefore, it is conceivable that differences in exemestane P450-mediated metabolism may contribute to the overall variability in clinical response experienced by patients.
According to the information provided by the manufacturer, exemestane undergoes oxidation of the methylene group in position 6 (to form 6-hydroxymethylexemestane) and reduction of the 17-keto group (to form 17-hydroexemestane) with subsequent formation of many secondary metabolites. After a single oral administration of radiolabeled exemestane to postmenopausal volunteers, 42% of the radioactivity was recovered in urine and feces over a 7-day period. Unchanged drug accounted for <1% of the administered dose (Clemett and Lamb, 2000). Despite this information, there are no published data that inform us about how these metabolic pathways were identified. Most importantly, the enzyme kinetic parameters of specific enzymes responsible for exemestane metabolism remain unknown, making prediction of factors controlling exemestane pharmacokinetics difficult. The aim of this study was to identify and assess the contribution of the specific cytochromes P450 involved in the in vitro primary metabolism of exemestane.
Materials and Methods
Chemicals.
Exemestane, nevirapine, and norgestrel were obtained from Toronto Research Chemicals Inc. (North York, ON, Canada). 17-Hydroexemestane was a kind gift of Pfizer (New York, NY). Glucose 6-phosphate, glucose-6-phosphate dehydrogenase, magnesium chloride, NADP, furafylline (CYP1A2 inhibitor), pilocarpine (CYP2A6 inhibitor), sulfaphenazole (CYP2C9 inhibitor), ticlopidine (CYP2B6 and CYP2C19 inhibitor), thioTEPA (CYP2B6 inhibitor), quercetin (CYP2C8 inhibitor), quinidine (CYP2D6 inhibitor), diethylthiocarbamate (CYP2E1 inhibitor), ketoconazole (CYP3A inhibitor), and troleandomycin (CYP3A inhibitor) were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals and solvents were of HPLC grade and were purchased from reliable commercial sources.
Microsomal Source.
Human liver microsomes (HLMs) were obtained from a commercial source (BD Gentest, Woburn, MA). BD Gentest states that the human materials were prepared from human liver donor tissues that were obtained with informed consent in conformance with the guidelines promulgated by the U.S. Uniform Anatomical Gift Act (1987). Each HLM was obtained from a single donor. Baculovirus insect cell-expressed human P450s (CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, and CYP4A11) were purchased from BD Gentest. All microsomal preparations were stored at −80°C until used. The total P450 content, protein concentrations, and specific activity of each P450 isoform were as supplied by the manufacturer.
Incubation Conditions.
Pilot experiments were carried out in HLMs to identify exemestane primary metabolites and to optimize conditions for incubation and HPLC analysis. Exemestane (500 μM, initial concentration) was prepared in methanol and serially diluted with methanol to the required concentration. Before the metabolic incubation, the methanol was evaporated to dryness under reduced pressure using a SpeedVac SC110 model RH40-12 (Savan Instruments, Holbrook, NY). The metabolism of exemestane was assessed in HLMs and a panel of recombinant P450 isoforms (CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, and CYP4A11). For studies conducted in HLMs, duplicate mixtures of exemestane reconstituted in a phosphate reaction buffer (pH, 7.4) and a NADPH-generating system (1.3 mM β-NADP+, 3.3 mM glucose 6-phosphate, 3.3 mM MgCl2, and 0.4 U/ml glucose 6-phosphate dehydrogenase) were preincubated at 37°C for 5 min. For studies conducted in expressed P450s, duplicate mixtures of exemestane and a NADPH-generating system (same composition as above) were used. After the reaction was initiated by adding 25 μl of HLMs (2.5 mg of protein/ml) or 25 μl of P450 (25 pmol) and incubated for 10 min at 37°C (final volume, 250 μl), the reaction was terminated by placing tubes on ice and immediately adding 500 μl of acetonitrile. Norgestrel (100 μl of 50 μM) was added as an internal standard to the incubation sample, vortexed for 30 s, and centrifuged at 14,000 rpm for 5 min in an Eppendorf model 5415D centrifuge (Brinkmann Instruments, Westbury, NY). The supernatant was removed to a clean tube and extracted using ethyl acetate under alkaline pH (0.5 ml of 0.5 M NaOH-glycine buffer, adjusted to pH 10). The sample was centrifuged for 15 min at 2500 rpm in a Beckman GS-6R centrifuge (Global Medical Instrumentation, Inc., Ramsey, MN). The organic layer was removed after centrifugation and evaporated to dryness by speed vacuum and reconstituted with 150 μl of the mobile phase from which 100 μl was injected into the HPLC system. Negative control incubations were run in parallel and included no incubation, no substrate, no cofactor, or no HLMs (bovine serum albumin was used instead). The formation of MI and MII was linear with time between 0 and 15 min and with protein over the range from 0 to 1 mg/ml (data not shown). The end concentrations of exemestane ranged from 0 to 300 μM. The substrate consumption was less than 15% over the incubation time (10 min).
HPLC Analysis.
An analytical liquid chromatography instrument combined with an ultraviolet detection system was used to quantify exemestane and its major metabolites MI and MII. In brief, ethyl acetate extracts of incubation samples were injected onto an Agilent Zorbax analytical C18 column (150 × 4.6-mm i.d., 5 μm, 100 A; Phenomenex, Torrance, CA) with a Luna 10 × 3-mm C18 guard column (Phenomenex) to a 245 UV detection lamp (Advanced Separation Technologies, Whippany, NJ) at a flow rate of 0.8 ml/min with a mobile phase composed of 40% acetonitrile and 10% methanol in 10 mM monobasic potassium phosphate. The HPLC system consisted of a model 515 pump, a model 717 plus autosampler, and a model 486 tunable absorbance detector (Waters, Milford, MA). The ratio of the area under the curve for the metabolite to the area under the curve for each internal standard was calculated. Concentrations of metabolites were measured by standard curves obtained with exemestane, because authentic samples of the synthetic metabolites were not available to us. The possibility that the ultraviolet intensity might differ between the metabolites and exemestane or between the metabolites themselves could not be excluded. Formation rates of metabolites normally presented as picomoles per milligram per minute (or picomoles per picomole of P450 per minute) should be viewed more appropriately as apparent velocities (arbitrary units) where an arbitrary unit = 1000 × (metabolite peak area/internal standard peak area)/slope of the exemestane standard curve. Intra- and interday coefficients of variation of the assays were less than 15%.
LC-MS/MS Assay Development and Identification of Exemestane Metabolites.
Two major primary exemestane metabolites (MI and MII) were detected in human liver microsomal incubates using the HPLC system as described above. To characterize these metabolites, we developed a new sensitive LC-MS/MS method. Reference standards of exemestane and 17-hydroexemestane were dissolved and serially diluted with methanol. After any methanol was removed by evaporation and reconstituted in a mobile phase, an aliquot was infused onto the MS/MS system, and a quantitative optimization was performed to optimize the compound-specific MS/MS conditions and to generate fragment ion patterns. Then, pilot experiments were performed to develop chromatographic conditions for the LC-MS/MS assay. Exemestane was injected onto an LC-MS/MS system that consisted of an LC-20AB pump and SIL-20A HT autosampler (Shimadzu, Addison, IL) and an API 2000 LC-MS/MS triple quadruple system (Applied Biosystems, Foster City, CA) with an electrospray ion source. Exemestane and 17-hydroexemestane were separated on a Luna 3μ C18(2) stainless steel column (100 × 2 mm, 100 A; Phenomenex). The components were eluted with 40% of a 0.1% formic acid in water and 60% methanol, which was degassed in a sonicator for 15 min and delivered at a flow rate of 0.2 ml/min. The LC eluate was introduced into the electrospray ionization source at the same flow rate. The mass spectrometer was operated using electrospray ionization with an ion spray voltage of +5000 V, and the temperature was set at 450°C. Both negative and positive ion modes were tested, and the positive ion multiple reaction monitoring (MRM) analysis was optimal and used in the subsequent experiments. The positive ion MRM mode analysis was performed using nitrogen as the collision gas. The nitrogen nebulizer gas and curtain gas were both set at 45 psi. The pressure in the collision cell was set at 2.20 mTorr. The orifice voltage and ring voltages were set at +35 and +400 V, respectively. Declustering potentials were 21 V for exemestane and 17-hydroexemestane. A dwell time of 400 ms and a pause time of 5 ms between scans were used to monitor the precursor/product ion pairs. Two positive transitions were used for exemestane (+297/120 and +297/148) and 17-hydroexemestane (+299/135 and +299/121). Exemestane and 17-hydroexemestane were measured with the quantifier MRM and confirmed with the qualifier MRM transition.
For the identification of MI and MII, microsomal incubates were injected onto the LC-MS/MS system described above. The following approaches were used to identify MI and MII metabolites in microsomal incubates of exemestane. Because we had no access to exemestane metabolites (except 17-hydroexemestane, which was commercially available), the fragmentation of exemestane was used as a guide for interpretation and identification of exemestane metabolites. Targeted precursor ion scans were performed as a first step of metabolite identification. As a guide, a list of expected metabolites was compiled, and possible metabolic pathways were reconstructed based on the available limited metabolism data in the product label of exemestane as well as predicted metabolic alterations. Suspected masses identified through the precursor ion scan were targeted for product ion analysis to further confirm the identity of these analytes as true metabolites derived from exemestane. HPLC with UV detection was run for retention time confirmation. On the basis of the data obtained from HPLC with UV detection and LC-MS/MS analysis, we identified two metabolite peaks that were consistent with 17-hydroexemestane and 6-hydroxymethylexemestane.
Kinetic Analyses in HLMs and Expressed P450s.
The enzyme kinetics of exemestane were characterized in three different HLMs. Exemestane (5–300 μM) was allowed to incubate for 10 min at 37°C at a protein concentration of 0.25 mg/ml. The enzyme kinetics of exemestane were also characterized in six different expressed P450s (CYP3A4, CYP3A5, CYP2B6, CYP2A6, CYP1A1, and CYP4A11). Exemestane (5–300 μM) was incubated for 10 min at 37°C at a protein concentration of 100 pmol of P450/ml. Concentrations of metabolites were measured by standard curves obtained with exemestane, because authentic samples of the synthetic metabolites were not available. Vmax normally presented as picomoles per milligram per minute (or picomoles per picomole of P450 per minute) should be viewed more appropriately as apparent velocities (arbitrary units), where an arbitrary unit = 1000 × (metabolite peak area/internal standard peak area)/slope of the standard curve.
Correlation Experiments in a Panel of HLMs.
Exemestane (10 μM) was incubated for 10 min at 37°C with microsomes from 15 different human livers (0.25 mg of protein/ml) and an NADPH-generating system. Values for the activity of each P450 were provided by the supplier of the HLMs studied (see http://www.gentest.com) and were detailed in an earlier publication (Ward et al., 2003).
Inhibition Experiments.
Formation rates of 17-hydroexemestane and 6-hydroxymethylexemestane from exemestane were evaluated in the absence (control) and presence of known P450 isoform-specific inhibitors using pooled HLMs. The inhibitors used were 20 μM furafylline (CYP1A2 inhibitor), 50 μM pilocarpine (CYP2A6 inhibitor), 50 μM sulfaphenazole (CYP2C9 inhibitor), 5 μM ticlopidine (CYP2B6, CYP2C19 inhibitor), 5 μM thioTEPA (CYP2B6 inhibitor), 20 μM quercetin (CYP2C8 inhibitor), 1 μM quinidine (CYP2D6 inhibitor), 50 μM diethylthiocarbamate (CYP2E1 inhibitor), 1 μM ketoconazole (CYP3A inhibitor), and 50 μM troleandomycin (CYP3A inhibitor). For competitive inhibitors, exemestane (10 μM) was prewarmed for 5 min at 37°C with or without the inhibitor and with an NADPH-generating system. HLMs (0.25 mg protein/ml) were added to initiate the reaction and incubated at 37°C for 15 min. For mechanism-based inhibitors (ticlopidine, troleandomycin, furafylline, and thioTEPA), the inhibitors and controls were first preincubated in the presence of an NADPH-generating system and HLMs at 37°C for 15 min before the reaction was initiated by addition of exemestane. The inhibitors were studied at concentrations chosen to be selective for the respective P450 isoforms (Bourrié et al., 1996). Percent inhibition of metabolite formation rate by isoform-specific inhibitors was calculated by comparing the inhibited activity with uninhibited controls (without inhibitors).
Data Analysis.
Formation of metabolite versus exemestane concentrations was fit to hyperbolic and nonhyperbolic enzyme kinetic models to estimate apparent kinetic parameters using the Enzyme Kinetics Module of SigmaPlot (Systat Software, Inc., San Jose, CA).
Correlation analysis was performed using a Spearman rank correlation test. p < 0.05 was considered statistically significant. All experiments were performed in duplicate. Data are presented as the mean of duplicate measurements.
Results
Identification of Exemestane Metabolites in Microsomal Incubates.
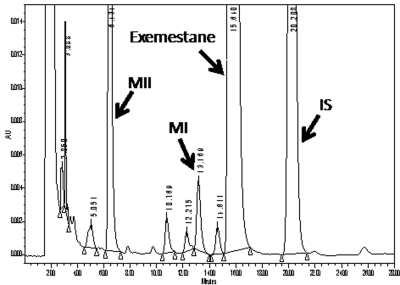
With use of HPLC with UV detection, two major chromatographic peaks that were unique to incubations consisting of exemestane, HLMs, and cofactors were noted. A typical chromatogram of exemestane and its two main metabolites (MI and MII) is shown in Fig. 1. The retention times for MII, MI, exemestane, and the internal standard (norgestrel) were 6.4, 13.1, 15.6, and 20.2 min, respectively. The formation of these two metabolites was not observed in the negative control experiments and depended on the NADPH-generating system, duration of incubation, microsomal protein concentration, and substrate concentration.
Fig. 1.
Representative chromatogram of primary exemestane metabolites (MI and MII) extracted from human liver microsomal incubates. Norgestrel was used as an internal standard (IS). The retention times of MII, MI, exemestane, and the internal standard norgestrel were 6.4, 13.1, 15.6, and 20.2 min, respectively. AU, arbitrary units.
To characterize MI and MII, a new LC-MS/MS method was developed. The parent drug fragmentation pattern and LC-MS/MS conditions were used as a guide to identify exemestane metabolites. First, precursor ion scanning identified two potential metabolites that exhibited 299.3 and 313.3 amu in the exemestane microsomal incubates that consisted of HLMs and cofactors (but not in negative control experiments) in addition to the 297.3 amu of exemestane. Second, to gain further structural information, MS/MS spectra of these potential metabolites were obtained by collusion-induced dissociation of their molecular ions. The MS/MS spectra detected two major characteristic fragment ions for each potential metabolite: m/z 135.1 and 121.1 and 109.0 and 91.0 were obtained from m/z 299.3 and m/z 313.3 spectra, respectively. These transition ions were selected in the MRM modes for identification and quantification of the metabolites: The metabolite with 299.3 amu was measured with the quantifier MRM m/z 299.3/135.1 and confirmed with the qualifier MRM transition 299.3/121.1 and the metabolite with 313.3 amu was measured with the quantifier MRM m/z 313.3/109.0 and confirmed with the qualifier MRM 313.3/91.0.
Kinetic Analysis in HLMs.
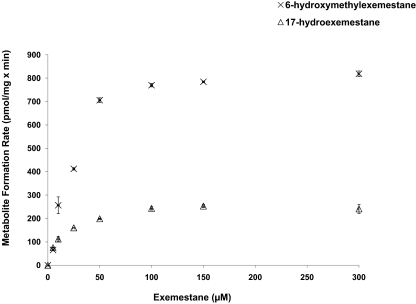
Kinetic analysis for the formation of MI and MII from exemestane was studied in three different HLM preparations. The kinetic profiles of exemestane metabolism to MI and MII in HLMs (HLM 452183, HLM 452165, and HLM 452172) are shown in Table 1. A representative Michaelis-Menten plot for MI and MII formation in HLM 452165 is shown in Fig. 2. The formation rate of MI and MII revealed Michaelis-Menten saturation curves. The Km (apparent) values of MI formation from exemestane for HLM 452183, HLM 452165, and HLM 452172 were 14.51, 15.65, and 10.92 μM, respectively. The Km (apparent) values of MII formation from exemestane for HLM 452183, HLM 452165, and HLM 452172 were 40.41, 31.50, and 39.34 μM, respectively. On average, the apparent hepatic intrinsic clearances [Clint app] of MI and MII formation were 16.23 μl/mg × min and 21.05 μl/mg × min, respectively.
TABLE 1.
Enzyme kinetic parameters of exemestane metabolite formation in three human liver samples
| HLMs | 17-Hydroexemestane |
6-Hydroxymethylexemestane |
||||
|---|---|---|---|---|---|---|
| Km app | Vmax app | Clint app | Km app | Vmax app | Clint app | |
| μM | pmol/mg × min | Vmax/Km | μM | pmol/mg × min | Vmax/Km | |
| HLM (452183) | 14.51 | 176.24 | 12.15 | 40.41 | 739.04 | 18.3 |
| HLM (452165) | 15.65 | 274.51 | 17.54 | 31.50 | 999.29 | 31.72 |
| HLM (452172) | 10.92 | 216.38 | 19.81 | 39.34 | 605.13 | 15.4 |
| Mean | 13.7 | 222.4 | 16.5 | 37.1 | 781.15 | 21.80 |
Km app, apparent affinity; Vmax app, apparent maximum reaction velocity; Clint app, apparent intrinsic clearance.
Fig. 2.
Representative enzyme kinetic plots of the velocities of 17-hydroexemestane (MI) and 6-hydroxymethylexemestane (MII) formation rates by HLM 452165 as a function of exemestane concentration (dose-response curves). Data represent means of duplicate determinations from a single experiment.
Correlation Analysis.
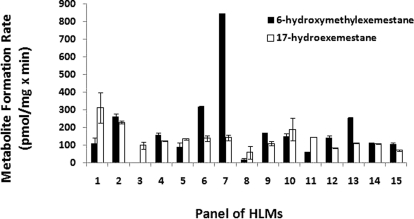
Figure 3 shows the rates of MI and MII formation from exemestane in a panel of 15 characterized HLMs. The average apparent formation rates of MI and MII at 10 μM exemestane were 137.32 ± 65.09 pmol/mg × min (range 59.45–312.14 pmol/mg × min, 5-fold) and 200.47 ± 202.08 pmol/mg × min (range 20.15–844.09 pmol/mg × min, 42-fold), respectively. As shown in Table 2, the MI formation rate correlated significantly with CYP1A2 (Spearman r = 0.60, p = 0.02) and CYP4A11 (Spearman r = 0.67, p = 0.01) isoform-specific activities, whereas the MII production rate correlated significantly with CYP2B6 (Spearman r = 0.57, p = 0.03) and CYP3A isoform-specific activities (Spearman r = 0.76, p = 0.005).
Fig. 3.
Formation rates of 17-hydroexemestane (MI) and 6-hydroxymethylexemestane (MII) from exemestane in a panel of well characterized human liver microsomal preparations. Incubations were performed with 0.25 mg/ml microsomal protein and 10 μM exemestane for 10 min at 37°C. Data represent means of duplicate determinations from a single experiment.
TABLE 2.
Correlation between formation rates of 17-hydroexemestane and 6-hydroxymethylexemestane from exemestane and individual P450 isoform-specific activities in a bank of HLMs
| P450s | 17-Hydroexemestane |
6-Hydroxymethylexemestane |
||
|---|---|---|---|---|
| Spearman r | p | Spearman r | p | |
| CYP1A2 | 0.60 | 0.02 | 0.0048 | 0.98 |
| CYP2A6 | 0.12 | 0.66 | 0.31 | 0.27 |
| CYP2B6 | 0.05 | 0.83 | 0.57 | 0.03 |
| CYP2C8 | 0.07 | 0.78 | 0.15 | 0.57 |
| CYP2C9 | 0.40 | 0.15 | 0.24 | 0.38 |
| CYP2C19 | 0.18 | 0.51 | 0.21 | 0.45 |
| CYP2D6 | 0.14 | 0.59 | 0.10 | 0.72 |
| CYP2E1 | 0.45 | 0.08 | 0.21 | 0.47 |
| CYP3A | 0.05 | 0.85 | 0.76 | 0.005 |
| CYP4A11 | 0.67 | 0.01 | 0.33 | 0.24 |
Metabolism of Exemestane by Recombinant P450s.
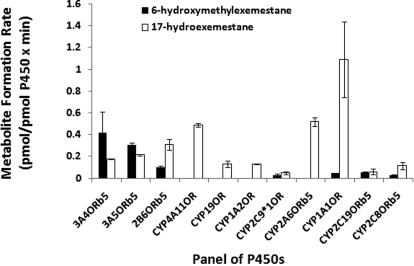
Because the lowest apparent Km in human liver microsomal preparations (Table 1) was 10.92 μM, exemestane was incubated with a panel of recombinant P450s at 10 μM at 37°C for 10 min. Formation rates of MI and MII from exemestane (10 μM) are shown in Fig. 4. CYP3A4, CYP3A5, and CYP2B6 were the major enzymes capable of MII formation from exemestane with rates of 0.42, 0.30, and 0.10 pmol/pmol P450 × min, respectively. CYP2C9, CYP1A1, CYP2C19, and CYP2C8 also contributed to MII formation, although their contributions were 5- to 20-fold lower. In contrast, CYP1A1, CYP2A6, and CYP4A11 were the predominant recombinant P450s involved in the formation of MI from exemestane with rates of 1.09, 0.51, and 0.48 pmol/pmol P450 × min, respectively. CYP3A4, CYP3A5, and CYP2B6 also catalyzed MI formation from exemestane with rates of 0.17, 0.21, and 0.30 pmol/pmol P450 × min, respectively. Other enzymes such as CYP19 and CYP1A2 also formed MI, but their rates were at least 10-fold lower than those of other recombinant P450s (Fig. 4).
Fig. 4.
Formation rates of 17-hydroexemestane (MI) and 6-hydroxymethylexemestane (MII) from exemestane in a panel of baculovirus-expressed P450 enzymes. Data represent means of duplicate determinations from a single experiment.
Because there was a strong correlation between the formation rate of MI and MII from 10 μM exemestane with CYP1A2, CYP4A11, CYP3A4/5, and CYP2B6 in a panel of 15 well characterized HLMs, full kinetic analyses were performed for these enzymes. In addition, CYP1A1 and CYP2A6 were also studied, because they showed strong involvement in the formation of MI in a panel of P450s (Fig. 4). The enzyme kinetic parameters of exemestane metabolism to MI and MII in the selected P450s are shown in Table 3. The formation rate of MI and MII revealed Michaelis-Menten saturation curves. The Km (apparent) of MI formation from exemestane was 28.62, 53, 55, 82.73, 98.66, and 177.2 μM for CYP1A1, CYP3A5, CYP2A6, CYP4A11, CYP3A4, and CYP2B6, respectively. The Km (apparent) of MII formation from exemestane was 9.24, 72.06, 93.46, and 104.56 μM for CYP2B6, CYP3A5, CYP3A4, and CYP1A1, respectively.
TABLE 3.
Enzyme kinetic parameters of exemestane metabolite formation in selected P450s
| P450s | 17-Hydroexemestane |
6-Hydroxymethylexemestane |
||||
|---|---|---|---|---|---|---|
| Km app | Vmax app | Clint app | Km app | Vmax app | Clint app | |
| μM | pmol/mg × min | Vmax/Km | μM | pmol/mg × min | Vmax/Km | |
| CYP3A4 | 98.66 | 0.78 | 0.008 | 93.46 | 78.57 | 0.84 |
| CYP3A5 | 53 | 1.89 | 0.036 | 72 | 9.76 | 0.13 |
| CYP2B6 | 177 | 3.9 | 0.022 | 9 | 1.93 | 0.21 |
| CYP2A6 | 55 | 2.33 | 0.042 | N.D. | N.D. | N.D. |
| CYP1A1 | 28.62 | 4.32 | 0.15 | 104.56 | 0.51 | 0.005 |
| CYP4A11 | 82.7 | 2.57 | 0.031 | N.D. | N.D. | N.D. |
Km app, apparent affinity; Vmax app, apparent maximum reaction velocity; Clint app, apparent intrinsic clearance; N.D., not detected.
Chemical Inhibition of Exemestane Metabolism.
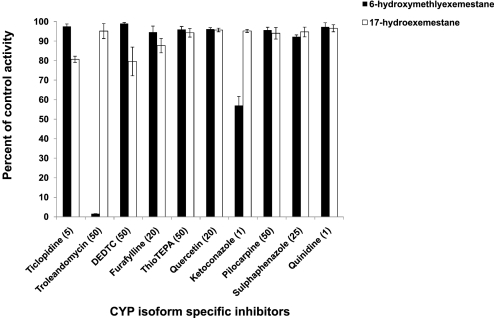
To identify the P450 isoforms participating in exemestane metabolism, 10 μM exemestane was incubated with P450 isoform-specific inhibitors with pooled HLMs and cofactors. As shown in Fig. 5, CYP3A inhibitors troleandomycin (50 μM) and ketoconazole (1 μM) showed a strong inhibitory effect on rates of 6-hydroxymethylexemestane formation (by 95 and 45%, respectively). The inhibitory effect of other isoform-specific inhibitors such as 20 μM furafylline (CYP1A2 inhibitor), 50 μM sulfaphenazole (CYP2C9 inhibitor), 5 μM ticlopidine (CYP2B6 and CYP2C19 inhibitor), 5 μM thioTEPA (CYP2B6 inhibitor), 20 μM quercetin (CYP2C8 inhibitor), 1 μM quinidine (CYP2D6 inhibitor), and 50 μM diethylthiocarbamate (CYP2E1 inhibitor) on the formation rates of either 6-hydroxymethylexemestane or 17-hydroexemestane was marginal (by <20%) (Fig. 5).
Fig. 5.
Chemical inhibition of 6-hydroxymethylexemestane and 17-hydroexemestane formation from exemestane by a panel of P450 isoform-specific inhibitors in pooled HLMs (see Materials and Methods for details). The final concentrations of the inhibitors used are indicated in parentheses. Data represent means of duplicate determinations from a single experiment. DEDTC, diethylthiocarbamate.
Discussion
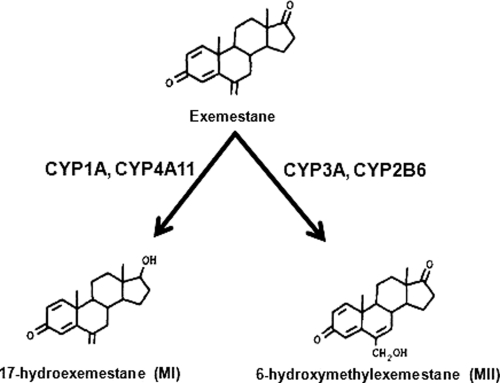
In this study, we used a bank of well characterized human liver microsomes with known isoform-specific activities, a panel of cDNA-expressed enzymes, and a panel of P450-specific chemical inhibitors to characterize the P450-mediated primary metabolism of exemestane in vitro. We show that exemestane is metabolized to 17-hydroexemestane (MI) by multiple P450s that include CYP4A11 and CYP1A1/2, whereas its oxidation to 6-hydroxymethylexemestane (MII) is primarily catalyzed by CYP3A (Fig. 6). These data serve as a basis to predict and estimate the contribution of specific drug-metabolizing enzymes to exemestane clearance.
Fig. 6.
Major primary metabolic pathways and P450 enzymes involved in exemestane metabolism in vitro.
Exemestane is extensively metabolized, with levels of the unchanged drug in plasma accounting for less than 10% of the total radioactivity after administration of radiolabeled exemestane to healthy postmenopausal women (Clemett and Lamb, 2000). Prescribing information provided by the manufacturer states that exemestane undergoes oxidation of the methylene group in position 6 (to form 6-hydroxymethylexemestane) and reduction of the 17-keto group (to form 17-hydroexemestane) with subsequent formation of many secondary metabolites. Our data concur with the manufacturer's label. In the present study, we were able to detect at least six metabolites of exemestane. Among those we have identified 1) 17-hydroexemestane (MI) as one of the main primary reduced metabolites of exemestane in vitro and 2) 6-hydroxymethylexemestane (MII) as the main primary oxidative metabolite of exemestane in vitro. The LC-MS/MS characteristics of MI (299.3/135.1) and MII (313.3/91.0) reported in this study indicate that these metabolites correspond to 17-hydroexemestane and 6-hydroxymethylexemestane, respectively. In fact, Corona et al. (2009) have recently shown that 17-hydroexemestane exhibits similar LC-MS/MS characteristics (m/z 299.1->134.9). Another group of investigators showed that 6-hydroxymethylandrosta-1,4,6-triene-3,7-dione (6-hydroxymethylexemestane) is a potential metabolite of exemestane (Buzzetti et al., 1993). Therefore, these data are consistent with the suggestion that exemestane undergoes reduction of the 17-keto group and oxidation of the methylene group in position 6.
Prescribing information provided by the manufacturer also states that exemestane undergoes extensive oxidation by CYP3A4 and reduction by aldoketoreductases. In this study, we did confirm that CYP3A4 is responsible for oxidation of the methylene group in position 6. The data supporting this conclusion include the facts that CYP3A isoform-specific activity (as measured by testosterone 6β-hydroxylation) correlated significantly with exemestane oxidation (MII formation) in a panel of 15 well characterized HLMs, that cDNA-expressed CYP3A4 catalyzed exemestane oxidation with an apparent catalytic efficiency (Vmax/Km) that was at least 4-fold higher than that of other P450s investigated, and that 6-hydroxymethylexemestane formation was inhibited (by 95 and 45%) by CYP3A-specific inhibitors troleandomycin and ketoconazole, respectively. On the other hand, we show for the first time that 17-hydroexemestane (MI) formation (exemestane reduction), which is the second major metabolic route, is mediated by multiple P450s that include CYP4A11 and CYP1A1/2. This is shown by the facts that CYP4A11 activity (as measured by lauric acid hydroxylase) correlated significantly with 17-hydroexemestane formation, that CYP1A2 activity (as measured by phenacetin O-deethylase) correlated significantly with 17-hydroexemestane formation in a panel of HLMs, and that cDNA-expressed CYP1A1 catalyzed 17-hydroexemestane formation with an efficiency that was at least 4-fold higher than those of other investigated P450s. To strengthen the conclusion that CYP3A is primarily responsible for MII formation and that multiple P450s contribute to MI formation from exemestane, we conducted a correlation analysis without outliers. In the absence of donor livers 6 and 7 (Fig. 3) that catalyzed the formation of MII at the highest rates (318 and 844 pmol/mg × min, respectively), a weaker but still significant correlation was found with CYP3A isoform-specific activity (Spearman r = 0.63, p = 0.035), compared with that found in the presence of outliers (Spearman r = 0.76, p = 0.005). However, upon elimination of donor livers 1 and 10 (Fig. 3), which exhibited the highest catalytic activities toward MI formation (312 and 230 pmol/mg × min, respectively), no significant correlation was found between CYP1A2 (Spearman r = 0.50, p = 0.08) or CYP4A11 (Spearman r = 0.51, p = 0.07) and MI formation rates. This analysis further suggests that CYP3A is the predominant enzyme responsible for the formation of MII, whereas not just one but rather multiple P450s are involved in the formation of MI. The involvement of multiple P450s in exemestane reduction is further confirmed by the fact that all of the tested chemical inhibitors marginally affected the formation of 17-hydroexemestane. According to the manufacturer's prescribing information, 17-hydroexemestane formation is thought to be primarily mediated by aldoketoreductases. The aim of this study was to identify and assess the contribution of P450s involved in the primary metabolism of exemestane. Therefore, we did not perform experiments with cytosolic hepatic fractions, which represent the main site of aldoketoreductase expression and are unable to comment on the relative contribution of P450s and aldoketoreductases toward 17-hydroexemestane formation. However, future studies focusing on the impact of non-P450 enzymes such as aldoketoreductases on the primary metabolism of exemestane are needed.
Taken together, our human liver microsomal data showed that 17-hydroexemestane (MI) formation and 6-hydroxymethylexemestane (MII) formation are the main metabolic routes with apparent catalytic efficiencies (Clint app) averaging 16.23 and 21.05 μl/mg × min, respectively. First, our correlation analysis suggested that CYP3A is the predominant P450 enzyme responsible for MII formation (Spearman r = 0.76), whereas CYP2B6 plays a minor role (Spearman r = 0.57). Second, we showed that CYP4A11 and CYP1A are the major catalysts of MI formation (Spearman r = 0.67 and r = 0.60, respectively). Because CYP4A11 is poorly expressed in the liver, CYP1A appears to be the major P450 responsible for MI formation in vivo. This conclusion is also supported by the similarities in apparent Km between HLMs and cDNA-expressed enzymes. On average, the Km of CYP3A4, CYP3A5, and CYP2B6 toward 6-hydroxymethylexemestane formation is 58 μM. The corresponding average Km in HLMs is 37 μM. Likewise, the Km of CYP1A1 toward 17-hydroexemestane formation is 28 μM. The corresponding average Km in HLMs is 14 μM. The kinetic parameters derived from HLMs and recombinant P450s do not entirely match because, unlike HLMs, recombinant P450s express artificially high and physiologically irrelevant levels of reductase and cytochrome b5, which give them different biochemical properties. Finally, using cDNA-expressed enzymes, we found that CYP3A and CYP1A, the major exemestane-metabolizing enzymes, catalyze the formation of MI and MII with apparent catalytic efficiencies of 150 and 840 nl/pmol P450 × min, respectively. Although cytochrome b5 was not uniformly present in all P450s tested, the use of two different microsomal systems (recombinant enzymes and a panel of well characterized human liver microsomes) predicts a major involvement of CYP3A and CYP1A in the primary metabolism of exemestane. Consistent with this conclusion, CYP3A and CYP1A have been shown to be involved in the metabolism of steroid-like compounds such as pregnenolone (Niwa et al., 1998), progesterone (Schwarz et al., 2000), estradiol (Ohe et al., 2000), testosterone (Kamdem et al., 2004), and estrone (Cribb et al., 2006). Furthermore, unpublished data provided by the prescribing manufacturer suggest that coadministration of ketoconazole (a potent CYP3A inhibitor) does not alter exemestane exposure in humans. This result suggests that alternative metabolic pathways exist. In fact, we have identified one of those alternative pathways, which is CYP1A-mediated formation of 17-hydroexemestane. Using HLMs, we found that the catalytic efficiency of 6-hydroxymethylexemestane (MII) formation (Clint app = 21.80 μl/min × mg) is almost similar to that of 17-hydroexemestane (MI) formation (Clint app = 16.5 μl/min × mg). Therefore, it is possible that upon coadministration of ketoconazole, exemestane metabolism shifts toward 17-hydroexemestane formation in vivo. It is also possible that the unknown role of aldoketoreductases in overall exemestane metabolism could lead to lack of drug-drug interaction with ketoconazole.
The clinical response to exemestane varies widely among individuals. For instance, some patients develop serious side effects [e.g., musculoskeletal pain (21%), arthralgia (18.6%), cardiovascular events (16.5% of patients), and bone fractures (7%)], whereas others do not (Robinson, 2009). The development of such vasomotor and/or joint symptoms, brought about not only by aromatase inhibitors but also by selective estrogen receptor modulators, has been shown to be associated with breast cancer recurrence rates (Cuzick, 2008; Mortimer et al., 2008). Therefore, the identification of predictive biomarkers of drug response and/or outcome would be of great clinical value. In this study, we made a step toward this goal. We showed that the formation rates of the major metabolites of exemestane (MI and MII) vary widely among human liver samples and that CYP3A4/5, CYP2B6, CYP1A, and CYP4A11 are involved in the biotransformation of the drug. Unlike CYP3A and to some extent CYP1A and CYP2B6, CYP4A11 is almost absent in the liver. Therefore, variable activity of CYP1A, CYP2B6, and CYP3A, brought about by genetic polymorphisms and drug interactions, may alter exemestane disposition and its antiestrogenic effect. It is well known that CYP3A4 and CYP3A5 are expressed in the liver and in extrahepatic tissues (e.g., gut and breast) (Kolars et al., 1994; Hellmold et al., 1998; Wojnowski and Kamdem, 2006). CYP3A5 is polymorphically expressed among different ethnic groups, with CYP3A5*3 (inactive gene form) allele frequencies of 30, 50, and 90% in Africans, Asians, and whites, respectively (Wojnowski and Kamdem, 2006). Unlike CYP3A5, CYP3A4 variability is mostly dictated by environmental factors, age, gender, and concomitant drug administration (Wilkinson, 2005). Wide interindividual variability in the expression of CYP2B6 has been seen at the level of mRNA (Chang et al., 2003), protein (Lang et al., 2001), and catalytic activity (Ekins et al., 1998), which is thought to be due to effects of genetic polymorphisms of CYP2B6 (Lang et al., 2001) or exposure to drugs that are inducers (Blizard et al., 2001) or inhibitors (Hesse et al., 2001) of CYP2B6. CYP1A1 is known to be involved in phase I xenobiotic and drug metabolism and is inhibited and induced by compounds such as macrolides and aromatic hydrocarbons, respectively (Smith et al., 1998). Several polymorphisms have been identified in CYP1A1, some of which lead to more highly inducible aryl hydrocarbon hydroxylase activity (Petersen et al., 1991; Cosma et al., 1993; Crofts et al., 1994). Although CYP1A represents a small fraction of the overall hepatic P450 pool, it is highly expressed in extrahepatic tissues. Because exemestane has a very large volume of distribution, it is conceivable that CYP1A-mediated extrahepatic metabolism contributes significantly to exemestane disposition in vivo. Further studies pertaining to the actual contribution of these pathways and enzymes as well as to the impact of drug-drug interaction and genetic polymorphisms on exemestane drug disposition in postmenopausal women with breast cancer are now warranted.
Acknowledgments
We thank Evan Ogburn for the technical support.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant 5U01GM061373-09].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.032276.
- P450
- cytochrome P450
- thioTEPA
- 1,1-phosphinothioylidynetrisaziridine
- HPLC
- high-performance liquid chromatography
- HLM
- human liver microsome
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- MRM
- multiple reaction monitoring
- amu
- atomic mass units.
Authorship Contributions
Participated in research design: Desta, Flockhart, and Kamdem.
Conducted experiments: Kamdem.
Contributed new reagents or analytic tools: Desta and Kamdem.
Performed data analysis: Kamdem.
Wrote or contributed to the writing of the manuscript: Desta, Flockhart, and Kamdem.
References
- Blizard D, Sueyoshi T, Negishi M, Dehal SS, Kupfer D. (2001) Mechanism of induction of cytochrome P450 enzymes by the proestrogenic endocrine disruptor pesticide-methoxychlor: interactions of methoxychlor metabolites with the constitutive androstane receptor system. Drug Metab Dispos 29:781–785 [PubMed] [Google Scholar]
- Bourrié M, Meunier V, Berger Y, Fabre G. (1996) Cytochrome P450 isoform inhibitors as a tool for the investigation of metabolic reactions catalyzed by human liver microsomes. J Pharmacol Exp Ther 277:321–332 [PubMed] [Google Scholar]
- Buzzetti F, Di Salle E, Longo A, Briatico G. (1993) Synthesis and aromatase inhibition by potential metabolites of exemestane (6-methylenandrosta-1,4-diene-3,17-dione). Steroids 58:527–532 [DOI] [PubMed] [Google Scholar]
- Chang TK, Bandiera SM, Chen J. (2003) Constitutive androstane receptor and pregnane X receptor gene expression in human liver: interindividual variability and correlation with CYP2B6 mRNA levels. Drug Metab Dispos 31:7–10 [DOI] [PubMed] [Google Scholar]
- Clemett D, Lamb HM. (2000) Exemestane: a review of its use in postmenopausal women with advanced breast cancer. Drugs 59:1279–1296 [DOI] [PubMed] [Google Scholar]
- Coombes RC, Hall E, Gibson LJ, Paridaens R, Jassem J, Delozier T, Jones SE, Alvarez I, Bertelli G, Ortmann O, et al. (2004) A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med 350:1081–1092 [DOI] [PubMed] [Google Scholar]
- Coombes RC, Kilburn LS, Snowdon CF, Paridaens R, Coleman RE, Jones SE, Jassem J, Van de Velde CJ, Delozier T, Alvarez I, et al. (2007) Survival and safety of exemestane versus tamoxifen after 2–3 years' tamoxifen treatment (Intergroup Exemestane Study): a randomised controlled trial. Lancet 369:559–570 [DOI] [PubMed] [Google Scholar]
- Corona G, Elia C, Casetta B, Diana C, Rosalen S, Bari M, Toffoli G. (2009) A liquid chromatography-tandem mass spectrometry method for the simultaneous determination of exemestane and its metabolite 17-dihydroexemestane in human plasma. J Mass Spectrom 44:920–928 [DOI] [PubMed] [Google Scholar]
- Cosma G, Crofts F, Taioli E, Toniolo P, Garte S. (1993) Relationship between genotype and function of the human CYP1A1 gene. J Toxicol Environ Health 40:309–316 [DOI] [PubMed] [Google Scholar]
- Cribb AE, Knight MJ, Dryer D, Guernsey J, Hender K, Tesch M, Saleh TM. (2006) Role of polymorphic human cytochrome P450 enzymes in estrone oxidation. Cancer Epidemiol Biomarkers Prev 15:551–558 [DOI] [PubMed] [Google Scholar]
- Crofts F, Taioli E, Trachman J, Cosma GN, Currie D, Toniolo P, Garte SJ. (1994) Functional significance of different human CYP1A1 genotypes. Carcinogenesis 15:2961–2963 [DOI] [PubMed] [Google Scholar]
- Cuzick J. (2008) Chemoprevention of breast cancer. Breast Cancer 15:10–16 [DOI] [PubMed] [Google Scholar]
- Ekins S, Vandenbranden M, Ring BJ, Gillespie JS, Yang TJ, Gelboin HV, Wrighton SA. (1998) Further characterization of the expression in liver and catalytic activity of CYP2B6. J Pharmacol Exp Ther 286:1253–1259 [PubMed] [Google Scholar]
- Hellmold H, Rylander T, Magnusson M, Reihnér E, Warner M, Gustafsson JA. (1998) Characterization of cytochrome P450 enzymes in human breast tissue from reduction mammaplasties. J Clin Endocrinol Metab 83:886–895 [DOI] [PubMed] [Google Scholar]
- Hesse LM, von Moltke LL, Shader RI, Greenblatt DJ. (2001) Ritonavir, efavirenz, and nelfinavir inhibit CYP2B6 activity in vitro: potential drug interactions with bupropion. Drug Metab Dispos 29:100–102 [PubMed] [Google Scholar]
- Jannuzzo MG, Poggesi I, Spinelli R, Rocchetti M, Cicioni P, Buchan P. (2004) The effects of degree of hepatic or renal impairment on the pharmacokinetics of exemestane in postmenopausal women. Cancer Chemother Pharmacol 53:475–481 [DOI] [PubMed] [Google Scholar]
- Kamdem LK, Meineke I, Koch I, Zanger UM, Brockmöller J, Wojnowski L. (2004) Limited contribution of CYP3A5 to the hepatic 6β-hydroxylation of testosterone. Naunyn Schmiedebergs Arch Pharmacol 370:71–77 [DOI] [PubMed] [Google Scholar]
- Kolars JC, Lown KS, Schmiedlin-Ren P, Ghosh M, Fang C, Wrighton SA, Merion RM, Watkins PB. (1994) CYP3A gene expression in human gut epithelium. Pharmacogenetics 4:247–259 [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM. (2001) Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11:399–415 [DOI] [PubMed] [Google Scholar]
- Mauras N, Lima J, Patel D, Rini A, di Salle E, Kwok A, Lippe B. (2003) Pharmacokinetics and dose finding of a potent aromatase inhibitor, aromasin (exemestane), in young males. J Clin Endocrinol Metab 88:5951–5956 [DOI] [PubMed] [Google Scholar]
- Meyer UA. (1994) The molecular basis of genetic polymorphisms of drug metabolism. J Pharm Pharmacol 46 (Suppl 1):409–415 [PubMed] [Google Scholar]
- Mortimer JE, Flatt SW, Parker BA, Gold EB, Wasserman L, Natarajan L, Pierce JP. WHEL Study Group (2008) Tamoxifen, hot flashes and recurrence in breast cancer. Breast Cancer Res Treat 108:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Yamazoe Y. (2002) Genetic polymorphism of human cytochrome p450 involved in drug metabolism. Drug Metab Pharmacokinet 17:167–189 [DOI] [PubMed] [Google Scholar]
- Niwa T, Yabusaki Y, Honma K, Matsuo N, Tatsuta K, Ishibashi F, Katagiri M. (1998) Contribution of human hepatic cytochrome P450 isoforms to regioselective hydroxylation of steroid hormones. Xenobiotica 28:539–547 [DOI] [PubMed] [Google Scholar]
- Ohe T, Hirobe M, Mashino T. (2000) Novel metabolic pathway of estrone and 17beta-estradiol catalyzed by cytochrome P-450. Drug Metab Dispos 28:110–112 [PubMed] [Google Scholar]
- Petersen DD, McKinney CE, Ikeya K, Smith HH, Bale AE, McBride OW, Nebert DW. (1991) Human CYP1A1 gene: cosegregation of the enzyme inducibility phenotype and an RFLP. Am J Hum Genet 48:720–725 [PMC free article] [PubMed] [Google Scholar]
- Rivera E, Valero V, Francis D, Asnis AG, Schaaf LJ, Duncan B, Hortobagyi GN. (2004) Pilot study evaluating the pharmacokinetics, pharmacodynamics, and safety of the combination of exemestane and tamoxifen. Clin Cancer Res 10:1943–1948 [DOI] [PubMed] [Google Scholar]
- Robinson A. (2009) A review of the use of exemestane in early breast cancer. Ther Clin Risk Manag 5:91–98 [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Kisselev P, Schunck WH, Chernogolov A, Boidol W, Cascorbi I, Roots I. (2000) Allelic variants of human cytochrome P450 1A1 (CYP1A1): effect of T461N and I462V substitutions on steroid hydroxylase specificity. Pharmacogenetics 10:519- 530 [DOI] [PubMed] [Google Scholar]
- Smith G, Stubbins MJ, Harries LW, Wolf CR. (1998) Molecular genetics of the human cytochrome P450 monooxygenase superfamily. Xenobiotica 28:1129–1165 [DOI] [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306:287–300 [DOI] [PubMed] [Google Scholar]
- Wilkinson GR. (2005) Drug metabolism and variability among patients in drug response. N Engl J Med 352:2211–2221 [DOI] [PubMed] [Google Scholar]
- Wojnowski L, Kamdem LK. (2006) Clinical implications of CYP3A polymorphisms. Expert Opin Drug Metab Toxicol 2:171–182 [DOI] [PubMed] [Google Scholar]