Abstract
Cytochrome P450 (P450)-mediated metabolism of arachidonic acid regulates inflammation in hepatic and extrahepatic tissue. CYP2C/CYP2J-derived epoxyeicosatrienoic and dihydroxyeicosatrienoic acids (EET+DHET) elicit anti-inflammatory effects, whereas CYP4A/CYP4F-derived 20-hydroxyeicosatetraenoic acid (20-HETE) is proinflammatory. Because the impact of inflammation on P450-mediated formation of endogenous eicosanoids is unclear, we evaluated P450 mRNA levels and P450 epoxygenase (EET+DHET) and ω-hydroxylase (20-HETE) metabolic activity in liver, kidney, lung, and heart in mice 3, 6, 24, and 48 h after intraperitoneal lipopolysaccharide (LPS) (1 mg/kg) or saline administration. Hepatic Cyp2c29, Cyp2c44, and Cyp2j5 mRNA levels and EET+DHET formation were significantly lower 24 and 48 h after LPS administration. Hepatic Cyp4a12a, Cyp4a12b, and Cyp4f13 mRNA levels and 20-HETE formation were also significantly lower at 24 h, but recovered to baseline at 48 h, resulting in a significantly higher 20-HETE/EET+DHET formation rate ratio compared with that for saline-treated mice. Renal P450 mRNA levels and P450-mediated eicosanoid metabolism were similarly suppressed 24 h after LPS treatment. Pulmonary EET+DHET formation was lower at all time points after LPS administration, whereas 20-HETE formation was suppressed in a time-dependent manner, with the lowest formation rate observed at 24 h. No differences in EET+DHET or 20-HETE formation were observed in heart. Collectively, these data demonstrate that acute activation of the innate immune response alters P450 expression and eicosanoid metabolism in mice in an isoform-, tissue-, and time-dependent manner. Further study is necessary to determine whether therapeutic restoration of the functional balance between the P450 epoxygenase and ω-hydroxylase pathways is an effective anti-inflammatory strategy.
Introduction
In addition to their role in xenobiotic metabolism, cytochromes P450 (P450) metabolize numerous endogenous substrates, including steroids, hormones, and fatty acids, to biologically active mediators (Roman, 2002). One such example is the oxidative metabolism of arachidonic acid to epoxyeicosatrienoic acids (EETs) and hydroxyeicosatetraenoic acids (HETEs). Olefin epoxidation of arachidonic acid to four EET regioisomers (5,6-EET, 8,9-EET, 11,12-EET, and 14,15-EET) is primarily catalyzed by CYP2C and CYP2J isoforms (Zeldin, 2001). Soluble epoxide hydrolase (sEH) (Ephx2) rapidly hydrolyzes EETs to dihydroxyeicosatrienoic acids (DHETs), which, in general, are less biologically active. In contrast, ω-hydroxylation of arachidonic acid by CYP4A and CYP4F isoforms produces 20-HETE (Roman, 2002).
EETs and 20-HETE regulate numerous biological processes, including vascular tone, angiogenesis, and the response to ischemia/reperfusion injury (Zeldin, 2001; Roman, 2002; Deng et al., 2010). Accumulating evidence has demonstrated that the P450 epoxygenase and ω-hydroxylase pathways also regulate inflammation. The EETs possess potent anti-inflammatory properties by attenuating cytokine-induced nuclear factor-κB (NF-κB) activation and leukocyte adhesion to the vascular wall (Node et al., 1999). In contrast, 20-HETE activates NF-κB signaling and induces expression of cellular adhesion molecules and cytokines, thereby promoting inflammation (Ishizuka et al., 2008).
Because of the divergent effects of the P450 epoxygenase and ω-hydroxylase pathways in the regulation of inflammation, alterations in the functional balance between these parallel pathways may contribute to the pathogenesis and progression of inflammatory diseases, such as sepsis, cancer, and cardiovascular disease. Although it is well established that acute inflammatory stimuli suppress hepatic P450 expression via a pretranslational mechanism, thereby decreasing xenobiotic metabolism and clearance in preclinical models and humans (Morgan, 2001; Riddick et al., 2004; Morgan et al., 2008), the effect on P450-mediated eicosanoid metabolism in hepatic and extrahepatic tissue has not been rigorously evaluated. Moreover, the biological properties of the P450 epoxygenase and ω-hydroxylase pathways in vivo are most commonly investigated in mouse models; however, the relative expression and function of each pathway across tissues in mice has not been well described to date. Therefore, we sought to characterize the 1) relative expression and metabolic activity of the P450 epoxygenase and ω-hydroxylase pathways across liver, kidney, lung, and heart in mice and the 2) impact of acute inflammation induced by systemic lipopolysaccharide (LPS) administration on P450 epoxygenase and ω-hydroxylase expression and metabolic activity in each tissue.
Materials and Methods
Reagents.
All reagents were purchased from Thermo Fisher Scientific (Waltham, MA) unless otherwise noted.
Experimental Protocol.
Male C57BL/6 mice (4–5 months of age) were treated with Escherichia coli LPS (1 mg/kg, serotype O111:B4, 1,000,000 endotoxin units/mg; Sigma-Aldrich, St. Louis, MO) or endotoxin-free saline by intraperitoneal injection and were euthanized by CO2 inhalation 3, 6, 24, or 48 h after treatment. Liver, kidney, lung, heart, and aorta were harvested and flash frozen in liquid nitrogen. Blood was collected in heparinized tubes, and plasma was separated by centrifugation. Tissue and plasma were stored at −80°C pending analysis. All studies were in accordance with principles outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, 1996) and were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
RNA Isolation, Reverse Transcription, and qRT-PCR.
Total RNA was isolated from whole tissue homogenates using the RNeasy Miniprep Kit (QIAGEN, Valencia, CA) per the manufacturer's instructions. Total RNA was reverse-transcribed to cDNA using an ABI High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) with a reaction temperature of 25°C for 10 min and then 37°C for 120 min. Expression of murine Cyp2c29, Cyp2c44, Cyp2j5, Cyp2j9, Cyp4a12a, Cyp4a12b, Cyp4f13, Cyp4f16, Ephx2, and GAPDH was quantified by qRT-PCR using commercially available TaqMan Assays on Demand (Applied Biosystems) (Supplemental Table 1). P450 isoforms were selected on the basis of known epoxygenase or ω-hydroxylase activity and/or expression in several of the tissues examined in an initial expression screen (Luo et al., 1998; Ma et al., 1999; Qu et al., 2001; DeLozier et al., 2004; Wang et al., 2004; Muller et al., 2007). The metabolic activity of murine Cyp4f isoforms has not been characterized, but these isoforms were included as CYP4F isoforms in other species and have been shown to catalyze 20-HETE formation (Powell et al., 1998; Christmas et al., 2001; Xu et al., 2004). Each reaction was performed in a 20-μl volume with 50 ng of cDNA, 20× Assay on Demand, and 2× TaqMan Universal PCR Master Mix. All reactions were performed in triplicate using the ABI Prism 7300 Sequence Detection System. The cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C followed by 60 s at 60°C. The efficiency of each RT-PCR probe was calculated over a range of cDNA amounts (1–100 ng), as described previously (Pfaffl, 2001) and was equivalent for all probes (data not shown). P450 mRNA levels were normalized to GAPDH and expressed relative to the saline-treated controls using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Microsome Isolation.
Microsomal fractions from liver, kidney, lung, and heart were isolated as described previously (Lee et al., 2007). In brief, frozen tissue was homogenized in 0.25 M sucrose-10 mM Tris-HCl buffer (pH 7.5) containing protease inhibitors. Liver and kidney homogenates were prepared from individual mice. Lung and heart homogenates were prepared from tissue pooled from two to four mice, because of the tissue size and low levels of P450 expression. Homogenates were centrifuged at 4°C at 2570g for 20 min and then at 10,300g for 20 min to remove cellular debris. The supernatants were then centrifuged at 100,000g at 4°C for 90 min. The resulting microsomal pellets were resuspended in 50 mM Tris-1 mM dithiothreitol-1 mM EDTA buffer (pH 7.5) containing 20% glycerol. Protein concentrations were quantified using a Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA), per the manufacturer's instructions.
Microsomal Incubations.
Incubations contained 300 μg (liver and kidney) or 350 μg (lung and heart) of microsomal protein and 50 μM (liver, kidney, and heart) or 150 μM (lung) arachidonic acid in a 1-ml volume of 0.12 M potassium phosphate incubation buffer containing 5 mM magnesium chloride, as described previously (Poloyac et al., 2004). In the presence of these saturating substrate concentrations, formation rates reflect the amount of metabolically active protein (Poloyac et al., 2004). The limited amount of tissue precluded assessment of P450 epoxygenase and ω-hydroxylase activity in aorta. Reactions were initiated by the addition of 1 mM NADPH and were performed at 37°C for 20 min (liver and kidney) or 60 min (lung and heart). In lung and heart incubations, an additional 1 mM NADPH was added after 30 min. Incubations were performed at saturating concentrations of substrate, and metabolite formation was linear with respect to incubation time and microsomal protein, as determined from preliminary incubations. The reactions were stopped by placing the samples on ice, and 12.5 ng of 20-HETE-d6 was added as an internal standard. Because of the high metabolite formation, liver incubations were diluted 20-fold in incubation buffer before addition of internal standard. Metabolites were extracted with diethyl ether, evaporated to dryness under nitrogen gas, and reconstituted in 80% methanol in deionized water for analysis.
UPLC-Tandem Mass Spectrometry.
Arachidonic acid metabolites (14,15-EET, 11,12-EET, 8,9-EET, 14,15-DHET, 11,12-DHET, 8,9-DHET, 5,6-DHET, and 20-HETE) in microsomal incubations and plasma were quantified by UPLC-tandem mass spectrometry as described previously (Miller et al., 2009). Analytes were separated on a UPLC BEH C-18, 1.7 μm (2.1 × 100 mm) reverse-phase column (Waters, Milford, MA) protected by a guard column (2.1 × 5 mm; Waters). Mobile phases consisted of 0.005% acetic acid-5% acetonitrile in deionized water (A) and 0.005% acetic acid in acetonitrile (B) at an initial mixture of 65% A and 35% B. Mobile phase B increased from 35 to 70% in a linear gradient over 4 min and then increased to 95% over 0.5 min where it remained for 0.3 min. This was followed by a linear return to the initial conditions over 0.1 min with a 1.5-min preequilibration period before the next sample run.
Mass spectrometric analysis was performed with a TSQ Quantum Ultra (Thermo Fisher Scientific) triple quadrupole mass spectrometer coupled with heated electrospray ionization operated in negative selective reaction monitoring mode. Unit resolutions at both Q1 and Q3 were set at 0.70 full width at half-maximum. Quantitation by selective reaction monitoring analysis on EETs, DHETs, and HETEs was performed by monitoring their m/z transitions. Parameters were optimized to obtain the highest [M − H]− ion abundance and were as follows: capillary temperature 400°C, spray voltage 3000 V, and a source collision-induced dissociation set at 0 V. Sheath gas, auxiliary gas, and ion sweep gas pressures were set at 65, 55, and 3, respectively. Scan time was set at 0.01 s, and collision gas pressure was set at 1.3 mTorr. Analytical data were acquired and analyzed using Xcaliber software version 2.0.6 (Thermo Fisher Scientific). Metabolite concentrations were calculated from a standard curve and are expressed as formation rates (picomoles per milligram of protein per min).
Statistical Analysis.
All data are expressed as the mean ± S.E.M. The sum formation rate of all EET and DHET regioisomers was calculated and used as an index of total P450 epoxygenase metabolic activity. The functional balance between the P450 ω-hydroxylase and epoxygenase pathways was assessed by the ratio of 20-HETE to total EET+DHET formation. Because the data were not normally distributed, mRNA and protein levels were transformed to ranks and metabolite formation rates were log-transformed before statistical analysis. Data from saline-treated mice at each time point were pooled to create a single control group for statistical comparisons. Data were analyzed by one-way analysis of variance followed by a post hoc Dunnett's test for comparison with the pooled saline control group. The relationship between P450 mRNA and protein levels and EET+DHET or 20-HETE formation was evaluated by Spearman rank correlation. Statistical analysis was performed using SAS software (version 9.1.3; SAS Institute, Cary, NC). P < 0.05 was considered statistically significant.
Results
Induction of Cytokine Expression.
Systemic LPS administration induced tumor necrosis factor-α (TNF-α) expression in a time-dependent manner in all tissues examined, consistent with acute activation of the innate immune response (Fig. 1). The most substantial increase was observed 3 and 6 h after LPS administration. TNF-α expression decreased over time but remained significantly elevated compared with that in saline-treated mice in most tissues at 48 h.
Fig. 1.
Effect of LPS administration (1 mg/kg i.p.) on TNF-α mRNA levels over 48 h in liver, kidney, lung, heart, and aorta. Data are expressed as mean ± S.E.M. fold change in expression, relative to the saline control group, using the 2−ΔΔCt method. n = 5–6 per time point. *, P < 0.05 versus saline control group.
Liver.
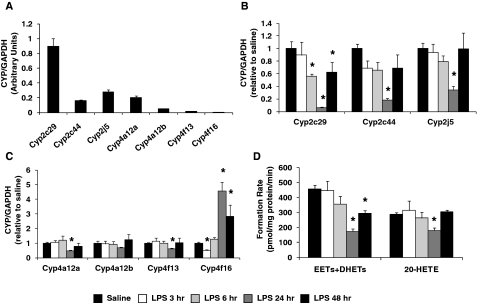
All P450 isoforms examined were expressed at high levels in liver, with Cyp2c29 and Cyp4a12a being the most abundant P450 epoxygenase and ω-hydroxylase, respectively (Fig. 2A). Hepatic Cyp2c29 (0.06 ± 0.02), Cyp2c44 (0.18 ± 0.03), and Cyp2j5 (0.34 ± 0.06) mRNA levels were markedly suppressed 24 h after LPS administration (Fig. 2B), with partial (Cyp2c29, 0.62 ± 0.16; Cyp2c44, 0.69 ± 0.22) or full (Cyp2j5, 1.00 ± 0.25) restoration of the expression to basal levels at 48 h. Hepatic CYP2C and CYP2J protein expression was also significantly suppressed 24 h after LPS administration (P < 0.05 versus saline) (Supplemental Fig. 1). At 48 h, CYP2C remained significantly suppressed, but CYP2J expression recovered to near-basal levels. Hepatic Cyp4a12a (0.47 ± 0.06), Cyp4a12b (0.67 ± 0.06), and Cyp4f13 (0.59 ± 0.08) mRNA levels were also significantly suppressed, but to a lesser degree, 24 h after LPS administration and returned to basal levels by 48 h (Cyp4a12a, 0.79 ± 0.22; Cyp4a12b, 1.21 ± 0.37; Cyp4f13, 1.02 ± 0.33). In contrast, Cyp4f16 mRNA levels were significantly higher 24 (4.56 ± 0.62) and 48 (2.82 ± 0.81) h after LPS administration compared with saline controls (Fig. 2C). No significant alterations in CYP4A or CYP4F protein expression were observed (Supplemental Fig. 1).
Fig. 2.
A, relative abundance of hepatic P450 mRNA was quantified in saline-treated mice (n = 20) and normalized to GAPDH. The time-dependent effect of LPS administration (1 mg/kg i.p.) on hepatic (B) Cyp2c29, Cyp2c44, and Cyp2j5 and (C) Cyp4a12a, Cyp4a12b, Cyp4f13, and Cyp4f16 mRNA levels was quantified by qRT-PCR and expressed relative to that for the saline control group [saline (pooled), n = 20; LPS 3 h, n = 6, LPS 6 h, n = 6; LPS 24 h, n = 15; LPS 48 h, n = 6]. D, effect of LPS administration on total P450 epoxygenase (EETs+DHETs) and ω-hydroxylase (20-HETE) metabolic activity in liver microsomes was determined [saline (pooled), n = 12; LPS 3 h, n = 4; LPS 6 h, n = 4; LPS 24 h, n = 12; LPS 48 h, n = 6]. *, P < 0.05 versus saline control group.
Total hepatic P450 epoxygenase metabolic activity was significantly lower 24 (173.9 ± 15.7 pmol/mg protein/min) and 48 (295.4 ± 18.8 pmol/mg protein/min) h after LPS administration compared with that for the saline control group (458.2 ± 24.0 pmol/mg protein/min), whereas no differences were observed at 3 and 6 h (Fig. 2D). Similar results were observed when each EET+DHET regioisomer was evaluated individually (Supplemental Fig. 2A). Total P450 epoxygenase metabolic activity was strongly correlated with hepatic Cyp2c29 (r = 0.74, P < 0.001), Cyp2c44 (r = 0.65, P < 0.001), and Cyp2j5 (r = 0.56, P < 0.001) mRNA levels, as well as CYP2C (r = 0.50, P = 0.006) and CYP2J (r = 0.37, P = 0.056) protein levels. Compared with saline-treated mice (286.7 ± 11.2 pmol/mg protein/min), 20-HETE formation was also significantly lower 24 h (180.8 ± 17.8 pmol/mg protein/min) after LPS administration, but returned to basal levels at 48 h (305.5 ± 11.7 pmol/mg protein/min) (Fig. 2D). No significant differences in hepatic 20-HETE formation were observed 3 or 6 h after LPS administration. 20-HETE formation was significantly correlated with hepatic Cyp4a12b mRNA levels (r = 0.51, P = 0.002).
Kidney.
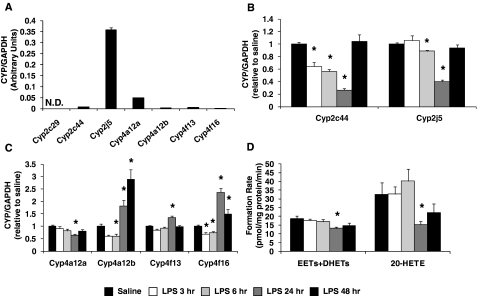
Cyp2j5 and Cyp4a12a were the most abundant P450 epoxygenase and ω-hydroxylase in kidney, respectively, whereas Cyp2c29 mRNA was undetectable (Fig. 3A). Renal Cyp2c44 mRNA levels were significantly suppressed 3 (0.64 ± 0.06), 6 (0.56 ± 0.03), and 24 (0.26 ± 0.03) h after LPS administration but returned to baseline at 48 h (1.04 ± 0.11). A similar profile was observed for Cyp2j5, but Cyp2j5 was suppressed to a lesser degree (Fig. 3B). Cyp4a12a expression was also significantly suppressed at 24 h (0.63 ± 0.06). In contrast, Cyp4a12b and Cyp4f16 mRNA levels were significantly lower at 6 h but were significantly higher 24 and 48 h after LPS administration (P < 0.05 versus saline) (Fig. 3C). Significant changes in Cyp4f13 mRNA levels were observed only at 24 h.
Fig. 3.
A, relative abundance of renal P450 mRNA was quantified in saline-treated mice (N.D., not detected; n = 20 for detected isoforms; n = 3 for undetectable isoforms) and normalized to GAPDH. The time-dependent effect of LPS administration (1 mg/kg i.p.) on renal (B) Cyp2c44 and Cyp2j5 and (C) Cyp4a12a, Cyp4a12b, Cyp4f13, and Cyp4f16 mRNA levels was quantified by qRT-PCR and expressed relative to that for the saline control group [saline (pooled), n = 20; LPS 3 h, n = 6; LPS 6 h, n = 6; LPS 24 h, n = 15; LPS 48 h, n = 6]. D, effect of LPS administration on total P450 epoxygenase (EETs+DHETs) and ω-hydroxylase (20-HETE) metabolic activity in kidney microsomes was determined [saline (pooled), n = 9; LPS 3 h, n = 6; LPS 6 h, n = 6; LPS 24 h, n = 9; LPS 48 h, n = 6]. *, P < 0.05 versus saline control group.
Total P450 epoxygenase metabolic activity was significantly lower in kidney microsomes 24 h (13.2 ± 0.7 pmol/mg protein/min) after LPS administration, compared with that for the saline control group (18.7 ± 1.6 pmol/mg protein/min). Formation of the 14,15- and 11,12- but not of the 8,9- or 5,6-regioisomers was significantly lower 24 h after LPS administration (Supplemental Fig. 2B). At 48 h, total EET+DHET formation was returning to basal levels (14.7 ± 1.4 pmol/mg protein/min; P = 0.12 versus saline) (Fig. 3D). Likewise, renal 20-HETE formation was significantly lower in LPS-treated mice at 24 h (15.3 ± 1.9 pmol/mg protein/min), compared with that for the saline control group (32.5 ± 6.7 pmol/mg protein/min), but was recovering toward baseline at 48 h (22.2 ± 4.9 pmol/mg protein/min; P = 0.45 versus saline) (Fig. 3D). No significant differences in renal EET+DHET or 20-HETE formation were observed 3 or 6 h after LPS administration. A significant correlation between total P450 epoxygenase and ω-hydroxylase metabolic activity and renal Cyp2j5 (r = 0.42, P = 0.012) and Cyp4a12a mRNA levels (r = 0.55, P < 0.001), respectively, was observed.
Lung.
Cyp2j9 was the most abundant P450 epoxygenase isoform expressed in lung (Fig. 4A). Cyp2j9 and Cyp2c44 mRNA levels were significantly lower in LPS-treated mice at 3, 6, 24, and 48 h (P < 0.05 versus saline) (Fig. 4B). Cyp2j5 also appeared to be suppressed at 24 h (0.20 ± 0.05; P = 0.11); however, because of substantial interanimal variability, these differences were not statistically significant (Fig. 4B). After LPS administration, Cyp4a12a, Cyp4a12b, Cyp4f13, and Cyp4f16 mRNA levels were significantly lower at almost every time point, although Cyp4f16 returned to basal levels at 48 h (Fig. 4C).
Fig. 4.
A, relative abundance of pulmonary P450 mRNA was quantified in saline-treated mice (n = 23 for detected isoforms; n = 3 for undetectable isoforms) and normalized to GAPDH. The time-dependent effect of LPS administration (1 mg/kg i.p.) on pulmonary (B) Cyp2c44, Cyp2j5, and Cyp2j9 and (C) Cyp4a12a, Cyp4a12b, Cyp4f13, and Cyp4f16 mRNA levels was quantified by qRT-PCR and expressed relative to that for the saline control group [saline (pooled), n = 23; LPS 3 h, n = 9; LPS 6 h, n = 9; LPS 24 h, n = 18; LPS 48 h, n = 6]. D, effect of LPS administration on total P450 epoxygenase (EETs+DHETs) and ω-hydroxylase (20-HETE) metabolic activity in lung microsomes (n = 2 mice/microsome preparation) was determined [saline (pooled), n = 6, LPS 3 h, n = 4; LPS 6 h, n = 3; LPS 24 h, n = 6; LPS 48 h, n = 3]. *, P < 0.05 versus saline control group. N.D., not detected.
Compared with saline-treated mice (54.1 ± 5.2 pmol/mg protein/min), total P450 epoxygenase metabolic activity in lung microsomes was lower, but not statistically significant, 3 (35.8 ± 3.0 pmol/mg protein/min, P = 0.063), 6 (38.4 ± 7.3 pmol/mg protein/min, P = 0.171), and 24 (38.7 ± 3.7 pmol/mg protein/min, P = 0.094) h after LPS treatment, whereas EET+DHET formation was significantly lower at the 48-h time point (28.9 ± 2.2 pmol/mg protein/min, P = 0.008) (Fig. 4D). Similar time-dependent changes in metabolism were observed for each regioisomer (Supplemental Fig. 2C). 20-HETE formation was significantly lower 6 (12.8 ± 5.9 pmol/mg protein/min) and 24 (9.4 ± 1.5 pmol/mg protein/min) h after LPS administration, compared with that for the saline control group (23.1 ± 1.4 pmol/mg protein/min), whereas no differences were observed at 3 or 48 h (Fig. 4D). No significant correlations between total P450 epoxygenase metabolic activity and Cyp2c44, Cyp2j5, or Cyp2j9 mRNA levels were observed. However, 20-HETE formation significantly correlated with pulmonary Cyp4a12a (r = 0.47, P = 0.026), Cyp4a12b (r = 0.46, P = 0.030), and Cyp4f13 (r = 0.46, P = 0.033) mRNA levels.
Heart/Aorta.
Overall, myocardial P450 mRNA levels were low, with Cyp4f13 and Cyp4f16 being the most abundant isoforms. Of the P450 epoxygenases examined, only Cyp2c44 was expressed at detectable levels (Fig. 5A). The P450 expression profile in aorta was similar; however, Cyp2c29 mRNA was expressed at detectable levels, and Cyp2c44 and Cyp4f13 mRNA levels were approximately 10-fold higher in aorta compared with heart (Supplemental Table 1). Myocardial Cyp2c44 mRNA levels were significantly lower 3 and 6 h after LPS administration (P < 0.05 versus saline) (Fig. 5B). Although it remained significantly suppressed at 24 h (0.51 ± 0.14), Cyp2c44 seemed to be returning to basal levels, with full recovery observed at 48 h (0.78 ± 0.13). In aorta, Cyp2c29 (0.38 ± 0.12, P = 0.128) and Cyp2c44 (0.26 ± 0.06, P = 0.128) mRNA appeared to be suppressed 24 h after LPS administration, relative to that for saline-treated mice, but this difference was not statistically significant because of substantial interanimal variability. In heart, both Cyp4f13 and Cyp4f16 mRNA levels were significantly lower in LPS-treated mice at 3 h, but significantly higher at 24 and 48 h (P < 0.05 versus saline) (Fig. 5C). Likewise, Cyp4f13 and Cyp4f16 mRNA levels in aorta were significantly higher in LPS-treated mice at 24 (Cyp4f13, 1.48 ± 0.12; Cyp4f16, 3.65 ± 0.44; P < 0.05 versus saline) and 48 h (Cyp4f13, 1.93 ± 0.18; Cyp4f16, 3.30 ± 0.61; P < 0.05 versus saline).
Fig. 5.
A, relative abundance of myocardial P450 mRNA was quantified in saline-treated mice (n = 24 for detected isoforms; n = 3 for undetectable isoforms) and normalized to GAPDH. The time-dependent effect of LPS administration (1 mg/kg i.p.) on myocardial (B) Cyp2c44 and (C) Cyp4f13 and Cyp4f16 mRNA levels was quantified by qRT-PCR and expressed relative to that for the saline control group [saline (pooled), n = 24; LPS 3 h, n = 9; LPS 6 h, n = 9; LPS 24 h, n = 18; LPS 48 h, n = 6]. D, effect of LPS administration on total P450 epoxygenase (EETs+DHETs) and ω-hydroxylase (20-HETE) metabolic activity in heart microsomes (n = 3–4 mice/microsome preparation) was determined [saline (pooled), n = 4; LPS 24 h, n = 4; LPS 48 h, n = 2]. *, P < 0.05 versus saline control group. N.D., not detected.
Total P450 epoxygenase metabolic activity (3.97 ± 0.44 pmol/mg protein/min) was approximately 8-fold higher than P450 ω-hydroxylase activity (0.51 ± 0.04 pmol/mg protein/min) in heart microsomes under basal conditions (Fig. 5D). Compared with saline, no significant differences in EET+DHET or 20-HETE formation were observed 24 h after LPS administration. Although EET+DHET formation appeared to be lower and 20-HETE formation appeared to be higher 48 h after LPS administration, the limited sample size (n = 2 incubations) at 48 h precluded statistical comparisons.
Ephx2 Expression.
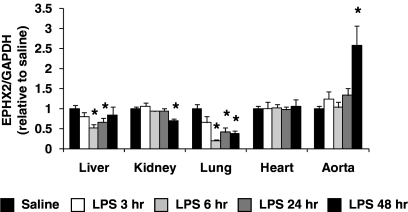
Ephx2 was abundantly expressed in all tissues examined. In liver, kidney, and lung, Ephx2 mRNA levels were similar to those for the P450 isoforms examined. In contrast, Ephx2 was approximately 100- and 10-fold more abundant than Cyp4f13 in heart and aorta, respectively (Supplemental Table 1). Tissue-specific alterations in Ephx2 expression were observed after LPS administration (Fig. 6). In liver, kidney, and lung, Ephx2 expression was suppressed by 30 to 80% 6 to 48 h after LPS administration, relative to the saline control. In contrast, no differences were observed in heart, and Ephx2 mRNA levels were significantly higher in aorta at 48 h (2.57 ± 0.49, P < 0.05 versus saline).
Fig. 6.
The time-dependent effect of LPS administration (1 mg/kg i.p.) on Ephx2 mRNA levels in liver, kidney, lung, heart, and aorta was quantified by qRT-PCR and expressed relative to that for the saline control group [saline (pooled), n = 18–24; LPS 3 h, n = 6–9; LPS 6 h, n = 6–9; LPS 24 h, n = 15–18; LPS 48 h, n = 6]. *, P < 0.05 versus saline control group.
Functional Balance between P450 Epoxygenase and ω-Hydroxylase Pathways across Tissues.
Under basal conditions, hepatic EET+DHET formation was higher than 20-HETE formation, resulting in a 20-HETE/EET+DHET formation rate ratio of 0.64 ± 0.03 in saline-treated mice (Fig. 7A). A similar 20-HETE/EET+DHET formation rate ratio was observed in lung (0.45 ± 0.06) (Fig. 7C) and heart (0.13 ± 0.02) (Fig. 7D), indicative of higher P450 epoxygenase metabolic activity relative to P450 ω-hydroxylase metabolic activity. In contrast, the renal ratio of P450 ω-hydroxylase to P450 epoxygenase metabolic activity was 1.69 ± 0.25 (Fig. 7B) because of higher 20-HETE compared with EET+DHET formation under basal conditions in kidney.
Fig. 7.
The time-dependent effect of LPS administration (1 mg/kg i.p.) on the ratio of 20-HETE to EET+DHET formation was determined in (A) liver, (B) kidney, (C) lung, and (D) heart microsomes. Saline, n = 4–12; LPS 3 h, n = 4–6; LPS 6 h, n = 3–6; LPS 24 h, n = 4–12; LPS 48 h, n = 2–6. *, P < 0.05 compared with saline-treated mice.
After LPS stimulation, the hepatic 20-HETE/EET+DHET formation rate ratio was significantly greater at 24 (1.02 ± 0.06) and 48 (1.05 ± 0.06) h compared with that in saline-treated mice (Fig. 7A). Although the ratio of P450 ω-hydroxylase to P450 epoxygenase metabolic activity in heart appeared higher 48 h after LPS treatment (0.23 ± 0.06), the limited sample size precluded formal statistical comparisons (Fig. 7D). In contrast, the pulmonary 20-HETE/EET+DHET formation rate ratio was significantly lower at 24 h (0.24 ± 0.02) compared with that for saline-treated mice (Fig. 7C). In kidney, no significant differences in the ratio of P450 ω-hydroxylase to P450 epoxygenase metabolic activity were observed after LPS treatment (Fig. 7B), which remained greater than 1.0 at all time points.
Compared with those in saline-treated mice, plasma 14,15-DHET levels were significantly higher 6 h after LPS administration; however, no differences in DHET levels were observed at 3, 24, or 48 h (Supplemental Fig. 2E). Plasma EETs and 20-HETE were below the limit of detection in the majority of mice (data not shown).
Discussion
Although it is well established that acute inflammatory stimuli suppress hepatic P450 expression and xenobiotic metabolism, the effect on P450-mediated eicosanoid metabolism in hepatic and extrahepatic tissues has not been characterized to date. To our knowledge, this is the first study demonstrating that acute activation of the innate immune response alters P450 epoxygenase and ω-hydroxylase metabolic activity in mice through pretranslational regulation of P450 expression and disrupts the functional balance between these parallel pathways in a tissue- and time-dependent manner. Collectively, these findings suggest that alteration of P450-mediated eicosanoid metabolism is an important consequence of the acute inflammatory response in vivo.
Under basal conditions, P450 epoxygenase and ω-hydroxylase metabolic activity in liver was approximately 10- to 20-fold greater than that in kidney and lung and 100-fold greater than that in heart. In contrast to levels in the other tissues examined, P450 mRNA levels in heart and aorta were low, with Cyp4f13 and Cyp4f16 being the most abundant isoforms. Myocardial EET+DHET formation was approximately 8-fold higher than 20-HETE formation despite the high levels of Cyp4f13 and Cyp4f16, suggesting that these isoforms play a minor role in 20-HETE biosynthesis.
Consistent with previous studies in human hepatocyte culture (Abdel-Razzak et al., 1993) and in rodents (Sewer et al., 1996; Barclay et al., 1999), hepatic P450 epoxygenase mRNA, protein, and metabolic activity was suppressed 24 h after LPS treatment, with partial (CYP2C) or full (CYP2J) restoration to basal levels at 48 h. The correlation between time-dependent changes in hepatic Cyp2c29, Cyp2c44, and Cyp2j5 mRNA levels and epoxygenase metabolic activity support the hypothesis that inflammation-mediated alterations in P450 metabolism are mediated primarily via pretranslational regulation of P450 expression (Morgan et al., 2008). Although Cyp2c29, Cyp2c44, and Cyp2j5 catalyze the formation of individual EET regioisomers in different proportions (Luo et al., 1998; Ma et al., 1999; DeLozier et al., 2004), the collective suppression of all EET+DHET regioisomers is consistent with the regioisomer profiles for these isoforms.
In contrast to the majority of P450s, hepatic CYP4A and CYP4F isoforms may be induced by inflammation, although this seems to be isoform- and species-specific (Sewer et al., 1996; Barclay et al., 1999; Kalsotra et al., 2003; Anwar-mohamed et al., 2010). The temporal profiles of hepatic Cyp4a12a, Cyp4a12b, and Cyp4f13 mRNA levels were similar to that for 20-HETE formation, which was also suppressed at 24 h, but to a lesser degree than EET+DHET formation. The significantly higher 20-HETE/EET+DHET formation rate ratio at 24 and 48 h suggests that after systemic activation of the innate immune response, the functional balance in liver is tipped in favor of the proinflammatory P450 ω-hydroxylase pathway.
Consistent with previous findings in rats (Anwar-mohamed et al., 2010), LPS suppressed renal Cyp2c44 and Cyp2j5 expression and epoxygenase metabolic activity, which correlated with Cyp2j5 mRNA levels. This result suggests that the mechanism is pretranslational and that Cyp2j5 is primarily responsible for renal EET formation. We observed lower Cyp4a12a but higher Cyp4a12b, Cyp4f13, and Cyp4f16 mRNA levels in kidney after LPS administration. Renal Cyp4a10 mRNA levels were also higher 24 and 48 h after LPS administration (data not shown), which is consistent with previous reports (Barclay et al., 1999). However, Cyp4a10 does not readily catalyze 20-HETE formation (Muller et al., 2007). Renal P450 ω-hydroxylase activity was suppressed at 24 h and significantly correlated with Cyp4a12a mRNA levels, suggesting that Cyp4a12a is primarily responsible for renal 20-HETE formation. In contrast to the other tissues examined, P450 ω-hydroxylase metabolic activity predominated under both basal and LPS-stimulated conditions, indicating that inhibition of 20-HETE biosynthesis may be a rational therapeutic strategy to attenuate renal inflammation.
In lung, our findings are consistent with studies demonstrating lower pulmonary CYP2C and CYP2J expression and epoxygenase metabolic activity in rat models of sepsis (Cui et al., 2004) and Pseudomonas pneumonia (Yaghi et al., 2003, 2004), providing further evidence that suppression of pulmonary EET formation is a key component of the pathological response to inflammation. Compared with basal conditions, 20-HETE formation and the 20-HETE/EET+DHET formation rate ratio were significantly lower 24 h after LPS administration, suggesting that the functional balance was shifted in favor of the anti-inflammatory P450 epoxygenase pathway. Although this could serve as a compensatory mechanism to facilitate resolution of the inflammatory response, additional studies are necessary to dissect the role of P450-mediated EET and 20-HETE biosynthesis in the regulation of pulmonary inflammation.
A recent study demonstrated that myocardial P450 epoxygenase activity was significantly suppressed, whereas P450 ω-hydroxylase activity was induced after LPS administration in rats (Anwar-mohamed et al., 2010). Although we observed no significant changes in P450 epoxygenase or ω-hydroxylase metabolic activity, the low arachidonic acid-metabolizing capacity of mouse heart limited our sample size. Further studies are needed to characterize the effect of inflammation on myocardial P450 epoxygenase and ω-hydroxylase metabolic activity.
Plasma EETs and 20-HETE were below the limit of detection in the majority of mice, consistent with previous reports (Schmelzer et al., 2005; Kubala et al., 2010). Plasma DHET levels appeared modestly elevated 6 h after LPS administration, consistent with previous studies (Schmelzer et al., 2005; Fife et al., 2008; Kubala et al., 2010); however, only elevation of 14,15-DHET was statistically significant. Of importance, the primary source of circulating EET, DHET, and 20-HETE levels in plasma has not been determined. A recent study demonstrated that LPS-induced elevations in plasma DHETs were attenuated in myeloperoxidase-deficient [Mpo(−/−)] mice, suggesting that lipid peroxidation by reactive oxygen species may serve as a P450-independent source of circulating eicosanoids (Kubala et al., 2010). Furthermore, because EETs and 20-HETE circulate at levels <1.0 ng/ml, it has been hypothesized that these mediators act predominantly in a paracrine manner (Roman, 2002). Our data demonstrate that systemic activation of the innate immune response significantly alters the formation of EETs and 20-HETE in tissue. Further studies to characterize the functional consequences of these local effects remain necessary.
It is believed that the primary mechanism by which P450 expression is down-regulated in response to LPS is cytokine-mediated suppression of gene transcription. However, identification of the specific nuclear receptors that mediate these effects has remained elusive (Morgan, 2001; Riddick et al., 2004; Morgan et al., 2008). For example, an initial study suggested that LPS-mediated alteration of hepatic and renal P450 expression was mediated by peroxisome proliferator-activated receptor-α (Barclay et al., 1999). However, subsequent experiments have demonstrated that LPS-mediated down-regulation of hepatic P450 expression is independent of peroxisome proliferator-activated receptor-α, pregnane X receptor, and signal transducer and activator of transcription-1 (Pan et al., 2003; Richardson and Morgan, 2005). Although we did not directly investigate the mechanism by which LPS alters P450 expression and metabolic activity, the observed isoform- and tissue-specific response suggests that the mechanism is complex and most likely involves multiple transcription factors that vary across isoforms and tissues. Moreover, the time-dependent changes observed further suggest a multifactorial mechanism. In addition, multiple murine P450 isoforms catalyze EET and 20-HETE formation, further complicating the ability to identify a single factor that underlies the net impact of the inflammatory response on P450-mediated eicosanoid metabolism in each tissue. Further studies are necessary to dissect these complex mechanisms.
P450-derived EETs possess anti-inflammatory properties via inhibition of NF-κB activation (Node et al., 1999), whereas 20-HETE activates NF-κB signaling and elicits proinflammatory effects (Ishizuka et al., 2008). These opposing effects on the regulation of inflammation suggest that inflammation-induced alterations in the functional balance between these parallel pathways may contribute to the pathologic consequences of the inflammatory response. A limitation of the current work is that we did not directly demonstrate that potentiation of the P450 epoxygenase pathway and/or inhibition of the P450 ω-hydroxylase pathway attenuates the acute inflammatory response to LPS in each tissue. However, our findings provide an important foundation to guide future studies evaluating these therapeutic strategies, particularly in tissues in which inflammatory stimuli tip the functional balance in favor of P450-mediated 20-HETE biosynthesis, such as liver. Indeed, the liver is the predominant source of cytokine production and drives the systemic inflammatory response, as observed during sepsis, via subsequent activation of extrahepatic inflammation and multiorgan dysfunction (Szabo et al., 2002). Inhibition of sEH-mediated EET hydrolysis has potent anti-inflammatory effects, including attenuation of endotoxemia-induced hypotension and mortality (Schmelzer et al., 2005; Luria et al., 2007; Liu et al., 2009). In contrast, a recent study demonstrated that LPS-induced hepatic inflammation was not attenuated in Ephx2(−/−) mice or by sEH inhibition (Fife et al., 2008). Although the mechanisms underlying these conflicting findings remain unclear, our data suggest that dual inhibition of 20-HETE biosynthesis and sEH-mediated EET hydrolysis may be a more effective means to attenuate hepatic inflammation. Further studies are necessary to evaluate these therapeutic strategies in disease models of local and systemic inflammation.
In conclusion, our findings demonstrate that acute activation of the inflammatory response with LPS alters P450 epoxygenase and ω-hydroxylase expression and metabolic activity in a tissue-, isoform-, and time-dependent manner. These results highlight the relative differences in P450-mediated eicosanoid metabolism across tissues under basal and inflammatory conditions and lay an important foundation to guide future studies that seek to determine whether therapeutic restoration of the functional balance between the P450 epoxygenase and ω-hydroxylase pathways is an effective anti-inflammatory strategy in vivo.
Supplementary Material
This work was supported in part by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant R01-NS052315]; the National Institutes of Health National Center of Research Resources [Grant S10-RR023461]; the National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM088199]; the American Foundation for Pharmaceutical Education (Predoctoral Fellowship); the University of North Carolina at Chapel Hill (Junior Faculty Development Award); and the American Heart Association [Beginning Grant-in-Aid].
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences, National Institute of Neurological Disorders and Stroke, or National Institutes of Health.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.035287.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- P450
- cytochrome P450
- EET
- epoxyeicosatrienoic acid
- 20-HETE
- 20-hydroxyeiocatetraenoic acid
- sEH
- soluble epoxide hydrolase
- DHET
- dihydroxyeicosatrienoic acid
- LPS
- lipopolysaccharide
- NF-κB
- nuclear factor-κB
- q
- quantitative
- RT
- reverse transcription
- PCR
- polymerase chain reaction
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- UPLC
- ultraperformance liquid chromatography
- TNF-α
- tumor necrosis factor-α.
Authorship Contributions
Participated in research design: Theken, Deng, Poloyac, and Lee.
Conducted experiments: Theken, Deng, Kannon, and Miller.
Contributed new reagents or analytic tools: Miller and Poloyac.
Performed data analysis: Theken and Lee.
Wrote or contributed to the writing of the manuscript: Theken, Poloyac, and Lee.
Other: Theken, Poloyac, and Lee acquired funding for the research.
References
- Abdel-Razzak Z, Loyer P, Fautrel A, Gautier JC, Corcos L, Turlin B, Beaune P, Guillouzo A. (1993) Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Mol Pharmacol 44:707–715 [PubMed] [Google Scholar]
- Anwar-mohamed A, Zordoky BN, Aboutabl ME, El-Kadi AO. (2010) Alteration of cardiac cytochrome P450-mediated arachidonic acid metabolism in response to lipopolysaccharide-induced acute systemic inflammation. Pharmacol Res 61:410–418 [DOI] [PubMed] [Google Scholar]
- Barclay TB, Peters JM, Sewer MB, Ferrari L, Gonzalez FJ, Morgan ET. (1999) Modulation of cytochrome P-450 gene expression in endotoxemic mice is tissue specific and peroxisome proliferator-activated receptor-α dependent. J Pharmacol Exp Ther 290:1250–1257 [PubMed] [Google Scholar]
- Christmas P, Jones JP, Patten CJ, Rock DA, Zheng Y, Cheng SM, Weber BM, Carlesso N, Scadden DT, Rettie AE, et al. (2001) Alternative splicing determines the function of CYP4F3 by switching substrate specificity. J Biol Chem 276:38166–38172 [DOI] [PubMed] [Google Scholar]
- Cui X, Wu R, Zhou M, Simms HH, Wang P. (2004) Differential expression of cytochrome P450 isoforms in the lungs of septic animals. Crit Care Med 32:1186–1191 [DOI] [PubMed] [Google Scholar]
- DeLozier TC, Tsao CC, Coulter SJ, Foley J, Bradbury JA, Zeldin DC, Goldstein JA. (2004) CYP2C44, a new murine CYP2C that metabolizes arachidonic acid to unique stereospecific products. J Pharmacol Exp Ther 310:845–854 [DOI] [PubMed] [Google Scholar]
- Deng Y, Theken KN, Lee CR. (2010) Cytochrome P450 epoxygenases, soluble epoxide hydrolase, and the regulation of cardiovascular inflammation. J Mol Cell Cardiol 48:331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fife KL, Liu Y, Schmelzer KR, Tsai HJ, Kim IH, Morisseau C, Hammock BD, Kroetz DL. (2008) Inhibition of soluble epoxide hydrolase does not protect against endotoxin-mediated hepatic inflammation. J Pharmacol Exp Ther 327:707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources (1996) Guide for the Care and Use of Laboratory Animals, 7th ed., Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council, Washington, DC [Google Scholar]
- Ishizuka T, Cheng J, Singh H, Vitto MD, Manthati VL, Falck JR, Laniado-Schwartzman M. (2008) 20-Hydroxyeicosatetraenoic acid stimulates nuclear factor-κB activation and the production of inflammatory cytokines in human endothelial cells. J Pharmacol Exp Ther 324:103–110 [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cui X, Antonovic L, Robida AM, Morgan ET, Strobel HW. (2003) Inflammatory prompts produce isoform-specific changes in the expression of leukotriene B4 ω-hydroxylases in rat liver and kidney. FEBS Lett 555:236–242 [DOI] [PubMed] [Google Scholar]
- Kubala L, Schmelzer KR, Klinke A, Kolarova H, Baldus S, Hammock BD, Eiserich JP. (2010) Modulation of arachidonic and linoleic acid metabolites in myeloperoxidase-deficient mice during acute inflammation. Free Radic Biol Med 48:1311–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Bottone FG, Jr, Krahn JM, Li L, Mohrenweiser HW, Cook ME, Petrovich RM, Bell DA, Eling TE, Zeldin DC. (2007) Identification and functional characterization of polymorphisms in human cyclooxygenase-1 (PTGS1). Pharmacogenet Genomics 17:145–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Tsai HJ, Hwang SH, Jones PD, Morisseau C, Hammock BD. (2009) Pharmacokinetic optimization of four soluble epoxide hydrolase inhibitors for use in a murine model of inflammation. Br J Pharmacol 156:284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Luo G, Zeldin DC, Blaisdell JA, Hodgson E, Goldstein JA. (1998) Cloning and expression of murine CYP2Cs and their ability to metabolize arachidonic acid. Arch Biochem Biophys 357:45–57 [DOI] [PubMed] [Google Scholar]
- Luria A, Weldon SM, Kabcenell AK, Ingraham RH, Matera D, Jiang H, Gill R, Morisseau C, Newman JW, Hammock BD. (2007) Compensatory mechanism for homeostatic blood pressure regulation in Ephx2 gene-disrupted mice. J Biol Chem 282:2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Qu W, Scarborough PE, Tomer KB, Moomaw CR, Maronpot R, Davis LS, Breyer MD, Zeldin DC. (1999) Molecular cloning, enzymatic characterization, developmental expression, and cellular localization of a mouse cytochrome P450 highly expressed in kidney. J Biol Chem 274:17777–17788 [DOI] [PubMed] [Google Scholar]
- Miller TM, Donnelly MK, Crago EA, Roman DM, Sherwood PR, Horowitz MB, Poloyac SM. (2009) Rapid, simultaneous quantitation of mono and dioxygenated metabolites of arachidonic acid in human CSF and rat brain. J Chromatogr B Analyt Technol Biomed Life Sci 877:3991–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan ET. (2001) Regulation of cytochrome P450 by inflammatory mediators: why and how? Drug Metab Dispos 29:207–212 [PubMed] [Google Scholar]
- Morgan ET, Goralski KB, Piquette-Miller M, Renton KW, Robertson GR, Chaluvadi MR, Charles KA, Clarke SJ, Kacevska M, Liddle C, et al. (2008) Regulation of drug-metabolizing enzymes and transporters in infection, inflammation, and cancer. Drug Metab Dispos 36:205–216 [DOI] [PubMed] [Google Scholar]
- Muller DN, Schmidt C, Barbosa-Sicard E, Wellner M, Gross V, Hercule H, Markovic M, Honeck H, Luft FC, Schunck WH. (2007) Mouse Cyp4a isoforms: enzymatic properties, gender- and strain-specific expression, and role in renal 20-hydroxyeicosatetraenoic acid formation. Biochem J 403:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Node K, Huo Y, Ruan X, Yang B, Spiecker M, Ley K, Zeldin DC, Liao JK. (1999) Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science 285:1276–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Xiang Q, Ball S, Scatina J, Kao J, Hong JY. (2003) Lipopolysaccharide-mediated modulation of cytochromes P450 in Stat1 null mice. Drug Metab Dispos 31:392–397 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloyac SM, Tortorici MA, Przychodzin DI, Reynolds RB, Xie W, Frye RF, Zemaitis MA. (2004) The effect of isoniazid on CYP2E1- and CYP4A-mediated hydroxylation of arachidonic acid in the rat liver and kidney. Drug Metab Dispos 32:727–733 [DOI] [PubMed] [Google Scholar]
- Powell PK, Wolf I, Jin R, Lasker JM. (1998) Metabolism of arachidonic acid to 20-hydroxy-5,8,11,14-eicosatetraenoic acid by P450 enzymes in human liver: involvement of CYP4F2 and CYP4A11. J Pharmacol Exp Ther 285:1327–1336 [PubMed] [Google Scholar]
- Qu W, Bradbury JA, Tsao CC, Maronpot R, Harry GJ, Parker CE, Davis LS, Breyer MD, Waalkes MP, Falck JR, et al. (2001) Cytochrome P450 CYP2J9, a new mouse arachidonic acid omega-1 hydroxylase predominantly expressed in brain. J Biol Chem 276:25467–25479 [DOI] [PubMed] [Google Scholar]
- Richardson TA, Morgan ET. (2005) Hepatic cytochrome P450 gene regulation during endotoxin-induced inflammation in nuclear receptor knockout mice. J Pharmacol Exp Ther 314:703–709 [DOI] [PubMed] [Google Scholar]
- Riddick DS, Lee C, Bhathena A, Timsit YE, Cheng PY, Morgan ET, Prough RA, Ripp SL, Miller KK, Jahan A, et al. (2004) Transcriptional suppression of cytochrome P450 genes by endogenous and exogenous chemicals. Drug Metab Dispos 32:367–375 [DOI] [PubMed] [Google Scholar]
- Roman RJ. (2002) P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev 82:131–185 [DOI] [PubMed] [Google Scholar]
- Schmelzer KR, Kubala L, Newman JW, Kim IH, Eiserich JP, Hammock BD. (2005) Soluble epoxide hydrolase is a therapeutic target for acute inflammation. Proc Natl Acad Sci USA 102:9772–9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer MB, Koop DR, Morgan ET. (1996) Endotoxemia in rats is associated with induction of the P4504A subfamily and suppression of several other forms of cytochrome P450. Drug Metab Dispos 24:401–407 [PubMed] [Google Scholar]
- Szabo G, Romics L, Jr, Frendl G. (2002) Liver in sepsis and systemic inflammatory response syndrome. Clin Liver Dis 6:1045–1066 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao Y, Bradbury JA, Graves JP, Foley J, Blaisdell JA, Goldstein JA, Zeldin DC. (2004) Cloning, expression, and characterization of three new mouse cytochrome P450 enzymes and partial characterization of their fatty acid oxidation activities. Mol Pharmacol 65:1148–1158 [DOI] [PubMed] [Google Scholar]
- Xu F, Falck JR, Ortiz de Montellano PR, Kroetz DL. (2004) Catalytic activity and isoform-specific inhibition of rat cytochrome P450 4F enzymes. J Pharmacol Exp Ther 308:887–895 [DOI] [PubMed] [Google Scholar]
- Yaghi A, Bend JR, Webb CD, Zeldin DC, Weicker S, Mehta S, McCormack DG. (2004) Excess nitric oxide decreases cytochrome P-450 2J4 content and P-450-dependent arachidonic acid metabolism in lungs of rats with acute pneumonia. Am J Physiol Lung Cell Mol Physiol 286:L1260–L1267 [DOI] [PubMed] [Google Scholar]
- Yaghi A, Bradbury JA, Zeldin DC, Mehta S, Bend JR, McCormack DG. (2003) Pulmonary cytochrome P-450 2J4 is reduced in a rat model of acute Pseudomonas pneumonia. Am J Physiol Lung Cell Mol Physiol 285:L1099–L1105 [DOI] [PubMed] [Google Scholar]
- Zeldin DC. (2001) Epoxygenase pathways of arachidonic acid metabolism. J Biol Chem 276:36059–36062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.