Abstract
Human ether-à-go-go-related gene 1 (hERG1) channels conduct the rapid delayed rectifier K+ current, IKr, an important determinant of action potential repolarization in mammals, including humans. Reduced IKr function caused by mutations in KCNH2 or drug block of hERG1 channels prolongs the QT interval of the electrocardiogram and increases the risk of ventricular fibrillation and sudden cardiac death. Several activators of hERG1 channels have been discovered in recent years. These compounds shorten the duration of cardiac action potentials and have been proposed as a new therapeutic approach for the treatment of acquired or congenital long QT syndrome. We defined previously the mechanism of action of 1,3-bis-(2-hydroxy-5-trifluoromethyl-phenyl)-urea (NS1643), a compound that increases hERG1 currents by shifting the voltage-dependence of inactivation to more positive potentials. Here, we use scanning mutagenesis of hERG1 and functional characterization of 56 mutant channels heterologously expressed in Xenopus laevis oocytes to define the molecular determinants of the binding site for NS1643. Most point mutations did not alter response to the drug; however, 10 mutant channels had reduced sensitivity, and F619A and I567A exhibited enhanced activation by the drug. Some of these residues form a cluster and, together with molecular modeling, suggest that NS1643 binds to a pocket near the extracellular ends of the S5/S6 segments of two adjacent hERG1 channel subunits. This putative binding site differs from the sites described previously for two other hERG1 activators, (3R,4R)-4-[3-(6-methoxy-quinolin-4-yl)-3-oxo-propyl]-1-[3-(2,3,5-trifluoro-phenyl)-prop-2-ynyl]-piperidine-3-carboxylic acid (RPR260243) and 2-(4-[2-(3,4-dichloro-phenyl)-ethyl]-phenylamino)-benzoic acid (PD-118057).
Introduction
Repolarization of the cardiac action potential is mediated by several K+ currents. In mammals, including humans, three K+ currents (IKr, IKs, and IK1) are predominantly responsible for terminating the action potential. hERG1 (Kv11.1) channel subunits coassemble to form the channels that conduct IKr (Sanguinetti et al., 1995; Trudeau et al., 1995). hERG1a is the original variant identified (Warmke and Ganetzky, 1994) and is the most well characterized in biophysical and pharmacological studies of heterologously expressed channels. Later, an alternatively spliced variant with a shorter N terminus was identified and named hERG1b (Lees-Miller et al., 1997; London et al., 1997). Heterologously expressed hERG1b channels deactivate faster than hERG1a channels, and coexpression of hERG1a plus hERG1b reveals a current with biophysical properties that most resembles native IKr (Lees-Miller et al., 1997; London et al., 1997; Larsen et al., 2008).
hERG1 channels have been extensively studied because of their unusual gating properties and association with congenital and acquired cardiac arrhythmias. Loss-of-function mutations in HERG1 (KCNH2) cause long QT syndrome type 2 (LQT2) (Curran et al., 1995), revealed on the body surface electrocardiogram as a prolonged QT interval and a small T-wave amplitude (Moss et al., 1995). The mechanism underlying prolonged QT intervals is delayed repolarization of ventricular action potentials. Serious arrhythmias (torsades de pointes, ventricular fibrillation) and sudden death associated with LQT2 emphasizes the importance of hERG1 channels for normal repolarization of ventricular action potentials. However, congenital long QT syndrome is relatively rare compared with acquired forms of cardiac repolarization disorders. Unintended block of hERG1 channels by noncardiac medications is a common cause of torsades de pointes and can sometimes lead to ventricular fibrillation and sudden cardiac death (Sanguinetti and Tristani-Firouzi, 2006). For this reason, in early stages of drug development, compounds are routinely assayed for their effects on hERG1 channel current. These assays have revealed the promiscuous nature of the hERG1 channel with regard to block by a wide spectrum of chemical entities (Redfern et al., 2003; Sanguinetti and Mitcheson, 2005). In addition, compounds have been discovered that enhance rather than inhibit hERG1 channel current. Discovered hERG1 channel activators include (3R,4R)-4-[3-(6-methoxyquinolin-4-yl)-3-oxo-propyl]-1-[3-(2,3,5-trifluoro-phenyl)-prop-2-ynyl]-piperidine-3-carboxylic acid (RPR260243) (Kang et al., 2005; Perry et al., 2007), 1,3-bis-(2-hydroxy-5-trifluoromethyl-phenyl)-urea (NS1643) (Casis et al., 2006; Hansen et al., 2006a), 2-[2-(3,4-dichloro-phenyl)-2,3-dihydro-1H-isoindol-5-ylamino]-nicotinic acid (PD-307243) (Gordon et al., 2008; Xu et al., 2008), 2-(4-[2-(3,4-dichloro-phenyl)-ethyl]-phenylamino)-benzoic acid (PD-118057) (Zhou et al., 2005; Perry et al., 2009), N-(4-bromo-2-(1H-tetrazol-5-yl)-phenyl)-N′-(3′-trifluoromethylphenyl)urea (NS3623) (Hansen et al., 2006b), {4-[4-(5-trifluoromethyl-1H-pyrazol-3-yl)-phenyl]-cyclohexyl}-acetic acid (A-935142) (Su et al., 2009), and 3-nitro-N-(4-phenoxyphenyl) benzamide (ICA-105574) (Gerlach et al., 2010).
The putative binding sites for hERG1 channel activators have been defined using site-directed mutagenesis, voltage-clamp assays, and molecular modeling. The binding site for RPR260243 was mapped to the intersection of the cytosolic ends of the S5 and S6 segments of a single hERG1 subunit (Perry et al., 2007). The binding site for PD-118057 was mapped to a hydrophobic pocket located between the S6 segment and pore helix of adjacent subunits of the channel (Perry et al., 2009). In contrast to RPR260243 and PD-118057, the binding site for NS1643 on hERG1 was proposed to be located on the outside of the channel (Xu et al., 2008). Several findings by Xu et al. (2008) support this hypothesis, including the lack of effect of NS1643 when injected into the cytoplasm of an oocyte, attenuation of agonist effect by mutations of residues located in the outer vestibule of the channel, and block of agonist activity by pretreatment of cells with BeKm-1, a peptide toxin that blocks hERG1 channels by binding to the outer mouth of the channel (Zhang et al., 2003). However, the specific mutations in the S5-P linker that reduced the effects of NS1643 (Xu et al., 2008) also significantly attenuate channel inactivation. Because the activation of hERG1 by NS1643 is mediated by altered channel inactivation, it would be expected that mutations that reduce inactivation would also reduce the agonist activity of the drug. For these reasons, the location of the binding site for NS1643 on the hERG1 channel remains uncertain. Here we use site-directed mutagenesis, functional voltage-clamp assay, and molecular modeling to identify the location of the binding site for NS1643 on hERG1.
Materials and Methods
Isolation and cRNA Injection of Oocytes.
Oocytes were collected from Xenopus laevis frogs anesthetized by immersion in a tricaine solution (2 g/l; Sigma-Aldrich, St. Louis, MO) according to guidelines approved by the Danish National Committee for Animal Studies and the University of Utah Institutional Animal Care and Use Committee. Isolated oocytes were incubated for 24 h at 19°C in Kulori medium consisting of 90 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM HEPES, pH 7.4 adjusted to with NaOH before injection with 50 nl of mRNA (approximately 5 ng) using a Nanoject microinjector (Drummond Scientific, Broomall, PA). Injected oocytes were incubated at 17 to 19°C occasionally in the presence of gentamicin and pyruvic acid for an additional 2 to 5 days before measurements were performed.
In Vitro Transcription.
Wild-type (WT) hERG1a (Kv11.1) cDNA was subcloned into the pXOOM and pSP64 expression vectors containing the 5′- and 3′-untranslated regions for X. laevis β-globin and a poly(A) segment, as described previously. Mutations in hERG1a pSP64 were introduced by site-directed mutagenesis using QuikChange (Stratagene, La Jolla, CA). Constructs were linearized with BamHI or EcoRI and in vitro transcribed with SP6 CapScribe (Roche Diagnostics, Indianapolis, IN) or mMessage mMachine kit (Ambion, Austin, TX). RNA concentrations were quantified by UV spectroscopy and RNA quality was checked by gel electrophoresis.
Two-Electrode Voltage-Clamp and Data Analysis.
Currents were monitored using a Dagan CA-1B (Dagan Corporation, Minneapolis, MN) or GeneClamp 500 (Molecular Devices, Union City, CA) two-electrode voltage-clamp amplifier. The pCLAMP 9 software suite was used for data acquisition and analysis. Electrodes were pulled from borosilicate glass capillaries and had tip resistance between 0.5 and 2.5 MΩ when filled with 1 M KCl. During the experiments oocytes were placed in a small chamber (volume, 200 μl) connected to a continuous flow system (flow, 2–3 ml/min). All recordings were performed at room temperature in Kulori solutions consisting of 89 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM HEPES, pH 7.4, with NaOH. Only oocytes with membrane potentials lower than −25 mV were used for current recordings.
To determine a standard current-voltage (I-V) relationship, the holding potential was −80 or −100 mV, and currents were activated with 1-s voltage steps to test potentials (Vt) ranging from −80 to +40 mV, applied in 10-mV increments. After each test pulse, the membrane was repolarized to −60 mV for 1.5 s to record deactivating (tail) currents. A plot of the peak tail current amplitude I versus Vt was fitted to a Boltzmann function to determine the voltage dependence of current activation: I = (Imax /{1 + e(V1/2 − Vt)/k}), where Imax is the maximal peak tail current amplitude, V1/2 is the voltage required for half-activation, and k is the slope factor. Fully activated I-V relationships were determined using a 1-s prepulse to +40 mV followed by a 3-s repolarizing step to a Vt that ranged from −120 to +40 mV. Between voltage steps, the oocyte was clamped at −80 or −100 mV for a minimum of 2 s.
Data analyses were performed using IGOR (Wavemetrics, Lake Oswego, OR) or Origin 8.1 (OriginLab Corp, Northampton, MA), pCLAMP 8 (Molecular Devices, Sunnyvale, CA) and GraphPad Prism (GraphPad Software, Inc., San Diego, CA) software. Data are presented as mean and S.E.M. values.
Molecular Modeling.
Modeling and docking were performed with the Insight II modules Homology, Builder, and Docking (version 8.2; Accelrys, San Diego, CA). The crystal structure of the mammalian Shaker Kv1.2 potassium channel (Protein Data Bank identification number 2A79) was used for creating a hERG1 homology model as described previously (Perry et al., 2009). A model of NS1643 structure was created and energy optimized. Its interaction with residues with the pore domains (from S4–S5 linker to the C-terminal end of S6) of two adjacent hERG1 subunits were determined by molecular docking at a maximal distance of 15 Å from residue Cys643 in hERG1. The docking was initiated from 200 random configurations of NS1643. Both the drug and hERG1 residues were flexible during the optimization procedure, and the consistent valence force field was used to calculate conformational energies. Optimal configurations were determined by simulated annealing with an initial temperature of 350 K and a final temperature of 300 K followed by final steepest descent minimization (steps = 1000).
Drugs and Solutions.
NS1643 was synthesized at NeuroSearch A/S (Ballerup, Denmark). The chemical structure is revealed in a previous publication (Hansen et al., 2006a). Unless otherwise noted, all other chemicals were of analytical grade and were obtained from Sigma-Aldrich. NS1643 was dissolved in dimethyl sulfoxide as a 100 mM concentrated stock solution and diluted directly into the superfusion solution to yield the final drug concentration. dimethyl sulfoxide concentration never exceeded 0.1% in final solutions. At this concentration no influence on any measurements were observed (data not shown).
Results
NS1643 Activates WT hERG1a Channels.
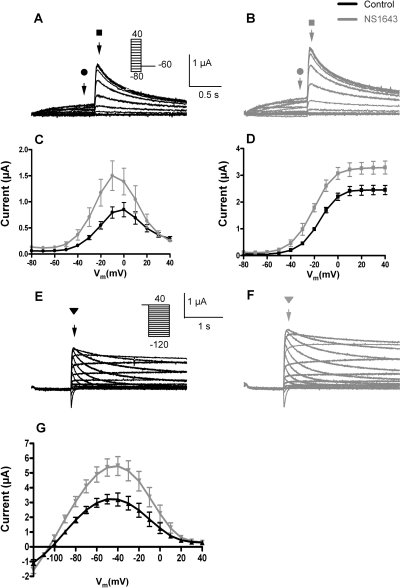
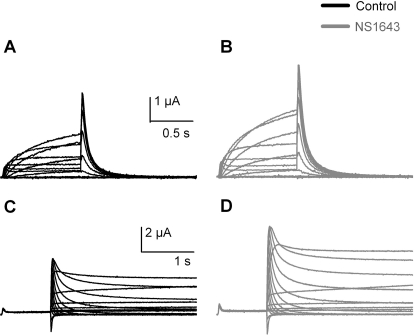
Currents conducted by hERG1a channels heterologously expressed in X. laevis oocytes were recorded using two different voltage-clamp protocols, and standard I-V and fully activated I-V currents are shown in Figs. 1A and 1E, respectively. As reported previously (Casis et al., 2006; Hansen et al., 2006a; Xu et al., 2008), NS1643 increased the magnitude of hERG1 channel current recorded with either pulse protocol and had relatively minor effects on the rate of current deactivation. At 30 μM, NS1643 enhanced currents at all test potentials (Fig. 1, A and B) and by ∼2-fold when measured at the peak of the standard I-V relationship (Fig. 1C). The peak value of tail currents measured at −60 mV was increased ∼1.5-fold (Fig. 1D), and the voltage-dependence of activation (V1/2) was shifted from −16.3 ± 1.3 to −21.3 ± 1.6 mV in the presence of NS1643 (n = 12). At 30 μM, NS1643 also enhanced the magnitude of currents recorded with the fully activated I-V protocol (Fig. 1, E and F), with a ∼1.7-fold change at the peak of the I-V (Fig. 1G).
Fig. 1.
Effect of NS1643 on WT hERG1 channels heterologously expressed in X. laevis oocytes. A and B, currents recorded from a single oocyte before (A) and after (B) treatment with 30 μM NS1643 using the standard I-V relationship voltage-clamp protocol. Step currents (indicated by circles) were elicited with 1-s pulses to test potentials (Vt) that ranged from −80 to +40 mV. Tail currents (indicated by squares) were measured at −60 mV. C, standard I-V relationships measured before (black circles) and after (gray circles) 30 μM NS1643. D, peak tail currents as a function of Vt measured before (black squares) and after (gray squares) 30 μM NS1643. Data were fitted to a Boltzmann function (smooth curves). For control, V1/2 = −16.3 mV; k = 8.1 mV (n = 13); for NS1643, V1/2 = −21.3 mV; k = 8.7 mV (n = 11). E and F, currents recorded from a single oocyte before (E) and after (F) treatment with 30 μM NS1643 using the fully activated I-V relationship voltage-clamp protocol. After a 1-s pulse to +40 mV, tail currents (triangles) were elicited with 3-s pulses to Vt that ranged from −120 to + 40 mV. G, fully activated I-V relationships measured before (black triangles) and after (gray triangles) 30 μM NS1643.
Scanning Mutagenesis of the Pore Domain of hERG1.
The putative binding site for two hERG1a activators, RPR260243 and PD-118057, were mapped previously using a combined site-directed mutagenesis, voltage-clamp assay, and molecular-modeling approach. A similar approach was used to identify specific residues in hERG1 that potentially interact with NS1643. We mutated 53 different residues in hERG1, including 21 residues in the S5 segment between Gly546 and Ile567, 25 residues in the S6 segment between Ser641 and Leu666, and 7 residues in the pore-helix between Thr613 and Thr623. Three of the mutant channels (T618A, S641C, and M645C) expressed poorly and were therefore studied using an extracellular solution that contained 20 mM KCl (with an equimolar reduction in [NaCl]). In addition, we evaluated the noninactivating S631A and the long QT syndrome-associated mutants R56Q and N470D. The effect of drug on channels harboring single point mutations were assayed with the same two voltage-clamp protocols used to characterize WT channels. Mutations that altered the response to NS1643, but not channel inactivation, were considered informative regarding residues that influence the binding of the drug to a site that mediates channel activation.
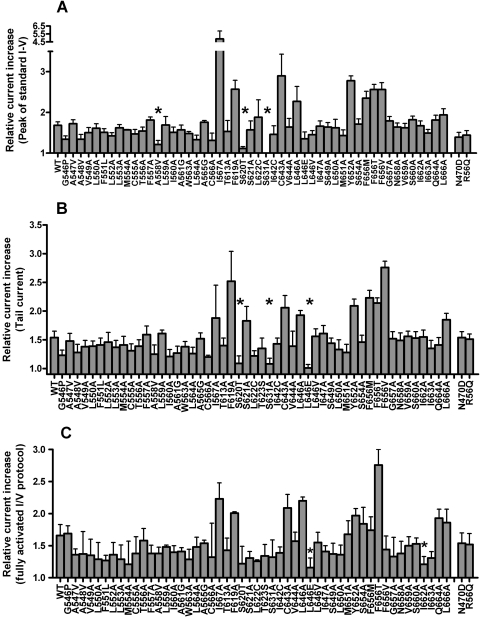
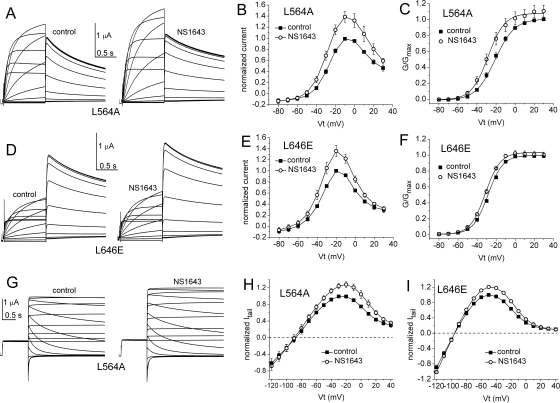
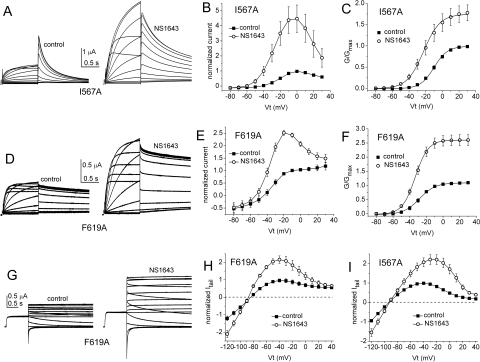
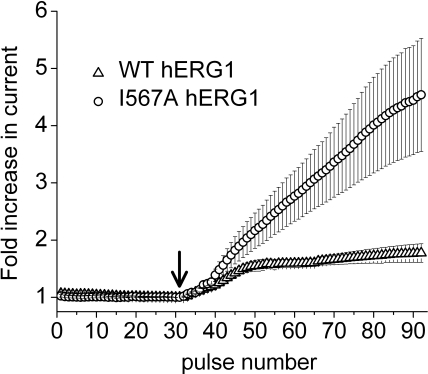
The results of the scanning mutagenesis experiments are summarized in Fig. 2. The fold increase in current magnitude induced by 30 μM NS1643, measured at the peak of the standard I-V relationship, is shown in Fig. 2A. The fold increase in the peak tail current measured at −60 mV after a 1-s test pulse is summarized in Fig. 2B, and the fold increase in the peak tail current of the fully activated I-V relationship is shown in Fig. 2C. Most mutations had no appreciable effect on the ability of 30 μM NS1643 to activate hERG1 currents. When addressing relative current increase from fully activated I-V curves (Fig. 2C), 11 point mutations (A547V, F551L, M554A, A558V, I560A, A561G, W563A, L622C, L646E, L650A, and I662A) reduced current activation by NS1643 compared with WT channels. On the other hand, only very few amino acid changes resulted in mutations being insensitive to activation by NS1643. These were A558V, S620T, and S631A for standard I-V curves, S620T, S631A, and L646E for tail current curves and L646E and I662A for fully activated I-V curves. Examples of currents recorded before and after treatment of oocytes expressing L564A or L646E hERG1 channels are shown in Fig. 3. Examples of I-V currents are shown in Fig. 3, A and D, whereas examples of fully activated I-V currents are shown in Fig. 3G. Plots of fold increase in current levels are shown in Fig. 3, B, C, E, F, H, and I. In contrast, I567A, F619A, C643A, Y652A, or mutations of Phe656 (to methionine, threonine, or valine) caused an enhanced response to the activating effects of NS1643. The almost complete lack of drug response to channels harboring mutations that eliminate inactivation (S620T, S631A) was reported previously, as was the enhanced drug response to channels containing mutations that disrupt the putative binding site for the pore-blocking effect of NS1643 (Y652A, F656M/T/V) (Casis et al., 2006; Xu et al., 2008). Currents for F619A and I567A channels are illustrated in Fig. 4. Unlike WT or other mutant channels, the increase in currents induced by NS1643 was very slow in onset for I567A hERG1 channels (Fig. 5). The mechanism responsible for this very slow enhancement of current is unknown but could be caused by an increase in trafficking of the mutant channel to the cell surface. For the three mutant channels recorded in oocytes bathed in an extracellular solution containing 20 mM KCl, the effect of drug was evaluated by determining the increase in peak tail currents elicited with the standard I-V pulse protocol. The fold increase in peak tail current by NS1643 (30 μM) for WT channels was 1.29 ± 0.016 (n = 3) and 1.45 ± 0.127 (n = 3) for T618C, 1.38 ± 0.036 (n = 5) for S641C and 1.16 ± 0.046 (n = 3) for M645C channels (data not shown). Only M645C channels were less sensitive to the drug compared with WT channels.
Fig. 2.
Summary of scanning mutagenesis of hERG1 channels. A, fold change in peak outward step current measured with the standard I-V protocol for indicated point mutant channels. Application of 30 μM NS1643 increased current significantly (P = 0.05, unpaired t test) for the majority of tested mutants. The only mutations that did not respond with significant current increase after exposure to NS1643 were A558V, S620T, and S631A. B, fold change in peak tail current at −60 mV measured with the standard I-V protocol. Application of 30 μM NS1643 increased current significantly (P = 0.05) for the majority of tested mutants. The only mutations that did not respond with significant current increase after exposure to NS1643 were S620T, S631A, and L646E compared with WT channels. C, fold change in peak outward tail current measured with the fully activated I-V protocol. Application of 30 μM NS1643 increased current significantly (P = 0.05) for most of the tested mutants. The only mutations that did not respond with significant current increase after exposure to NS1643 were L646E and I662A. For all summarized experiments, n = 4 to 9.
Fig. 3.
Effect of NS1643 on L564A and L646E hERG1 channel currents. A, L564A hERG1 currents recorded from a single oocyte before (control, left) and after (right) treatment with 30 μM NS1643 using the standard I-V relationship voltage-clamp protocol. Step currents were elicited with 1-s pulses to test potentials (Vt) that ranged from −80 to +30 mV; deactivating (tail) currents were measured at −60 mV. B, standard I-V relationship for L564A hERG1 channels, currents were normalized to peak outward current under control conditions (n = 5). C, normalized conductance-voltage (G/Gmax − V) relationship for L564A hERG1 channels. For control, V1/2 = −24.5 ± 1.2 mV; k = 8.2 ± 0.5 mV (n = 5); for NS1643, V1/2 = −28.6 ± 0.9 mV; k = 8.7 ± 0.5 mV (n = 5). D to F, effect of 30 μM NS1643 on L646E hERG1 channel currents (n = 3). For control, V1/2 = −27.6 ± 0.4 mV; k = 7.9 ± 0.2 mV (n = 3); for NS1643, V1/2 = −31.2 ± 1.4 mV; k = 7.3 ± 0.2 mV (n = 3). G, L564A hERG1 currents recorded from a single oocyte before (control, left) and after (right) treatment with 30 μM NS1643 using the fully activated I-V relationship voltage-clamp protocol. After a 1-s pulse to +40 mV, tail currents were elicited with pulses to test potentials (Vt) that ranged from −120 to +40 mV. H and I, effect of 30 μM NS1643 on fully activated I-V relationships for L564A (n = 5) and L646E (n = 3) hERG1 channels.
Fig. 4.
Effect of NS1643 on I567A and F619A hERG1 channel currents. A, I567A hERG1 currents recorded from a single oocyte before (control, left) and after (right) treatment with 30 μM NS1643 using the standard I-V relationship voltage-clamp protocol. Step currents were elicited with 1-s pulses to test potentials (Vt) that ranged from −80 to +30 mV; deactivating (tail) currents were measured at −60 mV. B, standard I-V relationship for I567A hERG1 channels; currents were normalized to peak outward current under control conditions (n = 9). C, normalized G-V relationship for I567A hERG1 channels. For control, V1/2 = −10.1 ± 1.6 mV; k = 8.4 ± 0.2 mV (n = 9); for NS1643, V1/2 = −20.0 ± 2.0 mV; k = 11.0 ± 0.8 mV (n = 9). D to F, effect of 30 μM NS1643 on F619A hERG1 channel currents (n = 7). For control, V1/2 = −28.1 ± 1.7 mV; k = 8.2 ± 0.3 mV (n = 7); for NS1643, V1/2 = −33.0 ± 0.8 mV; k = 7.9 ± 0.3 mV (n = 7). G, F619A hERG1 currents recorded from a single oocyte before (control, left) and after (right) treatment with 30 μM NS1643 using the fully activated I-V relationship voltage-clamp protocol. After a 1-s pulse to +40 mV, tail currents were elicited with pulses to Vt that ranged from −120 to +40 mV. H and I, effect of 30 μM NS1643 on fully activated I-V relationships for I567A (n = 9) and F619A (n = 7) hERG1 channels.
Fig. 5.
Increase of current by NS1643 is slow for I567A hERG1 channels. Currents were elicited every 10 s with a 1-s pulse to −10 mV. Outward current at the end of each pulse was plotted as a function of pulse number for a 5-min control period and during the initial 10 min after addition (marked by arrow) of 30 μM drug to the perfusate. WT currents reached an approximate steady-state within ∼5 min, whereas I567A mutant channel current increased monotonically throughout the 10-min period of measurement.
Some hERG1 channel activators, especially RPR260243 (Kang et al., 2005), slow the rate of channel deactivation. Although the mutations V549A, L550A, I662A, and L666A also cause a slowing of deactivation, the effects of NS1643 on current amplitude was comparable with the data obtained for WT hERG1 channels. To further substantiate these results, an additional deactivation perturbing mutation located in a complete different domain of the hERG1 channel was evaluated. The N terminus of hERG1a channels contains a Per-Arnt-Sim domain essential for the normal deactivation properties of hERG1 channels (Morais Cabral et al., 1998). The mutation R56Q has been identified in patients with LQT2 and, as shown previously (Chen et al., 1999), accelerates the deactivation of hERG1 currents (Fig. 6, A and C). NS1643 increased the current amplitude of R56Q hERG1a in a manner similar to results obtained with WT hERG channels with no apparent change in deactivation kinetics (Fig. 6, B and D).
Fig. 6.
The hERG1-R56Q mutant identified in patients with long QT type 2 is activated by NS1643. R56Q hERG1 channel current is characterized by fast deactivation (compared with WT channels shown in Fig. 1) revealed by I-V and fully activated I-V recordings (A and C). The response to 30 μM NS1643 (B and D) was comparable with data obtained with WT channels. Black traces represent control recordings and gray traces recordings in the presence of 30 μM NS1643.
Molecular Modeling.
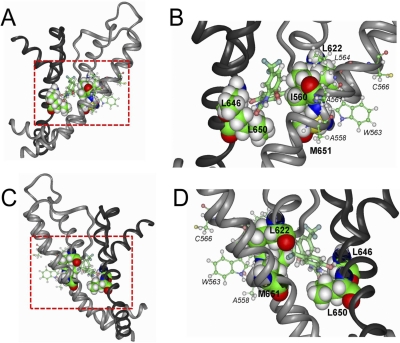
Molecular modeling was used to dock NS1643 to the Kv1.2-based homology model of the hERG1 channel pore. Figure 7 shows the preferred drug docking to the interstitial region of two adjacent subunits. In this figure, the pore helix–S6 region of one subunit and the S4–S5 linker to the end of S6 of an adjacent subunit are shown. Two views are illustrated; the configuration in Fig. A was rotated 180° relative to Fig. 7 C, and Fig. 7, B and D, show close-up views of the docking. Docking of NS1643 was initiated from 200 random configurations of the drug and allowed for binding positioned at a maximal distance of 15 Å from Cys643 of the S6 in one of the hERG1 subunits. NS1643 is in close proximity to Leu646 and Leu650 in the S6 helix of one subunit (colored dark gray), and to Ile560 (S5), Leu622 (pore helix), and Met651 (S6) of an adjacent subunit (colored light gray). Other residues identified as potentially important by scanning mutagenesis, including Ala558 and Trp563 in S5, do not apparently interact with the drug in this docking (Fig. 7, B and D). The spatial arrangement of identified residues makes it impossible that a single NS1643 molecule interacts with all of them. It is possible that mutation of these residues alters response to the drug by an allosteric effect and not by interfering with binding.
Fig. 7.
NS1643 docked to a homology model of two adjacent subunits of a hERG1 channel. A, side view of two adjacent hERG1 subunits highlighting the residues that were identified as important for drug activity based on the results of the voltage-clamp assay for hERG1 mutations as summarized in Fig. 2. NS1643 is portrayed in thick ball-and-stick mode and is located in the middle of the region delineated by the dashed box. The pore helix, selectivity filter, and S6 segment of one subunit is colored dark gray; the S4–S5 linker, S5, pore helix, selectivity filter, and S6 of an adjacent residue is colored light gray. The turret region (between pore helix and S6 segment) was not modeled and represents the structure of the Kv1.2 channel. B, amplified view of the region inside the boxed region shown in A. Residues identified by scanning mutagenesis as the most important for interaction with NS1643 are labeled. Boldface text was used for the residues shown in space-fill that are in close proximity to the drug. Smaller font and italicized text identify residues (thin ball-and-stick mode) that are not in close proximity to the drug. C and D, similar to A and B, but after 180° rotation.
Discussion
Multiple Binding Sites Identified for hERG1 Channel Activators.
The putative binding site on the hERG1 channel for two activators, RPR260243 and PD-118057, were mapped previously using a combined site-directed mutagenesis, voltage-clamp assay and molecular-modeling approach. The effects of activators on hERG1 channels harboring single point mutations and heterologously expressed in X. laevis oocytes were assayed using two microelectrode voltage-clamp techniques. Mutations that attenuated drug activity identified specific residues in hERG1 that directly or indirectly (via allosteric effects) interacted with the drug. Using this approach, distinct hydrophobic pockets in the pore domain (S5 and S6 segments and pore helices) of hERG1 were identified for RPR260243 (Perry et al., 2007) and PD-118057 (Perry et al., 2009). The most prominent effect of RPR260243 is a marked slowing of channel deactivation. The binding site for RPR260243 was mapped to the intersection of the cytosolic ends of the S5 and S6 segments of a single hERG1 subunit, a location that may explain how this compound profoundly affects the closure of the activation gate. In contrast, PD-118057 barely affects deactivation, but attenuates P-type inactivation and increases the open probability of hERG1 channels. The binding site for PD-118057 was mapped to a hydrophobic pocket located between the S6 segment and pore helix of adjacent subunits of the channel and its direct contact with the pore helix may explain how this compound alters both inactivation and single-channel open probability (Perry et al., 2009). Our findings reported here for NS1643 indicate that it binds to a site distinct from RPR260243 and PD-118057.
Putative Binding Site for NS1643.
The binding site for NS1643 on hERG1 was proposed previously to be located on the outside of the channel based on several findings (Xu et al., 2008). First, when injected into the cytoplasm of an oocyte, a high concentration of NS1643 did not affect current amplitude, whereas extracellular application of drug rapidly activated current (time constant of 3.3 min at 30 μM) in a concentration-dependent manner. Second, mutations of specific residues in the outer vestibule (S5–P linker) of hERG1 or extracellular application the peptide toxin BeKm-1 suppressed the agonist activity of NS1643. However, the specific mutations in the S5-P linker that reduced the effects of NS1643 also significantly attenuate channel inactivation. Because the increased activity of hERG1 by NS1643 is mediated by altered channel inactivation, it would be expected that mutations that reduce inactivation would also reduce the agonist activity of the drug. As reported previously, point mutations such as S631A or S620T (Casis et al., 2006; Xu et al., 2008) or N588K (Grunnet et al., 2008) that attenuate or remove inactivation also prevented the activation of current by NS1643. Thus, despite considerable effort of previous investigations, the location of the binding site for the activator activity of NS1643 remained undefined.
Mutation of Phe656 (to methionine, threonine, or valine) or Tyr652 (to alanine) enhanced the effect of NS1643 on hERG1 channels. We reported previously (Casis et al., 2006) that mutations of Phe656 (but not Y652A) enhanced activation by NS1643. Both Phe656 and Tyr652 are located in the S6 segment and face the central cavity of the channel and are key residues of the binding site for hERG1 blockers. NS1643 is a partial agonist, presumably because it binds to two different sites on the channel, one that mediates activation, and another mediating block. Disruption of the blocker binding site by mutation of Tyr652 or Phe656 reveals the undiminished agonist activity of the drug.
The molecular features of the NS1643 binding site are uncertain because none of the point mutations, with the possible exception of L646E, eliminated the effects of the drug and because the location of the key residues identified by mutagenesis are not clustered in a way consistent with identification of a binding pocket. However, drug docking to the homology model of the hERG1 pore suggests that five of the residues identified in the scan (Ile560, Leu622, Leu646, Leu650, and Met651) are in close proximity to the drug (Fig. 7) and that the putative binding site so identified is located away from the central cavity and close to the sites identified for two other hERG1 channel activators, RPR260243 (Perry et al., 2007) and PD-118057 (Perry et al., 2009). The potential importance of different amino acid changes to the same residue is illustrated by the mutations L646E, L646A, and L646V. Although L646E almost completely abolished the ability of NS1643 to activate the channels, this was not observed for L646A and L646V. A possible explanation for this phenomenon is that L646E induces steric or charge-based hindrance to drug binding, whereas this is not the situation for L646A and L646V.
As always for the combination of electrophysiological scanning mutagenesis experiments and molecular-docking models some limitations have to be emphasized. First, functional assays such as voltage clamp do not directly assess drug binding the way a binding assay can. Second, we cannot exclude the possibility that residues other than those identified here might contribute to the stabilization of drug binding. For example, relatively conservative mutations (e.g., valine to alanine) may not appreciably alter the affinity of drug binding and thus erroneously be overlooked. Third, some mutations may alter drug sensitivity by an allosteric effect and not by direct interference with drug binding. Fourth, the hERG1 homology model used here is based on the crystal structure of a related Kv channel and may not accurately reflect the specific conformation of the channel in its native environment. The model is also a static structure that does not take into account potentially important protein rearrangements associated with channel gating.
Antiarrhythmic and Proarrhythmic Activity of hERG1 Channel Activators.
Because hERG1 channel activators were initially described approximately 4 years ago, a number of investigations have addressed the possible antiarrhythmic effects of these compounds. This line of inquiry was prompted by the realization that although class III antiarrhythmic compounds are effective against atrial fibrillation, these drugs can also increase the risk for ventricular fibrillation and sudden cardiac death (Waldo et al., 1996). It is plausible that there are a higher propensity for a number of proarrhythmic biomarkers such as prolonged QT interval, increased action potential triangulation, reverse-use dependence, instability, and dispersion as a consequence of hERG1 channel inhibition. This will lead to increased risk for triggering events that can initiate arrhythmias and for creating substrates that are necessary to maintain the arrhythmia (Hondeghem, 2008). It was therefore expected that hERG1 channel agonists could decrease the proclivity for these biomarkers, and this assumption has been confirmed in a number of different experimental settings. hERG1 channel agonists decrease dispersion and triangulation, prevent bradypacing and pharmacological-induced arrhythmias, shorten QT intervals in vivo, and directly protect against arrhythmias (Kang et al., 2005; Hansen et al., 2007, 2008; Diness et al., 2008, 2009). However, hERG1 agonists can also be proarrhythmic under certain settings (Larsen et al., 2008; Lu et al., 2008), placing obvious restrictions on their potential therapeutic use. These compounds would theoretically be useful for treatment of long QT syndrome, heart failure, or cardiac remodelling as a consequence of hypertension. All known hERG1 channel activators have low potency, in part because most are partial agonists, causing both activation and block of hERG1 channels. A more detailed understanding of the molecular mechanisms of action for hERG1 channel activators will assist in the discovery of more potent compounds.
In conclusion, based on functional analysis of the effects of a large number of mutant channels, we have identified a putative binding site on hERG1 for the activator effects of NS1643. The key residues that seem to interact with NS1643 are located on the S5 and S6 segments of adjacent subunits and are situated near the pore helix. In all probability, drug bound to this site interferes with the subtle rearrangement of the pore helix/selectivity filter that is believed to underlie P-type inactivation.
Acknowledgments
We thank Camilla Irlind and Kam Hoe Ng for excellent technical assistance.
This work was supported by The Danish National Research Foundation Centre for Cardiac Arrhythmia; The Novo Nordisk Foundation; The Aase and Ejnar Danielsen Foundation; The National Danish Research Council; and the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL055236].
M.G. has minor-equity holdings in NeuroSearch A/S. The other authors have no potential conflicts of interest.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.067728.
- hERG1
- human ether-à-go-go-related gene type 1
- LQT2
- long QT syndrome type 2
- NS1643
- 1,3-bis-(2-hydroxy-5-trifluoromethyl-phenyl)-urea
- RPR260243
- (3R,4R)-4-[3-(6-methoxy-quinolin-4-yl)-3-oxo-propyl]-1-[3-(2,3,5-trifluoro-phenyl)-prop-2-ynyl]-piperidine-3-carboxylic acid
- PD-118057
- 2-(4-[2-(3,4-dichloro-phenyl)-ethyl]-phenylamino)-benzoic acid
- PD-307243
- 2-[2-(3,4-dichloro-phenyl)-2,3-dihydro-1H-isoindol-5-ylamino]-nicotinic acid
- NS3623
- N-(4-bromo-2-(1H-tetrazol-5-yl)-phenyl)-N′-(3′-trifluoromethylphenyl)urea
- ICA-105574
- 3-nitro-N-(4-phenoxyphenyl) benzamide
- A-935142
- {4-[4-(5-trifluoromethyl-1H-pyrazol-3-yl)-phenyl]-cyclohexyl}-acetic acid
- WT
- wild type.
Authorship Contributions
Participated in research design: Grunnet, Abbruzzese, Sachse, and Sanguinetti.
Conducted experiments: Grunnet, Abbruzzese, and Sachse.
Performed data analysis: Grunnet, Abbruzzese, and Sanguinetti.
Wrote or contributed to the writing of the manuscript: Grunnet, Abbruzzese, Sachse, and Sanguinetti.
References
- Casis O, Olesen SP, Sanguinetti MC. (2006) Mechanism of action of a novel human ether-a-go-go-related gene channel activator. Mol Pharmacol 69:658–665 [DOI] [PubMed] [Google Scholar]
- Chen J, Zou A, Splawski I, Keating MT, Sanguinetti MC. (1999) Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem 274:10113–10118 [DOI] [PubMed] [Google Scholar]
- Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. (1995) A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80:795–803 [DOI] [PubMed] [Google Scholar]
- Diness JG, Hansen RS, Nissen JD, Jespersen T, Grunnet M. (2009) Antiarrhythmic effect of IKr activation in a cellular model of LQT3. Heart Rhythm 6:100–106 [DOI] [PubMed] [Google Scholar]
- Diness TG, Yeh YH, Qi XY, Chartier D, Tsuji Y, Hansen RS, Olesen SP, Grunnet M, Nattel S. (2008) Antiarrhythmic properties of a rapid delayed-rectifier current activator in rabbit models of acquired long QT syndrome. Cardiovasc Res 79:61–69 [DOI] [PubMed] [Google Scholar]
- Gerlach AC, Stoehr SJ, Castle NA. (2010) Pharmacological removal of human ether-à-go-go-related gene potassium channel inactivation by 3-nitro-N-(4-phenoxyphenyl) benzamide (ICA-105574). Mol Pharmacol 77:58–68 [DOI] [PubMed] [Google Scholar]
- Gordon E, Lozinskaya IM, Lin Z, Semus SF, Blaney FE, Willette RN, Xu X. (2008) 2-[2-(3,4-dichloro-phenyl)-2,3-dihydro-1H-isoindol-5-ylamino]-nicotinic acid (PD-307243) causes instantaneous current through human ether-a-go-go-related gene potassium channels. Mol Pharmacol 73:639–651 [DOI] [PubMed] [Google Scholar]
- Grunnet M, Diness TG, Hansen RS, Olesen SP. (2008) Biophysical characterization of the short QT mutation hERG-N588K reveals a mixed gain-and loss-of-function. Cell Physiol Biochem 22:611–624 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Diness TG, Christ T, Demnitz J, Ravens U, Olesen SP, Grunnet M. (2006a) Activation of human ether-a-go-go-related gene potassium channels by the diphenylurea 1,3-bis-(2-hydroxy-5-trifluoromethyl-phenyl)-urea (NS1643). Mol Pharmacol 69:266–277 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Diness TG, Christ T, Wettwer E, Ravens U, Olesen SP, Grunnet M. (2006b) Biophysical characterization of the new human ether-a-go-go-related gene channel opener NS3623 [N-(4-bromo-2-(1H-tetrazol-5-yl)-phenyl)-N′-(3′-trifluoromethylphenyl)urea]. Mol Pharmacol 70:1319–1329 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Olesen SP, Grunnet M. (2007) Pharmacological activation of rapid delayed rectifier potassium current suppresses bradycardia-induced triggered activity in the isolated guinea pig heart. J Pharmacol Exp Ther 321:996–1002 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Olesen SP, Rønn LC, Grunnet M. (2008) In vivo effects of the IKr agonist NS3623 on cardiac electrophysiology of the guinea pig. J Cardiovasc Pharmacol 52:35–41 [DOI] [PubMed] [Google Scholar]
- Hondeghem LM. (2008) Use and abuse of QT and TRIaD in cardiac safety research: importance of study design and conduct. Eur J Pharmacol 584:1–9 [DOI] [PubMed] [Google Scholar]
- Kang J, Chen XL, Wang H, Ji J, Cheng H, Incardona J, Reynolds W, Viviani F, Tabart M, Rampe D. (2005) Discovery of a small molecule activator of the human ether-a-go-go-related gene (HERG) cardiac K+ channel. Mol Pharmacol 67:827–836 [DOI] [PubMed] [Google Scholar]
- Larsen AP, Olesen SP, Grunnet M, Jespersen T. (2008) Characterization of hERG1a and hERG1b potassium channels-a possible role for hERG1b in the I (Kr) current. Pflugers Arch 456:1137–1148 [DOI] [PubMed] [Google Scholar]
- Larsen AP, Olesen SP, Grunnet M, Poelzing S. (2010) Pharmacological activation of IKr impairs conduction in guinea pig hearts. J Cardiovasc Electrophysiol 21:923–929 [DOI] [PubMed] [Google Scholar]
- Lees-Miller JP, Kondo C, Wang L, Duff HJ. (1997) Electrophysiological characterization of an alternatively processed ERG K+ channel in mouse and human hearts. Circ Res 81:719–726 [DOI] [PubMed] [Google Scholar]
- London B, Trudeau MC, Newton KP, Beyer AK, Copeland NG, Gilbert DJ, Jenkins NA, Satler CA, Robertson GA. (1997) Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ Res 81:870–878 [DOI] [PubMed] [Google Scholar]
- Lu HR, Vlaminckx E, Hermans AN, Rohrbacher J, Van Ammel K, Towart R, Pugsley M, Gallacher DJ. (2008) Predicting drug-induced changes in QT interval and arrhythmias: QT-shortening drugs point to gaps in the ICHS7B Guidelines. Br J Pharmacol 154:1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. (1998) Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell 95:649–655 [DOI] [PubMed] [Google Scholar]
- Moss AJ, Zareba W, Benhorin J, Locati EH, Hall WJ, Robinson JL, Schwartz PJ, Towbin JA, Vincent GM, Lehmann MH. (1995) ECG T-wave patterns in genetically distinct forms of the hereditary long QT syndrome. Circulation 92:2929–2934 [DOI] [PubMed] [Google Scholar]
- Perry M, Sachse FB, Abbruzzese J, Sanguinetti MC. (2009) PD-118057 contacts the pore helix of hERG1 channels to attenuate inactivation and enhance K+ conductance. Proc Natl Acad Sci USA 106:20075–20080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M, Sachse FB, Sanguinetti MC. (2007) Structural basis of action for a human ether-a-go-go-related gene 1 potassium channel activator. Proc Natl Acad Sci USA 104:13827–13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern WS, Carlsson L, Davis AS, Lynch WG, MacKenzie I, Palethorpe S, Siegl PK, Strang I, Sullivan AT, Wallis R, et al. (2003) Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development. Cardiovasc Res 58:32–45 [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Jiang C, Curran ME, Keating MT. (1995) A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81:299–307 [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Mitcheson JS. (2005) Predicting drug-hERG channel interactions that cause acquired long QT syndrome. Trends Pharmacol Sci 26:119–124 [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Tristani-Firouzi M. (2006) hERG potassium channels and cardiac arrhythmia. Nature 440:463–469 [DOI] [PubMed] [Google Scholar]
- Su Z, Limberis J, Souers A, Kym P, Mikhail A, Houseman K, Diaz G, Liu X, Martin RL, Cox BF, et al. (2009) Electrophysiologic characterization of a novel hERG channel activator. Biochem Pharmacol 77:1383–1390 [DOI] [PubMed] [Google Scholar]
- Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. (1995) HERG, a human inward rectifier in the voltage-gated potassium channel family. Science 269:92–95 [DOI] [PubMed] [Google Scholar]
- Waldo AL, Camm AJ, deRuyter H, Friedman PL, MacNeil DJ, Pauls JF, Pitt B, Pratt CM, Schwartz PJ, Veltri EP. (1996) Effect of d-sotalol on mortality in patients with left ventricular dysfunction after recent and remote myocardial infarction. The SWORD Investigators. Survival With Oral d-Sotalol. Lancet 348:7–12 [DOI] [PubMed] [Google Scholar]
- Warmke JW, Ganetzky B. (1994) A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA 91:3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Recanatini M, Roberti M, Tseng GN. (2008) Probing the binding sites and mechanisms of action of two human ether-a-go-go-related gene channel activators, 1,3-bis-(2-hydroxy-5-trifluoromethyl-phenyl)-urea (NS1643) and 2-[2-(3,4-dichloro-phenyl)-2,3-dihydro-1H-isoindol-5-ylamino]-nicotinic acid (PD307243). Mol Pharmacol 73:1709–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Korolkova YV, Liu J, Jiang M, Grishin EV, Tseng GN. (2003) BeKm-1 is a HERG-specific toxin that shares the structure with ChTx but the mechanism of action with ErgTx1. Biophys J 84:3022–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Augelli-Szafran CE, Bradley JA, Chen X, Koci BJ, Volberg WA, Sun Z, Cordes JS. (2005) Novel potent human ether-a-go-go-related gene (hERG) potassium channel enhancers and their in vitro antiarrhythmic activity. Mol Pharmacol 68:876–884 [DOI] [PubMed] [Google Scholar]