Abstract
Red algal parasites are unusual because the vast majority of them parasitize species with which they share a recent common ancestor. This strategy has earned them the name “adelphoparasites,” from the Greek, adelpho, meaning “kin.” Intracellular adelphoparasites are very rare in nature, yet have independently evolved hundreds of times among the floridiophyte red algae. Much is known about the life history and infection cycle of these parasites but nearly nothing in known about their genomes. We sequenced the mitochondrial genomes of the free-living Gracilariopsis andersonii and its closely related parasite Gracilariophila oryzoides to determine what effect a parasitic lifestyle has on the genomes of red algal parasites. Whereas the parasite genome is similar to the host in many ways, the genes encoding essential proteins ATP8 and SDHC are pseudogenes in the parasite. The mitochondrial genome of parasite from a different class of red algae, Plocamiocolax puvinata, has lost the atp8 gene entirely, indicating that this gene is no longer critical in red algal parasite mitochondria.
Keywords: comparative genomics, mitochondria, red algae, parasitism, genome evolution, atp8
Introduction
Parasitism is a common evolutionary strategy that has arisen independently numerous times throughout the history of life and encompasses a wide variety of organismal interactions. Much is understood about parasite biology, but little is known about the method by which a free-living eukaryote adopts a parasitic lifestyle. To tease apart the stages of evolution resulting in a parasitic organism, a closely related, free-living taxon must be identified for comparative purposes in order to accurately access character change. Often, however, parasites are only distantly related to a free-living taxon. Red algae provide a unique model for understanding parasite evolution because most red algal parasites are sister species to their hosts, derived from a recent common ancestor. Therefore, direct genomic and molecular comparisons made between the host and parasite have the potential to unravel the fundamental genomic changes that must occur for an organism to become parasitic.
This study uses the host–parasite relationships that exist among red algae (Rhodophyta) as a model to investigate the evolution of parasites from a free-living ancestor. Multicellular red algae are ideal for understanding the evolution of parasitism because ∼10% of known species are parasites of free-living red algae, and approximately, 80% of these parasites have evolved from the most recent common ancestor with their hosts (Goff et al. 1996). These parasites are known as adelphoparasites (“adelpho” meaning kinsman) (Setchell 1918; Goff 1995). Morphologically reduced to a miniature, often colorless pustule, adelphoparasites are highly host dependent and occur exclusively on their sister-taxon (Goff 1982). The remaining 20% of red algal parasites are known as alloparasites (“allo” meaning other) because whereas they also diverged from free-living species, they have since radiated to exploit more distantly related red algal hosts (Goff et al. 1997). Although this terminology has somewhat fallen out of favor because a full spectrum of host/parasite relationships occur in nature (Zuccarello et al. 2004), the terms remain useful to designate differences in the biology of the organisms.
Red algal adelpho- and alloparasitism have arisen independently numerous times throughout the diversification of the florideophyte lineage, so a range of host–parasite relationships exists providing “windows” into the process of parasite evolution. For example, some parasites are not entirely obligate and have retained limited photosynthetic capabilities, whereas others are highly host specific to their sister species and others have multiple hosts. This range of relationships creates an evolutionary spectrum, allowing for comparative investigations into how these parasites evolve from a free-living species (Goff and Coleman 1987; Goff et al. 1997).
The mechanism by which red algal adelpho- and alloparasites infect host cells has been extensively studied (Setchell 1918; Goff and Coleman 1985, 1987; Goff 1995; Goff et al. 1997; Zuccarello et al. 2004). Upon contact with the host thallus, the nonmotile parasitic spore produces a short filament, or conjector cell, that fuses with a surface host cell. Minor variations in the subsequent steps from one species to the next are common, but in general, infection proceeds when the parasite deposits a nucleus, mitochondrion and dedifferentiated plastid, known as a proplastid, into a host cell. Once in the host cytoplasm, the parasite nuclei undergo DNA synthesis and the mitochondria divide rapidly. This cycle of host infection by the parasite results in the formation of nonphotosynthetic heterokaryotic tissue that appears as a gall or pustule on the exterior of the infected host thallus (Goff and Coleman 1985; Goff 1995; Zuccarello et al. 2004). The parasite cells in the pustule eventually give rise to gametes, 2n carpospores, or 1n tetraspores that are released allowing for the continued dispersal of the parasite. Spores packaged and produced by the parasite contain host-derived proplastids along with parasite nucleus and mitochondrion (Goff and Coleman 1987).
Although much is known about the life history strategies of red algae algal parasites, little work has been done assessing the molecular and genomic consequences of becoming a parasite. A paper published by Goff and Coleman (1995) nicely illustrates the fate of parasite organellar DNA during host cellular infection and transformation. By comparing the restriction fragment length polymorphisms derived from the nuclear, plastid, and mitochondrial genomes of both host and parasite, Goff and Coleman show that although the parasite plastid is lost in favor of co-opting the host version, the mitochondrial DNA (mtDNA) and nuclear DNA are maintained by the parasite. Because this pattern was confirmed in three different red algal parasites (representing three different red algal orders), it can be assumed that there is strong selection for the parasite maintaining its own genetically unique mitochondria rather than using the host version.
Here, we compare the mitochondrial genome sequences and architecture of Gracilariopsis andersonii and its adelphoparasite Gracilariophila oryzoides, as well as alloparasite Plocamiocolax pulvinata, to shed light on the evolutionary mechanisms that have resulted in the differences between a parasitic and free-living lifestyle. Because of the close evolutionary relationships between the red algal host and parasite, direct comparisons can be made between the mitochondrial genomes of Gr. andersonii and G. oryzoides with differences attributed to their respective lifestyles. The mitochondrial genome of the slightly more divergent alloparasite, P. pulvinata, provides further insight into general trends that may be observed among red algal parasites.
Materials and Methods
Sample Collection, DNA Isolation, and Genome Sequencing
Host, Gr. andersonii, and parasite, G. oryzoides, were collected in December 2008 and June 2009 from Pigeon Point, Pescadero, CA, USA. Noninfected Gr. andersonii were found growing along sandy substrata in the lower intertidal region. Plants were removed by hand and placed directly into a two gallon Zip-Lock bag filled with seawater. Within an hour, freshly collected, noninfected Gr. andersonii plants were desiccated on silica gel. Gracilariophila oryzoides material was isolated from freshly collected infected host Gr. andersonii by excising the white pustules formed by the mature parasites. Parasitic tissue was removed using forceps and placed directly into a 1.5-ml eppie filled with silica.
Dried Gr. andersonii and G. oryzoides were immersed in liquid nitrogen and ground to a fine powder using a mortar and pestle. Total DNA (genomic and organellar) was isolated from Gr. andersonii and G. oryzoides using an optimized red algal extraction buffer (Saunders 1993) and a standard phenol/chloroform genomic DNA extraction (Saunders 1993). mtDNA from Gr. andersonii was amplified by polymerase chain reaction (PCR) and long range-PCR using primers designed in Geneious Pro 4.8.5 (Drummond et al. 2009) from known red algal mitochondrial genomes (Leblanc et al. 1995; Ohta et al. 1998; Burger et al. 1999). Primer sequences are listed in supplementary table 1 (Supplementary Material online).
Conserved genes were amplified by PCR using cycling parameters that included an initial denaturation cycle of 94 °C for 30 s followed by 38 cycles of 94 °C for 30 s, 50 °C for 45 s, and 72 °C for 2 min. A final extension step was performed at 72 °C for 10 min followed by a 10 °C hold. For larger amplicons (between conserved genes), a high-fidelity taq (Takara LA Taq Polymerase) was employed to reduce amplification errors. The thermal profile for long range-PCR amplification included an initial denaturation cycle of 94 °C for 30 s followed by 36 cycles of 94 °C for 30 s, 50 °C for 45 s, and 68 °C for 4–8 min depending on the estimated sequence length (approximately 1 min extension time for every 1,000 bp). A final extension step was performed at 68 °C for 10 min followed by a hold at 10 °C until the samples were processed. Amplified products were purified and sent for sequencing on an Applied Biosystems 3130xl Genetic Analyzer at the Rhode Island Genomics and Sequencing Center. Individual sequences were assembled into contigs and edited based on quality scores using Geneious Pro 4.8.5. Primers built from assembled Gr. andersonii sequences were used to “primer-walk” gaps and close sequence holes in the host mitochondrial genome.
The mtDNA sequence of P. pulvinata was acquired from Lynda Goff that was sequenced previously using the same methods described above for Gr. andersonii. The 25, 894 bps contig was imported into Geneious Pro 4.8.5 for open-reading frame (ORF) identification and genome annotation.
Total genomic and organellar DNA were isolated from dried G. oryzoides using the same methods described above for Gr. andersonii. Five milligrams of parasite total genomic DNA was sequenced using the 454 Genome Sequencer FLX (Roche) at the McGill University and Genome Quebec Innovation Centre. A ∼25, 000 bp contig resulting from the assembly was imported into Geneious Pro 4.8.5 for further annotation. Contig coverage was approximately 30-fold for the mitochondrial contig. Regions of ambiguity or where frameshifts were detected were resequenced using PCR and Sanger Sequencing.
Genome Annotation
ORFs and annotations of Gr. andersonii, G. oryzoides, and P. pulvinata mtDNA were performed using Geneious Pro 4.8.5. Genes were identified using the BlastN and BlastX algorithms (Altschul et al. 1997) to compare the predicted ORFs to the NCBI GenBank database. Frameshift mutations resulting in pseudogenes were confirmed by manual assessment of 454-sequence alignments, and Sanger sequencing was employed in areas of homopolymer runs, low coverage, or frameshifts. Small and large ribosomal rRNA subunits were identified in each genome by alignment to the mtDNA sequences to previously sequenced red algal rRNA subunits (Leblanc et al. 1995; Burger et al. 1999). Transfer RNAs were determined using tRNAscan-SE version 1.21 (Lowe and Eddy 1997).
Analysis: dN/dS Ratio, AT Content
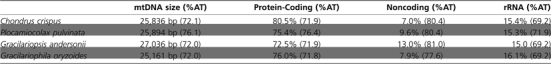
For each mtDNA genome, all protein-coding genes and pseudogenes were extracted and aligned at the nucleotide and amino acid level with the only published floridiophyte mitochondrial genome, Chondrus crispus (NC001677), using the Geneious Pro software package. To estimate the ratio of nonsynonymous to synonymous substitutions (dN/dS), a nucleotide alignment and Neighbor-Joining tree were constructed for each protein-coding gene and imported into the CODEML program from the PAML package (Yang 2007). Analyses were performed with the following parameters specified in the default control file from PAML for the CODMEL package: runmode = 0 (sequence comparisons take into account phylogenetic history); Seqtype = 1, and Codonfreq = 2 (average nucleotide frequencies at the three codon positions); Model = 0; and omega (measuring dN/dS ratio) and kappa (measuring transitions/transversions) were estimated, not fixed. To generate a pairwise comparison between each species, all the parameters were kept constant except runmode, which was denoted as a −2. AT content for each genome was determined by the extraction and concatenation of noncoding, protein-coding, and rRNA sequence, respectively (table 1).
Table 1.
Red Algal Mitochondrial Genome Comparisons
 |
Genome Size Estimates: Pulsed-Field Gel Electrophoresis and Southern Hybridization
Using pulsed-field gel electrophoresis (PFGE), mitochondrial genome size was estimated for both Gr. andersonii and G. oryzoides. Whole cells extracted from freshly collected host and parasite material were embedded in agarose plugs and digested in a buffer (10 mM Tris/HCl, 400 mM EDTA, 1% lauroylsarcosine, 0.25 mg Proteinase K). Plugs were run in a PFGE, on a 1% agarose TBE gel in 0.5% TBE buffer and prestained in 1X SYBR Safe. The gel was run with the MidRange I PFGE (BioLabs) ladder for 11–24 h at 14 °C with a switch time of 1–6 s at a 120° angle to optimize the migration of the mitochondrial genome.
Results
Genome Size and Architecture
The entire mitochondrial genomes of Gr. andersonii (27,036 bps, GenBank accession HQ586060), G. oryzoides (25,161 bps, HQ586059), and P. pulvinata (25,894 bps, HQ586061) were sequenced and mapped to a circular model based on our ability to amplify the genomes in entirely overlapping fragments. Whereas this does not preclude the possibility that the genome may be a linear molecule with varying termination points between different copies, we have no evidence to support this alternative. The overall AT composition of the three mitochondrial genomes was 72.0, 72.0, and 76.1%, respectively. Coding regions of the three genomes had lower AT content and higher GC content than intergenic or noncoding regions (table 1). Consistent with previously sequenced red algal mitochondrial genomes (Leblanc et al. 1995; Ohta et al. 1998; Burger et al. 2000), Gr. andersonii, G. oryzoides, and P. pulvinata have densely packed mitochondrial genomes that are 72.5, 75.4, and 76.0% coding, respectively. Genes are encoded on both the positive and negative strand of all three genomes and there is considerable sequence synteny across the three red algal species sequenced in this study and in mitochondrial genomes previously sequenced; C. crispus, Porphyra purpurea, and Cyanidioschyzon merolae.
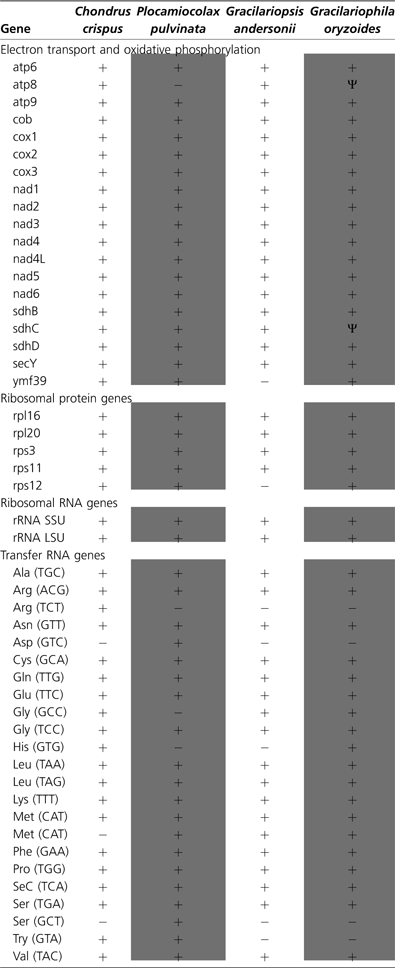
The mitochondrial genes in C. crispus, Gr. andersonii, G. oryzoides, and P. pulvinata that code for proteins, rRNA, and tRNAs are listed in table 2. All three genomes code for both 26s RNA and 16s RNA. The number and content of tRNAs varies among Gr. andersonii, G. oryzoides, and P. pulvinata mtDNA. Consistent with C. crispus, P. purpurea, and C. merolae, these genomes do not contain a full set of tRNAs needed to complete translation (table 2). The protein-coding genes found in all three genomes are virtually identical, and the content similarity extends to C. crispus as well. The few exceptions to this trend will be discussed below.
Table 2.
Mitochondrial Genome Comparisons: Protein-Coding Genes, rRNA, and tRNA Content
 |
PFGE Genome Size Estimates
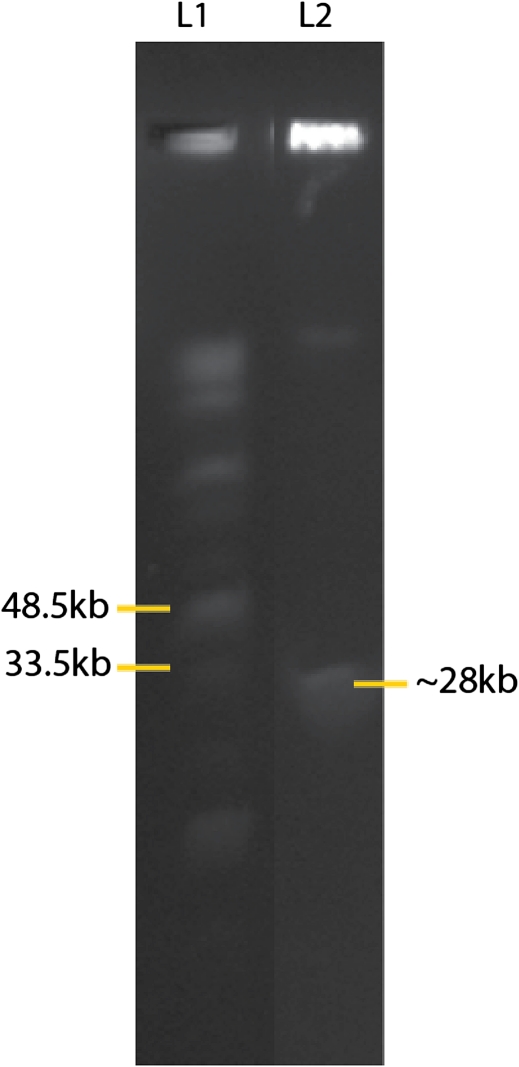
To ensure our assemblies were consistent with estimated genome size, PFGE was performed for Gr. andersonii. The mtDNA of Gr. andersonii was estimated to be ∼28 Kb (fig. 1), indicating that our sequence is not missing data nor are assembly artifacts present that might change artificially the sequence length.
FIG. 1—
PFGE of total DNA from Gracilariopsis andersonii, stained with SYBR Safe. A midrange size ladder was run in lane one (L1) and the relevant sized ladder fragments are labeled. The ∼28 Kb fragment in lane two (L2) is similar in size to the sequenced mitochondrial genome of Gr. andersonii and is the only DNA band in the likely size range of red algal mitochondrial genomes.
Mitochondrial Gene Content and Order in G. andersonii, G. oryzoides, and P. pulvinata in Comparison with Each Other and Previously Sequenced Red Algal mtDNA
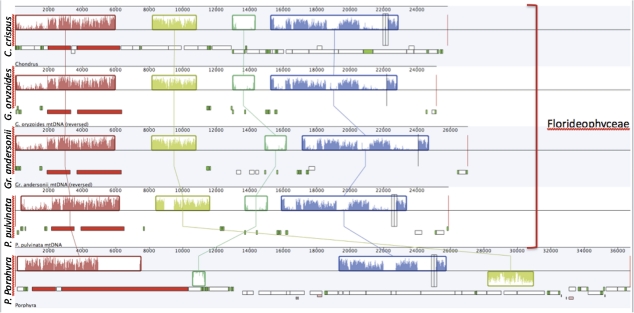
Similar to the mitochondrial genomes of P. purpurea and C. crispus, the mt genomes of Gr. andersonii, G. oryzoides, and P. pulvinata can be broken into three conserved gene clusters. The longest cluster is comprised of atp6-atp8-nad5-nad4-sdhD-nad2-nad1-nad3. In P. pulvinata, there has been a deletion of atp8 but the neighboring genes are unaffected. The cox1-cox2-cox3 genes form the second well-conserved gene cluster among sequenced red algal mtDNA. Finally, the cluster SSU-nad4L-LSU occurs in all sequenced red algal mtDNA except for a slight difference in C. merolae where the gene yeiU exists between nad4L and LSU. Overall, gene order and content are largely static in red algae despite the age of the lineage.
Sequence analysis of Gr. andersonii, G. oryzoides, and P. pulvinata mtDNA indicates that all three genomes code for three small ribosomal proteins (rps3, rps11, and rps12) and two large ribosomal proteins (rpl16 and rpl20) (table 2). Genes involved in electron transport and oxidative phosphorylation are well conserved among newly sequenced red algal mtDNA and C. crispus. NADH dehydrogenase subunits nad1, nad2, nad3, nad4, nad4L, nad5, and nad6 are transcribed by all red algal mtDNA sequenced so far. Protein-coding genes involved in cytochrome C oxidase (cox1, cox2, cox3) and cytochrome C oxidoreductase (cob) are also found among all red algal mtDNA. Subunits sdhB, sdhC, and sdhD involved in respiratory chain complex II (succinate dehydrogenase) are identified in all red algal mtDNA except for in G. oryzoides, where sdhC is now a pseudogene. Genes involved in ATP synthesis, atp6, atp8, atp9, are found throughout red algal mtDNA, except atp8 is not found in P. pulvinata and appears to be a pseudogene in G. oryzoides.
Host/Parasite and Parasite/Parasite Comparisons
Protein-coding gene content and order are identical between the mitochondrial genomes of Gr. andersonii and G. oryzoides with a few exceptions; rps11 is in the same position but in different directions (figs. 2 and 3); the presence of ymf (a gene without identified function) following cox3 in G. oryzoides (fig. 3); the development of two pseudogenes in G. oryzoides (fig. 3); and differences in unique ORFs. Additionally, Gr. andersonii contains an intergenic space of ∼2,000 bps between cob and nad6 that is not found in G. oryzoides. This intergenic space is unique to G. andersonii and is not found in other red algal mtDNA available for comparison.
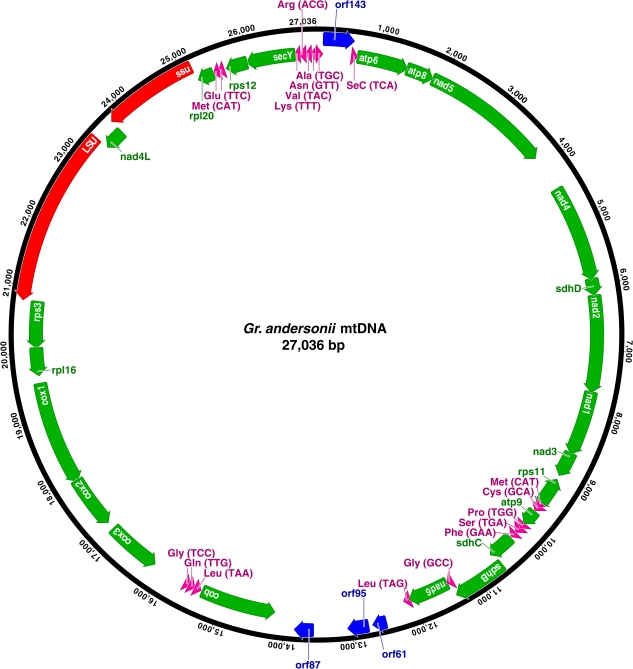
FIG. 2.—
Gene map of the mitochondrial genome of Gracilariopsis andersonii (GenBank accession HQ586060). Protein-coding genes are indicated by green bars with arrowheads showing the direction of the coding sequence. tRNAs are displayed in purple, rRNA in red, and unidentified ORFs in blue.
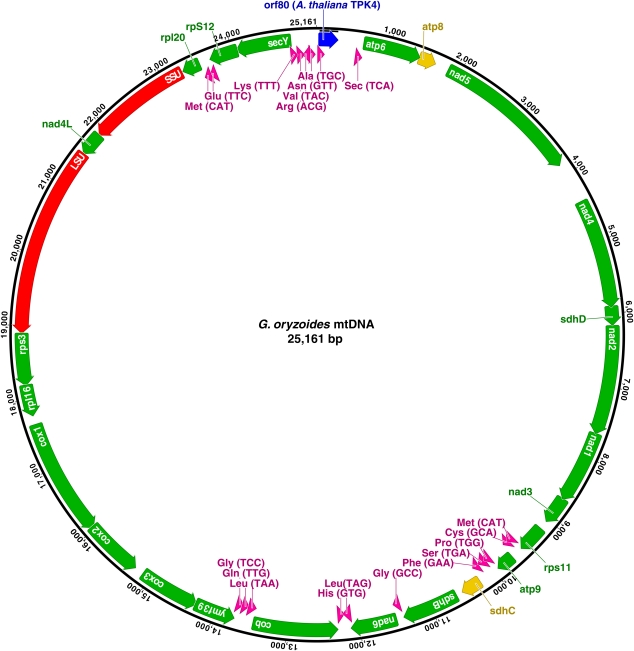
FIG. 3.—
Mitochondrial genome map of the parasite Gracilariophila oryzoides (GenBank accession HQ586059). Colors of all ORFs are the same as figure 2, with the addition of yellow indicating pseudogenes.
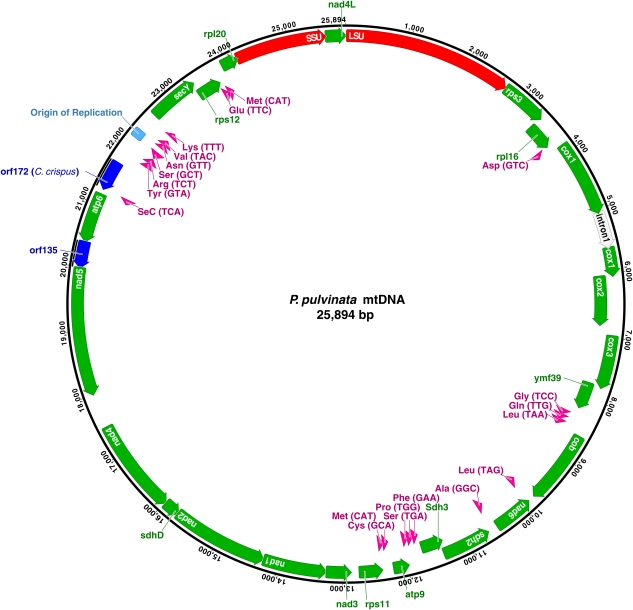
Parasites P. pulvinata and G. oryzoides also have very similar gene content (figs. 3 and 4). Both parasites, unlike Gr. andersonii, encode ymf39 (also found in C. crispus and P. purpurea) following cox3 and have either lost or developed a frameshift mutation in their atp8 gene. The two notable differences between the parasite mitochondrial genomes are the presence of an intron (557 bps) within cox1 in P. pulvinata (fig. 4) and the absence of a truncated sdhC gene in P. pulvinata.
FIG. 4.—
Genome map from the Plocamiocolax pulvinata mithochondrion (GenBank accession HQ586061). Colors are the same as figure 2, except an intron in the cox1 gene is shown in gray. The predicted origin of replication is also indicated.
Pseudogenes
Due to frameshift mutations and the development of premature stop codons, G. oryzoides contains two pseudogenes; atp8 and sdhC. The remaining portion of the atp8 gene in G. oryzoides is ∼200 bps shorter than the atp8 gene found in Gr. andersonii and C. crispus, which are 411 and 407 bps long, respectively. The sdhC pseudogene has undergone multiple deletions resulting in a frameshift mutation and development of a premature stop codon. The sdhC pseudogene in G. oryzoides is 255 bps, which is ∼120 bps shorter than the sdhC gene found in Gr. andersonii, P. pulvinata, and C. crispus. Despite being pseudogenes, the genes are still being transcribed as parts of multigene transcripts based on Illumina sequenced cDNA libraries (Lane CE, unpublished).
tRNAs
Dispersed throughout the Gr. andersonii mtDNA are 18 tRNA genes (fig. 2 and table 2). All tRNA gene sequences show standard cloverleaf secondary structures and are between 72 and 88 nucleotides long. Transfer RNA sequences are found on both the negative and positive strand and are typically single or in groups of two or three. Gracilariophila oryzoides encodes 19 tRNA genes distributed on both strands throughout the mtDNA (fig. 3 and table 2), which also show conventional size and structure. Transfer RNAs coding for the same amino acids are found in the mtDNA of Gr. andersonii and G. oryzoides with the exception of HIS (GTG), which is only in the G. oryzoides mtDNA. Twenty tRNAs are found throughout the mtDNA of P. pulvinata (fig. 4 and table 2). Unlike Gr. andersonii and G. oryzoides, P. pulvinata mtDNA does not contain Gly (GCC) but does encode Ser (GCT) and Try (GTA) (table 2). Plocamiocolax pulvinata, like Gr. andersonii, does not encode tRNA HIS (GTG). No pseudo tRNAs are found in any of the three mitochondrial genomes sequenced.
ORFs
Four unique ORFs (orf61, orf87, orf95, and orf143) are present in the mtDNA of Gr. andersonii and based on predictions using Geneious Pro 4.8.5, all four ORFs exhibit hydrophobic characteristics indicative of membrane embedded proteins. One unique ORF (orf80) is found in the mtDNA of G. oryzoides. Blast results indicate weak homology between orf80 and tpk4 (e value: 5.89 × 10−1); a protein found in Arabidopsis thaliana that is involved in the plasma membrane K+-channel (Gobert et al. 2007). Two unique ORFs (orf172 and orf135) are found in the mtDNA of P. pulvinata. Orf152 shows weak sequence similarity (E value: 7.62 × 10−1) with orf172 in C. crispus. Similar to C. crispus orf172, orf152 is highly hydrophobic and most likely membrane bound, if it codes for a functional protein. Orf135 shows characteristics typically observed in transmembrane proteins. Protein predictions of ymf39, present in both P. pulvinata and G. oryzoides, indicate that this hypothetical protein has both hydrophobic and hydrophilic properties that may be associated with a transmembrane protein.
dN/dS Ratios
Table 3 shows the ratio of nonsynonymous to synonymous substitutions (dN/dS) for all the protein-coding genes shared by C. crispus, P. pulvinata, Gr. andersonii, and G. oryzoides. A low ratio (dN/dS < 1) indicates purifying or stabilizing selection, whereas a high ratio (dN/dS > 1) signifies positive selection. Because mitochondrial genomes show AT skew, each codon position was considered when calculating both the synonymous and nonsynonymous substitutions (see Materials and Methods). Cox1, cox2, and cob have some of the lowest dN/dS ratios (table 3), which is expected considering these genes are typically used in phylogenetics due to their low rate of evolutionary divergence. Mitochondrial genes encoding ribosomal proteins (rps3, rps11, rps12, rpl16, and rpl20) have some of the higher dN/dS ratios along with sdhB, sdhC, and sdhD. On the whole, genes involved in electron transport (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) and ATP synthase (atp6, atp8, and atp9) have lower dN/dS ratios. The dN/dS ratio of rps11 (1.72) and secY (1.19) imply that these genes are under positive selection. In the case of the atp8 and sdhC G. oryzoides pseudogenes, the 5′ end of each gene was included in the analysis and in pairwise comparisons. In both cases, the ratio was similar to that found among functional copies of the genes.
Table 3.
Table 3dN/dS Ratios for the 24 Protein-Coding Genes Shared between C. crispus (CC), P. pulvinata (PP), Gr. andersonii (GA), and G. oryzoides (GO)
| Gene | Average dN/dS | CC/GA | GA/GO | GO/PP | CC/GO | CC/PP |
| atp6 | 0.14923 | 0.1222 | 0.1948 | 0.1463 | 0.1211 | 0.1464 |
| atp8 | 0.17942 | 0.1144 | 0.2086 | NA | 0.2092 | NA |
| atp9 | 0.031 | 0.0318 | 0.0581 | 0.04 | 0.0419 | 0.0229 |
| cob | 0.12321 | 0.091 | 0.0913 | 0.1686 | 0.0868 | 0.1444 |
| cox1 | 0.09423 | 0.0744 | 0.0573 | 0.1107 | 0.0724 | 0.1152 |
| cox2 | 0.13519 | 0.0996 | 0.1626 | 0.1501 | 0.1176 | 0.1328 |
| cox3 | 0.14909 | 0.1245 | 0.1074 | 0.1862 | 0.1347 | 0.1673 |
| nad1 | 0.10879 | 0.0911 | 0.1051 | 0.1433 | 0.0862 | 0.088 |
| nad2 | 0.42864 | 0.3198 | 0.6045 | 0.7016 | 0.4609 | 0.2944 |
| nad3 | 0.18078 | 0.1788 | 0.0938 | 0.258 | 0.1591 | 0.1729 |
| nad4 | 0.16722 | 0.1181 | 0.1165 | 0.1956 | 0.1159 | 0.2021 |
| nad4L | 0.18756 | 0.1918 | 0.2048 | 0.2109 | 0.2444 | 0.1646 |
| nad5 | 0.35287 | 0.2902 | 0.1675 | 0.5156 | 0.293 | 0.5299 |
| nad6 | 0.34997 | 0.2493 | 0.1919 | 0.6968 | 0.2177 | 0.6968 |
| rpl16 | 0.3923 | 0.419 | 0.2002 | 1.1754 | 0.3767 | 1.8547 |
| rpl20 | 0.32286 | 0.8087 | 0.1827 | 0.8081 | 0.5991 | 0.3551 |
| rps3 | 0.66532 | 1.5084 | 0.3131 | 0.5835 | 1.4929 | 1.3762 |
| rps11 | 1.72201 | 3.261 | 2.6059 | 4.427 | 1.3158 | 1.8863 |
| rps12 | 0.33198 | 0.6998 | 0.4949 | 1.4743 | 0.2276 | 0.4949 |
| sdhB | 0.12757 | 0.1358 | 0.1094 | 3.3678 | 0.1435 | 3.1396 |
| sdhC | 0.52405 | 0.2853 | 0.4833 | 0.4903 | 0.2246 | 1.2409 |
| sdhD | 0.44802 | 0.3186 | 0.3684 | 0.6144 | 0.37 | 0.4201 |
| secY | 1.19583 | 1.7927 | 0.317 | 2.5772 | 1.5177 | 8.0768 |
| ymf | 0.83624 | NA | NA | 1.5478 | 1.0369 | 0.4958 |
Note.—Average values and pairwise comparisons are given, with values >1 in bold. NA represents missing genes and in cases where pseudogenes were present, sequence preceding a frameshifting indel was used.
Discussion
Because of the medical implications of parasitic organisms and their tendency toward smaller genomes, they were among the first eukaryotes to have their genomes sequenced (e.g., Katinka et al. 2001; Gardner et al. 2002; Berriman et al. 2005; El-Sayed et al. 2005; Ivens et al. 2005; Haas et al. 2009). As a result, much is known about parasite genome architecture and construction. Parasite genomes, like those of most symbionts, tend to follow certain evolutionary trends such as: an increase in AT content (Moran 1995; Cavalier-Smith 2005); decreased coding capacity due to the loss of biosynthetic and metabolic pathways (Corradi et al. 2007); and organellar genome reduction (Rocha and Danchin 2002; Keeling and Slamovits 2004; Keeling and Slamovits 2005; Moya et al. 2008).
On initial comparison between the adelphoparasite and its host it would appear that, much like the nuclear genome, organellar genome reduction appears to be a common characteristic among parasites. The mtDNA of adelphoparasite G. oryzoides is 1,873 bp smaller than host Gr. andersonii. However, difference in genome size can mostly be accounted for by the deletion of a large intergenic space in the host, with ORFs of unknown function (orf61, orf87, and orf95) between cob and nad6 that accounts for most of the difference (fig. 3). Due to the lack of a closely related “out group” sequence, parasite genome reduction is indistinguishable from genome expansion in the host. Although the mitochondrial genome of the alloparasite, P. pulvinata (25,894 bps), is also smaller than that of Gr. andersonii, direct genome size comparisons are difficult to make because the parasites are not sister species. Both parasite genomes fall well within the range of already sequenced red algal mtDNA (Leblanc et al. 1995; Ohta et al. 1998; Burger et al. 2000) and do not appear to be significantly reduced in size. What the mtDNA sequence of P. pulvinata does allow for is parasite/parasite comparisons and the identification of genomic trends among red algal parasites.
Typically parasite genomes have a higher A/T content because the energetic demand to produce the GTP nucleotide and CTP nucleotide is more expensive than to produce ATP and UTP (Rocha and Danchin 2002). The overall A/T content of G. oryzoides mtDNA is the same as Gr. andersonii at 72%; an A/T percentage typically observed in mitochondrial genomes (Gray et al. 2004). When the genomes are separated into protein coding, noncoding, and rRNA, the A/T content percentage stays almost the same for G. oryzoides and Gr. andersonii (table 1), indicating similar evolutionary pressure across both host and parasite genomes. The overall A/T composition of P. pulvinata mtDNA is higher than that of the three other floridiophyte genomes at 76.1% (table 1). Additionally, the protein-coding regions of P. pulvinata are more A/T rich (76.4%) than that of other red algal mtDNA sequenced thus far.
The trend in A/T content is noteworthy because it is conventionally believed that red algal parasites evolve in a linear fashion, starting as adelphoparasites before diversifying and attacking novel hosts as an alloparasite (Zuccarello et al. 2004). If this theory is correct, the alloparasite P. pulvinata diverged earlier than G. oryzoides and had a greater amount of time to accrue sequence change, such as transversions. Combined with the outright loss of atp8 in P. pulvinata, as opposed to a recognizable pseudogene, our data are consistent with the notion that alloparasites are more evolutionarily divergent than adelphoparasites.
The development of pseudogenes atp8 and sdhC suggest that some essential genes are being rendered unnecessary in the parasite. This is interesting considering 1) overall mt genome conservation observed among diverse lineages of red algae, 2) the impact of mitochondrial dysfunction on cellular energetics, and 3) the compatibility between host and parasite nuclear and organellar DNA.
Red Algal mtDNA Conservation
Prior to this study, three red algal mitochondrial genomes representing three divergent orders were sequenced; C. crispus (Gigartinales), P. purpurea (Bangiales), and C. merolae (Cyanidiales). The mitochondrial genomes of Gr. andersonii and G. oryzoides (Gracilariales) and P. pulvinata (Plocamiales) fall into the same class (Florideophyceae) as C. crispus, whereas P. purpurea belongs to the Bangiophyceae and C. merolae to the Cyanidiophyceae. The mitochondrial genomes sequenced represent a broad spectrum of evolutionary time and species divergence (Le Gall and Saunders 2007). As noted by Ragan et al. (1994, p. 7278) “Rhodophyta are more divergent among themselves than are 1) fungi or 2) green algae and green plants together.” Despite this evolutionary divergence, mitochondrial genomes of sequenced red algae are incredibly well conserved in both content and genome architecture (Gray et al. 2004). Gene content and order uniformity are seemingly the product of a low mutation rate—a characteristic to consider when comparing red algal hosts with their respective red algal adelpho—and alloparasites (figs. 2–5, Leblanc et al. 1997; Gray et al. 2004). Mitochondrial genome differences between sister taxa Gr. andersonii and G. oryzoides are noteworthy considering the only changes over the ∼600 My since the divergence between the Bangiophyceae and the Florideophyceae (Yoon et al. 2004) are minor rearrangements of the conserved gene blocks (fig. 5). On the other hand, it is remarkable that such a dramatic change in lifestyle has resulted in only the loss of one (in the case of P. pulvinata) or two (G. oryzoides) genes from these mitochondria and almost no other detectable changes. The conserved dN/dS ratios across all taxa (table 3), independent of their lifestyle, is an additional indication of strong sequence constraint on both free living and parasite mitochondria. This low rate of red algal mitochondrial evolution indicates that the changes seen in parasite mtDNA over short time scales are a response to lifestyle change.
FIG. 5.—
Genome synteny of all sequenced floridiophyte mitochondrial genomes and that of the bangiophyte Porphyra purpurea. Among all of these genomes, coding genes fall into four highly conserved blocks. Within the floridiophytes, these blocks are maintained in the same order across divergent taxa.
Mitochondrial Dysfunction
The development of atp8 and sdhC as pseudogenes is perplexing considering the primary function of the mitochondria is to support aerobic respiration and produce cellular ATP. The atp8 gene encodes the ATP8 subunit of the F1F0-ATP synthase complex. This complex is composed of two main structural domains: the hydrophilic F1 domain and the hydrophobic F0 domain. Mitochondrial encoded atp6 and atp9, along with atp8, make up the F0 domain of the complex. Embedded in the inner membrane of the mitochondrion, the F0 domain anchors the ATP synthase complex and functions in proton translocation allowing for the F1 portion of the complex to complete ATP synthesis (Jesina et al. 2004; Jonckheere et al. 2008). Mutations in the atp6 gene have been shown to disturb the function of the ATPase proton channel. Without the ATP6 subunit, the ATP-synthase complex does not assemble correctly resulting in a decrease in ATP production (Funes et al. 2001). Mutations in the atp8 gene are less common but have been noted to occur in humans (Jonckheere et al. 2008), where data are extensive.
To further support the link between the loss of mitochondrial atp8 and a parasitic lifestyle, the atp8 gene is entirely absent from the mtDNA of P. pulvinata. The absence of atp8 in a mitochondrial genome is not entirely surprising considering organellar genes are often transferred to the nucleus (Dreyer and Steiner 2006; Pesaresi et al. 2007). The genes atp8, atp6, and atp9 are encoded by all red algal mtDNA sequenced to date but atp8 has been transferred to the nucleus in other lineages, including ciliates (Burger et al. 2000), apicomplexans and dinoflagellates (Slamovits et al. 2007), and even green algae (Denovan-Wright et al. 1998). Among all red algal mtDNA, atp8 is found between atp6 and nad5 in a highly conserved cluster of genes (atp6-atp8-nad5-nad4-sdhD-nad2-nad1-nad3). It remains unclear as to whether the atp8 gene was transferred to the nuclear genome or has simply been lost, but we have searched for a nuclear copy of atp8 in ∼373 Mb of nuclear sequence from G. oryzoides and ∼1,292 Mb of expressed sequence tag sequence (Lane Lab, unpublished) and were unable to locate a nuclear sequence with any homology. Although far from a definitive answer, the lack of any hint of a nuclear atp8 in data that represents >2X coverage of the genome (based on estimates in Kapraun 2005) is consistent with the outright loss of the gene.
The sdhC gene, like atp8, encodes a membrane bound subunit involved in the succinate dehydrogenase complex (complex II). More specifically, this complex catalyzes the oxidation of succinate to fumarate in the citric acid cycle while generating electrons from succinate for mitochondrial respiration (Bayley et al. 2005). Complex II is made up of four subunits; two are hydrophilic and involved in the catalytic portion of the complex (SDHA and SDHB) and the other two, SDHC and SDHD, are hydrophobic and act as anchors to the entire complex (Elorza et al. 2004; Bayley et al. 2005). Typically sdhB, sdhC, and sdhD are mitochondrially encoded, whereas sdhA has been transferred to the nuclear genome and is imported in from the cytosol. Mutations in SDH subunits are well studied and have been shown to have detrimental effects on cellular function and are most often tumorigenic (Bayley et al. 2005). It has been hypothesized that improper assembly of complex II results in either an accumulation of succinate, which then moves out of the mitochondria causing a hypoxic response, or leads to the production of reactive oxygen species (ROS) (Szeto et al. 2007). A frameshift mutation causing the development of a premature stop codon in the sdhC gene raises questions about the functionality of complex II in the parasite mitochondrial genome.
Mitochondria-encoded genes function in electron transport and oxidative phosphorylation. These pathways generate the majority of ATP for the cell, which is then used as a source of chemical energy to carry out energetic demands. Mutations in these genes have been shown to have broader implications on cell growth and longevity and are associated with a wide array of human neuromuscular and neurodegenerative diseases, including: Parkinson’s disease (VanItallie 2008), multiple sclerosis (Geurts and Barkhof 2008), various cancers (Ishikawa, Koshikawa, et al. 2008; Ishikawa, Takenaga, et al. 2008), Leigh syndrome (Pronicki et al. 2008), ROS production (Ishikawa, Koshikawa, et al. 2008), and Alzheimer’s disease (Reeve et al. 2008). The severity of these diseases is due to the cell’s inability to make energy and is often the result of single basepair insertions or deletions resulting in frameshift mutations (He et al. 2002; DiMauro and Schon 2008).
A recent study by Ishikawa, Takenaga, et al. (2008) demonstrated the acute impact of minor basepair changes in mitochondrial genes on metastatic capability of mouse tumor cells. When the resident mitochondria of mouse tumor cells with poor metastatic capability were replaced with those from a highly metastatic tumor cell line, metastatic potential was transferred from one cell type to the other. High metastasis resulted from two basepair mutations in the NADH dehydrogenase subunit 6 causing an overall deficiency in respiratory complex I activity. Seemingly, minor basepair changes in mtDNA can therefore have enormous consequences on cell function.
If these essential genes, atp8 and sdhC, are truncated and do not encode the entire protein, the parasite mitochondria must at least be less efficient at generating cellular energy. Based on our sequencing of RNA transcripts from host and parasite, it is clear that these genes are transcribed as part of multigene transcripts, much like other mitochondrial genes. It is unlikely given the degree of sequence divergence after the stop codon, however, that posttranscriptional modifications result in functional proteins. If the genes code for any functional protein, the more likely scenario (based on the 5′ sequence conservation) is that a truncated version of the protein is performing some function. If, instead, ATP8 and SDHC are not encoded by the parasite and it is still generating ATP efficiently, there are two possible explanations: 1) these proteins are coming from the parasite nuclear genome or the host mitochondrial genome or 2) ATP8 and SDHC are being compensated for by other proteins involved in their respective complexes. Both scenarios are interesting to consider because they would provide insight into host/parasite nuclear and organellar compatibility. Thus far, however, nuclear copies of either gene have not surfaced in either DNA or RNA sequencing efforts (Lane lab, unpublished).
Host/Parasite Nuclear and Organellar Compatibility
Work done by Goff and Coleman (1995) showed that upon host infection red algal parasites G. oryzoides and P. pulvinata maintain genetically unique nuclear and mtDNA while co-opting host plastids. Because host plastids are utilized by the parasite and incorporated into parasite spores, the nuclear genome of the parasite must be able to, in some way, communicate with the host plastid. Additionally, as cited by Goff and Coleman (Goff 1995), P. pulvinata, although colorless during its vegetative stage, has pigmented mature carpospores that appear to be partially photosynthetic (Kugrens and Delivopoulos 1986; Goff 1995). This, along with the occurrence of other photosynthetically capable alloparasites such as Choreocolax (Callow et al. 1979), indicates that the parasite nuclei have many of the genes needed for protein synthesis required by the plastid. Interestingly, in contrast, we observe the conservation of unique parasite mitochondria. This again raises the inevitable question of why the parasite maintains its own mitochondria. Are the parasite nuclei and host mitochondria incompatible? This question becomes increasingly interesting now that the mtDNA of both a red algal host and adelphoparasite have been sequenced. Although there are differences between the two genomes, they are similar overall.
An abstract published by Goff and McLaughlin (1997) describes the mitochondrial genome sequence of parasite P. pulvinata and its host Plocamium cartilagineum. The most notable difference between these two genomes is the retention of a tRNA cluster in the parasite mtDNA. The authors suggest that the additional tRNAs present in the parasite genome function in host cellular transformation. Essentially, these tRNAs are needed by the parasite to effectively complete spore germination, filament growth, and host infection. Interestingly, the mtDNA of G. oryzoides has 19 tRNAs, whereas Gr. andersonii has 18 tRNAs. The tRNA content of G. oryzoides and Gr. andersonii is identical expect for the presence of the histidine tRNA in G. oryzoides. The histidine tRNA is most likely a tRNA loss in Gr. andersonii rather than a gain in G. oryzoides because it is present in the mtDNA of C. crispus, a more divergent red alga. A tRNA gain would indicate a duplication event of an existing tRNA in the mtDNA of G. oryzoides that then underwent a mutation in the anticodon resulting in a GUG. Although possible, alignments of the G. oryzoides tRNAs show little sequence similarity to the G. oryzoides histidine tRNA. While in contrast, an alignment between the G. oryzoides histidine and C. crispus histidine tRNA shows considerable sequence similarity.
Interestingly, the expression of the histidyl-tRNA in the bloodstream-form of Trypanosoma brucei has a significant effect on its growth. Through RNAi silencing Merritt et al. (2010) were able to knockdown the histidine tRNA gene, consequently suppressing cell growth by a factor of >103 over 4 days (Merritt et al. 2010). The role of the histidyl-tRNA in G. oryzoides may not be as crucial to its overall growth success as it is in T. brucei, however, such findings add insight into the role of various genes on overall parasite success. The significance of tRNA content among red algal host/parasite pairs remains unknown. Although Goff and McLaughlin (1997) suggest that tRNAs are retained by the parasite so that spores function more efficiently during the initial stages of host infection, it is unclear after sequencing G. oryzoides, whether this really is the case.
Red algal adelpho- and alloparasites provide a unique model for understanding mitochondrial genome evolution. Host /parasite and parasite/parasite mtDNA comparisons reveal the effect of a parasitic lifestyle on the genomic architecture and sequence of the mitochondrion. Results indicate that parasite mtDNA is undergoing reduction and compaction through the loss of coding and noncoding sequences. No single feature, however, points to a reason why the mitochondrial genome is retained by red algal parasites. The overall efficiency of the parasite mitochondria would appear to be reduced considering the loss of atp8 in P. pulvinata and existence of atp8 and sdhC as pseudogenes in G. oryzoides. The trend toward the loss of atp8 is significant considering rapid rates of evolution are not occurring among the majority of mitochondrial genes (table 3). This creates a bit of a paradox because genes considered to be vital for function are being lost while, at the same time, there are clearly selective forces maintaining the genome sequence from further losses and accelerated mutational rates. Further analysis of host and parasite nuclear DNA may elucidate some fundamental genomic shifts and molecular changes that have resulted in a parasitic existence.
Supplementary Material
Supplementary table 1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank John Archibald and two anonymous reviewers for critically reading the manuscript as well as Megan O'Brien for producing data for the project and Ian Misner for assistance with data manipulation. We also acknowledge the Gordon and Betty Moore Foundation sponsored MEGAMER facility for resources and use of space at UCSC and particularly Brandon Carter for help and logistical facilitation.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley JP, Devilee P, Taschner PE. The SDH mutation database: an online resource for succinate dehydrogenase sequence variants involved in pheochromocytoma, paraganglioma and mitochondrial complex II deficiency. BMC Med Genet. 2005;6:39. doi: 10.1186/1471-2350-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Burger G, Saint-Louis D, Gray MW, Lang BF. Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea. Cyanobacterial introns and shared ancestry of red and green algae. Plant Cell. 1999;11:1675–1694. doi: 10.1105/tpc.11.9.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, et al. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J Mol Biol. 2000;297:365–380. doi: 10.1006/jmbi.2000.3529. [DOI] [PubMed] [Google Scholar]
- Callow JA, Callow ME, Evans LV. Nutritional studies of the parasitic red alga Choreocolax polysiphoniae. New Phytol. 1979;83:451–462. [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, et al. Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS One. 2007;2:e1277. doi: 10.1371/journal.pone.0001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denovan-Wright EM, Nedelcu AM, Lee RW. Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol Biol. 1998;36:285–295. doi: 10.1023/a:1005995718091. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G. The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica—and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 2006;3:13. doi: 10.1186/1742-9994-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, et al. Geneious v4.5 [Internet] 2009. Available from: http://www.geneious.com/ default, 1103, citations.sm. [Google Scholar]
- El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Elorza A, et al. Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiol. 2004;136:4072–4087. doi: 10.1104/pp.104.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S, et al. The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0-ATPase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J Biol Chem. 2001;277:6051–6058. doi: 10.1074/jbc.M109993200. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts JJG, Barkhof F. Grey matter pathology in multiple sclerosis. Lancet Neurol. 2008;7:841–851. doi: 10.1016/S1474-4422(08)70191-1. [DOI] [PubMed] [Google Scholar]
- Gobert A, Isayenkov S, Voelker C, Czempinski K, Maathuis FJM. The two-pore channel TPK1 gene encodes the vacuolar K conductance and plays a role in K homeostasis. Proc Natl Acad Sci U S A. 2007;104:10726–10731. doi: 10.1073/pnas.0702595104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff L, Ashen J, Moon D. The evolution of parasites from their hosts: a case study in the parasitic red algae. Evolution. 1997;51:1068–1078. doi: 10.1111/j.1558-5646.1997.tb03954.x. [DOI] [PubMed] [Google Scholar]
- Goff L, et al. The evolution of parasitism in the red algae: molecular comparisons of adelphoparasites and their hosts. J Phycol. 1996;32:297–312. [Google Scholar]
- Goff LJ. The biology of parasitic red algae. Phycol Res. 1982;1:289–370. [Google Scholar]
- Goff LJ. Fate of parasite and host organelle DNA during cellular transformation of red algae by their parasites. Plant Cell. 1995;7:1899–1911. doi: 10.1105/tpc.7.11.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LJ, Coleman AW. The role of secondary pit connections in red algal parasitism. J Phycol. 1985;21:483–508. [Google Scholar]
- Goff LJ, Coleman AW. Nuclear transfer from parasite to host—a new regulatory mechanism of parasitism. Ann N Y Acad Sci. 1987;503:402–423. [Google Scholar]
- Goff LJ, Coleman AW. Fate of parasite and host organelle DNA during cellular transformation of red algae by their parasites. Plant Cell. 2005;7:1899–1911. doi: 10.1105/tpc.7.11.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LJ, McLaughlin IJ. The complete sequence of the mitochondrial genome of the red algal parasite Plocamiocolax pulvinata (Plocamiales) and comparison to its host, Plocamium cartilagineum, and two other Plocamium non-host species. Phycologia. 1997;36:35. [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- He LP, et al. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res. 2002;30:e68. doi: 10.1093/nar/gnf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Koshikawa N, Takenaga K, Nakada K, Hayashi JI. Reversible regulation of metastasis by ROS-generating mtDNA mutations. Mitochondrion. 2008;8:339–344. doi: 10.1016/j.mito.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:4. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Ivens AC, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesina P, et al. Diminished synthesis of subunit a (ATP6) and altered function of ATP synthase and cytochrome c oxidase due to the mtDNA 2 bp microdeletion of TA at positions 9205 and 9206. Biochem J. 2004;383:561–571. doi: 10.1042/BJ20040407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere AI, et al. A novel mitochondrial ATP8 gene mutation in a patient with apcial hypertrophic cardiomyopathy and neuropathy. J Med Genet. 2008;45:129–133. doi: 10.1136/jmg.2007.052084. [DOI] [PubMed] [Google Scholar]
- Kapraun D. Nuclear DNA content estimates in multicellular green, red and brown algae: phylogenetic considerations. Ann Bot. 2005;1:7–44. doi: 10.1093/aob/mci002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Slamovits CH. Simplicity and complexity of microsporidian genomes. Eukaryot Cell. 2004;3:1363–1369. doi: 10.1128/EC.3.6.1363-1369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Slamovits CH. Cause and effects of nuclear genome reduction. Sci Direct. 2005;15:601–608. doi: 10.1016/j.gde.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Kugrens P, Delivopoulos SG. Ultrastructure of the carposporophyte and carposorogenesis in the parasitic red alga Plocamiocolax pulvinata SETCH. (Gigartinales, Plocamiaceae) J Phycol. 1986;22:8–21. [Google Scholar]
- Le Gall L, Saunders GW. A nuclear phylogeny of the Florideophyceae (Rhodophyta) inferred from combined EF2, small subunit and large subunit ribosomal DNA: establishing the new red algal subclass Corallinophycidae. Mol Phylogenet Evol. 2007;34:1118–1130. doi: 10.1016/j.ympev.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Leblanc C, et al. Complete sequence of the mitochondrial DNA of the rhodophyte Chondrus crispus (Gigartinales)—gene content and genome organization. J Mol Biol. 1995;250:484–495. doi: 10.1006/jmbi.1995.0392. [DOI] [PubMed] [Google Scholar]
- Leblanc C, et al. Origin and evolution of mitochondria: what have we learnt from red algae? Curr Genet. 1997;31:193–207. doi: 10.1007/s002940050196. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, et al. Crystal structures of trypanosomal histidyl-tRNA synthetase illuminate differences between eukaryotic and prokaryotic homologs. J Mol Biol. 2010;397:481–494. doi: 10.1016/j.jmb.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1995;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Ohta N, Sato N, Kuroiwa T. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 1998;26:5190–5198. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Curr Opin Plant Biol. 2007;10:600–606. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Pronicki M, et al. Light and electron microscopy characteristics of the muscle of patients with SURF1 gene mutations associated with Leigh disease. J Clin Pathol. 2008;61:460–466. doi: 10.1136/jcp.2007.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan, et al. A molecular phylogeny of the marine red algae (Rhodophyta) based on the nuclear small-subunit rRNA gene. Proc Nat Acad Sci. 1994;91:7276–7280. doi: 10.1073/pnas.91.15.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve AK, Krishnan KJ, Turnbull DM. Age related mitochondrial degenerative disorders in humans. Biotechnol J. 2008;3:750–756. doi: 10.1002/biot.200800066. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- Saunders GW. Gel purification of red algal genomic DNA—an inexpensive and rapid method for the isolation of polymerase chain reaction-friendly DNA. J Phycol. 1993;29:251–254. [Google Scholar]
- Setchell WA. Parasitism among the red algae. Proc Amer Philos Soc. 1918;57:155–172. [Google Scholar]
- Slamovits CH, Saidarriaga JF, Larocque A, Keeling PJ. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol. 2007;372:356–368. doi: 10.1016/j.jmb.2007.06.085. [DOI] [PubMed] [Google Scholar]
- Szeto SSW, Reinke SN, Sykes BD, Lemire BD. Ubiquinone-binding site mutations in the Saccharomyces cerevisiae succinate dehydrogenase generate superoxide and lead to the accumulation of succinate. J Biol Chem. 2007;282:27518–27526. doi: 10.1074/jbc.M700601200. [DOI] [PubMed] [Google Scholar]
- VanItallie TB. Parkinson disease: primacy of age as a risk factor for mitochondrial dysfunction. Metab Clin Exp. 2008;57:S50–S55. doi: 10.1016/j.metabol.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Zuccarello GC, Debra M, Goff LJ. A phylogenetic study of parasitic genera placed in the family Choreocolacaceae (Rhodophya) J Phycol. 2004;40:937–945. [Google Scholar]