Summary
A-Kinase Anchoring Proteins (AKAPs) coordinate cell-signaling events. AKAP79 brings together different combinations of enzyme binding partners to customize the regulation of effector proteins. In neurons muscarinic agonists mobilize an AKAP79-anchored pool of PKC that phosphorylates the KCNQ2 subunit of the M channel. This inhibits potassium permeability to enhance neuronal excitability. Using a dual fluorescent imaging/patch-clamp technique, we visualized AKAP79-anchored PKC phosphorylation of the kinase activity reporter CKAR concurrently with electrophysiological changes in KCNQ2 channels to show that AKAP79 synchronizes both signaling events to optimize the attenuation of M currents. AKAP79 also protects PKC from certain ATP competitive inhibitors. Related studies suggest that context dependent protein-protein interactions alter the susceptibility of another protein kinase, PDK1, to ATP analog inhibitors. This implies that intracellular binding partners not only couple individual molecular events in a cell signaling process but can also change the pharmacological profile of certain protein kinases.
Introduction
Non-catalytic regulatory proteins profoundly influence the action of protein kinases and phosphatases (Scott and Pawson, 2009; Tasken and Aandahl, 2004). A-Kinase Anchoring Proteins (AKAPs) are signal-organizing molecules that tether these enzymes in subcellular environments to control the phosphorylation state of neighboring substrates (Wong and Scott, 2004). A prototypic example is AKAP79/150: a family of three orthologs (human AKAP79, murine AKAP150, and bovine AKAP75) that were initially discovered as binding proteins for the type II regulatory subunit of the cAMP dependent protein kinase (PKA) (Carr et al., 1992). AKAP79/150 also anchors the calcium/phospholipid dependent kinase (PKC), and the calcium/calmodulin dependent phosphatase (PP2B) (Coghlan et al., 1995; Klauck et al., 1996). These signaling complexes reside on the inner face of the plasma membrane where they respond to the generation of intracellular second messengers such as cAMP, calcium and phospholipid (Dell’Acqua et al., 1998).
Molecular and cellular approaches have demonstrated that AKAP79/150 directs its cohort of anchored enzymes towards selected transmembrane proteins to facilitate their efficient regulation. Functional studies in multiple cell types have confirmed this notion showing that different AKAP79/150 complexes regulate the activity of ion channels including AMPA receptors, L-type calcium channels, M-type potassium channels, and heat-activated TRPV1 channels (Gao et al., 1997; Hoshi et al., 2005; Tavalin et al., 2002; Zhang et al., 2008). AKAP79/150 has been implicated in cardiovascular signaling through PKA mediated phosphorylation of β-adrenergic receptors and suppression of adenylyl cyclase 5/6 activity (Bauman et al., 2006; Fraser et al., 2000). In addition AKAP79/150 influences the onset of angiotensin II-induced hypertension (Navedo et al., 2008). AKAP79/150 also participates in the modulation of the muscarine-sensitive M current, a voltage-gated potassium current that counteracts neuronal excitability (Hoshi et al., 2003). The KCNQ2 subunit of the M channel binds AKAP79/150, while C-terminal regions of the anchoring protein interact with the m1 muscarinic receptor (Hoshi et al., 2005). This maintains PKC where it can optimally respond to activating signals from the m1 receptor and preferentially phosphorylate the KCNQ2 subunit. Such a receptor-AKAP-channel complex is believed to enhance the suppression of M currents (Tunquist et al., 2008). In this report, we delve more deeply into how AKAP79/150 augments this signaling pathway. We have discovered that the anchoring protein modifies the activity of anchored PKC in a manner that changes the pharmacological profile of the enzyme. Related studies on another protein kinase PDK1 suggest that context dependent protein-protein interactions alter its sensitivity to ATP analog inhibitors.
Results
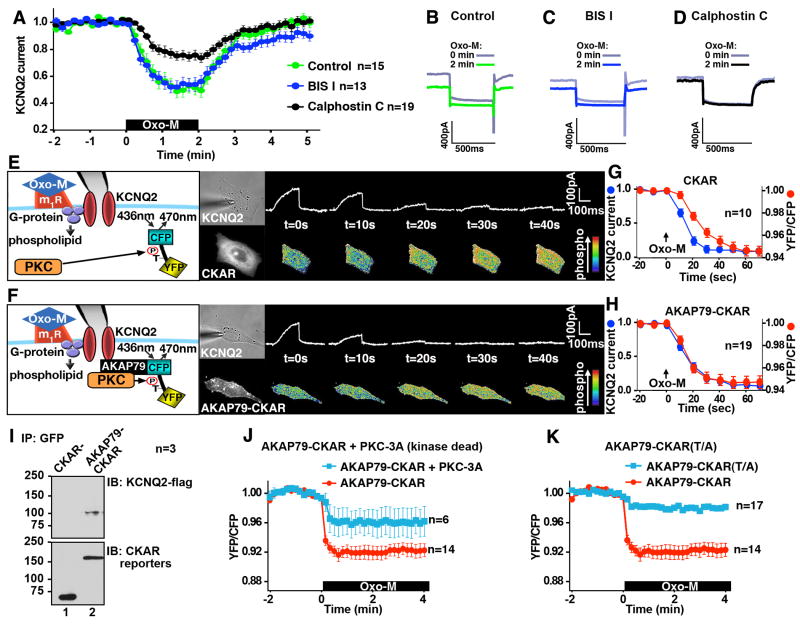
Muscarinic agonists such as acetylcholine mobilize an anchored pool of PKC that phosphorylates the KCNQ2 subunit of the M channel on Ser 541 to decrease potassium permeability (Hoshi et al., 2003). Yet paradoxically, muscarinic receptor operated M channels are insensitive to some PKC inhibitors (Bosma and Hille, 1989; Suh and Hille, 2002). Whole cell patch-clamp electrophysiology experiments in cultured Sympathetic Cervical Ganglion (SCG) neurons confirmed this observation. Application of the muscarinic agonist oxotremorine-M (Oxo-M) promoted suppression of M currents (n=15; Fig 1A & B; green). Similar results were obtained when these experiments were repeated in the presence of bisindolylmaleimide I (BIS I) a general inhibitor of PKCs that targets the ATP binding pocket of the enzyme (n=13; Fig 1A & C; blue). In contrast, Oxo-M induced suppression of M currents was reduced when neurons were treated with calphostin C, a PKC inhibitor that targets the diacylglycerol (DAG) binding site of the kinase (n=19; Fig 1A & D; black). Although AKAP79/150 has been implicated in this important signaling event, little is known about how this anchoring protein synchronizes individual steps in this process or how AKAP79-anchored PKC exhibits a differential sensitivity to pharmacological inhibitors. To address this we configured a patch-clamp apparatus to allow fluorescent imaging of AKAP79-anchored PKC activity and simultaneous electrophysiological recording of the ion channel. A Chinese Hamster Ovary (CHO) cell line that stably expresses the m1 muscarinic receptor (Selyanko et al., 2000) was used to ensure optimal expression of the ion channel and the fluorescent reporter.
Figure 1. AKAP79 synchronizes muscarinic activation of PKC with KCNQ2 current suppression.
A) Electrophysiological recording of the M current from SCG neurons. The M current suppression induced by a muscarinic agonist, 1μM Oxo-M, was attenuated by 100 nM calphostin C but not by 100 nM BIS I. Representative traces at 0 and 2 min after the application of 1μM Oxo-M in B) untreated SCG neurons, C) 100 nM BIS I, and D) 100 nM Calphostin C treated neurons. E) Simultaneous recording of KCNQ2 channel activity and PKC phosphorylation. Diagram of experimental setup: the patch-clamp and FRET based PKC activity reporter (CKAR) system. CHO cells co-expressing CKAR and KCNQ2 were simultaneously analyzed for changes in whole cell currents and PKC activity after 3 μM Oxo-M. Phosphorylation of CKAR induces conformational changes that decrease the YFP/CFP ratio. Micrographs of a single cell show the patch-pipet (bright-field, upper middle) and CKAR reporter (fluorescent, lower middle). Primary data: KCNQ2 currents (top) and pseudocolor images depicting FRET changes (bottom) at specified times after Oxo-M. F) Simultaneous recording of KCNQ2 channel and AKAP79-CKAR. Model of an AKAP79-CKAR reporter and M channel configuration (left). Micrographs of a single cell show the patch-pipet (bright field, upper middle) and AKAP79-CKAR reporter (fluorescent, lower middle). Primary data: Time course of KCNQ currents (top) and pseudocolor images of FRET changes (bottom; YFP/CFP ratio). G & H) Time course response to Oxo-M: amalgamated data from simultaneous patch-clamp with CKAR (G) or AKAP79-CKAR (H). KCNQ2 currents (blue) and respective reporter FRET ratio: YFP/CFP (red). N values and mean +/- SEM are indicated. I) Co-immunoprecipitation of flag-tagged KCNQ2 channel with AKAP79-CKAR. J) Disruption of AKAP79-CKAR response by overexpression of kinase-dead PKCβII (PKC-3A). Co-expression of PKC-3A, blunted the AKAP79-CKAR response to 3 μM Oxo-M (blue) compared to AKAP79-CKAR alone (red). K) Effect of removal of phosphorylation acceptor threonine. Thr/Ala mutant at the PKC phosphorylation acceptor in AKAP79-CKAR did not respond to 3 μM Oxo-M (blue), suggesting relevant threonine residue is the only site that is detected by AKAP79-CKAR. (See also Figures S1 and S2.)
Agonist-dependent activation of PKC was detected using cytoplasmic CKAR, a genetically encoded fluorescence resonance energy transfer (FRET) based PKC activity reporter (Violin et al., 2003). Cells were excited at 436nm to elicit FRET and patch-clamped to measure channel activity (Fig 1E & F, Supplemental Fig S1). A montage of current amplitudes (top) and FRET data (bottom) at specified times after activation of the m1 receptor are presented. To facilitate comparisons of current activity with simultaneous FRET measurements of PKC activity we chose to portray increases in PKC phosphorylation as a decrease in the ratio of YFP/CFP. These data show that Oxo-M induces changes in the cytoplasmic CKAR signal and evokes suppression of the KCNQ2 currents (Fig 1G). However, these events were not synchronized. Half-maximal responses (T½) of KCNQ2 currents and cytoplasmic CKAR fluorescence were observed 13.5+/- 2.2 sec (Fig 1G blue, n=10) and 24.2+/-1.4 sec (Fig 1G red, n=10, P<0.001) seconds after application of Oxo-M respectively.
Fusion with AKAP79 confers a new property on CKAR; namely it targets the reporter to the ion channel thereby anchoring PKC close to its substrate (Fig 1F). Control experiments demonstrated that the AKAP79-CKAR protein interacts with the KCNQ2 subunits inside cells (Fig 1I). Under this configuration, the time course of the AKAP79-CKAR response was nearly identical to that for the suppression of the KCNQ2 current (Fig 1F & H). KCNQ2 current and AKAP79-CKAR fluorescence T½ occurred at 14.4+/- 0.8 sec (n=19; Fig 1H, blue) and 13.7+/- 1.2 sec (n=19; Fig 1H, red) after application of Oxo-M respectively. Cells co-expressing a catalytically inactive kinase (PKC-3A) that competes with endogenous PKC for interaction with AKAP79-CKAR exhibited blunted FRET responses (n=6; Fig 1J, blue). Other controls showed that AKAP79-CKAR Thr/Ala (T/A), a non-phosphorylatable form of the reporter, was refractory to Oxo-M stimulation (n=17; Fig 1K, blue). Attempts to further examine this effect using myristoylated-CKAR that non-specifically localizes the reporter to membranes were unsuccessful. Comparable FRET responses were obtained with the membrane-tethered versions of CKAR and the T/A mutant (Supplemental Fig S2). In addition, cells overexpressing cytoplasmic CKAR and AKAP79 did not show any difference in their responses from cells expressing CKAR alone (data not shown). Collectively these data in show that AKAP79 optimizes the relay of muscarinic signals by synchronizing molecular events that contribute to the down-regulation of KCNQ2 channels.
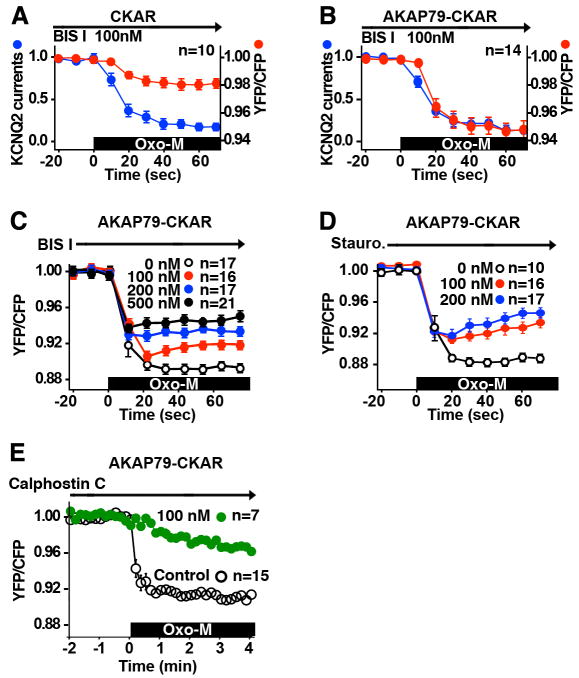
Next we explored anchored PKC action. Membrane permeable compounds such as bisindolylmaleimides or staurosporine are frequently used to inhibit PKCs although they also affect other kinases (Davies et al., 2000). These small-molecule inhibitors compete with ATP. Striking differences were observed between the cytoplasmic- and AKAP79- associated pools of PKC when we used these inhibitors with our simultaneous FRET and patch-clamp apparatus. BIS I, (100nM) blocked PKC activity 76.3 ± 7.6% in cells expressing cytoplasmic-CKAR when compared to controls (n=10; Fig 2A; red). Yet surprisingly, muscarinic suppression of the KCNQ2 currents persisted in the presence of this inhibitor (n=10; Fig 2A, blue). One interpretation of this observation is that CHO cells harbor a pool of PKC close to the channel that is resistant to BIS I.
Figure 2. AKAP79-anchored PKC activity shows resistance to ATP competitive inhibitors.
A & B) BIS I inhibition of CKAR or AKAP79-CKAR and KCNQ2 Oxo-M stimulated responses. Cells coexpressing KCNQ2 and CKAR (A) or AKAP79-CKAR (B) were pretreated with 100 nM BIS I prior to 3 μM Oxo-M stimulation. BIS I inhibited CKAR (A) response but not KCNQ2 response as compared with Fig. 1G. BIS I failed to inhibit AKAP79-CKAR (B) or KCNQ2 responses compared with Fig. 1H. C) Dose-response of BIS I inhibition of AKAP79-CKAR. D) Dose-response of another ATP competitive inhibitor, staurosporine. Anchored PKC was resistant to a wide range of BIS I and staurosporine concentrations. E) Calphostin C inhibition of AKAP79-CKAR. 100 nM Calphostin C, a PKC inhibitor that targets the DAG binding site attenuated the Oxo-M stimulated AKAP79-CKAR activity.
This was further explored in cells expressing the AKAP79-CKAR reporter. Surprisingly, the proximity of anchored PKC to the AKAP abrogated the inhibitory action of BIS I (n=14; Fig 2B, red). More detailed analysis revealed that cells expressing AKAP79-CKAR and KCNQ2 show marked resistance to BIS I and staurosporine over a range of concentrations (Fig 2 C & D). In contrast, calphostin C, a small molecule inhibitor that targets the DAG-binding site of PKC reduced phosphorylation of the AKAP79-CKAR reporter (n=7; Fig 2E; green).
These observations indicate that AKAP79/150 not only controls access to substrates but may also influence how binding partners such as PKC respond to certain pharmacological inhibitors. This provides an explanation as to why PKC was initially discounted as a modulator of KCNQ2 channel function since early reports concluded that muscarinic suppression of M channels was unaffected by staurosporine (Bosma and Hille, 1989).
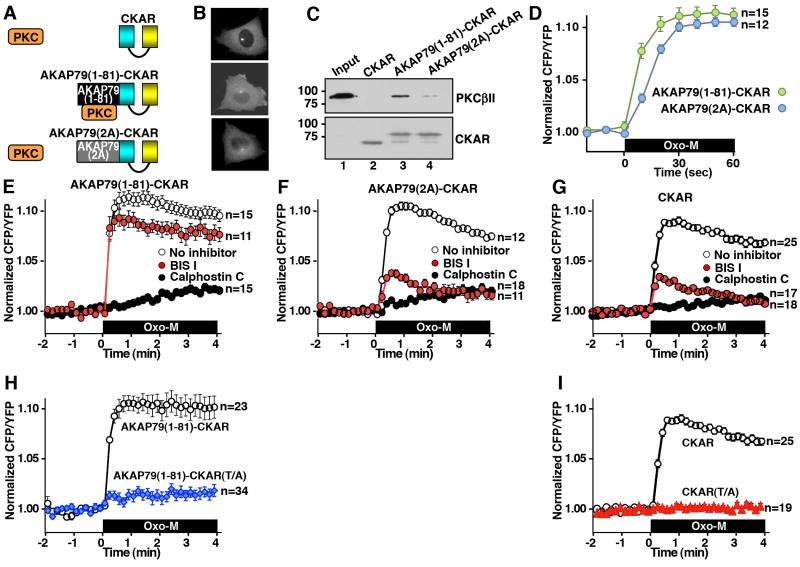
The first 81 amino acids of AKAP79 encompass a “pseudosubstrate-like” domain that is sufficient for PKC binding in vitro (Faux and Scott, 1997). Fusion of this region to CKAR created a cytoplasmic reporter that sequesters PKC but does not interact with either PKA or PP2B (Fig 3 A & B, middle). We replaced arginines at positions 40 and 41 of the AKAP79 1-81 fragment with alanines to produce a PKC-anchoring deficient reporter called AKAP79(2A)-CKAR (Fig 3A & B, bottom). Both reporters were found to be predominantly cytoplasmic and binding assays demonstrated that PKCβII associates with AKAP79(1-81)-CKAR but not with AKAP79(2A)-CKAR or CKAR alone (Fig 3C). Control experiments confirmed that a) these modified CKAR reporters did not interact with PKA or phosphatases and b) KCNQ2 channels were not sensitive to PP2B inhibitors (data not shown). In addition, it has been documented that CKAR is sensitive to attenuation by cellular phosphatase activity (Violin et al., 2003). The AKAP79(1-81)-CKAR reporter responded more rapidly to PKC activation (Fig 3D, green; T½ = 8.3 +/- 1.0 sec, n=15) than cells expressing AKAP79(2A)-CKAR (Fig 3D, blue; T½ = 14.6 +/- 0.5 sec, n=12) or CKAR alone (T½ = 12.1 +/- 0.8 sec, n = 25). This lends further credence to the notion that anchoring of PKC enables preferential phosphorylation of this reporter. Although we acknowledge it is formally possible that submembrane tethering of the anchored enzyme could minimally contribute to the activation of the reporter it is important to note that these data support a model where PKC-AKAP79 interaction in the cytosol is sufficient to promote preferential phosphorylation of the reporter. Data in figure 3D and the remainder of this report PKC activation is now represented as an increase in the CFP/YFP ratio.
Figure 3. PKC binding to a region of AKAP79 affects susceptibility to small molecule inhibitors.
A) Cartoon of PKC interaction with either CKAR, AKAP79(1-81)-CKAR or the PKC binding-deficient AKAP79(2A)-CKAR reporters. B) Micrographs showing expression and subcellular localization of these constructs in CHO cells. C) Input and co-immunoprecipitation of PKCβII with FRET reporters using anti-GFP antibody assessed by immunoblot (top). Immunoblot control (bottom). D) Time course of PKC response to Oxo-M enhanced in AKAP79(1-81)-CKAR (green) FRET reporter relative to AKAP79(2A)-CKAR. Phosphorylation of the reporter = increase in the CFP/YFP ratio. E-G) Amalgamated data from cells expressing the E) AKAP79(1-81)-CKAR, (F) AKAP79(2A)-CKAR or (G) CKAR reporters were pretreated with either the ATP competitive inhibitor, BIS I (red, 200 nM) or a PKC inhibitor that targets the DAC binding site, calphostin C (black, 100 nM) or no PKC inhibitor (white) prior to 3 μM Oxo-M application. H) A Thr/Ala mutant, AKAP79(1-81)-CKAR(TA), did not respond to 3 μM Oxo-M. I) CKAR(T/A) did not respond to 3 μM Oxo-M. Phosphorylation of the reporter = increase in the CFP/YFP ratio. N values and mean +/- SEM are indicated. (See also Figure S3.)
Different classes of PKC inhibitor were then used to investigate the activity state of the AKAP79 bound enzyme. Application of the ATP-competitive inhibitor BIS I and staurosporine had no effect on Oxo-M mediated activation of anchored PKC as measured with the AKAP79(1-81)-CKAR reporter (Fig 3E, red; n=11; Supplemental Fig S3). In contrast, application of calphostin C that does not bind in the ATP binding site blocked activation of anchored PKC (n=15; Fig 3E, black). Thus, association of PKC with the first 81 residues of AKAP79 confers a resistance to ATP competitive inhibitors. Control experiments demonstrated that both BIS I and calphostin C were effective PKC inhibitors when the AKAP79(2A)-CKAR or CKAR reporters were used to monitor enzyme activity (Figs 3F & G). Additional controls showed that the AKAP79(1-81)-CKAR T/A and CKAR T/A reporters were refractory to Oxo-M stimulation (Fig 3H & I). Taken together the data in figure 3 imply that interactions with the “pseudosubstrate like” site on AKAP79 alters the conformation of the catalytic core of PKC, thereby restricting the accessibility of certain ATP competitive inhibitors. This raises an intriguing possibility that the intracellular pharmacology of certain small molecule inhibitors may be influenced by the molecular context of their target enzyme.
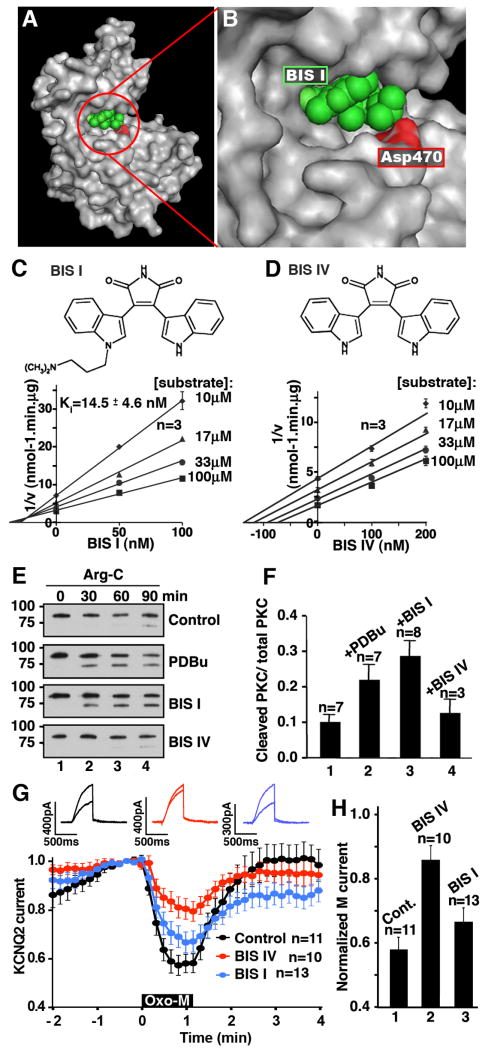
X-ray crystallography studies have shown that BIS I (and staurosporine) occupy an ATP binding pocket located in a cleft formed between the two lobes of the kinase catalytic core (Grodsky et al., 2006, & Fig 4A). Closer inspection reveals that the dimethylamino group of BIS I forms a hydrogen bond with aspartate 470 in PKCβII, a conserved residue in the substrate recognition groove of all PKC subtype (Fig 4B & C). Previous studies have shown that the “pseudosubstrate-like” region of AKAP79 also binds within this region of PKC (Klauck et al., 1996). BIS I and the PKC binding peptide from AKAP79 are competitive inhibitors of PKCβII with inhibition constants (Ki) of 14.5 ± 4.6 nM, (n=6; Fig 4C) and 1.41 ± 0.28 μM (Klauck et al., 1996). In contrast, BIS IV, a bisindolylmaleimide derivative that lacks the dimethylamino group is an uncompetitive inhibitor with respect to the substrate peptide (Fig 4D). This latter finding suggests that BIS IV acts by a different mechanism of action than that observed with BIS I or AKAP79.
Figure 4. Certain ATP inhibitor competitors stabilize PKC in an open conformation.
A & B) Molecular model of PKCβII catalytic core in gray modified from the PDB file (2I0E). BIS I (green) in the ATP binding pocket. Aspartate 470 of PKCβII (red). C & D) Chemical structures of BIS I and BIS IV. Dixon plots of PKCβII activity against peptide substrate with inhibitor BIS I (C) or BIS IV (D). Substrate peptide concentrations are indicated. E) Conformational sensitivity of PKCβII to proteolysis. Purified PKCβII was preincubated with either BIS I, BIS IV, or PDBu before proteolysis with Arg-C for 0-90 min and SDS-PAGE of reaction products. Immunoblot detection with an antibody that recognizes the catalytic core of PKC showed proteolytic products. 75kDa band indicates increased sensitivity to proteolysis due to open conformation of PKC. No pretreatment control shows resistance to proteolysis. F) Amalgamated data from multiple Arg-C protease experiments, n values and SEM are indicated. G) Differential actions of 10 μM BIS I and 10 μM BIS IV on the muscarinic suppression of KCNQ2 currents. H) Amalgamated data from experiments shown in (G). N values and mean +/- SEM are indicated.
Biochemical and electrophysiological experiments provided support for this concept (Figs 4E-H). Purified PKCβII was pre-incubated with BIS I or BIS IV prior to digestion with the Arginine-C terminal (Arg-C) proteinase. The appearance of a faster migrating band on SDS PAGE gels was indicative of PKC in the open conformation, as the exposed amino-terminal regulatory domain of the enzyme is sensitive to proteolysis at Arg 19 (Orr, 1992). Control experiments with or without a PKC activator, 100 nM PDBu, confirmed that only activated PKC is sensitive to Arg-C treatment (Fig 4E top two panels). Also, we have previously demonstrated that the PKC binding peptide from AKAP79 stabilized PKC at the open state rendering the pseudosubstrate domain more sensitive to proteolysis (Faux et al., 1999). Preincubation with BIS I resulted in the accumulation of the 75 kDa fragment whereas incubation with BIS IV had almost no effect on proteolytic stability (Fig 4E bottom two panels). Quantification of multiple experiments showed that with BIS I there was a 2.9 ± 0.4 (n=7) fold enhancement of the 75kDa band over controls (Fig 4F). It appears that BIS I binding stabilizes the open conformation of PKC, whereas BIS IV has a different effect on the enzyme. Likewise BIS I and BIS IV modulated KCNQ2 channels differently. Pretreatment with BIS IV blocked Oxo-M mediated suppression of KCNQ2 currents (n=10; Fig 4G, red & 4H bar 2) whereas pretreatment with BIS 1 had little effect (n=13; Fig 4G, blue & 4H bar 3).
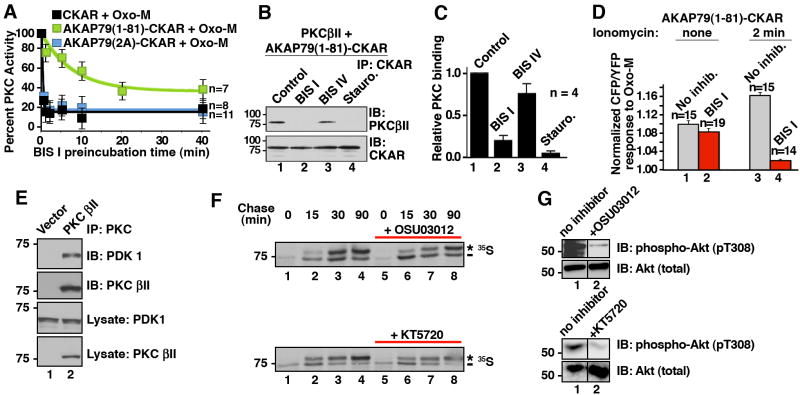
Our working hypothesis was that both AKAP79 and BIS I target the substrate-binding loop of PKC to force the enzyme into an open conformation. Therefore we tested if BIS I could compete with AKAP79 for binding to PKC. CHO cells were pre-incubated with BIS I for various times up to 40 min. Oxo-M was applied to the cells and 30 sec later cellular PKC activity was measured using our FRET based assay. In the AKAP79(1-81)-CKAR cells inhibition of PKC by BIS I took a considerable time to reach equilibrium (n=7; Fig 5A, green, T½ = 9.2 ± 4.0 min). In contrast, BIS I treatment immediately inhibited PKC in cells expressing AKAP79(2A)-CKAR or CKAR (Fig 5A; blue T½= 0.28 ± 0.05 min, n=8; & black T½=0.27 ± 0.04 min, n=11, respectively). When these results are considered with the data presented in figure 2B, it appears that AKAP79 confers some form of temporal constraint on BIS I action towards PKC. This could occur by one of two mechanisms: 1) it may take additional time for BIS I to enter the AKAP79 signaling complex and bind to the catalytic core of anchored PKC; or 2) PKC binds AKAP79 with a micromolar dissociation constant such that the enzyme exists in equilibrium between the bound and free states. Furthermore, competition between inhibitor and the endogenous pseudosubstrate domain may retard binding of inhibitors. Only the free enzyme is susceptible to ATP competitors such as BIS I. PKC binding studies demonstrated that pre-incubation with either BIS I or staurosporine for 30 minutes reduced PKCβII interaction with AKAP79(1-81)-CKAR as assessed by immunoblot of CKAR immune complexes (Fig 5B & C, lanes 2 & 4, n=4). Pretreatment with BIS IV did not disrupt PKC interaction with the anchoring protein (Fig 5B & C, lane 3). These data support the second postulate that BIS I can only act on PKC when it is released from AKAP79. Additional cell based experiments were conducted to develop this notion. We previously found that an influx of calcium across the synaptic membranes recruits Ca2+/calmodulin to AKAP79. One consequence is the release of PKC from the AKAP79 signaling complex (Faux and Scott, 1997). Therefore, cells expressing AKAP79(1-81)-CKAR were preincubated with the calcium ionophore ionomycin for 2 mins prior to stimulation with Oxo-M. Calcium influx not only increased the magnitude of the PKC response in the absence of inhibitors (Fig 5D, bars 1 & 3) but also restored PKC sensitivity to inhibition by BIS I (Fig 5D, bar 4). Activation of calcium dependent proteinases or calmodulin kinases could also contribute to this cellular event. Nevertheless these findings show that PKC is only available for inhibition by the ATP competitive inhibitor BIS I following release from interaction with the anchoring protein.
Figure 5. AKAP79 binding protects PKC from BIS I inhibition.
A) AKAP79 interaction alters PKC inhibition. Time course of FRET response to 3 μM Oxo-M: amalgamated data of cellular PKC activity measured from cells incubated with BIS I (500nM) for indicated times. Different reporters were used; CKAR (black, n=8), AKAP79(1-81)-CKAR (green, n=7) or AKAP79(2A)-CKAR (blue, n=11). Time points show peak PKC activity (CFP/YFP) 30 seconds after application of Oxo-M as percent of the response observed in the absence of BIS I treatment. B & C) Purified AKAP79(1-81)-CKAR was treated with PKC inhibitors (30 min) prior to addition of purified PKCβII. The binding is assessed by co-precipitating with AKAP79(1-81)-CKAR and PKCβII was identified by immunoblot. C) Amalgamated data from four co-IP experiments. PKC inhibitor competition with AKAP79 is shown as a decrease in relative PKC binding. D) In situ release of PKC from AKAP79(1-81)-CKAR was done by pretreatment of cells with ionomycin (2 min) prior to BIS I (500 nM, 5 min) and Oxo-M application. PKC activity was measured 30 sec after stimulation with Oxo-M. E) Co-immunoprecipitation of PDK1 with PKCβII. F) In pulse-chase experiments with 35S Met/Cys the ATP competitive PDK inhibitors, 10 μM OSU03012 or 1 μM KT5720, could not inhibit PDK-dependent phosphorylation of associated PKCβII. The asterisk indicates the mature, fully phosphorylated PKC and the dash indicates the newly synthesized, faster migrating PKC. G) The same concentration of OSU03012 and KT5720 inhibited PDK phosphorylation of a non-associated substrate, Akt1. Control immunoblots show cellular Akt protein levels are unchanged in the presence of inhibitors. N values and mean +/- SEM are indicated. (See also Figure S4.)
An important extension to this study was to establish if other protein kinases became resistant to pharmacological inhibitors when they were associated with binding partners. We tested this notion with PDK1, another binding partner for PKC (Dutil et al., 1998; Le Good et al., 1998). It has been previously established that PDK1 and newly synthesized PKCβII form a stable complex inside cells (Gao et al., 2001; Keranen et al., 1995). COS7 cells were transfected with empty vector or PKCβII (Fig 5E). PKC was immunoprecipitated from detergent-solubilized lysates, and endogenous PDK1 was detected by immunoblot (Fig 5E, top panel, lane 2).
Phosphorylation by PDK1 at the activation loop initiates the processing of PKCβII into a fully-active enzyme that is phosphorylated at two key sites in the C-terminus (Dutil et al., 1998). Although PDK1 is required for the maturation of PKC, once PKC is fully-phosphorylated at these C-terminal sites, the phosphorylation of the PKC activation loop becomes dispensable. Thus PDK1 does not affect the steady-state level of mature PKC, rather, it is required to initiate the processing of newly synthesized PKC (Newton, 2003). Two different ATP competitive inhibitors that selectively target PDK-1, OSU03012 and KT5720 were tested to see if pharmacological inhibition of PDK1 activity would ablate the processing of its binding partner PKC. COS7 cells transfected with PKCβII were incubated with Met/Cys-deficient media and 0.5mCi ml-1of 35S Met/Cys tracer for 7 min at 37 °C. The media was replaced and at specified times points after the chase (15, 30 & 90 min) PKC was immunoprecipitated. Immune complexes were analyzed for processing of PKCβII by 35S autoradiography. In the absence of PDK1 inhibitors, the newly synthesized PKC migrated with a faster mobility (Fig 5F; lane 1) but shifted to a slower migrating band with a half-life of approximately 30 mins (Fig 5F; lanes 2-4). This mobility shift reflects the two tightly coupled C-terminal phosphorylations at Thr-641 and Ser-660 in PKC βII1. Surprisingly, the rate of PKC processing was similar to controls in the presence of two different PDK1 inhibitors. Neither OSU03012 nor KT5720 altered the kinetics of appearance of the fully-phosphorylated species of PKC. In the presence of OSU03012 53% of PKC was fully phosphorylated after 30 min (Fig 5F, top blot, lanes 3 vs. 4). In the absence of this drug 55% of PKC migrated as the fully phosphorylated species (Fig 5F, top blot, lanes 7 vs. 8). Similar results were obtained when the PKC-PDK1 complex was treated with KT5720 where 60% of PKC was processed at 15 min whether or not KT5720 was present (Fig. 5F, bottom blot, lanes 2 vs. 4 and 6 vs. 8). This suggests that these ATP competitive inhibitors are ineffective within the framework of the PDK1-PKCβII complex. Importantly, when phosphorylation of phosphoinositide-dependent kinase, Akt1, a well characterized, non-associated PDK1 substrate was measured in the same cell extracts, OSU03012 or KT5720 potently blocked PDK1 activity (n=3; Fig 5G, lane 2). Note also that in vitro this same concentration of OSU03012 inhibited the phosphorylation of pure and unscaffolded PKC by PDK1 (Supplemental figure S4). This PKC, purified from insect cells which yield a population of PKC that is completely phosphorylated at C-terminal sites but only half the PKC population is phosphorylated at the activation loop, has a considerably lower affinity for PDK1, thus serving as an unscaffolded control (Gao et al., 2001; Keranen et al., 1995). Together these studies suggest that these ATP competitive inhibitors remain effective only in the context of if the unscaffolded enzymes.
Discussion
The two principle findings of this study refine and broaden our understanding of anchored kinase function. First, it has been assumed that AKAPs couple sequential signaling events. We now have experimental evidence to support this concept. Second, we have discovered that interaction with AKAPs or other interacting proteins can modify kinase susceptibility to pharmacological agents. This offers a surprising explanation to a long-standing conundrum – namely why muscarinic receptor operated M channels are insensitive to PKC inhibitors (Bosma and Hille, 1989). Our discovery that another anchored kinase PDK1 is also rendered insensitive to ATP competitive inhibitors consolidates this new concept that binding partners influence the susceptibility of AGC kinases to pharmacological inhibitors. A broader implication is that intracellular pharmacology cannot be reliably inferred from in vitro enzyme assays.
Suppression of muscarinic sensitive currents primes neurons for generating the next action potential. This is such a critical aspect of neuronal function that multiple signaling pathways are involved in its regulation, including an AKAP79/150-dependent pathway that involves PKC phosphorylation (Hoshi et al., 2003). The coincident monitoring of PKC activity and KCNQ channel function in a single cell has revealed previously unappreciated roles of AKAP action. Cytoplasmic PKC activation lagged 10.7 seconds behind suppression of the channel (Fig 1G). However, in the presence of the AKAP79-CKAR reporter we were able to monitor PKC activation and changes in ion channel function simultaneously (Fig 1H). This may be because AKAP79/150 serves as a molecular bridge linking the m1 receptor to the KCNQ2 subunit (Hoshi et al., 2005). The binding studies presented in figure 1I suggest that the AKAP79-CKAR reporter can effectively substitute for the endogenous anchoring protein in this capacity. The utility of such a GPCR-AKAP-channel ternary complex is evident in AKAP150 -/- mice where muscarinic suppression of the M current is blunted 40% when compared to their +/+ littermates (Tunquist et al., 2008). Unfortunately, technical issues including low transfection efficiency and the instability of the AKAP79-CKAR viral vector prevented us from using the AKAP79-CKAR reporter in AKAP150 -/- neurons.
Data presented in figure 1 argues that AKAP79 efficiently couples PKC phosphorylation events with suppression of channel activity. A variation on this theme is the ability of AKAP79/150 to synchronize individual molecular reactions. Although it is challenging to calculate the precise temporal advantage gained with AKAP79, data with another anchoring protein suggest an approximate twofold increase in the overall rate of signaling. AKAP-Lbc integrates signals from α1-adrenergic and endothelin receptors to mobilize a protein kinase cascade involved in pathological cardiac hypertrophy. In the presence of this AKAP the rate of nuclear export of the DNA effector protein HDAC5 was doubled and gene re-programming accelerated (Carnegie et al., 2008). Our observations with these two anchoring proteins support a refined anchoring hypothesis, namely that components in an enzyme cascade are synchronized by AKAPs to increase signaling efficiency.
Small molecule kinase inhibitors are valuable research tools that are often used to identify biologically relevant phosphorylation events. A select few of these compounds have been developed into therapeutics (Druker et al., 2001). Although the therapeutic benefit, pharmacokinetics, and side effects associated with these “kinase inhibitor drugs” has been rigorously investigated, much less is known about their mechanism of action. In vitro and cellular profiling has revealed that few if any of these drugs target individual kinases. For example, Gleevec a highly effective anticancer drug blocks the action of the Abl tyrosine kinase to manage the progression of acute myeloid leukemia. Yet, this ATP competitive inhibitor also targets the related Arg kinase, the PDGF receptor kinase, and the c-Kit tyrosine kinase (Druker, 2006). Given the concern about specificity of kinase inhibitors one broad implication of our study is that protein-protein interactions change the pharmacology of these well-defined inhibitors. Our observation that AKAP79/150 protects PKC from the ATP competitor class of PKC inhibitors creates a seemingly paradoxical circumstance. Association with AKAP79 alters PKC to impair the action of ATP competitive inhibitors yet still allows the enzyme to utilize ATP for catalysis. This fits with the conclusions of two elegant chemical biology studies that have appeared while this manuscript was under review (Cameron et al., 2009; Okuzumi et al., 2009). Both propose that ATP competitive inhibitors promote phosphorylation dependent maturation of certain AGC kinases independent of inherent kinase activity. Shokat and colleagues envisage that this so-called “intrinsic activation” or “kinase hijacking” may involve endogenous kinase-binding partners that replicate the role of ATP analogs inside cells (Okuzumim et al., 2009). This prediction could explain our data in figure 5 showing that PKCβII maturation is resistant to OSU03012 or KT5720 when the kinase is in complex with PDK1. Moreover, the manner in which PKC binds to AKAP79 may shed further light on this “kinase hijacking” model. We have previously shown that a “pseudosubstrate like” sequence on AKAP79 binds at the catalytic core of PKC (Faux and Scott, 1997). Consequently, the enzyme adopts an open conformation but remains inactive. Interruption of this protein-protein interaction is required for PKC activation. Thus, when the enzyme is liberated from AKAP79, it is in the open conformation, primed for catalysis, and in proximity to target substrates such as the KCNQ2 channel. However the phosphotransfer reaction cannot proceed without ATP as a second substrate. It is at this stage that ATP and ATP competitive inhibitors act. The phosphotransfer reaction would be favored at this location since the intracellular concentration of ATP is millimolar and thus approaching 100 fold greater than the highest concentration of BIS I in any of our experiments.
Although AKAP79/150 interacts with all classes of PKC (Faux et al., 1999), our in vitro studies have been limited to analysis of the conventional PKCβII isoform. Therefore, other PKC isoforms may respond differently to ATP competitors when bound to AKAP79/150. PKCs are also compartmentalized through interactions with other AKAPs (gravin/AKAP220 and yotiao/AKAP350) and other classes of PKC interacting proteins. Likewise a growing body of evidence suggests that PDK1 interacts with a range of intracellular binding partners including the brain and muscle selective anchoring protein mAKAP (Michel et al., 2005). It will be interesting to evaluate how interaction with other binding partners may affect the susceptibility of PKC and PDK1 to ATP competitive inhibitors.
In conclusion AKAP79 not only integrates signals from cell surface receptors to aid in the rapid attenuation of ion channels but also enhances the precision of cell signaling by providing both spatial and temporal control of a multistep cellular process. Evidence that interaction with AKAP79 protects PKC from certain pharmacological inhibitors such as BIS I and staurosporine sheds new light on how protein-protein interactions may modulate specific phosphorylation events. Our hypothesis that binding to AKAP79 locks PKC in the open conformation is consistent with the notion that anchoring proteins are allosteric modifiers of their enzyme-binding partners. This explains how some anchored kinases acquire reduced sensitivities to small molecule inhibitors to create pockets of active kinase in situ. The resistance of PDK1 to OSU03012 or KT5720 when in complex with PKCβII could represent another example of what may come to be recognized as a common mechanism. These findings should have important consequences for drug discovery and research projects predicated on the selectivity of pharmacological protein kinase inhibitors.
Experimental Procedures
Expression plasmids
KCNQ2 cDNA has been described previously (Hoshi et al., 2005). Cytosolic version of CKAR cDNA has been described previously (Gallegos et al., 2006). AKAP79 and AKAP79-fragment CKAR fusion proteins were created by recombinant PCR. Sequencing verified PCR-derived constructs.
Cell cultures
HEK and COS7 cells grown in DMEM with 10% fetal bovine serum (FBS). CHO hm1 cells grown in alpha MEM with 10% FBS and 500μg/ml G415. Rat SCG culture described previously (Hoshi et al., 2005). Transfections done with Fugene6 reagent (Roche) or TransIT-LT1 reagent (Mirus).
Biochemicals
Bisindolylmaleimide I (BIS I), bisindolylmaleimide IV (BIS-IV), staurosporine, calphostin C and phorbol 12,13-dibutyrate (PDBu) purchased from Sigma–Aldrich and/or EMD Chemical.
Live-cell imaging
Cells cultured on coverslips for 48-72h post-transfection were washed (2X10 min) in external solution consisted of 144 mM NaCl, 5 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, 10 mM HEPES pH7.4 prior to imaging to minimize activation of PKC by serum. Fluorescence emission was acquired using an Axiovert 135 microscope (Zeiss), Cascade II:512 camera (Roper) and a lambda 10-2 filter wheel (Sutter) controlled by MetaMorph 6.3 (Universal Imaging). Dual-emission images were obtained simultaneously through a dual-view module (Roper) with S470/30 and S535/30 emission filters and 505 dcxr dichroic mirror (Chroma). Illumination was 75 W Xenon lamp through D436/10 (12.5% ND) or HQ500/20 (1.25% ND) filter. Exposure time was 100-300 ms; image interval 10 sec. FRET changes were calculated as ratio of fluorescent intensities from two channels (Mdonor, MindirectAcceptor) after subtraction of background signal. The third channel (MdirectAcceptor) was used to monitor photobleaching during experiments, which was not detected. FRET ratio (YFP/CFP and CFP/YFP) changes are shown (normalized to the average FRET ratio value before stimulation).
Electrophysiology
Whole-cell recordings for CHO cells performed on isolated cells using an Axopatch 200B (Molecular Devices). For simultaneous recordings, signals were sampled at 2 kHz, filtered at 1 kHz, and acquired using pClamp software (version 9, Molecular Devices) that was triggered by Metamorph. The patch pipette with resistance of 4-8 MΩ was filled with pipette solution containing 135 mM potassium aspartate, 2 mM MgCl2, 3 mM EGTA, 1 mM CaCl2, 5 mM Mg ATP, 0.1 mM GTP, 10 mM HEPES. Fluorescent signals from MdirectAcceptor were stable during experiments, suggesting that the pipette solution did not affect CKAR fluorescence. External solution is described above. KCNQ2 currents evoked by 500-ms test pulses to -10 mV from a holding potential of -70 mV. Linear leak and capacitance transient were subtracted by P/4 protocol.
Measurement of the M current from SCG neurons: amphotericin B based perforated patch technique as described previously (Hoshi et al., 2005). Amplitude of the M currents measured as deactivating currents during 500-ms test pulses to -50 mV from a holding potential of -20 mV. Data are presented as mean +/- SEM.
KCNQ2-FLAG, CKAR binding assay
Immunoprecipitation was performed as described (Hoshi et al., 2005). Details can be found in Supplemental Experimental Procedures S1.
PKC binding assay
Purified CKAR was prepared by immunoprecipitation and high salt wash. CKAR attached to beads were incubated with 100 ng PKCβII (Calbiochem) in 10 μl buffer (1% Triton X-100, 150 mM NaCl, 1mM CaCl2, 10μg/ml PS/DAG, complete mix) for 1hr 4°C. After 3X wash with T-HBS containing 0.1% Triton X-100, 150 mM NaCl, 20mM HEPES (pH7.4), PKC binding measured by Western blot. To evaluate effect of ATP competitors, washed beads were incubated for 30 min with a binding buffer containing 0.1% Triton X-100, 150 mM NaCl, 250 nM PDBu, 1mMDTT, 10 mM HEPES (pH7.4) for control, or 500 nM ATP competitors (BIS I, BIS IV, ST). Then, 60 ng PKCβII (Calbiochem) added and further incubated for two minutes. After 3X wash with binding buffer plus PKC inhibitors, associated proteins were eluted by SDS-loading buffer and analyzed by Western blot.
PKC kinase assay
PKC kinase assay was performed using 32P PKC assay kit (Millipore) using a substrate peptide (QKRPSQRSKYL) and 32P-ATP. ATP and concentrations were 200 μM.
Arg-C assay
Proteolysis of PKC by Arg-C performed as described (Orr, 1992). Purified PKCβII (Calbiochem) 650 ng incubated at 37°C with 10 ng of Arg-C (Sigma-Aldrich, Arg-C to PKC molar ratio of 1:50) in buffer (0.1% Triton X-100, 750 μM CaCl2, 1 mM DTT, 10 mM Tris, pH 7.5) containing indicated PKC inhibitors. 10 μl samples at each time point were analyzed by Western.
Pulse-chase assay
COS7 cells transfected with WT PKCβII for 24 -30 h, treated with 10 μM OSO03012 (Calbiochem) or 1 μM KT5720 (Calbiochem) for 3 h at 37 °C. Cells were then pulse-labeled with 0.5 mCi ml-1 [35S]Met/Cys (P.E.) in Met/Cys-deficient DMEM, 7 min at 37 °C, media removed, and cells chased with DMEM as previously described (Sonnenburg et al., 2001). At indicated times cells were lysed in Buffer A (50 mM Tris, pH 7.5, 1% TritonX-100, 50 mM NaF, 10 mM Na4P2O7, 100 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, and 1 mM PMSF), centrifuged at 16,000 × g for 5 min at 22 °C, and PKCβII in supernatant immunoprecipitated with PKCα monoclonal antibody (cross-reactive with PKCβII, B.D.) overnight at 4 °C. Immune complexes were collected with Ultra-Link protein A/G beads (Pierce), washed with Buffer A and analyzed by SDS-PAGE and autoradiography. Lysates from these experiments were analyzed for phospho-Akt (Thr-308, Cell Signaling). Prolonged incubation of cells with PDK1 inhibitors for up to 24 hours did not affect cellular levels of PKCβII.
Immunoprecipitation of PKC/Akt1
COS7 cells were transfected with either PKCβII or HA-Akt1. Approximately 24 h post-transfection, cells were lysed in Buffer A, centrifuged at 16,000 × g for 5 min at 22 °C and detergent solubilized supernatants incubated with either a monoclonal PKCα antibody (to IP PKCβII) or a monoclonal HA antibody (Roche, to IP Akt) 16hr at 4 °C. Immune complexes were collected with Ultra-Link protein A/G beads, washed with Buffer A, and analyzed by SDS-PAGE/Western for interaction with endogenous PDK1 (polyclonal anti-PDK1, Cell Signaling). Expression of PKCβII and HA-Akt1 was assessed by a monoclonal antibody to PKCβII (B.D.) and a monoclonal HA antibody (Roche).
Supplementary Material
Acknowledgments
We thank Alyssa Wu Zhang for experiments in supplemental figure S4 and colleagues at the University of Washington for critical evaluation of this manuscript. This work was supported in part by NIH grant GM48231 to JDS. ACN and LKL are supported by NIH program grant DK54441.
Footnotes
Unfortunately we were unable to directly monitor changes in the phosphorylation of T500 on PKC because this covalent modification, which initiates the maturation process is not accompanied a mobility shift in the enzyme (Newton, A.C. 2003).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauman AL, Soughayer J, Nguyen BT, Willoughby D, Carnegie GK, Wong W, Hoshi N, Langeberg LK, Cooper DM, Dessauer CW, et al. Dynamic regulation of cAMP synthesis through anchored PKA-adenylyl cyclase V/VI complexes. Mol Cell. 2006;23:925–931. doi: 10.1016/j.molcel.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma MM, Hille B. Protein kinase C is not necessary for peptide-induced suppression of M current or for desensitization of the peptide receptors. Proc Natl Acad Sci USA. 1989;86:2943–2947. doi: 10.1073/pnas.86.8.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AJ, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat Struct Mol Biol. 2009;16:624–630. doi: 10.1038/nsmb.1606. [DOI] [PubMed] [Google Scholar]
- Carnegie GK, Soughayer J, Smith FD, Pedroja BS, Zhang F, Diviani D, Bristow MR, Kunkel MT, Newton AC, Langeberg LK, et al. AKAP-Lbc mobilizes a cardiac hypertrophy signaling pathway. Mol Cell. 2008;32:169–179. doi: 10.1016/j.molcel.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr DW, Stofko-Hahn RE, Fraser IDC, Cone RD, Scott JD. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins: characterization of AKAP79. J Biol Chem. 1992;24:16816–16823. [PubMed] [Google Scholar]
- Coghlan VM, Perrino BA, Howard M, Langeberg LK, Hicks JB, Gallatin WM, Scott JD. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–112. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5- bisphosphate. EMBO J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ. Circumventing resistance to kinase-inhibitor therapy. N Engl J Med. 2006;354:2594–2596. doi: 10.1056/NEJMe068073. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Dutil EM, Toker A, Newton AC. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1) Curr Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- Faux MC, Rollins EN, Edwards AS, Langeberg LK, Newton AC, Scott JD. Mechanism of A-kinase-anchoring protein 79 (AKAP79) and protein kinase C interaction. Biochem J. 1999;343:443–452. [PMC free article] [PubMed] [Google Scholar]
- Faux MC, Scott JD. Regulation of the AKAP79-protein kinase C interaction by Ca2+/calmodulin. J Biol Chem. 1997;272:17038–17044. doi: 10.1074/jbc.272.27.17038. [DOI] [PubMed] [Google Scholar]
- Fraser ID, Cong M, Kim J, Rollins EN, Daaka Y, Lefkowitz RJ, Scott JD. Assembly of an A kinase-anchoring protein-beta(2)-adrenergic receptor complex facilitates receptor phosphorylation and signaling. Curr Biol. 2000;10:409–412. doi: 10.1016/s0960-9822(00)00419-x. [DOI] [PubMed] [Google Scholar]
- Gallegos LL, Kunkel MT, Newton AC. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem. 2006;281:30947–30956. doi: 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- Gao T, Toker A, Newton AC. The carboxyl terminus of protein kinase c provides a switch to regulate its interaction with the phosphoinositide-dependent kinase, PDK-1. J Biol Chem. 2001;276:19588–19596. doi: 10.1074/jbc.M101357200. [DOI] [PubMed] [Google Scholar]
- Gao T, Yatani A, Dell’Acqua ML, Sako H, Green SA, Dascal N, Scott JD, Hosey MM. cAMP-dependent regulation of cardiac L-type Ca2+ channels requires membrane targeting of PKA and phosphorylation of channel subunits. Neuron. 1997;19:185–196. doi: 10.1016/s0896-6273(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Grodsky N, Li Y, Bouzida D, Love R, Jensen J, Nodes B, Nonomiya J, Grant S. Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry. 2006;45:13970–13981. doi: 10.1021/bi061128h. [DOI] [PubMed] [Google Scholar]
- Hoshi N, Langeberg LK, Scott JD. Distinct enzyme combinations in AKAP signalling complexes permit functional diversity. Nat Cell Biol. 2005;7:1066–1073. doi: 10.1038/ncb1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi N, Zhang JS, Omaki M, Takeuchi T, Yokoyama S, Wanaverbecq N, Langeberg LK, Yoneda Y, Scott JD, Brown DA, et al. AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists. Nat Neurosci. 2003;6:564–571. doi: 10.1038/nn1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keranen LM, Dutil EM, Newton AC. Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr Biol. 1995;5:1394–1403. doi: 10.1016/s0960-9822(95)00277-6. [DOI] [PubMed] [Google Scholar]
- Klauck TM, Faux MC, Labudda K, Langeberg LK, Jaken S, Scott JD. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- Michel JJ, Townley IK, Dodge-Kafka KL, Zhang F, Kapiloff MS, Scott JD. Spatial restriction of PDK1 activation cascades by anchoring to mAKAPalpha. Mol Cell. 2005;20:661–672. doi: 10.1016/j.molcel.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Navedo MF, Nieves-Cintron M, Amberg GC, Yuan C, Votaw VS, Lederer WJ, McKnight GS, Santana LF. AKAP150 is required for stuttering persistent Ca2+ sparklets and angiotensin II-induced hypertension. Circ Res. 2008;102:e1–e11. doi: 10.1161/CIRCRESAHA.107.167809. [DOI] [PubMed] [Google Scholar]
- Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Inhibitor hijacking of Akt activation. Nat Chem Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr JW, Keranen LM, Newton AN. Reversible exposure of the pseudosubstrate domain of protein kinase C by phosphatidylserine and diacylglycerol. J Biol Chem. 1992;267:15263–15266. [PubMed] [Google Scholar]
- Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selyanko AA, Hadley JK, Wood IC, Abogadie FC, Jentsch TJ, Brown DA. Inhibition of KCNQ1-4 potassium channels expressed in mammalian cells via M1 muscarinic acetylcholine receptors. J Physiol. 2000;522(Pt 3):349–355. doi: 10.1111/j.1469-7793.2000.t01-2-00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Gao T, Newton AC. The phosphoinositide-dependent kinase, PDK-1, phosphorylates conventional protein kinase C isozymes by a mechanism that is independent of phosphoinositide 3-kinase. J Biol Chem. 2001;276:45289–45297. doi: 10.1074/jbc.M107416200. [DOI] [PubMed] [Google Scholar]
- Suh BC, Hille B. Recovery from muscarinic modulation of M current channels requires phosphatidylinositol 4,5-bisphosphate synthesis. Neuron. 2002;35:507–520. doi: 10.1016/s0896-6273(02)00790-0. [DOI] [PubMed] [Google Scholar]
- Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- Tavalin SJ, Colledge M, Hell JW, Langeberg LK, Huganir RL, Scott JD. Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J Neurosci. 2002;22:3044–3051. doi: 10.1523/JNEUROSCI.22-08-03044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunquist BJ, Hoshi N, Guire ES, Zhang F, Mullendorff K, Langeberg LK, Raber J, Scott JD. Loss of AKAP150 perturbs distinct neuronal processes in mice. Proc Natl Acad Sci U S A. 2008;105:12557–12562. doi: 10.1073/pnas.0805922105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Scott JD. AKAP Signalling complexes: Focal points in space and time. Nature Reviews Molecular Cell biology. 2004;5:959–971. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li L, McNaughton PA. Proinflammatory mediators modulate the heat-activated ion channel TRPV1 via the scaffolding protein AKAP79/150. Neuron. 2008;59:450–461. doi: 10.1016/j.neuron.2008.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.