Abstract
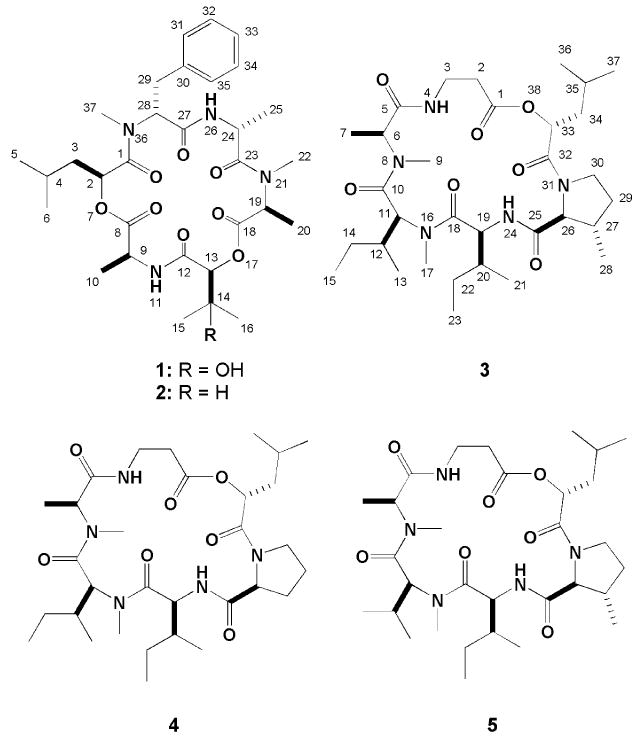
Two novel cyclic depsipeptides, guangomides A (1) and B (2), together with a new destruxin derivative (3) were isolated from the cytotoxic extract obtained from the saltwater culture of an unidentified sponge-derived fungus. The new structures were elucidated based on analysis of extensive 1D and 2D NMR data sets, and the absolute configurations of 2S, 9S, 13S, 19S, 24R, 28R of 1 was determined based on the combined X-ray and Marfey's method structure analysis. Identical absolute configurations were assumed for 2. The cytotoxicity of the extract was found to be due to brefeldin A, while 1 and 2 showed weak antibacterial activity against Staphylococcus epidermidis and Enterococcus durans.
A few marine invertebrates are known to be a source of structurally fascinating and biologically active peptides and depsipeptides. Some most noteworthy examples include the peptide anticancer drug candidates dolastatin 10,i from the sea hare Dolabella auricularia (but also the cyanophyte Symploca sp VP642ii) and kahalalide Fiii from the herbivorous marine mollusk Elysia rufescens (and also its dietary alga Bryopsis sp.). Significantly, both of these chemotypes are continuing subjects of clinical evaluation.iv Two closely related PKS/NRPS cyclidepspeptides — jasplakinolide from spongesv and chondramide C from myxobacteriavi — are of much interest because they are both potent in causing the hperassembly of G-actin into F-actin.vii Our attempts to date to discover marine-derived fungi as a source of unusual peptides have been somewhat successful. These include the isolation of the cytotoxic bicyclic peptide malformin C from Aspergillus niger,viii and highly N-methylated linear peptides of the RHM family from an unidentified fungal strain.ix Recently we began a project stimulated by the observation that extracts of an unidentified fungal strain separated from an Inathella sponge possessed potent cytotoxicity and selectivity in our disk diffusion assay system.x Most importantly its 13C NMR spectra displayed clusters of peaks centered at δ175. The initial bioassay-guided dereplication efforts showed that brefeldin Axi was a non-peptide major component responsible for the cytotoxicity. Subsequently, deeper evaluation of crude extract fractions employing bioassay and LCMS data led to discovery of three novel cyclic depsipeptides named guangomides A (1), B (2) and homodestcardin (3). We now describe the structure elucidation and the biological activity of these depsipeptides.
The molecular formula of C31H46N4O9 (m/z = 641.3152 [M+Na]+) was established for guangomide A (1) by the HRESIMS data. The low field 13C NMR resonances noted above were confirmed to be associated with peptidic functionality that was further validated by 1H NMR data showing the presence of two amide protons, six α-protons, and two N-methyl groups. There were a total of six carbonyl carbons in the 13C NMR spectrum (Table 1). Four of those carbonyl carbons were assigned as amide carbons and consistent with the four nitrogen atoms in the molecular formula. Therefore, the assumption that two ester carbons were present was verified by the HMBC correlations from two α-oxy methines (δH 5.00, δC 70.5 and δH 5.22, δC 76.5) to these carbonyl carbons (δC 172.6, 169.0).
Table 1. 1H and 13C NMR data for guangomide A (1) and B (2) in CDCl3a.
| Structural unit | 1 | 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 13C (δ, mult) | 1H (δ, mult., J, Hz) | gCOSY | gHMBC | NOESY | 13C (δ, mult) | 1H (δ, mult., J, Hz) | gHMBC | ||||
| S -Hic | 1 | 172.4 (s) | 172.4 (s) | ||||||||
| 2 | 70.5 (d) | 5.00 (dd, 8.7, 6.2) | 3a, 3b | 1, 3, 4, 8 | 3a, 3b, 4, 5, 6, 37 | 70.3 (d) | 5.00 (dd,8.9, 6.4) | 1, 3, 4, 8 | |||
| 3 | 38.8 (t) | a | 0.98 (ddd, 14.2, 6.2, 5.3) | 2, 3b | 1, 4, 5, 6 | 2, 3b, 4, 5, 6, 37 | 38.8 (t) | a | 0.96 (ddd, 14.2, 6.4, 5.3) | 2, 4, 5, 6 | |
| b | 1.55 (ddd, 14.2, 8.7, 6.7) | 2, 3a, 4 | 1, 2, 4, 5, 6 | 2, 3a, 4, 5, 6 | b | 1.54 m | 4, 5, 6 | ||||
| 4 | 23.8 (d) | 1.15 m | 3a, 5, 6 | 3, 5, 6 | 2, 3a, 3b, 5, 6, 31, 32, 34, 35, 37 | 23.8 (d) | 1.19 (hept, 6.7) | ||||
| 5 | 22.8 (q) | 0.78 (d, 6.6) | 4 | 3, 4, 6 | 2, 3a, 3b, 4, 31, 32, 34, 35, 37 | 22.9 (q) | 0.78 (d, 6.6) | 3, 4, 6 | |||
| 6 | 22.3 (q) | 0.72 (d, 6.6) | 4 | 4, 5, 6 | 2, 3a, 3b, 4, 31, 32, 34, 35, 37 | 22.2 (q) | 0.72 (d, 6.6) | 4, 5, 6 | |||
| l-Ala | 8 | 172.6 (s) | 172.9 (s) | ||||||||
| 9 | 47.1 (d) | 4.97 (dq, 8.7, 7.1) | 11 | 8, 10, 12 | 10, 11 | 47.0 (d) | 4.98 (dq, 8.9, 7.4) | 8, 10 | |||
| 10 | 19.3 (q) | 1.53 (d, 7.1) | 8, 9 | 9, 11 | 19.3 (q) | 1.52 (d, 7.4) | 8, 9 | ||||
| 11 | 8.22 (d, 8.7) | 9 | 12 | 9, 10, 13, 15 | 7.89 (d, 8.5) | ||||||
| S -Dhiv | 12 | 169.8 (s) | 168.5 (s) | ||||||||
| (S -Hiv) | 13 | 76.5 (d) | 5.22 s | 12, 14, 15, 16, 18 | 11, 15, 16 | 78.0 (d) | 5.23 (d, 2.5) | 12, 14, 15, 16 | |||
| 14 | 71.8 (s) | 30.0 (d) | 2.61 (hept.d, 6.7, 2.5) | ||||||||
| 15 | 26.7 (q) | 1.17 s | 12, 13, 14, 16 | 11, 13, 16, OH | 19.2 (q) | 0.93 (d, 6.7) | 13, 14, 16 | ||||
| 16 | 24.1 (q) | 1.26 s | 13, 14, 15 | 13, 15, OH | 15.8 (q) | 0.92 (d, 6.7) | 13, 14, 15 | ||||
| OH | 5.27 br.s | 15, 16 | |||||||||
| l-N-MeAla | 18 | 169.0 (s) | 169.7 (s) | ||||||||
| 19 | 60.5 (d) | 3.69 (q, 6.8) | 20 | 18, 20, 22, 23 | 20, 22 | 60.7 (d) | 3.69 (q, 6.8) | 18, 20, 22, 23 | |||
| 20 | 13.5 (q) | 1.52 (d, 6.8) | 19 | 18, 19 | 19, 22 | 13.6 (q) | 1.54 (d, 6.9) | 18, 19 | |||
| 22 | 36.9 (q) | 3.20 s | 19, 23 | 19, 20, 24, 25 | 36.9 (q) | 3.19 s | 19, 23 | ||||
| d-Ala | 23 | 171.3 (s) | 171.1 (s) | ||||||||
| 24 | 46.2 (d) | 4.85 (quint, 7.1) | 25, 26 | 23, 25, 27 | 22, 25, 26 | 46.3 (d) | 4.84 (quint, 7.1) | 23 | |||
| 25 | 18.1 (q) | 1.41 (d, 6.8) | 24 | 23, 24 | 22, 26, 37 | 18.1 (q) | 1.42 (d, 6.7) | 23, 24 | |||
| 26 | 7.09 (d, 7.3) | 24 | 27 | 24, 25, 28, 29a | 7.12 (d, 7.4) | ||||||
| d-N-Me-Phe | 27 | 168.5 (s) | 168.5 (s) | ||||||||
| 28 | 56.6 (d) | 5.76 (dd, 11.9, 5.3) | 29a, 29b | 1, 27, 29, 37 | 26, 29a, 29b, 31, 35 | 56.5 (d) | 5.76 (dd, 11.8, 5.3) | 1, 27, 29, 37 | |||
| 29 | 33.2 (t) | a | 2.92 (dd, 15.3, 11.8) | 28, 29b | 27, 28, 30, 31, 35 | 26, 29b, 28, 31, 35 | 33.2 (t) | a | 2.92 (dd, 15.1, 11.8) | 27, 30, 31, 35 | |
| b | 3.49 (dd, 15.3, 5.3) | 28, 29a | 30, 30, 31, 35 | 28, 29a, 31, 35 | b | 3.47 (dd, 15.1, 5.3) | 27, 30, 31 | ||||
| 30 | 137.1 (s) | 137.1 (s) | |||||||||
| 31,35 | 128.6 (d) | 7.16 m | 32, 34 | 29, 30, 32, 33, 34 | 5, 6, 28, 29a, 29b | 128.6 (d) | 7.16 m | 29, 30, 32, 33, 34 | |||
| 32,34 | 128.4 (d) | 7.24 m | 31, 33, 35 | 30, 31, 33, 35 | 5, 6 | 128.4 (d) | 7.24 m | 30, 31, 33, 35 | |||
| 33 | 126.6 (d) | 7.19 m | 32, 34 | 31, 32, 34, 35 | 126.6 (d) | 7.19 m | 31, 32, 34, 35 | ||||
| 37 | 30.2 (q) | 2.92 s | 1, 28 | 2, 3a, 4, 5, 6, 25 | 30.1 (q) | 2.92 s | 1, 28 | ||||
Measured at 500 MHz (1H) and 125 MHz (13C).
There were six carbonyl containing substructures envisioned based on the 2D NMR data and they are shown in Figure 1. Further, as expected, the gCOSY spectrum of 1 revealed six distinct spin systems. The first included the subset associated with the 2-hydroxyisocaproic acid (A). Another cluster included the three sets of overlapping resonances for the alanine residues (B, D and E). The protons associated with the phenylalanine residue (F) were identified by HMBC correlations (H28/C30 and H29/C31, C35) shown in Figure 1. Finally, the remaining subunit, 2,3-dihydroxyisovaleric acid (C), was established by the HMBC correlations (H13/C14, C15, C16, H15, H16/C13) also shown in Figure 1. Next, the locations of the N-methyl groups (δH 3.21, δC 37.1 and δH 2.94, δC 30.3) were affirmed as connected to the alanine (D) and the phenylalanine (F) based on the HMBC correlations (H22/C19, H37/C28). The final task of sequencing the six subunits was accomplished based on the HMBC correlations from the α-protons, the amide protons and the N-methyl groups to the carbonyl carbons (H2/C8, H9, NH11/C12, H13/C18, H19, H22/C23, H24, NH26/C27, H28, H37/C1) as detailed in Figure 2.
Figure 1. Structural units for 1.
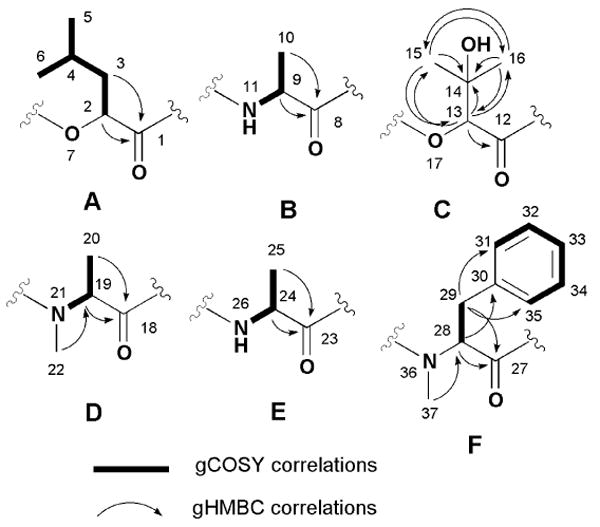
Figure 2. Selected gHMBC correlations for 1.
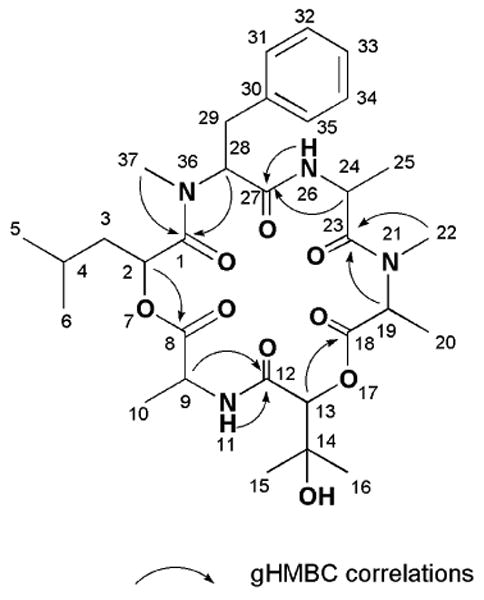
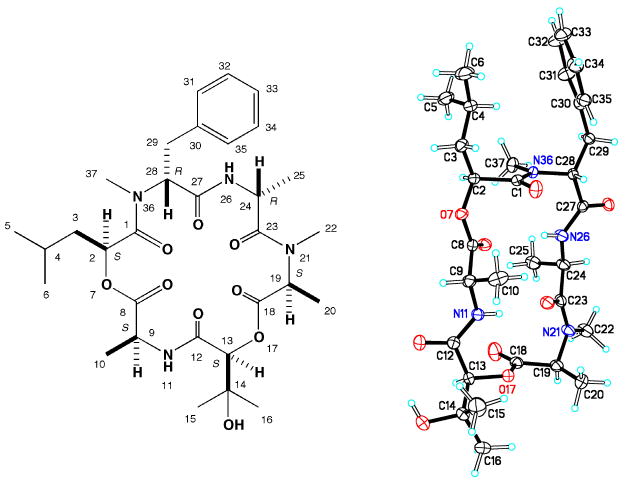
Assignment of the absolute configuration for the amino acid residues was accomplished using complimentary approaches. A d-N-methyl-phenylalanine was assigned based on results derived from HPLC-analysis of the products obtained from the Marfey's acid hydrolysate.xii Though our attempts to determine the relative configurations of chiral centers using NOESY data were unsuccessful, positive results were obtained through X-ray analysis structure of 1 shown in Figure 3. xiii Combining the relative configurations deduced from the X-ray data and using the d-N-methyl-phenylalanine as an anchor point supported the final absolute stereostructure as 2S, 9S, 13S, 19S, 24R, 28R.
Figure 3. Absolute structure of 1 including X-ray crystal structure.
The next compound to be analyzed was guangomide B (2), whose molecular formula of C31H46N4O8 differed from that of 1 by just a single oxygen atom. Not surprisingly, its 13C and 1H NMR spectra (Table 1) were almost identical with those of 1 The differences included shifted 13C resonances for the C13/C14/C15/C16 of 2 versus 1 consistent with the proposal that an H group replaced the OH group at C14 of substructure C (Figure 1). Likewise the 1H NMR spectrum of 1 showed diastereotopic H315 and H316 as doublets. Thus, these data were in firm support of substructure D as 2-hydroxyisovaleric acid, which was further confirmed by the COSY and the HMBC measurements. The stereochemistry assignments shown for 2 are based on a biogenetic analogy to 1 plus the observations that both compounds have the same sign of the optical rotation and both have parallel NMR shifts at each of the chiral centers.
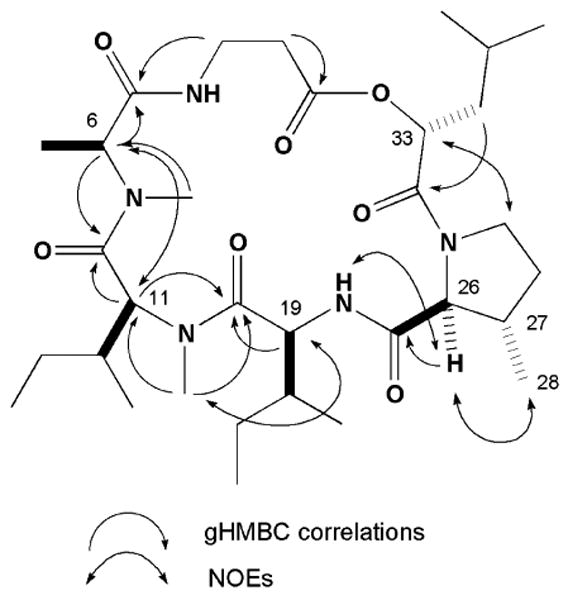
The last new compound, homodestcardin (3) had the molecular formula C32H55N5O7 established by the HRESIMS data. This compound was concluded to be a member of the destruxin family based on the similar profile of 1H and 13C (Table 2) to that of homodestruxin B (4)xiv and roseocardin (5).xv Detail analysis of the 2D NMR data of 3 pinpointed the β-methyl proline and N-methyl leucine residues versus the proline residue on 4 and N-methyl valine residue on 5, respectively. Our proposed planar structure derived by analogy to 4 and 5 was confirmed from the 2D NMR data (Figure 4). Extending the comparisons of the NMR data between this trio further revealed that 3 and 4 possessed identical shifts at the western chiral centers (C6, C11, C12, C19 and C20). Alternatively 3 and 5 possessed parallel shifts at the eastern chiral centers (C26, C27 and C33) were almost identical with those of 5. The significant NOE observed between H26 and H28 was also consistent with these stereochemical conjectures. The final relative stereoassignments proposed for 3 are shown in Figure 4.
Table 2. 1H and 13C NMR data for homodestcardin (3) in CDCl3a.
| Structural unit | 3 | ||||||
|---|---|---|---|---|---|---|---|
| 13C (δ, mult) | 1H (δ, mult., J, Hz) | gCOSY | gHMBC | NOESY | |||
| β-Ala | 1 | 173.8 (s) | |||||
| 2 | 34.5 (t) | a | 2.56 (ddd, 18.5, 5.0, 1.8) | 2b, 3b | 1, 3 | 2b, 3a | |
| b | 2.68 (ddd, 18.5, 11.5, 2.1) | 2a, 3a | 2a, 3a, 4 | ||||
| 3 | 33.2 (t) | 3.08 (m) | 2b, 3b, 4 | 2a, 2b, 3b, 4 | |||
| 4.05 (m) | 2a, 3a, 4 | 3a | |||||
| 4 | 8.27 (d, 8.5) | 3a, 3b | 2b, 3a, 6, 20, 22b, 24 | ||||
| N-Me-Ala | 5 | 169.7 (s) | |||||
| 6 | 55.5 (d) | 5.16 (q, 6.8) | 7 | 5, 8 | 4, 7, 11 | ||
| 7 | 15.3 (q) | 1.30 (d, 6.9) | 6 | 5, 6 | 6, 9 | ||
| 9 | 28.1 (q) | 2.73 (s) | 6, 10 | 7 | |||
| N-Me-Ile | 10 | 171.1 (s)b | |||||
| 11 | 56.8 (d) | 5.03 (d, 10.9) | 12 | 9, 10, 12, 14, 18 | 6, 12, 14a, 14b, 15 | ||
| 12 | 33.5 (d) | 2.06 (m) | 11, 13 | 11, 13, 14b, 15, 17 | |||
| 13 | 16.2 (q) | 0.85 (m)c | 11 | 11, 12, 14 | 12 | ||
| 14 | 25.8 (t) | a | 0.98 (m) | 14b, 15 | 11 | ||
| b | 1.43 (m) | 14a, 15 | 11, 12, 14b, 17 | ||||
| 15 | 11.1 (q) | 0.91 (t, 7.2) | 14a, 14b | 12, 14 | 11, 12, 14a | ||
| 17 | 31.0 (q) | 3.20 (s) | 11, 18 | 12, 14b, 19, 20 | |||
| Ile | 18 | 173.6 (s) | |||||
| 19 | 53.5 (d) | 4.83 (dd, 9.2, 6.8) | 20, 24 | 18, 20 | 17, 20, 21, 22a, 22b, 24 | ||
| 20 | 37.5 (d) | 1.92 (m) | 19, 21 | 4, 17, 19, 21, 22b, 24 | |||
| 21 | 15.3 (q) | 0.84 (m)c | 20 | 19, 20 | 19, 20 | ||
| 22 | 24.6 (t) | a | 1.29 (m) | 22b, 23 | 19, 23, 24 | ||
| b | 1.44 (m) | 22a, 23 | 4, 19, 20, 22b, 23, 24 | ||||
| 23 | 11.4 (q) | 0.85 (m)c | 22a, 22b | 19, 20 | 22a, 22b | ||
| 24 | 7.07 (d, 8.8) | 19 | 4, 19, 20, 22a, 22b, 26, 30b | ||||
| 3-Me-Pro | 25 | 171.0 (s)b | |||||
| 26 | 67.1 (d) | 4.27 (d, 0.3) | 27 | 25, 27, 28, 29 | 24, 27, 28 | ||
| 27 | 36.2 (d) | 2.78 (m) | 26, 28, 29b | 26, 28, 29a, 29b | |||
| 28 | 19.0 (q) | 1.11 (d, 7.1) | 27 | 26, 27, 29 | 26, 27, 29a, 30a | ||
| 29 | 30.9 (t) | a | 1.70 (m) | 29b, 30a, 30b | 28, 29b, 30a, 27 | ||
| b | 2.09 (m) | 27, 29a, 30a, 30b | 27, 29a, 30b | ||||
| 30 | 44.9 (t) | a | 3.56 (dt, 9.5, 7.4) | 29a, 29b, 30b | 28, 29a, 30b, 33, 34a | ||
| b | 3.86 (td, 9.5, 3.0) | 29a, 29b, 30a | 24, 29b, 30a, 33 | ||||
| Hica | 32 | 169.9 (s) | |||||
| 33 | 71.9 (d) | 4.90 (dd, 10.4, 3.3) | 34a, 34b | 30a, 30b, 34a, 34b, 37 | |||
| 34 | 39.3 (t) | a | 1.37 (ddd, 14.5, 9.0, 3.4) | 34b, 35 | 32, 35, 36 | 30a, 33, 34b, 35 | |
| b | 1.97 (m) | 34a | 32, 35, 36 | 33, 34a, 35, 36 | |||
| 35 | 24.4 (d) | 1.85 (m) | 34a, 36, 37 | 34a, 34b, 36, 37 | |||
| 36 | 23.4 (q) | 1.00 (d, 6.6) | 35 | 35, 37 | 34b, 35 | ||
| 37 | 21.7 (q) | 0.96 (d, 6.6) | 35 | 35, 36 | 33, 35 | ||
Measured at 500 MHz (1H) and 125 MHz (13C).
Assignment may be interchangeable.
Coupling constant could not be measured due to signal overlap.
Figure 4. Selected 2D NMR correlations and the relative structure of 3.
The literature, aside from that noted above of cyclic depsipetides, including those with significant bioactivity properties isolated from marine-derived fungi is rather sparse. One set of compounds includes 15G256γ, 156G256δ and 15G256ε which are lipodepsipeptides isolated from a Hypoxylon oceanium separated from mangrove wood.xvi Exumolides A and B isolated from a marine plant-derived Scytaladium sp. are cytostatic cyclic hexadepsipetides.xvii Sansalvamidexviii and N-methylsansalvamidexix isolated from two different marine alga-derived Fusarium spp. are cytotoxic cyclic pentadepsipeptides. Those previous examples contain one ester group in the molecule. By contrast, the guangomides (1 and 2) isolated in this study are different from known cyclic depsipetides from marine-derived fungi because of the presence of two ester groups in the molecule. In addition, homodestcardin (3) is the first example of destruxin analog from marine-derived fungi. The biological properties of 1 and 2 were investigated and deserve brief comment. Guangomide A (1) was inactive against murine and human tumor cell lines in a disk diffusion assay. However, 1 and 2 showed weak antibacterial activity against Staphylococcus epidermidis (MIC=100μg/mL, each) and Enterococcus durans (MIC=100μg/mL, each).
Experimental Section
General Experimental Procedures
UV/vis measurements were recorded on HP 8453 diode array spectrometer. Optical rotations were obtained on a JASCO DIP-370 digital polarimeter. The NMR spectra were recorded on a Varian UNITY INOVA-500 spectrometer, operating at 500 and 125.7 MHz for 1H and 13C, respectively. High resolution mass measurements were obtained on a bench-top Mariner ESI-TOF mass spectrometer. HPLC was performed with a column of 4μm ODS.
Biological Materials
The fungus (strain no. 001314c) was isolated from an Inathella sp. (coll. no. 00314) collected using SCUBA off the coast of Guango, Papua New Guinea on December 2000 by the same techniques described previously.xx Attempts to identify this fungal strain by the alignment of the D2 region of the 25S ribosomal DNA sequence and its fruiting body were unsuccessful (the closest fungus: Fusarium graminearum with a genetic distance of 9.75%). This fungus is maintained in a cryopreserved state at UCSC.
Culture conditions
The fungal strain was grown in a liquid medium (20 L) containing 1.5 % malt extract broth in filtered Monterey Bay seawater adjusted to pH 7.4 at 180 rpm for 28 days at room temperature (25°C).
Disk Diffusion Soft Agar Colony Formation Assay
An in vitro cell-based assay was employed to identify solid tumor selectivity for original extracts, extract partition fractions, and pure compounds. The differential cytotoxicity is expressed by observing a zone differential between any solid tumor cell (Colon 38, Colon H116, Lung H125) and either leukemia cells (L1210 or CEM) or normal cells (CFU-GM). The sample is designated as “solid tumor selective” if the zone units of solid tumor – normal cell or leukemia cells is greater than 250 units.
Extraction and Isolation
The culture was filtered under suction and the broth was extracted with equal volumes of EtOAc three times. The EtOAc extract was partitioned by Kupchan type extraction reported previously.xxi The CH2Cl2 extract (EFD; 2.6 g) contained crystals, which were purified by washing away with MeOH and were identified brefeldin A (560 mg) by comparison of its spectral data to the published data.8 The CH2Cl2 soluble portion was separated with a flash silica gel chromatography using CH2Cl2-MeOH stepwise gradient as the eluent. B3 (170 mg) obtained from MeOH-CH2Cl2 (1:99) was purified by a reverse phase HPLC with MeOH-H2O (3:7 up to 1:0) to furnish 2 (1.5 mg). B4 (190 mg) obtained from MeOH-CH2Cl2 (1:49) was purified by a reverse phase HPLC with MeOH-H2O (2:3 up to 9:1) to furnish 1 (11.0 mg) and 3 (1.2 mg).
Guangomide A (1)
Colorless crystals from Hexane/EtOAc/MeOH (1/1/1). Mp. 255-257°C. [α]28D −44.6 (c 0.8, CHCl3); λmax (MeOH)/nm 203 (log ε 4.38); 1H and 13C NMR data are listed in Table 1; HRESIMS: m/z 641.3152 [M+Na]+ (calcd for C31H46N4O9Na, 641.3157).
Guangomide B (2)
White amorphous powder. [α]28D −18.1 (c 0.9, CHCl3); λmax (MeOH)/nm 204 (log ε 4.38); 1H and 13C NMR data are listed in Table 1; HRESIMS: m/z 625.3208 [M+Na]+ (calcd for C31H46N4O8Na, 625.3208).
X-ray crystallography of 1
The single crystal X-ray analysis was conducted as follows. Suitable crystals were obtained from Hexane/EtOAc/MeOH (1/1/1) by the vapor diffusion method. This crystal (0.60 × 0.40 × 0.30 mm3) was mounted on a Bruker SMART diffractometer (MoKα; -100°C). A hemisphere of data was taken using a narrow-scan routine (1406 frames, 0.3° steps ω-scan, exposure time was 30 s/frame, 2θmax = 63.62°). Raw data was integrated with the Bruker SAINT+ programxxii to yield a total 33908 reflections, of which 10117 were independent (Rint = 2.58 %, completeness 94.9 %) and 8852 with I > 2σ(I). Data were collected for absorption using SADABS program (min. and max. transmission are 0.9479 and 0.9735, respectively).xxiii The structure was solved by direct methods and refined by full matrix least square on F2 techniques using anisotropic displacement parameters for all non-hydrogen atoms.xxiv All hydrogen atoms were found in the difference Fourier map and refined isotropically. At final convergence, R1 = 3.88 % and GOF = 1.022 for 581 parameters.
Amino acid analysis of 1 using Marfey's method
Guangomide A (1) (1.4 mg) and 6N HCl (2 mL) were added to MeOH (0.2mL) and heated at 110°C for 14hr in a sealed vial. The cooled reaction mixture was evaporated to dryness. To the residue was added 1M NaHCO3 (0.1 mL) and 1% Marfey's reagent (FDAA) in acetone (0.1 mL), incubated at 37°C for 0.5 hr. The reaction mixture was quenched with 2N HCl (50 μL), analyzed by reverse phase HPLC. The analysis was performed by the following condition: Alltech altima C18 column (5μ, 250mm × 10mm id.), solvent system acetonitrile/H2O (4:1) up to (1:1) over 60 min with 1 mL/min flow rate, UV detection at 340 nm. Separately N-methyl-d and l-phenylalanine were derivatized with FDAA in the same manner as that of 1. The HPLC conditions gave the distinguishable retention times for d and l forms (51.0 min and 50.3 min, respectively). The configuration of the N-methylphenylalanine was determined to be d-form based on co-injection of each form of the standard amino acids and 1.
Homodestcardin (3)
White amorphous powder. [α]27D −143.8 (c 0.9, CHCl3); λmax (MeOH)/nm 204 (log ε 3.75); 1H and 13C NMR data are listed in Table 2; HRESIMS: m/z 622.4170 [M+H]+ (calcd for C32H56N5O7, 622.4174).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 CA 47135). Additional financial support, at UCSC, was provided by equipment grant from NSF BIR-94-19409 (NMR) and a supplement to NIH CA 52955 (ESI-TOF-MS). The work, at Harvard, was supported by the National Institutes of Health (R01 CA24487 JC). We thank Dr. W. Inmann (Tularik) for taking the NMR data of guangomide A. We thank Mr. M. Waddington, Accugenix, a division of Acculabs, INS and Dr. D. A. Sutton, Department of Pathology, University of Texas Health Science Center at San Antonio for taxonomic identification of the fungal strain. We thank Dr. M. C. Diaz (UCSC, IMS) for the identification of the sponge taxonomy. We also thank Mr. P. Ralifo and Ms. C. M. Boot of UCSC for the antibacterial assay.
Footnotes
Supporting Information Available: 1H and 13C NMR spectra of 1 – 3, bioassay data and X-ray data of 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Note
- 1.Pettit GR, Kamano Y, Herald CL, Tuinmann AA, Boettner FE, Kizu H, Schmidt JM, Baczynkyji L, Tomer KB, Bontems RJ. J Am Chem Soc. 1987;109:6883–6885. [Google Scholar]
- 2.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J Nat Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 3.Original structure: Hamann MT, Scheuer PJ. J Am Chem Soc. 1993;115:5825–5826. Revised structure: Bonnard I, Manzanares I, Rinehart KL. J Nat Prod. 2003;66:1466–1470. doi: 10.1021/np030334c.
- 4.Newman D, Cragg GM. J Nat Prod. 2004;67:1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 5.Sonnenschein RN, Farias JJ, Tenney K, Mooberry SL, Lokovosky E, Clardy J, Crews P. Org Lett. 2004;6:779–782. doi: 10.1021/ol036446c. [DOI] [PubMed] [Google Scholar]
- 6.Sasse F, Kunze B, Gronewold TM, Reichenbach H. J NCI. 1998;90:1559–1563. doi: 10.1093/jnci/90.20.1559. [DOI] [PubMed] [Google Scholar]
- 7.(a) Bai R, Covell DG, Liu C, Ghosh AK, Hamel E. J Bio Chem. 2002;277:32165–32171. doi: 10.1074/jbc.M205076200. [DOI] [PubMed] [Google Scholar]; (b) Holzinger A, Lütz-Meindl U. Cell Mot Cytosk. 2001;48:87–95. doi: 10.1002/1097-0169(200102)48:2<87::AID-CM1000>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]; J Nat Prod. 2004;67:1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 8.Isol: Varoglu M, Corbett TH, Valariote FA, Crews P. J Org Chem. 1997;62:7078–7079. doi: 10.1021/jo970568z. Syn: Govek SP, Overman LE. J Am Chem Soc. 2001;123:9468–9469. doi: 10.1016/j.tet.2007.05.127.
- 9.Boot CM, Tenny K, Valeriote FA, Crews P. J Nat Prod. 2006;69:83–92. doi: 10.1021/np0503653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Sperry S, Valeriote FA, Corbett TH, Crews P. J Nat Prod. 1998;61:241–247. doi: 10.1021/np970467w. [DOI] [PubMed] [Google Scholar]; (b) Valeriote FA, Grieshaber CK, Media J, Pietraszkewicz H, Pan M, McLaughlin S. J Exper Ther Oncol. 2002;2:228–236. doi: 10.1046/j.1359-4117.2002.01038.x. [DOI] [PubMed] [Google Scholar]
- 11.Cole RJ. Handbook of Toxic Fungal Metobolites. Academic Press; New York: 1981. pp. 844–847. [Google Scholar]
- 12.Marfey P. Carlsberg Res Commun. 1984;49:591–596. [Google Scholar]
- 13.Crystallographic data for guangomide A (1) have been deposited at the Cambridge Crystallographic Data Center. Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK [fax; +44-(0)1223-336033 or e-mail; deposit@ccdc.cam.ac.uk or http://www.ccdc.cam.ac.uk/].
- 14.Isol; Krasnoff SB, Gibson DM, Belofsky GN, Gloer KB, Gloer JB. J Nat Prod. 1996;59:485–489. Syn; Ward DE, Gai Y, Lazny R, Pedras MSC. J Org Chem. 2001;66:7832–7840. doi: 10.1021/jo015953+.
- 15.Tsunoo A, Kamijo M, Taketomo N, Sato Y, Ajisaka KJ. Antibiot. 1997;50:1007–1013. doi: 10.7164/antibiotics.50.1007. [DOI] [PubMed] [Google Scholar]
- 16.Schlingmann G, Milne L, Williams DR, Carter GT. J Antibiot. 1998;51:303–316. doi: 10.7164/antibiotics.51.303. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins KM, Renner MK, Jensen PR, Fenical W. Tetrahedron Lett. 1998;39:2463–2466. [Google Scholar]
- 18.Belofski GN, Jensen PR, Fenical W. Tetrahedron Lett. 1999;40:2913–2916. [Google Scholar]
- 19.Cueto M, Jensen PR, Fenical W. Phytochemistry. 2000;55:223–226. doi: 10.1016/s0031-9422(00)00280-6. [DOI] [PubMed] [Google Scholar]
- 20.Varoglu M, Crews P. J Nat Prod. 2000;63:41–43. doi: 10.1021/np9902892. [DOI] [PubMed] [Google Scholar]
- 21.Amagata T, Rath C, Rigot JF, Tarlov N, Tenney K, Valeriote FA, Crews P. J Med Chem. 2003;46:4342–4350. doi: 10.1021/jm030090t. [DOI] [PubMed] [Google Scholar]
- 22.SMART & SAINT Software Reference manuals, version 5.042. Bruker Analytical X-ray Systems; Madison WI: 1998. [Google Scholar]
- 23.Sheldrick GM. SADABS, a software for empirical absorption correction. University of Gottingen; Germany: 2000. [Google Scholar]
- 24.Sheldrick GM. SHELXTL Reference Manual, version 5.1. Bruker Analytical X-ray Systems; Madison WI: 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


