Abstract
Whooping cough is caused by Bordetella pertussis and still constitutes one of the top five causes of death in young children, particularly in developing countries. The calmodulin-activated adenylyl cyclase (AC) toxin CyaA substantially contributes to disease development. Thus, potent and selective CyaA inhibitors would be valuable drugs for the treatment of whooping cough. However, it has been difficult to obtain potent CyaA inhibitors with selectivity relative to mammalian ACs. Selectivity is important for reducing potential toxic effects. In a previous study we serendipitously found that bis-methylanthraniloyl (bis-MANT)-IMP is a more potent CyaA inhibitor than MANT-IMP (Mol Pharmacol 72:526–535, 2007). These data prompted us to study the effects of a series of 32 bulky mono- and bis-anthraniloyl (ANT)-substituted nucleotides on CyaA and mammalian ACs. The novel nucleotides differentially inhibited CyaA and ACs 1, 2, and 5. Bis-ANT nucleotides inhibited CyaA competitively. Most strikingly, bis-Cl-ANT-ATP inhibited CyaA with a potency ≥100-fold higher than ACs 1, 2, and 5. In contrast to MANT-ATP, bis-MANT-ATP exhibited low intrinsic fluorescence, thereby substantially enhancing the signal-to noise ratio for the analysis of nucleotide binding to CyaA. The high sensitivity of the fluorescence assay revealed that bis-MANT-ATP binds to CyaA already in the absence of calmodulin. Molecular modeling showed that the catalytic site of CyaA is sufficiently spacious to accommodate both MANT substituents. Collectively, we have identified the first potent CyaA inhibitor with high selectivity relative to mammalian ACs. The fluorescence properties of bis-ANT nucleotides facilitate development of a high-throughput screening assay.
Introduction
Whooping cough is caused by the Gram-negative bacterium Bordetella pertussis (Guiso, 2009; Carbonetti, 2010). Although vaccinations against whooping cough are available and the disease can be treated with antibiotics, it is still one of the five leading causes of death in young children, particularly in countries of the developing world (Crowcroft and Pebody, 2006). Thus, novel strategies for the treatment of whooping cough are urgently needed.
B. pertussis secretes two virulence factors that substantially contribute to the pathogenesis of whooping cough. Pertussis toxin ADP-ribosylates Gi protein α-subunits and, thereby, blocks the coupling of chemoattractant receptors to Gi proteins and cellular effector systems in phagocytes that kill invading bacteria (Carbonetti, 2010). This mechanism is complemented by the AC toxin CyaA, a protein consisting of 1706 amino acids. After secretion from the bacteria, CyaA inserts into the plasma membrane of host cells. CyaA then binds calmodulin (CaM), stimulating its AC activity and resulting in massive production of cAMP (Ladant and Ullmann, 1999; Vojtova et al., 2006). cAMP, like pertussis toxin, blunts the host-defense function of phagocytes. Accordingly, the synergistic actions of pertussis toxin and CyaA facilitate colonization of the respiratory tract with bacteria (Ladant and Ullmann, 1999; Vojtova et al., 2006; Carbonetti, 2010). As a result, the infection becomes more severe and lasts longer.
Based on the pathophysiological function of CyaA, it is a logical approach to develop CyaA inhibitors. In fact, several noncompetitive so-called P-site inhibitors for mammalian ACs also inhibit the catalytic activity of CyaA (Johnson and Shoshani, 1990). However, the potency of these inhibitors is rather low compared with mammalian ACs, rendering them unsuitable as a starting point for drug development. High-throughput screening studies yielded a low-potency (Ki, 20 μM) CyaA inhibitor (Soelaiman et al., 2003). To this end, 9-[2-(phosphonomethoxy)ethyl]adenine diphosphate (PMEApp), the active metabolite of the antiviral drug adefovir, is the most potent CyaA inhibitor known so far (Ki in the presence of Mn2+, 1 nM) (Guo et al., 2005). However, little is known about the selectivity of PMEApp for CyaA relative to mammalian ACs.
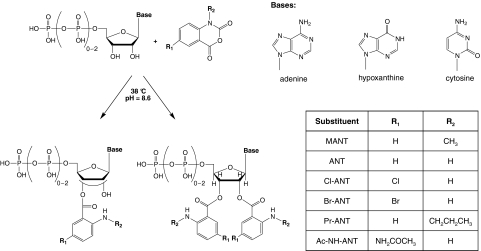
In other studies we have shown that 2′(3′)-O-MANT-substituted NTPs are moderately potent competitive CyaA inhibitors with Ki values in the range of 0.2 to 10 μM (Gille et al., 2004; Göttle et al., 2007). CyaA exhibits a broad base specificity for purine and pyrimidine nucleotides (Göttle et al., 2007, 2010), indicating that the catalytic site possesses substantial conformational flexibility and is spacious. Figure 1 provides an overview of structures of MANT nucleotides. MANT nucleotides are fluorescent and can be used to monitor the binding of nucleotides to CyaA and conformational changes in CyaA upon binding of CaM. Interaction of the MANT group with Phe306 results in a CaM-dependent increase in fluorescence (Göttle et al., 2007).
Fig. 1.
Overview of the structures and synthesis of mono- and bis-(M)ANT-substituted nucleotides. Nucleotides differ from each other in terms of base, phosphate chain length, and substitution of the 2′- and 3′-O-ribosyl group. In mono-(M)ANT-substituted nucleotides, the substituent can isomerize spontaneously between the 2′ and 3′ positions (Suryanarayana et al., 2009). In bis-(M)ANT-substituted nucleotides, this isomerization cannot occur anymore. In a single step, nucleotides and diversified isatoic anhydrides are reacted to the corresponding acylated nucleotides.
In an effort to obtain potent and selective CyaA inhibitors, in the present study, we examined the effects of 32 substituted bis- and mono-(M)ANT nucleotides. The starting point for this study was the serendipitous finding that bis-MANT-IMP was an at least 5-fold more potent inhibitor of CyaA than mono-MANT-IMP (Göttle et al., 2007). However, the Ki value of bis-MANT-IMP is still very low, i.e., in the range of 20 to 40 μM, depending on the experimental conditions. Because the length of the phosphate chain is important for high potency of AC inhibitors (Mou et al., 2005, 2006; Göttle et al., 2007; Taha et al., 2009), we focused our synthesis effort on NTPs. The ANT group was also modified by halogens and an acetylated amino group at the 5-position of the phenyl ring system. Moreover, the amino function of the ANT group was modified with propyl groups. Figure 1 provides an overview fn the structural properties of the newly synthesized compounds. For comparison with CyaA, we studied ACs 1, 2, and 5 as representative membranous mammalian ACs (mACs). We also conducted fluorescence studies with CyaA and representative nucleotides and modeled the interaction of CyaA with bis-MANT nucleotides. Here, we report that bis-Cl-ANT-ATP is a very potent CyaA inhibitor with substantial selectivity relative to mACs.
Materials and Methods
Materials.
MANT-UTP, MANT-UDP, MANT-CTP, MANT-CDP, MANT-ITP, MANT-IDP, MANT-IMP, ANT-IMP, ANT-ATP, and ANT-ADP were synthesized according to Hiratsuka (1983) with modifications described in Taha et al. (2009). Methylisatoic anhydride, isatoic anhydride, chloroisatoic anhydride, bromoisatoic anhydride, aminoisatoic anhydride, ATP, ITP, CTP, IMP, and bovine serum albumin, fraction V, highest quality, were purchased from Sigma-Aldrich (Seelze, Germany). MnCl2 tetrahydrate (highest quality) and aluminum oxide 90 active, (neutral, activity 1; particle size, 0.06–0.2 mm) were from MP Biomedicals (Eschwege, Germany). PMEApp was supplied by Gilead Sciences (Foster City, CA). The catalytic domain of B. pertussis AC protein (CyaA, amino acids 1–373) was purified as described previously (Shen et al., 2002). [α-32P]ATP (800 Ci/mmol) was purchased from PerkinElmer Life and Analytical Sciences (Rodgau Jügesheim, Germany). Lyophilized calmodulin from bovine brain was from Calbiochem (Darmstadt, Germany). Forskolin was supplied by LC Laboratories (Woburn, MA). For all experiments double-distilled water was used. Sources of all other biochemical reagents have been described previously (Gille et al., 2004; Göttle et al., 2007; Taha et al., 2009).
Mono- and Bis-MANT Nucleotide Synthesis General Procedure.
Synthesis of new substituted mono- and bis-(M)ANT nucleotides followed the general reaction scheme shown in Fig. 1 to obtain (bis-)Cl-ANT-ATP, (bis-)Cl-ANT-ITP, (bis-)Br-ANT-ATP, (bis-)Br-ANT-ITP, (bis-)Br-ANT-ADP and (bis-)Pr-ANT-ATP, (bis-)Pr-ANT-ITP and (bis-)Ac-NH-ANT-ATP, and (bis-)Ac-NH-ANT-ITP. Furthermore, we generated the bis-(M)ANT derivatives of known mono-(M)ANT nucleotides, i.e., bis-MANT-ATP, bis-MANT-ITP, bis-MANT-CTP, bis-MANT-ADP, bis-MANT-ADP, bis-MANT-IMP, and bis-ANT-IMP. Detailed synthesis procedures and chemical analysis of compounds, chemical structures, and their purity are documented in Supplementary Information 1. Under the basic reaction conditions mono- and bis-(M)ANT-NTPs partially decomposed to the corresponding NDPs. Those compounds were isolated as well.
During the synthesis of MANT-IMP, we observed a large new peak at later retention times, when the crude reaction mixture was analyzed by reversed-phase HPLC. Because of the long retention time of the unknown peak, a more lipophilic compound with additional nonpolar groups was expected. Thus, substitution of a second MANT group was hypothesized. The analysis of LC/MS online coupling corroborated the hypothesis. The esterfication of an additional MANT group was identified by the mass-per-charge ratio of 613.2 Da for the negative electrospray ionization measurement. The chromatogram of the crude reaction mixture displayed the typical two-peak system for the expected N-methyl-2′- and 3′-O-anthraniloyl nucleotide isomers at a retention time of 21.4 and 22.0 min, respectively. However, despite separation by LC, purified isomers cannot be used experimentally because of rapid reisomerization (Jameson and Eccleston, 1997). Seven minutes later, the peak for bis-MANT-ITP appeared (Rt = 29 min). The high polarity of nonreacted IMP resulted in a fast elution directly after the dead time (minor peak at Rt < 2 min). Further peaks were identified for the excess of methylisatoic anhydride (Rt ∼17 min) and decomposition product of the nucleoside of hypoxanthine (Rt ∼23 min). The assignment of all signals was achieved by LC/MS online coupling. For the purification of IMP derivatives, only size-exclusion chromatography was required, yielding MANT nucleotides of high purity of approximately 99%. Confirmation for production of bis-MANT nucleotides was provided by NMR spectroscopy. Structure determination of bis-MANT-IMP by one- and two-dimensional NMR measurements confirmed our assumption of the second acylated free hydroxyl group. The proton spectrum showed two sets of signals for the two MANT groups. Furthermore, the heteronuclear multiple bond coherence spectrum definitely identified 3J correlation between protons and quaternary carbons to ensure no substitution in the purine system of the nucleobase.
At the beginning of our in-house MANT-NTP synthesis program, we did not observe formation of bis-substituted MANT-NTPs. However, after the serendipitous discovery of bis-MANT-IMP (Göttle et al., 2007), we addressed the question of whether bis-MANT-NTPs were produced as well. The standard purification procedure was performed by size-exclusion chromatography for separation of starting materials. Nonreacted nucleotide and isatoic anhydride were removed by this method as precleaning. Unfortunately, bis-MANT-NTPs were lost by this separation as well. When we performed HPLC analysis of the crude reaction mixture without the precleaning, similar peaks as for bis-MANT-IMP appeared in the chromatogram. Diode array detection and fluorescence detection for HPLC analysis supported the assignment of bis-(M)ANT nucleotide signals because of the similarity of spectroscopic properties to mono-substituted (M)ANT nucleotides, especially of UV absorption. Furthermore, the formation of bis-MANT nucleotides was also confirmed by LC/MS online coupling. Under the basic reaction conditions, degradation of the labile γ-phosphate always occurred for mono- and bis-(M)ANT nucleotides. Thus, preparative HPLC was applied for the separation of (M)ANT-NTPs and (M)ANT-NDPs. In general, the purification by this method offered the possibility to obtain four compounds simultaneously.
Cell Culture and Membrane Preparation.
Cell culture and membrane preparations (Gille et al., 2004) were performed as described previously. In brief, Sf9 cells were cultured in SF 900 II medium (Invitrogen, Carlsbad, CA) supplemented with 5% (v/v) fetal bovine serum and 0.1 mg/ml gentamicin. High-titer baculoviruses for ACs 1, 2, and 5 were generated through two sequential amplification steps as described previously (Gille et al., 2004). In each amplification step the supernatant fluid was harvested and stored under light protection at 4°C. For membrane preparation Sf9 cells (3.0 × 106 cells/ml) were infected with corresponding baculovirus encoding different ACs (1:100 dilutions of high-titer virus) and cultured for 48 h. Membranes expressing each construct and membranes from uninfected Sf9 cells were prepared as described. Cells were harvested, and cell suspensions were centrifuged for 10 min at 1000g at 4°C. Pellets were resuspended in 10 ml of lysis buffer (1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptine, and 10 μg/ml benzamide, pH 7.4). Thereafter, cells were lysed with 20 to 25 strokes by using a Dounce homogenizer. The resultant cell fragment suspension was centrifuged for 5 min at 500g and 4°C to sediment nuclei. The cell membrane-containing supernatant suspension was transferred into 30-ml tubes and centrifuged for 20 min at 30,000g and 4°C. The supernatant fluid was discarded, and cell pellets were resuspended in buffer consisting of 75 mM Tris/HCl, 12.5 mM MgCl2, and 1 mM EDTA, pH 7.4. Membrane aliquots of 1 ml were prepared and stored at −80°C, and protein concentration for each membrane preparation was determined with the Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Hercules, CA).
AC Activity Assay.
AC activity in Sf9 membranes expressing ACs 1, 2, or 5 was determined essentially as described previously (Göttle et al., 2009). Before starting experiments, membranes were sedimented by a 15-min centrifugation at 4°C and 15,000g and resuspended in 75 mM Tris/HCl, pH 7.4. Reaction mixtures (50 μl, final volume) contained 20 to 40 μg of membrane protein, 40 μM ATP/Mn2+ plus 5 mM MnCl2, 100 μM forskolin, 10 μM GTPγS, and (M)ANT nucleotides at concentrations from 0.1 nM to 1 mM as appropriate to obtain saturated inhibition curves. After a 2-min preincubation at 37°C, reactions were initiated by adding 20 μl of reaction mixture containing (final) 1.0 to 1.5 μCi/tube [α-32P]ATP and 0.1 mM cAMP. AC assays were conducted in the absence of an NTP-regenerating system to allow for the analysis of (M)ANT-NDPs that could otherwise be phosphorylated to the corresponding (M)ANT-NTPs (Gille et al., 2004). Reactions were conducted for 20 min at 37°C and terminated by adding 20 μl of 2.2 N HCl.
For the determination of CyaA AC activity, assay tubes contained 10 μl of inhibitor at final concentrations from 1 nM to 100 μM and 20 μl of CyaA protein (final concentration, 10 pM) in 75 mM HEPES/NaOH, pH 7.4, containing 0.1% (mass/v) bovine serum albumin. After a 2-min preincubation at 25°C reactions were initiated by the addition of 20 μl of reaction mixture consisting of (final) 100 mM KCl, 10 μM free Ca2+, 5 mM free Mn2+, 100 μM EGTA, 100 μM cAMP, 100 nM calmodulin, 40 μM ATP, and [α-32P]ATP (0.2 μCi/tube). For the determination of Km and Vmax values in kinetic studies, 10 μM to 2 mM ATP/Mn2+ plus 5 mM free Mn2+ were added. To ensure linear reaction progress, tubes were incubated for 10 min at 25°C, and reactions were stopped by adding 20 μl of 2.2 N HCl. Denatured protein was precipitated by a 1-min centrifugation at 25°C and 15,000g. Sixty microliters of the supernatant fluid was applied onto disposable columns filled with 1.3 g of neutral alumina. [32P]cAMP was separated from [α-32P]ATP by elution of [32P]cAMP with 4 ml of 0.1 M ammonium acetate, pH 7.0. Recovery of [32P]cAMP was ∼80% as assessed with [3H]cAMP as standard. Blank values were approximately 0.02% of the total added amount of [α-32P]ATP; substrate turnover was <3% of the total added [α-32P]ATP. Samples collected in scintillation vials were filled up with 10 ml of double-distilled water and Cerenkov radiation was measured in a Tri-Carb 2800TR liquid scintillation analyzer (PerkinElmer Life and Analytical Sciences, Waltham, MA). Free concentrations of divalent cations were calculated with Win-MaxC (http://www.stanford.edu/∼cpatton/downloads.htm). Competition isotherms were analyzed by nonlinear regression using Prism 4.0 software (GraphPad Software Inc., San Diego, CA). Km values were 120 μM (AC1), 100 μM (AC2), 70 μM (AC5) and were taken from Gille et al. (2004). The Km value for CyaA was 45 μM and was determined in our previous study (Göttle et al., 2007).
Fluorescence Spectroscopy.
Experiments were conducted by using a quartz UV ultra-microcuvette from Hellma (Müllheim, Germany) (light path length, 3 mm; center, 15 mm; total volume, 70 μl; type 105.251-QS). Measurements were carried out in a Cary Eclipse fluorescence spectrometer (Varian Inc., Palo Alto, CA) at a constant temperature of 25°C (scan rate, 120 nm/min; averaging time, 0.5 s; photomultiplier voltage, 700 V; data interval, 1 nm; slit width, 5 nm) (Göttle et al., 2007). Initially, the cuvette contained 64 μl of 75 mM HEPES/NaOH buffer, pH 7.4, 100 μM CaCl2, 100 mM KCl, and 5 mM MnCl2. Next, nucleotide, CyaA, and CaM were added successively. The cuvette content was mixed after each addition to end up with a total volume of 70 μl. (M)ANT nucleotides were excited at 350 nm, and steady-state emission spectra were recorded from 380 to 550 nm at low speed. The final concentrations of CyaA and CaM were 2.4 μM each. Basal fluorescence in the presence of buffer alone was subtracted. (M)ANT nucleotides were displaced from CyaA by PMEApp at concentrations of 100 nM to 3 μM. For an estimation of the responsiveness of the MANT group to a hydrophobic environment, fluorescence of mono- and bis-(M)ANT nucleotides was determined with dimethyl sulfoxide ranging from 0 to 100% (v/v). Fluorescence recordings were analyzed with the spectrum package of Cary Eclipse software (Varian).
Modeling of Nucleotide Binding Mode to mAC and CyaA.
Docking studies were performed with the molecular modeling package SYBYL 7.3 (Tripos, St. Louis, MO) on an Octane workstation (SGI, Mountain View, CA). Initial computer models of bis-Br-ANT-ITP in complex with mAC and bis-Br-ANT-ATP in complex with CyaA were generated from the PDB crystal structures 3g82 (3′-MANT-ITP bound to mAC) and 1zot (PMEApp bound to CyaA) (Guo et al., 2005), respectively. In the case of CyaA, the starting conformation of bis-Br-ANT-ATP was derived from complexes of 3′-MANT-ATP and TNP-ATP with mAC (PDB structures 2gvz and 2gvd, respectively) (Mou et al., 2005, 2006). An initial docking position resulted from superposition of roughly optimized conformations with PMEApp, allowing the modification of rotatable bonds, and from consideration of the fluorescence data (interaction of the 3′-MANT group with Phe306). In both cases, the ribosyl conformations were adjusted to avoid clashes of the second Br-ANT group with the cores of mAC and CyA, respectively. Hydrogens were added and charges were assigned (proteins and water molecules, AMBER_FF99; ligands, Gasteiger-Hueckel). The ions (Mn2+ or Mg2+) received formal charges of 2. Each complex was refined in a stepwise approach. First, ∼50 minimization cycles with fixed ligand using the AMBER_FF99 force field (Cornell et al., 1995) (steepest descent method); second, ∼100 minimization cycles of the ligand and the surrounding (distance up to 6 Å) protein residues (Tripos force field) (Clark et al., 1989), and third, ∼100 minimization cycles with fixed ligand (AMBER_FF99 force field, Powell conjugate gradient) were performed. The second and third steps were repeated with a larger number of cycles until a root mean square force of 0.01 kcal/mol × Å−1 was approached. To avoid overestimation of electrostatic interactions, a distance-dependent dielectric constant of 4 was applied. Molecular surfaces and lipophilic potentials (protein variant with the new Crippen parameter table) (Heiden et al., 1993; Ghose et al., 1998) were calculated and visualized by the program MOLCAD (MOLCAD, Darmstadt, Germany) contained within SYBYL.
Results
Structure-Activity Relationships of Mono-(M)ANT Nucleotides for mACs and CyaA.
We examined the inhibitory effects of 16 mono-MANT nucleotides on the catalytic activity of mammalian ACs 1, 2, and 5 and bacterial CyaA (Table 1). Nucleotides differed from each other in base (adenine, hypoxanthine, and cytosine) and phosphate chain length (monophosphate, diphosphate, or triphosphate). Mono-substituted compounds (1–16) undergo spontaneous isomerization under physiological pH between the 2′- and 3′-O-ribosyl positions (Suryanarayana et al., 2009). The ANT moiety differed by hydrogen, chlorine, bromine, and an acetylamino group in the 5-position of the phenyl ring (Fig. 1; R1). Moreover, substitution at the amino function of the ANT-group led to methyl (MANT) and propyl (Pr-ANT) derivatives (Fig. 1; R2).
TABLE 1.
Inhibition of catalytic activity of recombinant mammalian ACs 1, 2, and 5 and bacterial AC toxin CyaA by (M)ANT nucleotides
Activity of ACs 1, 2, and 5 and CyaA was determined in the presence of (M)ANT nucleotides at increasing concentrations. Ki values were calculated by nonlinear regression. Data are the mean values ± S.D. of four to five independent experiments performed in triplicates with at least two different membrane preparations (for mACs).
| (M)ANT Nucleotide | AC 1 | AC 2 | AC 5 | CyaA | |
|---|---|---|---|---|---|
| nM | |||||
| 1 | MANT-ATP | 150 ± 40 | 330 ± 100 | 100 ± 30 | 4300 ± 400 |
| 2 | MANT-ITP | 2.8 ± 0.9 | 13.5 ± 0.5 | 1.2 ± 0.1 | 600 ± 100 |
| 3 | MANT-CTP | 150 ± 30 | 690 ± 20 | 150 ± 30 | 1100 ± 100 |
| 4 | Cl-ANT-ATP | 80 ± 4 | 490 ± 20 | 45 ± 1 | 350 ± 40 |
| 5 | Cl-ANT-ITP | 3.3 ± 0.3 | 8 ± 1 | 2.0 ± 0.2 | 700 ± 200 |
| 6 | Br-ANT-ATP | 120 ± 20 | 450 ± 40 | 70 ± 20 | 330 ± 30 |
| 7 | Br-ANT-ITP | 7.0 ± 0.1 | 22 ± 4 | 4.6 ± 0.4 | 920 ± 50 |
| 8 | Pr-ANT-ATP | 440 ± 20 | 1100 ± 100 | 360 ± 60 | 820 ± 250 |
| 9 | Pr-ANT-ITP | 22 ± 1 | 68 ± 12 | 10 ± 2 | 3800 ± 500 |
| 10 | Ac-NH-ANT-ATP | 6800 ± 200 | 11,000 ± 1000 | 3400 ± 40 | 710 ± 40 |
| 11 | Ac-NH-ANT-ITP | 140 ± 30 | 390 ± 80 | 37 ± 7 | 4800 ± 900 |
| 12 | MANT-ADP | 1300 ± 200 | 2900 ± 500 | 800 ± 200 | 12,000 ± 2000 |
| 13 | MANT-IDP | 39 ± 12 | 86 ± 9 | 31 ± 12 | 11,000 ± 3000 |
| 14 | Br-ANT-ADP | 560 ± 10 | 1700 ± 100 | 280 ± 20 | 8700 ± 1300 |
| 15 | MANT-IMP | 4600 ± 400 | 8200 ± 800 | 3400 ± 200 | >100,000 |
| 16 | ANT-IMP | 7400 ± 1200 | 7500 ± 1,400 | 4300 ± 600 | >100,000 |
| 17 | Bis-MANT-ATP | 700 ± 200 | 2100 ± 600 | 430 ± 50 | 360 ± 10 |
| 18 | Bis-MANT-ITP | 310 ± 20 | 1100 ± 200 | 140 ± 40 | 3000 ± 400 |
| 19 | Bis-MANT-CTP | 620 ± 40 | 7800 ± 100 | 750 ± 40 | 2500 ± 500 |
| 20 | Bis-Cl-ANT-ATP | 1700 ± 100 | 2400 ± 100 | 1600 ± 100 | 16 ± 1 |
| 21 | Bis-Cl-ANT-ITP | 66 ± 1 | 200 ± 10 | 65 ± 3 | 15 ± 1 |
| 22 | Bis-Br-ANT-ATP | 670 ± 50 | 1100 ± 100 | 900 ± 90 | 12.6 ± 0.3 |
| 23 | Bis-Br-ANT-ITP | 21 ± 1 | 71 ± 2 | 15 ± 2 | 20 ± 2 |
| 24 | Bis-Pr-ANT-ATP | 18,000 ± 3000 | 36,000 ± 3000 | 18,000 ± 5000 | 700 ± 200 |
| 25 | Bis-Pr-ANT-ITP | 440 ± 10 | 1400 ± 100 | 250 ± 20 | 2100 ± 100 |
| 26 | Bis-Ac-NH-ANT-ATP | 22,000 ± 1000 | 7000 ± 1000 | 6100 ± 1300 | 280 ± 20 |
| 27 | Bis-Ac-NH-ANT-ITP | 1700 ± 100 | 5600 ± 300 | 480 ± 30 | 7500 ± 100 |
| 28 | Bis-MANT-ADP | 700 ± 300 | 1600 ± 300 | 510 ± 70 | 6500 ± 800 |
| 29 | Bis-MANT-IDP | 1000 ± 100 | 1400 ± 100 | 700 ± 200 | 6600 ± 900 |
| 30 | Bis-Br-ANT-ADP | 1600 ± 300 | 3300 ± 200 | 1800 ± 200 | 91 ± 5 |
| 31 | Bis-MANT-IMP | >100,000 | >100,000 | >100,000 | 20,000 ± 3000 |
| 32 | Bis-ANT-IMP | >100,000 | >100,000 | >100,000 | 12,000 ± 3000 |
Recombinant ACs 1, 2, and 5 showed different sensitivity to inhibition by (M)ANT nucleotides (Table 1). In accordance with previous studies (Gille et al., 2004; Göttle et al., 2009), AC2 was the AC isoform with the lowest inhibitor sensitivity because of the exchange of A409P and V1108I in AC1 and AC5 versus AC2 (Mou et al., 2005). (M)ANT-NTPs with the purine base hypoxanthine had the highest potency. Specifically, MANT-ITP is the most potent inhibitor known so far for AC1 and AC5 with Ki values of 1 to 3 nM (Göttle et al., 2009). The exchange of adenine with cytosine had only marginal impact on potency (1 and 3). The halogenated ANT-ATP derivatives (4 and 6), in comparison with MANT-ATP (1), showed slightly more potent inhibition of ACs 1 and 5. The corresponding inosine compounds (5 and 7) exhibited lower potency than MANT-ITP on a still high level. Substitution of bromine with chlorine in ANT-ITP yielded 2-fold more potent inhibition of mACs (7 → 5). Elongation of the alkyl residue from N-methylated to N-propylated ANT nucleotides lowered Ki values 3- to 8-fold (1 → 8, 2 → 9). A further decrease in potency was observed for the acetylated aminoanthraniloyl nucleotides. The inhibition effect of Ac-NH-ANT-ATP (10) dropped into the micromolar range, and the corresponding inosine derivative (11) was 30- to 50-fold less potent than MANT-ITP (2). It is noteworthy that Ac-NH-ANT-ITP (11) displayed the highest 4-fold selectivity for AC5 compared with AC1. Deletion of the γ-phosphate reduced inhibitor affinity 3- to 26-fold (1 → 12, 2 → 13, 6 → 14) and is in accordance with previous data. Crystallographic studies showed that the Mn2+ ion in the B-site coordinates with the γ-phosphate of MANT-NTPs (Mou et al., 2005, 2006). The lack of this phosphate group destabilizes the diphosphate chain in its binding site. The deletion of the β-phosphate group of the MANT-NMP reduced inhibitor potency 120-fold (13 → 15). Exchange of the MANT group with an ANT group had only little effect on inhibitor affinity (15 → 16).
Generally, the inhibitor potency of mono-(M)ANT-NTPs at CyaA was lower than at mACs or maximally reached the affinity range of AC2. MANT-ATP (1) was less potent than MANT-ITP (2), but at all other combinations of adenosine and inosine derivatives the corresponding ANT-ATPs exhibited higher inhibition potency (4 → 5, 6 → 7, 8 → 9, 10 → 11). MANT-CTP (3) is the most potent MANT nucleotide inhibitor of edema factor AC toxin from Bacillus anthracis with a Ki value of 100 nM (Taha et al., 2009). This preference for cytosine was not observed for CyaA (Göttle et al., 2007). Insertion of halogens in ANT-ATPs reduced Ki values by 12- to 13-fold (1 → 4, 1 → 6). However, the Ki values of the corresponding inosine derivatives remained almost constant (2 → 5, 2 → 7). Pr-ANT-ATP (8) and Ac-NH-ANT-ATP (10) revealed still stable inhibitory potency, but Pr-ANT-ITP (9) and Ac-NH-ANT-ITP (11) lost 6- to 8-fold inhibitor affinity compared with MANT-ITP (1). Deletion of γ-phosphate reduced inhibitor potency 3- to 20-fold, and omission of γ- and β-phosphates further decreased inhibition up to the high micromolar range.
Structure-Activity Relationships of Bis-Substituted (M)ANT Nucleotides for mACs and CyaA.
We examined the inhibitory effects of 16 bis-MANT nucleotides on the catalytic activity of mACs 1, 2, and 5 and bacterial CyaA (Table 1). Overall, bis-(M)ANT nucleotides exhibited lower inhibition potencies for ACs 1, 2, and 5 than the corresponding mono-substituted derivatives. However, the reduction of the potency does not consistently depend on the nucleobase. In the case of bis-MANT and bis-Ac-NH-ANT derivatives, hypoxanthine led to the strongest decrease, whereas the affinity of bis-halogen-ANT nucleotides was more reduced with adenine. Comparing mono- and bis-(M)ANT nucleotides, the potency ratio of hypoxanthine versus adenine analogs was not markedly changed by the introduction of a second (M)ANT group. Exceptions were bis-MANT and bis-Ac-NH-ANT derivatives with a significantly lower ratio for the bis-substituted compounds. For the highest inhibition affinities of bis-(M)ANT-NTPs inosine was still required. Bis-MANT-ATP (18) and bis-MANT-CTP (19) inhibited AC1 and AC5 with almost equal potency. Bis-halogen-ANT-ITPs (21 and 23) showed Ki values between 15 and 66 nM for AC1 and AC5, but the corresponding adenine nucleotides (20 and 22) exhibited 25- to 60-fold lower potency. N-propylated instead of N-methylated ANT nucleotides lost affinity dramatically only for bis-Pr-ANT-ATP (24) (17- to 42-fold) but not for bis-Pr-ANT-ITP (25). Bis-Ac-NH-ANT-ATP (26) exhibited low inhibition potency, but possessed 3-fold higher sensitivity for AC2 and AC5 compared with AC1. Usually, the remaining inhibitors showed higher potencies at ACs 1 and 5 than AC2. It is noteworthy that bis-MANT-ADP (28) was as potent as bis-MANT-ATP (17). Bis-MANT-IDP (29) and bis-Br-ANT-ADP (30) were only 1.3- to 5-fold less potent than the corresponding (M)ANT-NTPs. Bis-MANT-IMP (31) and bis-ANT-IMP (32) showed only exceedingly low affinity for mACs.
At CyaA, bis-substituted (M)ANT-NTPs exhibited a broad variety in potency with Ki values from the nanomolar up to the micromolar range. In general, inhibition potencies of adenine nucleotides were the highest. Bis-MANT-CTP (19) and bis-MANT-ITP (18) exhibited 7- and 8-fold lower inhibition potency than bis-MANT-ATP (17). It is noteworthy that halogenated bis-ANT-nucleotide derivatives (20–23) increased the inhibition potency 20- to 240-fold compared with bis-MANT nucleotides (16–19). Bis-Br-ANT-ATP (22) showed the lowest Ki value of 12.6 nM for CyaA, but the remaining halogen-ANT-NTPs were only slightly less potent (Ki values 15–20 nM). The affinity of bis-Pr-ANT-ATP (24) decreased only 2-fold compared with bis-MANT-ATP (17), whereas the corresponding propylated inosine derivative (25) exhibited a slight increase in potency compared with bis-MANT-ITP (18). Again, deletion of the γ-phosphate reduced potency [bis-(M)ANT-NDPs 28–30]. For most (M)ANT-NMPs and (M)ANT-NTPs and all (M)ANT-NDPs the inhibitor affinity at CyaA was increased or kept constant by introduction of a second (M)ANT group. In the case of bis-halogen-ANT nucleotides, the increase was 18- to 73-fold (4–7 → 20–23). Bis-Ac-NH-ANT-ATP (26) was only 2.5-fold more and bis-Ac-NH-ITP (27) even slightly less potent than the corresponding mono-substituted derivatives (10, 11). A second Pr-ANT substitution did not significantly change affinity (8, 9 → 24, 25). An exception to the rule was bis-MANT-ITP (17), exhibiting 5-fold lower inhibitory potency compared with MANT-ITP (2). In most cases of (M)ANT-NTPs, the affinity order with respect to the nucleobase (adenine > hypoxanthine) was not altered by the introduction of a second (M)ANT group, but for halogen derivatives, the increase was greater with ITP than with ATP. Again, the bis-MANT-NTPs were outlier, because the affinity order of the mono-MANT-NTPs (hypoxanthine > adenine) was reversed.
Analysis of the Enzyme Kinetics of CyaA.
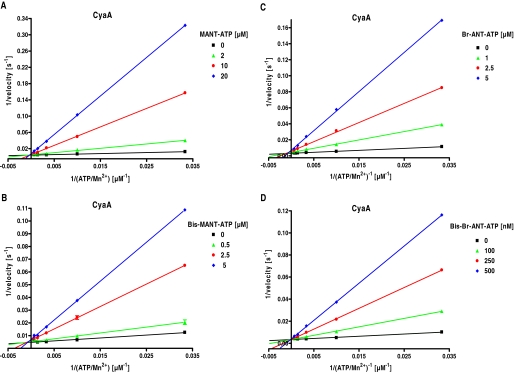
Two classes of AC inhibitors are known, i.e., so-called P-site inhibitors that are noncompetitive or uncompetitive, and competitive inhibitors. Prototypical P-site inhibitors are adenine nucleotide analogs with a 3′-phosphate chain (Tesmer et al., 2000). Most P-site inhibitors require pyrophosphate as a cofactor and bind to an AC-pyrophosphate conformation (Tesmer et al., 2000). MANT nucleotides are competitive mAC inhibitors and possess a 5′- phosphate chain (Gille and Seifert, 2003). Based on these data, for our newly synthesized mono- and bis-substituted (M)ANT nucleotides we expected identical inhibition properties as for known mono-substituted MANT nucleotides. To prove our hypothesis, enzyme kinetics of CyaA were determined with two pairs of compounds, i.e., MANT-ATP (1)/bis-MANT-ATP (17) and Br-ANT-ATP (6)/bis-Br-ANT-ATP (22) (Fig. 2). In fact, Lineweaver-Burk double-reciprocal plotting of CyaA inhibition kinetics displayed competitive inhibition pattern for mono- and bis-(M)ANT nucleotides. The linear regression lines intersected at the y-axis, i.e., Vmax remained constant, whereas Km increased with rising inhibitor concentrations.
Fig. 2.
Enzyme kinetics of CyaA: inhibition by mono- and bis-substituted (M)ANT nucleotides. AC activity of CyaA was determined with the indicated concentrations of MANT-ATP (0, 2, 10, and 20 μM) (A), bis-MANT-ATP (0, 0.5, 2.5, and 5.0 μM) (B), Br-ANT-ATP (0, 1.0, 2.5, and 5.0 μM) (C) or bis-Br-ANT-ATP (0, 100, 250, and 500 nM) (D). Reaction mixtures contained 10 pM CyaA, 100 mM KCl, 10 μM free Ca2+, 5 mM free Mn2+, 100 μM EGTA, 100 μM cAMP, 100 nM calmodulin, 0.2 μCi/tube [α-32P]ATP, and unlabeled ATP/Mn2+ concentrations as indicated. Data were plotted double-reciprocally and analyzed by linear regression according to Lineweaver-Burk. The r2 values of the regression lines were 0.97 to 0.99. Shown are the results of a representative experiment performed in triplicate. Similar results were obtained in at least two independent experiments.
Fluorescence Experiments.
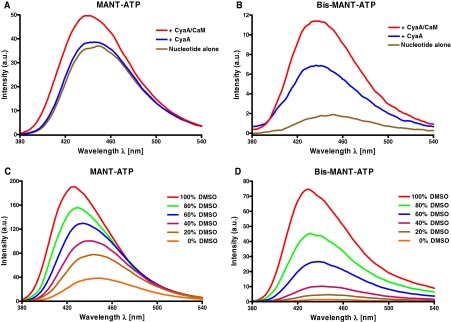
Although the present nucleotides differ in mono/bis substitution and in diversified (M)ANT groups, emission spectra were similar. Nucleotides were excited at 350 nm, and emission was scanned from 380 to 550 nm. Figure 3, A and B shows representative fluorescence emission spectra for the MANT-ATP/bis-MANT-ATP pair. MANT-ATP was added first to the buffer, displaying high autofluorescence at λem = 449 nm (brown lines in Fig. 3). Addition of CyaA did not change the intrinsic fluorescence significantly (blue lines in Fig. 3). However, upon addition of CaM, fluorescence increased by 36%. This result is in accordance to our previous fluorescence resonance energy transfer studies (Göttle et al., 2007). It is noteworthy that bis-MANT-ATP revealed a nearly 20-fold lower autofluorescence (brown lines in Fig. 3) compared with MANT-ATP. The addition of CyaA into the cuvette containing bis-MANT-ATP increased fluorescence by 4-fold. Moreover, the emission maximum was shifted to shorter wavelengths (blue shift). Thus, CyaA bound bis-MANT-ATP even without the activator CaM. Likewise, 2′,3′-O-(2,4,6-trinitrophenyl)-substituted NTPs bind to CyaA in the absence of CaM (Göttle et al., 2007). In contrast, for binding of mono-substituted MANT-NTPs to CyaA, interaction with CaM is required (Göttle et al., 2007). Upon addition of CaM, bis-MANT-ATP displayed an even larger fluorescence increase (6-fold higher compared with basal nucleotide fluorescence).
Fig. 3.
Fluorescence properties of MANT-ATP and bis-MANT-ATP: binding of nucleotides to CyaA and the effect of dimethyl sulfoxide. The assay buffer contained 75 mM HEPES/NaOH, pH 7.4, 100 μM CaCl2, 100 mM KCl, and 5 mM MnCl2, pH 7.4. A and B, MANT-ATP (A) and bis-MANT-ATP (B) were added to the buffer to yield a final concentration of 2 μM, and emission was scanned at an excitation wavelength of 350 nm. CyaA and CaM were added successively to yield a final concentration of 2.4 μM each. Shown are superimposed recordings of a representative experiment. Similar data were obtained in three independent experiments. C and D, MANT-ATP (C) and bis-MANT-ATP (D) were added to water-dimethyl sulfoxide (DMSO) mixtures ranging from 0 to 100% (v/v) to yield a final concentration of 2 μM. Nucleotides were directly excited at λex = 350 nm to mimic binding of the MANT group to a hydrophobic binding pocket. Shown are superimposed recordings of a representative experiment. Similar data were obtained in two independent experiments. Fluorescence intensities are given in arbitrary units (a.u.).
We also studied the responsiveness of MANT-ATP and bis-MANT-ATP to a hydrophobic environment (Göttle et al., 2007). Emission spectra were recorded with dimethyl sulfoxide ranging from 0 to 100% (v/v). Fluorescence of MANT-ATP increased from water environment to pure dimethyl sulfoxide just 5-fold (Fig. 3C). In marked contrast, bis-MANT-ATP exhibited a ∼40-fold fluorescence increase upon exposure to pure dimethyl sulfoxide (Fig. 3D). Thus, independently of the 12-fold higher potency for CyaA compared with MANT-ATP, bis-MANT-ATP displayed an improved signal-to-noise ratio for the fluorescence analysis of CyaA. Blue shifts were detected for both compounds (MANT-ATP: λmax = 448 nm → 426 nm; bis-MANT-ATP: λmax = 445 nm → 429 nm).
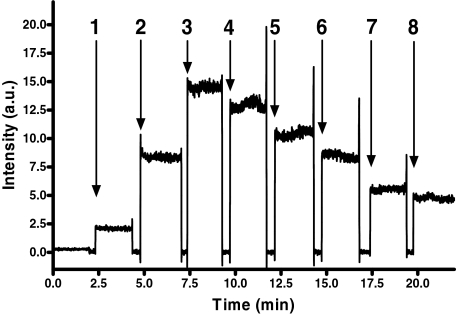
In kinetic experiments CyaA/CaM-evoked fluorescence was inhibited by the nonfluorescent nucleotide analog PMEApp in a concentration-dependent manner (Fig. 4). Because of the higher potency of PMEApp, half-maximal displacement of 2 μM bis-MANT-ATP occurred at a PMEApp concentration of approximately 1 μM. Fluorescence changes in CyaA stimulated by CaM and inhibition by PMEApp occurred within mixing time, i.e., a few seconds. These data show that the fluorescence changes monitored were rapid, specific, and reversible. Because fluorescent nucleotides, e.g., bis-MANT-ATP, were competitively displaced from CyaA by PMEApp, the affinity of nonlabeled inhibitors may also be estimated using this approach. Experiments with two additional compound pairs, i.e., MANT-ITP/bis-MANT-ITP and Br-ANT-ATP/bis-Br-ANT-ATP, showed similar fluorescence properties as with MANT-ATP/bis-MANT-ATP (data not shown). Control experiments were conducted to rule out false-positive fluorescence signals, i.e., combination of CaM and fluorophore without CyaA caused no signal and denatured CyaA (10 min at 95°C) reduced fluorescence to basal response (data not shown).
Fig. 4.
Time-resolved activation of CyaA by CaM and stepwise abolishment of fluorescence by PMEApp. Excitation wavelength was 350 nm, and emission was detected at 440 nm over time. Bis-MANT-ATP (2 μM) (1), 2.4 μM CyaA (2), 2.4 μM CaM (3), and PMEApp in the given concentrations (addition steps: 4, 100 nM; 5, 500 nM; 6, 1 μM; 7, 2 μM; 8, 3 μM) were added in sequence. A recording of a representative experiment is shown. Similar data were obtained in three independent experiments. Fluorescence intensities are given in a.u.
Modeling of Nucleotide Binding to mAC and CyaA.
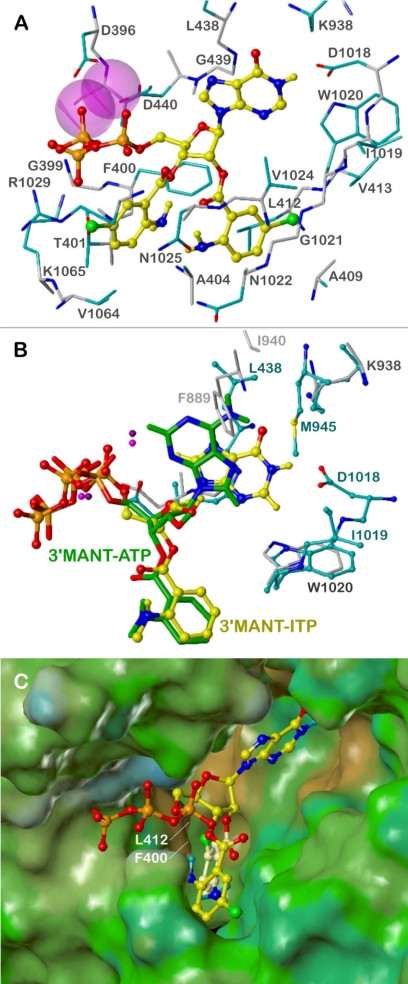
Models of binding modes of bis-(M)ANT nucleotides on mAC were based mainly on the crystal structure of mAC in complex with 3′-MANT-ITP. In this structure, the N- and C-terminal halves of the catalytic core (VC1 and IIC2) originate from AC isoforms 5 and 2, respectively. 3′-MANT-ITP occupies the same binding pocket at the C1:C2 interface as 3′-MANT-GTP and 3′-MANT-ATP in previous crystal structures (Mou et al., 2005, 2006). The ribosyl moiety adopts a 2′-endo envelope conformation, leading to an equatorial orientation of the 2′-hydroxyl incompatible with binding of a 2′ (M)ANT group (strong sterical clashes with VC1). However, the binding modes of mono- and bis-(M)ANT nucleotides should be similar, because an exchange of hypoxanthine by adenine leads to a related drop in potency in most cases of mono- and bis-substituted derivatives (see Table 1). On the base of the “mono-(M)ANT-NTP mode,” docking of bis-(M)ANT-NTPs to mAC is possible in a 3′-endo envelope ribosyl conformation with an axial 2′- and an equatorial 3′-substituent. This enables to align the 2′-(M)ANT group with the 3′-(M)ANT moiety of mono-(M)ANT nucleotides and provides an open channel for binding of the 3′-(M)ANT group.
Figure 5A shows the suggested mAC binding mode of bis-Br-ANT-ITP (23), the most potent bis-(M)ANT nucleotide from the present series. The binding pocket is formed by amino acids of VC1 (396–440) and IIC2 (938–1065). The first Mn2+ coordinates Asp440 and the α- and β-phosphates, whereas the second Mn2+ stabilizes the ligand by octahedral coordination with Asp396, Ile397, Asp440, and the β- and γ-phosphates, which additionally interact with the backbone of Gly399 and the side chains of Thr401 and Lys1065. The ribosyl oxygen is involved in a weak hydrogen bond with the backbone NH of Asp440. The hypoxanthine moiety interacts with Lys938 and Asp1018 by two charge-assisted hydrogen bonds of the 6-oxygen and the 1-NH functions, respectively.
Fig. 5.
Interactions of (M)ANT nucleotides with mAC. The models are based on the crystal structures of mAC in complex with MANT-ITP, PDB 3g82, and with MANT-ATP, PDB 2gvz (Mou et al., 2006). Colors of atoms, unless otherwise indicated, are: orange, phosphorus; red, oxygen; blue, nitrogen; yellow, carbon, hydrogen; green, bromine; purple, Mn2+. A, suggested binding mode of bis-Br-ANT-ITP; amino acids within a sphere of ∼3 Å around the ligand are shown and labeled: carbon atoms of the backbone, gray; carbon atoms of side chains, cyan; Mn2+ ions represented by van der Waal radii. B, comparison of the binding modes of 3′-MANT-ITP (ball and stick model, yellow C atoms) and 3′-MANT-ATP (stick model, green C atoms); amino acids within a sphere of ∼3 Å around the nucleobases are shown and labeled (3g82, ball and stick model, cyan labels and C atoms; 2gvz, stick model, light gray labels and C atoms; both structures, dark gray labels). C, binding site of bis-Br-ANT-ITP, represented by the lipophilic potential mapped onto a MOLCAD Connolly surface (brown, hydrophobic areas; green and blue, polar areas); bis-Br-ANT-ITP in ball and stick model, the 2′-Br-ANT group is highlighted by light yellow carbon atoms.
All interactions described so far correspond to those of 3′-MANT-ITP in the crystal structure (PDB 3g82). Obvious differences to the complex of 3′-MANT-ATP with mAC (PDB 2gvz) may account for the higher inhibitory potency of ITP derivatives (Fig. 5B). The hypoxanthine base adopts an anti conformation with respect to the ribosyl moiety, which enables the hydrogen bonds with Lys936 and Asp1018. This conformation is impossible in the case of 3′-MANT-ATP, probably because of electrostatic repulsion with Asp1018. In the alternative syn conformation, the adenine base forms no hydrogen bonds, but only some weak van der Waals contacts with mAC.
The 2′-Br-ANT group binds in close analogy to the 3′-MANT moiety of 3′-MANT-ITP in the same groove between β1-α1-α2 of VC1 and α4′-β5′ of IIC2 (compare Fig. 5, A and B). The model predicts a hydrogen bond of the ortho-amino substituent with the side-chain amide oxygen of Asn1025. In the case of 3′-MANT-ITP, the methyl group prevents the formation of this hydrogen bond by sterical hindrance (clash with Asn1022), but otherwise contributes to the high potency of MANT-ITP by hydrophobic binding to Ala404. There are significant hydrophobic interactions of the 2′-Br-ANT group with the VC1 site: a face-to-edge approach of the phenyl ring to Phe400 and contacts of the bromine with Leu412 and Val413. With respect to the IIC2 site, the 2′-Br-ANT moiety is aligned to the backbone of β5′ (Trp1020, Gly1021, Asn1022), forming only van der Waals contacts. This amphiphilic nature of the 2′-Br-ANT binding site (hydrophobic VC1, hydrophilic IIC2) becomes also obvious from the lipophilic potential map of mAC (Fig. 5C).
Interactions of the 3′-Br-ANT group with mAC are rather weak compared with the 2′-Br-ANT moiety. Van der Waals contacts occur mainly with the side chains of Thr401, Asn1025, Arg1029, Val1064, and Lys1065. Figure 5C shows that the hydrophobic Br-phenyl fragment projects into a polar environment, which may in part explain the drop in potency of bis-(M)ANT nucleotides compared with their mono-(M)ANT analogs. Other possible reasons are conformational strain caused by restricted space in the binding site and electrostatic repulsion of the ester function in 3′-position with the γ-phosphate, leading to insufficient fit of the phosphate groups.
The crystal structure of CyaA in complex with CaM and PMEApp (Guo et al., 2005) served as a base for predictions of the binding mode of mono- and bis-substituted (M)ANT-ATP derivatives. The AC domain of CyaA includes the catalytic site at the interface of two structural domains, CA (Met1-Gly61, Ala187-Ala364) and CB (Val62-Thr186). Compared with PMEApp, the substrate ATP and the fluorescent nucleotides are conformationally constrained because of the ribosyl moiety. However, the spacious cavity between CA and CB may accommodate the different scaffolds so that an alignment of the adenine bases and the terminal phosphates is possible. Hydrophobic interactions, especially of the 3′-MANT-group with Phe306, significantly contribute to the binding of MANT-ATP to CyaA (Göttle et al., 2007). In support of this model, the CyaA-F306A mutant failed to increase the fluorescence signal of 3′-ANT-2′dATP (Guo et al., 2005).
In our previous study (Göttle et al., 2007), we observed relatively high inhibition potency of 3′-MANT-2′-dATP and 2′-MANT-3′-dATP for CyaA. 3′-MANT-2′-dATP adopted an ideal position for π-stacking of the phenyl ring of the inhibitor and Phe306. This stacking also accounts for the substantial fluorescence increases. Docking of 2′-MANT-3′-dATP reproduced the interaction pattern of 3′-MANT-2′-dATP. The only difference is a 3′-endo conformation of the desoxyribosyl moiety as is present, e.g., in A-DNA.
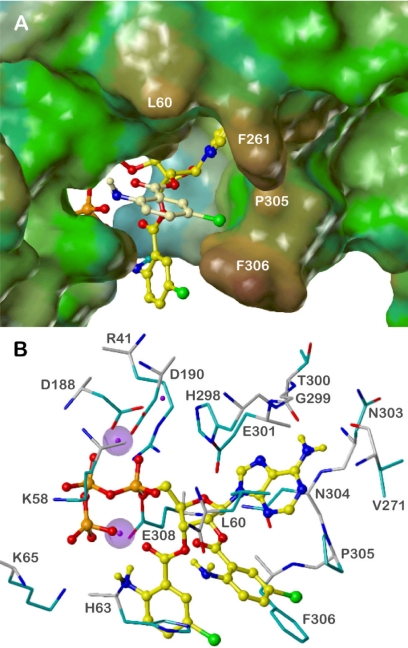
Figure 6 shows bis-Br-ANT-ATP in complex with CyaA. Docking was based on 3′-MANT-2′-dATP, because the equatorial position of the 2′-H atom to be replaced by the second Br-ANT group enables this substituent to be accommodated outside of the active site and to form additional specific interactions with CyaA. The ribosyl moiety adopts a half-chair (2′-endo, 3′-exo) conformation. The initial hypothesis that the second ANT group may interact with Phe261 (Göttle et al., 2007) was not confirmed because of conformational restrictions of the ester moiety (clash with the adenine base). Instead, an energetically favorable conformation of bis-Br-ANT-ATP is possible where the 2′-Br-ANT substituent can easily expand to Phe306. The binding mode of the rest of the molecule is largely the same as in the case of 3′-MANT-2′-dATP, but the ideal π-stacking with Phe306 is not retained. The Br-ANT moieties rather enclose Phe306 from both sides (Fig. 6A). Obviously, both Br (or Cl) substituents increase the hydrophobic interactions with Phe306, thus accounting for the high inhibitory potency of bis-halogen-ANT-ATPs compared with their dehalogenated derivatives. Additional hydrophobic contacts are formed between the 2′-Br-ANT group and the side chains of Leu60 (phenyl ring) and Pro305 (Br).
Fig. 6.
Docking of bis-Br-ANT-ATP to CyaA. The models are based on the crystal structure of CyaA in complex with PMEApp, PDB 1zot (Guo et al., 2005). Colors of atoms, unless otherwise indicated, are: orange, phosphorus; red, oxygen; blue, nitrogen; yellow, carbon, hydrogen; green, bromine; purple, Mg2+. A, overview of the binding site, represented by the lipophilic potential mapped onto a MOLCAD Connolly surface (brown, hydrophobic areas; green and blue, polar areas); bis-Br-ANT-ATP in ball and stick model. The 2′-Br-ANT group is highlighted by light yellow carbon atoms. B, detailed binding mode. Amino acids within a sphere of ∼3 Å around the ligand are shown and labeled: carbon atoms of the backbone, gray; carbon atoms of side chains, cyan. For the two Mg2+ ions interacting with the inhibitor, the van der Waal radii are shown.
Figure 6B presents the putative binding mode of bis-Br-ANT-ATP in more detail. The adenine base, the ribosyl nucleus, and the phosphate groups form similar interactions as described previously for 3′-MANT-2′-dATP (Göttle et al., 2007). Two of the three Mg2+ ions are in positions like those in the CyaA-PMEApp complex. Both are coordinated with Asp188 and Asp190, one additionally with His298, and the other with the α- and β-phosphate. The third Mg2+ ion interacts with Glu308, the carbonyl oxygen of the 3′-ANT group, and the γ-phosphate, which also contacts Lys58 and Lys65. These interactions account for the higher inhibitory potencies of the NTPs compared with NDPs and NMPs (Table 1). The adenine moiety is sandwiched between the side chains of Leu60 and Asn304 whose amide NH2 group may form an additional hydrogen bond with the ribosyl ring oxygen. Hydrogen bonds are also possible between the 6-amino substituent of the nucleobase and the backbone oxygens of Val271 and Thr300. However, the loop between Gly299 and Asn304 may interact with different nucleobases. In the case of ITP derivatives the backbone NH of Gly299 may serve as a hydrogen donor for the carbonyl oxygen in the 6-position. The diversity of possible interactions may account for inconsistent potency differences resulting from base substitution (Table 1). In addition, the affinity of each nucleotide may be affected by a specific arrangement of water molecules that cannot be simply transferred from the PMEApp-bound CyaA structure (Guo et al., 2005).
Hydrophobic interactions with the phenyl ring of Phe306 are formed mainly by both Br substituents. Not only the side chain, but also the backbone oxygen of Leu60, may interact with the 2′-Br-ANT group by forming a hydrogen bond with the amine. The amino group of the 3′-Br-ANT moiety contacts the γ-phosphate and is involved in a charge-assisted hydrogen bond with the carboxylate of Glu308. All of these interactions may be weaker or impossible in the case of secondary amino groups caused by sterical hindrance. This may explain subtle potency differences between ANT, MANT, and Pr-ANT derivatives.
Discussion
In previous studies we found MANT nucleotides to be highly potent mAC inhibitors (Ki values in the 1- to 20-nM range), but their affinity for CyaA was considerably lower (Ki values in the 0.2- to 10-μM range) (Gille et al., 2004; Göttle et al., 2007, 2009). In other previous studies, it was also difficult to identify highly potent and selective CyaA inhibitors (Johnson and Shoshani, 1990; Soelaiman et al., 2003). In this study we found, for the first time, not only highly potent (M)ANT nucleotides for CyaA inhibition with Ki values in the range of 10 to 20 nM, but also inhibitors with substantial selectivity for CyaA versus ACs 1, 2, and 5 (Table 1). The starting point for our systematic analysis of 25 novel mono- and bis-(M)ANT-substituted nucleotides was the unexpected finding that bis-MANT-IMP showed at least 5-fold higher potency at CyaA than mono-MANT-IMP (Göttle et al., 2007). Although the absolute potency of bis-MANT-IMP was still very low, i.e., in the 20- to 40-μM range, depending on the experimental conditions, we nonetheless decided to follow up on this observation and aim at optimizing inhibitor affinity. In fact, we achieved a 1000-fold increase in affinity with a relatively small number of novel compounds.
Bis-halogen-ANT-adenine nucleotides possess the best selectivity ratio. Our prediction of facile accommodation of bulky bis-substituted (M)ANT nucleotides in the large cavity of the catalytic site of CyaA, causing an increase in affinity (Göttle et al., 2007), was confirmed experimentally. Bis-Cl-ANT-ATP (20) is the most interesting compound with a 100- to 150-fold selectivity for CyaA relative to mACs. The equipotent CyaA inhibitor bis-Br-ANT-ATP (22) exhibited high selectivity as well (50- to 90-fold). Generally, the CyaA selectivity of the four bis-halogen-ANT derivatives is only caused by different affinities for mACs. Bis-halogen-ANT-ITPs (21 and 23) displayed only marginal selectivity because of higher mAC potency, probably depending on two specific hydrogen bonds of the hypoxanthine base with Lys936 and Asp1018 (Fig. 5). Bis-Cl-ANT-ATP is 3- to 4-fold less potent at mACs than its bis-Br analog and therefore the most CyaA-selective inhibitor. Docking of both structures on the mAC model confirms this difference. The calculated interaction energy (steric and electrostatic potentials) of bis-Br-ANT-ATP with mAC is by ∼3 kcal/mol greater than that of bis-Cl-ANT-ATP, probably because of better fit of the 3′-Br-ANT group, because the 2′-substituents bind at the site of the 2′- or 3′-(M)ANT group of mono-substituted derivatives (Fig. 5). However, in this case Cl leads to 2-fold higher potency than Br (compounds 4 and 5 versus 6 and 7).
It is noteworthy that the more space-filling propyl and acetylated amino group of ANT-ATPs implemented also selectivity aspects. Bis-propyl-ANT-ATP (24) was 26- to 52-fold more selective for CyaA compared with mAC, and bis-Ac-NH-ANT-ATP (26) was 20- to 80-fold more selective. For acylated amino ANT nucleotides even the mono-substituted derivative (10) yielded a 5- to 15-fold selectivity. Docking of bis-substituted halogen anthraniloyl derivatives to CyaA (Fig. 6) highlighted the critical importance of Phe306 for both high inhibitor potency (Table 1) and large fluorescence increases upon binding of inhibitor (Fig. 3B). The second halogen-ANT group fits to CyaA via strong hydrophobic interactions with Phe306, but aligns to a polar channel of mAC, forming only van der Waals contacts and inducing conformational strain. The resulting increase of CyaA and decrease of mAC potency is the first contributing factor for CyaA selectivity, but not effectual on its own. The second factor is caused mainly by weak interactions of the adenine base with mAC (only van der Waals contacts, no hydrogen bonds; see Fig. 5) (Mou et al., 2006). Both factors together account for the high CyaA selectivity of bis-halogen-ANT-ATPs. In accordance with the very low mAC potency of bis-Pr-ANT-ATP, nucleotides substituted with a bulky boron-dipyrromethene group are less potent mAC inhibitors than mono-MANT nucleotides (Gille et al., 2004). Compared with the catalytic site of mACs, the catalytic site of CyaA is more spacious and possibly more flexible, offering novel and unexpected opportunities for inhibitor design and selectivity.
Pro-drugs based on NMPs with protected phosphate could be feasible drug candidates for inhibition of CyaA and treatment of whooping cough (Laux et al., 2004). The lipophilic NMP pro drug has first to be activated by esterases and then phosphorylated by specific kinases to yield the active NTP derivative. Moreover, it is conceivable that high-affinity bis-(M)ANT nucleotides injected intravenously could bind to extracellularly circulating CyaA and, thereby, block catalytic and cytotoxic activity when the toxin inserts into the membrane of host cells. The compounds characterized in this study can also serve as pharmacophore for the rational development of non-nucleotide-based CyaA inhibitors that could be used in the treatment of whooping cough (Soelaiman et al., 2003).
Bis-substituted (M)ANT nucleotides offer the advantage of excellent signal-to-noise ratio in fluorescence spectroscopy, compared with mono-substituted (M)ANT nucleotides (Fig. 3). Probably, intramolecular interactions of the two MANT-groups with each other quench endogenous fluorescence, which is then unmasked upon binding to a hydrophobic amino acid in the target protein (Phe306 in case of CyaA). Considering then the fact that CyaA can be purified in large quantities (Guo et al., 2005), this fluorescence assay offers interesting opportunities for high-throughput screening studies aimed at the identification of novel non-nucleotide-based CyaA inhibitors (Fig. 4).
Noncompetitive/uncompetitive P-site inhibitors preferentially bind to the catalytically active enzyme (Tesmer et al., 2000). Although an increase in catalytic activity also increases the affinity of (M)ANT nucleotides for AC (Gille and Seifert, 2003; Gille et al., 2004), it is also clear that certain (M)ANT nucleotides can bind to mammalian and bacterial ACs in the absence of an activator. For example, MANT-GTP binds to the purified catalytic subunits of mAC in the absence of the activator forskolin, with the diterpene further enhancing the fluorescence increase (Mou et al., 2005, 2006). The effective binding of bis-MANT-ATP to CyaA in the absence of CaM reported in this study is a particularly impressive illustration for activator-independent interaction of inhibitors with ACs. Crystallographic studies are required to precisely define differences in structure of CyaA bound to bis-ANT-NTPs in the absence and presence of CaM. An approach to explore such potential conformational differences is the examination of the effects of CyaA inhibitors in the absence and presence of CaM using the fluorescence assay with bis-MANT-ATP. Thus, in a broader mechanistic perspective, our data support the concept of multiple activation-dependent AC conformations stabilized by nucleotide inhibitors.
There are both similarities and dissimilarities between CyaA and edema factor in terms of interaction with (M)ANT nucleotides. Both toxins possess a phenylalanine residue in the catalytic site (Phe306 in the case of CyaA and Phe586 in the case of edema factor), being responsible for the fluorescence increases upon interaction with CaM (Göttle et al., 2007; Taha et al., 2009). Moreover, MANT nucleotides inhibit the AC activities of both toxins competitively (Fig. 2) (Taha et al., 2009). Nonetheless, the inhibitory profiles of edema factor and CyaA are different. Most notably, MANT-ATP, defined 2′-MANT- and 3′-MANT-ATP isomers, and MANT-CTP inhibit catalytic activity of CyaA with approximately 10-fold lower potency than catalysis by edema factor, respectively (Göttle et al., 2007; Suryanarayana et al., 2009; Taha et al., 2009). Based on these differences it is also reasonable to assume that the enzymological analysis of edema factor with our new compound library will give different results than the analysis of CyaA. Beyond whooping cough and anthrax infection, it will be very interesting to examine the effects of the new compounds on the AC toxin ExoY from Pseudomonas aeruginosa (Yahr et al., 1998) as well as mycobacterial ACs (Linder and Schultz, 2008).
Since their initial description by Hiratsuka (2003), (M)ANT nucleotides have been broadly used for the analysis of conformational changes in various proteins involved in signal transduction, including GTP-binding proteins, protein kinases, phosphodiesterases, and ACs (Rensland et al., 1991; Jameson and Eccleston, 1997; Ni et al., 2000; Bessay et al., 2008). To this end, attention in the field was paid only to mono-(M)ANT nucleotides, although mono- and bis-(M)ANT nucleotides are produced simultaneously during the synthetic procedure. Because purification procedures have focused only on mono-(M)ANT nucleotides and bis-(M)ANT nucleotides have quite different physiochemical properties, most notably higher lipophilicity, formation of the latter nucleotides has largely gone undetected. In this article, we describe methods that allow for the convenient isolation of both mono- and bis-(M)ANT nucleotides. Based on the results of this study, we anticipate that beyond the field of ACs, bis-(M)ANT nucleotides may have broader applications in signal transduction research, specifically in cases where there are no large spacious constraints in the nucleotide-binding site. Small GTP-binding proteins are suitable candidates (Rensland et al., 1991).
Collectively, in our present study, we have developed a novel series of mono- and bis-(M)ANT-substituted nucleotides. Some of these nucleotides possess surprisingly high selectivity for CyaA relative to mACs, showing that the development of selective CyaA inhibitors for the improved treatment of whooping cough is not an elusive goal. Our data show that numerous chemical modifications such as halogenation and introduction of acetyl and propyl groups can be implemented in the (M)ANT group, resulting in substantial changes in pharmacological properties of compounds. Future studies will have to explore in detail the chemical opportunities offered by the 2′- and 3′-O-ribosyl substituent of nucleotides, facing a spacious cavity in CyaA.
Supplementary Material
Acknowledgments
We thank Dr. M. Göttle for helpful discussions, Mrs. S. Brüggemann for expert technical assistance, and the reviewers for their constructive critique.
This work was supported by the Deutsche Forschungsgemeinschaft [Grant 529/5–2] (to R. S.) and the National Institutes of Health National Institute of General Medical Sciences [Grant GM 062558] (to W.-J.T.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.174219.
 The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- AC
- adenylyl cyclase
- mAC
- mammalian membranous AC
- ANT
- anthraniloyl
- CaM
- calmodulin
- CyaA
- Bordetella pertussis adenylyl cyclase toxin
- HPLC
- high-pressure liquid chromatography
- MANT
- methylanthraniloyl
- MS
- mass spectroscopy
- NDP
- nucleoside 5′-diphosphate
- NMP
- nucleoside 5′-monophosphate
- PMEApp
- 9-[2-(phosphonomethoxy)ethyl]adenine diphosphate
- Pr
- propyl
- Rt
- retention time
- PDB
- Protein Data Bank
- DMSO
- dimethyl sulfoxide
- a.u.
- arbitrary units.
Authorship Contributions
Participated in research design: Geduhn, Dove, König, and Seifert.
Conducted experiments: Geduhn.
Contributed new reagents or analytic tools: Geduhn, Shen, Tang, and König.
Performed data analysis: Geduhn, Dove, and Seifert.
Wrote or contributed to the writing of the manuscript: Geduhn, Dove, Shen, Tang, König, and Seifert.
Other: König provided funding and laboratory space. Dove conducted molecular modeling. Seifert directed and coordinated study among collaborators and provided funding and laboratory space.
References
- Bessay EP, Blount MA, Zoraghi R, Beasley A, Grimes KA, Francis SH, Corbin JD. (2008) Phosphorylation increases affinity of the phosphodiesterase-5 catalytic site for tadalafil. J Pharmacol Exp Ther 325:62–68 [DOI] [PubMed] [Google Scholar]
- Carbonetti NH. (2010) Pertussis toxin and adenylate cyclase toxin: key virulence factors of Bordetella pertussis and cell biology tools. Future Microbiol 5:455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M, Cramer RD, III, Van Opdenbosch N. (1989) Validation of the general purpose Tripos 5.2 force field. J Comput Chem 10:982–1012 [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KMJ, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. (1995) A second generation force field for the simulation of proteins and nucleic acids. J Am Chem Soc 117:5179–5197 [Google Scholar]
- Crowcroft NS, Pebody RG. (2006) Recent developments in pertussis. Lancet 367:1926–1936 [DOI] [PubMed] [Google Scholar]
- Ghose AK, Viswanadhan VN, Wendoloski J. (1998) Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: an analysis of ALOGP and CLOGP. J Phys Chem A 102:3762–3772 [Google Scholar]
- Gille A, Seifert R. (2003) 2′(3′)-O-(N-methylanthraniloyl)-substituted GTP analogs: a novel class of potent competitive adenylyl cyclase inhibitors. J Biol Chem 278:12672–12679 [DOI] [PubMed] [Google Scholar]
- Gille A, Lushington GH, Mou TC, Doughty MB, Johnson RA, Seifert R. (2004) Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides. J Biol Chem 279:19955–19969 [DOI] [PubMed] [Google Scholar]
- Göttle M, Dove S, Kees F, Schlossmann J, Geduhn J, König B, Shen Y, Tang WJ, Kaever V, Seifert R. (2010) Cytidylyl and uridylyl cyclase activity of Bacillus anthracis edema factor and Bordetella pertussis CyaA. Biochemistry 49:5494–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttle M, Dove S, Steindel P, Shen Y, Tang WJ, Geduhn J, König B, Seifert R. (2007) Molecular analysis of the interaction of Bordetella pertussis adenylyl cyclase with fluorescent nucleotides. Mol Pharmacol 72:526–535 [DOI] [PubMed] [Google Scholar]
- Göttle M, Geduhn J, König B, Gille A, Höcherl K, Seifert R. (2009) Characterization of mouse heart adenylyl cyclase. J Pharmacol Exp Ther 329:1156–1165 [DOI] [PubMed] [Google Scholar]
- Guiso N. (2009) Bordetella pertussis and pertussis vaccines. Clin Infect Dis 49:1565–1569 [DOI] [PubMed] [Google Scholar]
- Guo Q, Shen Y, Lee YS, Gibbs CS, Mrksich M, Tang WJ. (2005) Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin. EMBO J 24:3190–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden W, Moeckel G, Brickmann J. (1993) A new approach to analysis and display of local lipophilicity/hydrophilicity mapped on molecular surfaces. J Comput Aided Mol Des 7:503–514 [DOI] [PubMed] [Google Scholar]
- Hiratsuka T. (1983) New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim Biophys Acta 742:496–508 [DOI] [PubMed] [Google Scholar]
- Jameson DM, Eccleston JF. (1997) Fluorescent nucleotide analogs: synthesis and applications. Methods Enzymol 278:363–390 [DOI] [PubMed] [Google Scholar]
- Johnson RA, Shoshani I. (1990) Inhibition of Bordetella pertussis and Bacillus anthracis adenylyl cyclases by polyadenylate and “P”-site agonists. J Biol Chem 265:19035–19039 [PubMed] [Google Scholar]
- Ladant D, Ullmann A. (1999) Bordetella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol 7:172–176 [DOI] [PubMed] [Google Scholar]
- Laux WH, Pande P, Shoshani I, Gao J, Boudou-Vivet V, Gosselin G, Johnson RA. (2004) Pro-nucleotide inhibitors of adenylyl cyclases in intact cells. J Biol Chem 279:13317–13332 [DOI] [PubMed] [Google Scholar]
- Linder JU, Schultz JE. (2008) Versatility of signal transduction encoded in dimeric adenylyl cyclases. Curr Opin Struct Biol 18:667–672 [DOI] [PubMed] [Google Scholar]
- Mou TC, Gille A, Fancy DA, Seifert R, Sprang SR. (2005) Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2′(3′)-O-(N-methylanthraniloyl)-guanosine 5′-triphosphate. J Biol Chem 280:7253–7261 [DOI] [PubMed] [Google Scholar]
- Mou TC, Gille A, Suryanarayana S, Richter M, Seifert R, Sprang SR. (2006) Broad specificity of mammalian adenylyl cyclase for interaction with 2′,3′-substituted purine- and pyrimidine nucleotide inhibitors. Mol Pharmacol 70:878–886 [DOI] [PubMed] [Google Scholar]
- Ni Q, Shaffer J, Adams JA. (2000) Insights into nucleotide binding in protein kinase A using fluorescent adenosine derivatives. Protein Sci 9:1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensland H, Lautwein A, Wittinghofer A, Goody RS. (1991) Is there a rate-limiting step before GTP cleavage by H-ras p21? Biochemistry 30:11181–11185 [DOI] [PubMed] [Google Scholar]
- Shen Y, Lee YS, Soelaiman S, Bergson P, Lu D, Chen A, Beckingham K, Grabarek Z, Mrksich M, Tang WJ. (2002) Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins. EMBO J 21:6721–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soelaiman S, Wei BQ, Bergson P, Lee YS, Shen Y, Mrksich M, Shoichet BK, Tang WJ. (2003) Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough. J Biol Chem 278:25990–25997 [DOI] [PubMed] [Google Scholar]
- Suryanarayana S, Wang JL, Richter M, Shen Y, Tang WJ, Lushington GH, Seifert R. (2009) Distinct interactions of 2′- and 3′-O-(N-methyl)anthraniloyl-isomers of ATP and GTP with the adenylyl cyclase toxin from Bacillus anthracis, edema factor. Biochem Pharmacol 78:224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha HM, Schmidt J, Göttle M, Suryanarayana S, Shen Y, Tang WJ, Gille A, Geduhn J, König B, Dove S, et al. (2009) Molecular analysis of the interaction of anthrax adenylyl cyclase toxin, edema factor, with 2′(3′)-O-(N-methyl)anthraniloyl)-substituted purine and pyrimidine nucleotides. Mol Pharmacol 75:693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Dessauer CW, Sunahara RK, Murray LD, Johnson RA, Gilman AG, Sprang SR. (2000) Molecular basis for P-site inhibition of adenylyl cyclase. Biochemistry 39:14464–14471 [DOI] [PubMed] [Google Scholar]
- Vojtova J, Kamanova J, Sebo P. (2006) Bordetella adenylate cyclase toxin: a swift saboteur of host defense. Curr Opin Microbiol 9:69–75 [DOI] [PubMed] [Google Scholar]
- Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. (1998) ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA 95:13899–14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.