Abstract
The copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction, optimized for biological molecules in aqueous buffers, has been shown to rapidly label mammalian cells in culture with no loss in cell viability. Metabolic uptake and display of the azide derivative of N-acetylmannosamine developed by Bertozzi, followed by CuAAC ligation using sodium ascorbate and the ligand tris(hydroxypropyltriazolyl)methylamine (THPTA), gave rise to abundant covalent attachment of dye-alkyne reactants. THPTA serves both to accelerate the CuAAC reaction and to protect the cells from damage by oxidative agents produced by the Cu-catalyzed reduction of oxygen by ascorbate, which is required to maintain the metal in the active +1 oxidation state. This procedure extends the application of this fastest of azide-based bioorthogonal reactions to the exterior of living cells.
Keywords: click chemistry, cell labeling, metabolic labeling, bioconjugation
Introduction
To understand the roles of proteins, DNA, RNA, lipids, and glycans in a wide variety of cellular processes, imaging of biomolecules in their native environment using a spectroscopic probe has become increasingly popular (1-3). For proteins, genetically encoded tags such as GFP are routinely used. When smaller and less structurally perturbing labels are required, the target protein can be endowed with a peptide sequence that is selectively addressed with a small-molecule probe by either chemical or enzymatic reaction (4-9). These and other examples of chemoselective labeling of biomolecules in vivo are furthered by the development of bioorthogonal ligation reactions (10).
Cycloaddition reactions are attractive in this context, since they usually involve weakly polarized reactants, minimizing undesired side reactions with biomolecules. Recent examples include the reactions of strained olefins with nitrile oxides (11) or tetrazines (12), as well as unstrained olefins with photogenerated nitrile imines (13). Azide-based click reactions (14) are particularly applicable, since the azide group is stable and easy to introduce into biomolecules or probe molecules without dramatically changing their functional properties. The Staudinger ligation with phosphine-esters and cycloaddition reactions with strained cyclic alkynes have been used to excellent effect, led by Bertozzi and coworkers (15-19), and augmented by others (20,21).
While ligand-accelerated catalysis allows the copper-mediated azide-alkyne cycloaddition (CuAAC) reaction to achieve very high rates for in vitro bioconjugation reactions (22-24), the Cu(I) catalyst is regarded as toxic and therefore incompatible with living cells. Since both azide and alkyne groups (25-27) can be appended to biomolecules without altering their function or metabolic processing, it would be very useful if the CuAAC reaction could be adapted for this purpose. As a first step toward this goal, we describe here the application of optimized CuAAC reaction conditions (24) to the rapid and efficient labeling of cell-surface glycans on mammalian cells in culture.
Experimental Procedures
Cell-surface labeling of azido glycans on HeLa and CHO cells and imaging by confocal microscopy
Cells were seeded at 1×105 cells/mL on glass bottom petri dishes (35 mm) and grown overnight at 37 °C and 5 % CO2 in growth medium (MEM medium containing 10% fetal calf serum, 1% glutamine, and 1% penstrep) with or without 50 μM Ac4ManNAz for 2 days. The medium was gently aspirated, and the cells were washed two times with 1 mL of DPBS. In an eppendorf tube, CuSO4 and THPTA in a 1:5 molar ratio were added to DPBS at 4 °C containing dye-alkynes 1 or 2 (final conc. 25 μM) and aminoguanidine (final conc. 1 mM). A freshly-prepared stock solution of sodium ascorbate (100 mM) was added to establish a final ascorbate concentration of 2.5 mM. This reaction mixture was incubated on ice for 10 minutes at 4 °C before adding to the cells. After incubation at 4 °C for 1 or 5 minutes, the cells were washed and fixed with a mixture of 3% paraformaldehyde, 0.3% glutaraldehyde and 1 mM MgCl2 in DBPS for 10 min at room temperature. Cell nuclei were stained by adding 4′,6-diamidino-2-phenylindole (DAPI). In between each step the slides were rinsed three times with DPBS. Slides were mounted using Vecta Shield mounting medium (Vector Laboratories, Burlingame, CA). Sections were imaged using a Biorad 2100 confocal microscope with a 60× oil objective. Data were analyzed and images were created using ImageJ (http://rsbweb.nih.gov/ij/). For dual labeling studies, the cells were washed twice with 1 mL of growth medium after the labeling reaction and returned to medium containing 50 μM Ac4ManNAz for another 20 hours. Optimized conditions for cell-surface labeling were 25 μM alkyne-488, CuSO4 (50 μM), THPTA (250 μM), aminoguanidine (1 mM), and sodium ascorbate (2.5 mM) for 1 to 5 min in medium at 4°C.
Cell-Surface labeling of azido glycans on Jurkat cells with biotinylated conjugates
Jurkat cells were grown in RPMI medium containing 10% fetal calf serum, 1% glutamine, and 1% penstrep with or without 10 μM Ac4ManNAz. Cells were collected using Enzyme-free Hank's based Cell Dissociation Buffer, distributed in 200 μL portions at a concentration of 5×106 cells/mL in the wells of a 96-well V-bottom shaped microtiter plate, pelleted (1,500 × g, 3 min), and washed twice with 200 μL of labeling buffer DPBS. On a separate 96-well plate, premixed CuSO4 and THPTA at a 1:5 molar ratio were added to DPBS at 4°C containing biotin-alkyne 3 (final conc. 50 μM) and aminoguanidine (final conc. 1 mM). A freshly-prepared stock solution of sodium ascorbate (100 mM, 2.5 μL) was added to establish a final ascorbate concentration of 2.5 mM. The reaction mixture was incubated on ice for 60 minutes before adding to the cells. After incubation for 5 minutes at 4 °C, the cells were washed two times with DPBS buffer containing 1 mM EDTA pH 8.0, 25 mM HEPES pH 7.5 and 1% fetal bovine serum, fixed with 2% (v/v) formaldehyde in DPBS for 10 min at room temperature, and resuspended in the same buffer containing FITC-streptavidin (1:250 dilution) for 20 minutes at 4 °C. Cells were analyzed using a FACS Calibur instrument (BD Biosciences, Franklin Lakes, NJ). At least 10,000 events were collected. Experiments were repeated at least twice, and triplicates of each sample were measured and data analyzed using FlowJo 8.7.1 software (Tree Star, Inc, Ashland, OR).
Cell viability assay
Cells were incubated for 2 days in untreated medium or medium containing 10μM-50 μM Ac4ManNAz on 96-well plates. The medium was gently aspirated, and the cells were washed twice with 200 μL of DPBS. In an eppendorf tube, premixed CuSO4 and THPTA at 5:1 ratio were added to DPBS at 4 °C containing aminoguanidine (1 mM) to the desired copper concentrations. Sodium ascorbate (100 mM) was added to the final concentration of 2.5 mM. The reaction mixture was incubated on ice for 10 minutes at 4 °C before adding to the cells. After incubation for 5 minutes, the cells were washed twice with the labeling buffer, and returned to growing medium for 24 hours. An equal volume of premixed Cell Titer-Glo Reagent from Promega was added, plates were shaken for 10 minutes at room temperature, and luminescence was monitored.
Synthesis of Alexa Fluor derivatives
To a solution of the appropriate Alexa Fluor cadaverine (1.0 equivalent) in DMSO (final concentration of 0.05 M) was added a solution of N-(4-pentynoyloxy)succinimide (10.0 equiv) and then N,N-diisopropylethylamine (10.0 equiv). The solution was stirred at room temperature overnight in the dark, diluted in 9:1 water:acetonitrile, purified by reversed-phase HPLC using water and acetonitrile, and lyophilized to a fine powder. Electrosprayionization mass spectrometry (negative ion detection): Alexa 488-alkyne parent ion [1•H+]− (C30H27N4O11S2)−, calcd, 683.1, found, 683.1; Alexa 568-alkyne parent ion [2•H+]− (C42H43N4O11S2)−, calcd, 843.2, found, 843.2.
Results and Discussion
While many enzymes require copper, including cytochrome oxidase, Cu/Zn superoxide dismutase, and lysyl oxidase (28,29), copper ions can be harmful to cells, mostly because they catalyze the production of reactive oxygen species (ROS) from atmospheric oxygen. Therefore, free Cu is essentially absent inside cells of aerobic organisms (30), but the overall intracellular copper concentration is approximately 70 μM (31), all bound up as CuI among proteins that constitute a well-developed and highly conserved system for importing and distributing this essential ion (32).
The CuI oxidation state required for the CuAAC reaction is most often achieved and/or maintained by the reduction of CuII with ascorbate. The Cu/ascorbate system is a prodigious generator of oxygen radicals and other reactive species in air (33-38). We have previously shown that a water-soluble derivative of the tris(triazolylmethyl)amine family of ligands (39) intercepts reactive oxygen species generated in the coordination sphere of the metal and accelerates Cu-mediated peroxide degradation (24). The additive aminoguanidine was also shown to capture dehydroascorbate and its decomposition products before they can react with protein side chains. The combined use of these two additives gave rise to a robust protocol to label proteins, polynucleotides, and other biomolecules in aqueous buffers with high yield and efficiency in the presence of air (24).
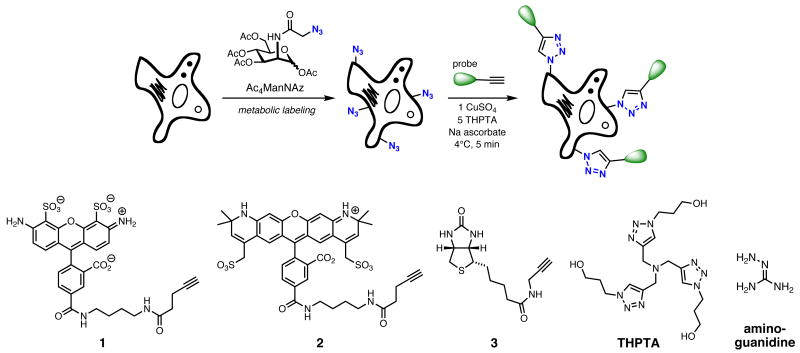
The applicability of this protocol to live cells was tested by metabolic labeling using peracetylated N-azidoacetylmannosamine (Ac4ManNAz), which is taken up and incorporated into cell-surface sialylated (SiaNAz) glycans (Figure 1) (40-42). In order to gauge the effect of THPTA on eliminating the toxicity of copper and sodium ascorbate, HeLa, Chinese hamster ovary (CHO), and Jurkat cells were incubated at 4 °C with these click reaction components. Preliminary exploratory experiments showed 5 minutes to be an incubation period in which significant protective effects from the ligand could be observed, and we knew from previous studies that the CuAAC reaction is fast enough to provide extensive labeling in this amount of time. Longer incubations gave rise to increasing cytotoxicity under most conditions. Thus, variations in Cu and ligand concentrations were explored for five-minute incubations, after which the cells were washed, placed back in media for 24 hours at 37 °C and 5% CO2, and then assessed for viability (Figure 2). In each case, the Cu/ligand/ascorbate mixture was allowed to stand for 10 minutes before introduction to the cells in order to allow the catalyst to quench the ROS and peroxide species generated in the reduction of dissolved oxygen (24).
Figure 1.
(Top) Cell labeling steps. (Bottom) alkynyl probe reagents and catalyst additives.
Figure 2.
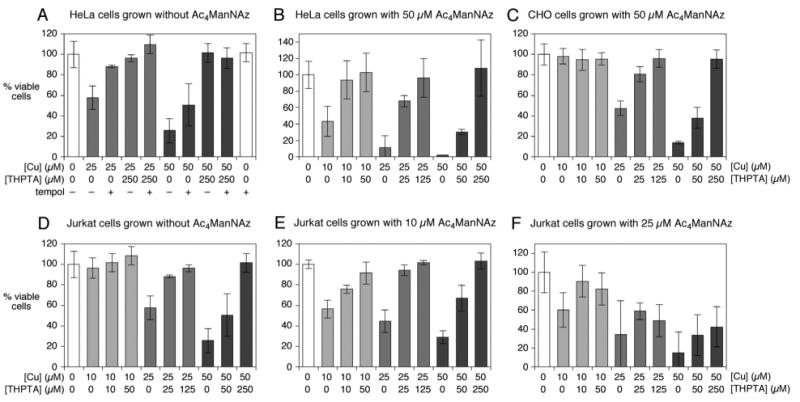
Viability of mammalian cells after Cu-ascorbate treatment ([CuSO4] and [THPTA] as indicated, [Na ascorbate] = 2.5 mM, DPBS, 4 °C, 5 minutes), followed by washing in buffer and incubation in growth media for 24 h. Cell viability was determined by CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI).(43) (B,C,E) Cells were grown for two days in the presence or absence of Ac4ManNAz, as indicated, before Cu treatment. Error bars indicate standard deviation from the averaging of results from at least three independent experiments.
As expected, in the absence of ligand, increasing copper concentration induced increased cell death for each cell type. Growth in the presence of the azido-sugar made the cells significantly more sensitive to Cu-ascorbate (Figure 2A vs. B, D vs. E,F), presumably because the health of the cells or integrity of the membrane is compromised by the incorporation of the non-natural glycan, or because the added Cu complex somehow engages the surface azide groups to the cell's detriment. The latter possibility is unlikely given the fact that organic azides are not consumed by treatment with CuI complexes in the absence of terminal alkynes. The ligand proved to be largely protective: THPTA reversed the toxic effect of Cu-ascorbate effect in a dose-dependent manner, a 5:1 ligand:Cu ratio preserving the viability of all cell types at each Cu concentration tested (Figure 2). This is consistent with the previous observations concerning the ability of the ligand to intercept the formation of oxygen radicals with maximum efficiency (24).
Treatment of the Ac4ManNAz-treated cells with dye-alkynes under the above CuAAC conditions resulted in extensive labeling, as shown in Figures 3 and 4. We chose to perform the CuAAC reactions at 4°C to eliminate internalization of cell-surface species during this process and inhibit energy-dependent internalization of reactants. As has been previously described for in vitro bioconjugation reactions (24), the CuI•THPTA system functions best at 50 μM Cu or greater, showing much greater labeling of SiaNAz-bearing CHO and HeLa cells than 25 μM (Figure 3C vs. B, G vs.F). A reaction time of just one minute gave substantial labeling but five minutes was far better (Figure 3G vs. H). In each case, evenly-distributed labeling over the cell surface was observed, consistent with the expected distribution of the azido sugar throughout the exterior of the cell membrane. The viability of the cells 24 hours after labeling was confirmed as above, with the same results of complete protection against ROS-induced toxicity in the presence of 5 equivalents of THPTA ligand. As shown in panel D, CHO cells returned to the growth medium for 24 hours after labeling showed the dye distributed in the cell interior, reflecting the expected internalization of the sialylated proteins.
Figure 3.
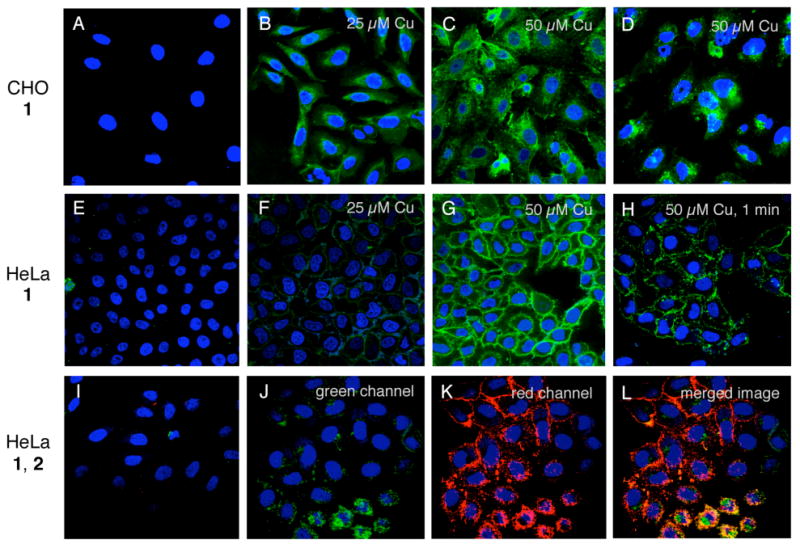
Representative labeling of cells (grown in the presence of 50 μM Ac4ManNAz for 48 h) with the indicated dye-alkynes (25 μM), in the presence of Cu catalyst (5 equiv. THPTA, 2.5 mM Na ascorbate, 1 mM aminoguanidine), in PBS pH 7.4 at 4 °C. The CuAAC reaction was conducted for 5 minutes unless otherwise noted, followed by washing in buffer and fixation for confocal microscopy imaging after incubation in fresh media for 15 minutes (except for panel D, for which the cells were incubated for 24 h before fixation and imaging). Nuclei were stained with DAPI (blue). Panels A, E, and I show the result of CuAAC labeling treatment with 1 on cells lacking the azido sugar, showing that non-specific adsorption or labeling with dye does not occur. The procedure for labeling of cells shown in panels J-L is described in the text. All images are 155 by 155 microns.
Figure 4.
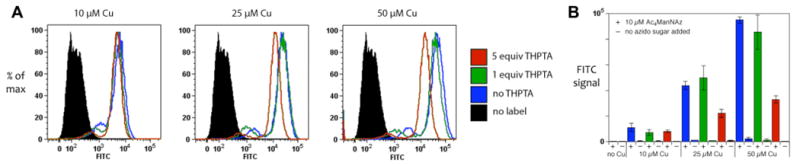
Flow cytometry analysis of Jurkat cells, pre-incubated for 48 h in growth medium containing 10 μM Ac4ManNAz, and then labeled with 3 (50 μM) under standard conditions (CuSO4 and THPTA at the indicated concentrations, 2.5 mM Na ascorbate, 1 mM aminoguanidine, in DPBS, 4 °C, 5 min). (A) All samples were analyzed in triplicate; 10,000 events each were recorded in each case, and representative histograms are shown. (B) Mean fluorescence intensities with error bars representing standard deviations (statistical analysis performed using FlowJo software 8.7.1).
The same pattern was observed in HeLa cells, which were further characterized by a two-color sequential labeling experiment. HeLa cells grown for 48 h in the presence of Ac4ManNAz were labeled with Alexa-488 alkyne 1 at 4 °C, washed, and returned to growth media containing the azido sugar for 24 h to install additional SiaNAz residues. The cells were then labeled a second time with Alexa-568 alkyne 2, and imaged, with the results shown in Figure 3J-L. After 24 h, the initially-labeled (green) sialic acid residues had trafficked largely to the inside of the cells, whereas the freshly-labeled (red) sugars were distributed on the cell surface. These results suggest that the labeling protocol does not significantly perturb glycan trafficking on the time scale of the experiment.
The non-adherent Jurkat cells were analyzed by flow cytometry after the same labeling procedure described above. Ac4ManNAz proved to be toxic to these cells at 50 μM, and cells grown in the presence of 25 μM of the sugar were more sensitive to copper than others (Figure 2F). Labeling studies where therefore performed after growth in the presence of 10 μM Ac4ManNAz. The cells were labeled by CuAAC reaction with biotin alkyne 3, and the label was subsequently detected with a fluorescein-streptavidin conjugate (see Experimental Section). As shown in Figure 4, labeling was most effective with 50 μM Cu, and, surprisingly, the ligand could either be omitted or used in equimolar amounts with respect to Cu to achieve maximum signal. In the case of this two step assay, however, the relationship between output signal and the degree of surface labeling may not be direct. In any case, cell labeling and viability were both excellent.
We demonstrate here a robust and general method for the surface labeling of live cells in culture using the CuAAC click reaction. The toxicity commonly attributed to copper results from the production of reactive oxygen species by Cu, ascorbate, and atmospheric oxygen. The important features of the cell labeling protocol are the use of a water-soluble accelerating ligand (THPTA) that also acts as a sacrificial reductant for oxidative species as they are produced in the coordination sphere of the metal and aminoguanidine to intercept strongly electrophilic byproducts of dehydroascorbate. The rapid nature of the reaction that allows for efficient labeling in a short time, preserving cell viability.
Acknowledgments
This work was supported by the NIH (R01 CA112075 to M.M. and M.G.F.; K99 EB009105 to N.F.S.), an American Heart Association Postdoctoral Fellowship (N.F.S.), the Skaggs Institute for Chemical Biology, and the W.M. Keck Foundation.
References
- 1.Foley TL, Burkart MD. Site-specific protein modification: advances and applications. Curr Opin Chem Biol. 2007;11:12–19. doi: 10.1016/j.cbpa.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 2.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Applied Microbiology and Biotechnology. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 3.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 4.Tsukiji S, Nagamune T. Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. ChemBioChem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 5.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. and references therein. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z, Cironi P, Lin AJ, Xu Y, Hrvatin S, Golan DE, Silver PA, Walsh CT, Yin J. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka F, Fuller R, Asawapornmongkol L, Warsinke A, Gobuty S, Barbas CF., III Development of a small peptide tag for covalent labeling of proteins. Bioconjugate Chem. 2007;18:1318–1324. doi: 10.1021/bc070080x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Suárez M, Baruah H, Martínez-Hernández L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duckworth BP, Zhang Z, Hosokawa A, Distefano MD. Selective labeling of proteins by using protein farnesyltransferase. ChemBioChem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 10.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutsmiedl K, Wirges CT, Ehmke V, C T. Copper-free “click” modification of DNA via nitrile oxide-norbornene 1,3-dipolar cycloaddition. Org Lett. 2009;4:2405–2408. doi: 10.1021/ol9005322. [DOI] [PubMed] [Google Scholar]
- 12.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song W, Wang Y, Lin Q. Selective functionalization of a genetically encoded alkene-containing protein via ‘photo-click chemistry’ in bacterial cells. J Am Chem Soc. 2008;130:9654–9655. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- 14.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 16.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 18.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proc Natl Acad Sci U S A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast Huisgen cycloadditions. Angew Chem Int Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J Am Chem Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen Gupta S, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Accelerated bioorthogonal conjugation: a practical method for the ligation of diverse functional molecules to a polyvalent virus scaffold. Bioconjugate Chem. 2005;16:1572–1579. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- 23.Rodionov VO, Presolski S, Díaz DD, Fokin VV, Finn MG. Ligand-accelerated Cu-catalyzed azide-alkyne cycloaddition: a mechanistic report. J Am Chem Soc. 2007;129:12705–12712. doi: 10.1021/ja072679d. [DOI] [PubMed] [Google Scholar]
- 24.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang P, Chen X, Smyrniotis C, Hu T, Bertozzi CR, Wu P. Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew Chem Int Ed. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci U S A. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yap MC, Kostiuk MA, Martin DD, Perinpanayagam MA, Hak PC, Siddam A, Majjigapu JR, Rajaiah G, Keller BO, JA P, Wu P, Bertozzi CR, Falck JR, Berthiaume LG. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J Lipid Res. 2010;51:1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaim W, Rall J. Copper - a “modern” bioelement. Angew Chem Int Ed. 1996;35:43–60. [Google Scholar]
- 29.Rosenzweig AC, O'Halloran TV. Structure and chemistry of the copper chaperone proteins. Curr Opin Chem Biol. 2000;4:140–147. doi: 10.1016/s1367-5931(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni CJ. Imaging of the intracellular topography of copper with a fluorescent sensor and by synchrotron X-ray fluorescence microscopy. Proc Natl Acad Sci U S A. 2005;102:11179–11184. doi: 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 32.Luk E, Jensen LT, Culotta VC. The many highways for intracellular trafficking of metals. J Biol Inor Chem. 2003;8:803–809. doi: 10.1007/s00775-003-0482-3. [DOI] [PubMed] [Google Scholar]
- 33.Houghton EA, Nicholas KM. In vitro reactive oxygen species production by histatins and copper(I,II) J Biol Inorg Chem. 2009;14:243–251. doi: 10.1007/s00775-008-0444-x. [DOI] [PubMed] [Google Scholar]
- 34.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 35.Khossravi M, Borchardt RT. Chemical pathways of peptide degradation: IX. Metal-catalyzed oxidation of histidine in model peptides. Pharm Res. 1998;15:1096–1102. doi: 10.1023/a:1011946631197. [DOI] [PubMed] [Google Scholar]
- 36.Serrano J, Jové M, Boada J, Bellmunt MJ, Pamplona R, Portrero-Otín M. Dietary antioxidants interfere with Amplex Red-coupled-fluorescence assays. Biochem Biophys Res Commun. 2009;388:443–449. doi: 10.1016/j.bbrc.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 37.Liu PY, Jiang N, Zhang J, Wei XH, Lin HH, Y XQ. The oxidative damage of plasmid DNA by ascorbic acid derivatives in vitro: the first research on the relationship between the structure of ascorbic acid and the oxidative damage of plasmid DNA. Chemistry & Biodiversity. 2006;3:958–965. doi: 10.1002/cbdv.200690104. [DOI] [PubMed] [Google Scholar]
- 38.Khossravi M, Borchardt RT. Chemical pathways of peptide degradation. X: Effect of metal-catalyzed oxidation on the solution structure of a histidine-containing peptide fragment of human relaxin. Pharm Res. 2000;17:851–858. doi: 10.1023/a:1007564410491. [DOI] [PubMed] [Google Scholar]
- 39.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 40.Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Constructing azide-labeled cell surfaces using polysaccharide biosynthetic pathways. Methods Enzymol. 2003;362:249–272. doi: 10.1016/S0076-6879(03)01018-8. [DOI] [PubMed] [Google Scholar]
- 41.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 42.Dube DH, Bertozzi CR. Metabolic oligosaccharide engineering as a tool for glycobiology. Curr Opin Chem Biol. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Niles AL, Moravec RA, Riss TL. Update on in vitro cytotoxicity assays for drug development. Exp Opin Drug Discov. 2008;3:655–669. doi: 10.1517/17460441.3.6.655. [DOI] [PubMed] [Google Scholar]