Abstract
The high mortality rate associated with cardiovascular disease is partially due to the lack of proliferative cells in the heart. Without adequate repair following myocardial infarction, progressive dilation can lead to heart failure. Stem cell therapies present one promising option for treating cardiovascular disease, though the specific mechanisms by which they benefit the heart remain unclear. Before stem cell therapies can be used safely in human populations, their biology must be investigated using innovative technologies such as multi-modality molecular imaging. The present review will discuss the basic principles, labeling techniques, clinical applications, and drawbacks associated with four major modalities: radionuclide imaging, magnetic resonance imaging, bioluminescence imaging, and fluorescence imaging.
Keywords: Stem cells, cell transplantation, imaging, radionuclide, magnetic resonance, bioluminescence, fluorescence
Cardiovascular disease, the leading cause of morbidity and mortality in the United States, ends one life every minute. Many of these deaths occur in individuals under age 65, far below the average life expectancy of 78 years (1). One major reason for the high morbidity and mortality is that the heart has an inadequate regenerative response following ischemia caused by myocardial infarction (MI) or other chronic cardiovascular diseases; the reasons for such limited regenerative ability are unclear (2, 3). Cell death from ischemic damage can lead to progressive remodeling and ventricular dilation, though the processes of vascular remodeling are complex and not fully understood (4). Following MI, ventricular remodeling can result in heart failure, for which the main end-stage treatment is transplantation; however, cardiac transplantation is not an option for most patients because of its high cost and the chronic shortage of suitable organs. Novel regenerative therapies that can promote neovascularization and neomyogenesis, and attenuate apoptotic cell death in the critical post-infarct period are therefore in urgent need. In order to evaluate the effectiveness of stem cell therapies, it is necessary to develop and test innovative noninvasive imaging technologies. Together, advances in stem cell biology and molecular imaging present a multi-disciplinary approach to cardiac disease management, fusing basic and clinical research.
STEM CELL THERAPY
Human and animal studies are adding to the growing body of research investigating the role of stem cell therapies in cardiac disease. With inconsistent results emerging from clinical studies over the last decade, a close examination of cellular mechanisms via imaging is critical.
Skeletal myoblasts (SKMs) were one of the earliest cell types investigated for their applications in cardiac regeneration; however, their potential for arrhythmogenicity and failure to differentiate into cardiomyoctyes led the field to seek out other cell types (5).
Embryonic stem cells (ESCs) are, theoretically, ideal candidates for cardiac regeneration because of their ability for unlimited self-renewal and pluripotency (6). Human embryonic stem cell-derived cardiomyocytes have been transplanted into the murine heart following ischemia-reperfusion injury and these pre-differentiated cells can promote short-term functional recovery (7). However, ESC use has been hindered by teratoma formation in vivo, with intramyocardial teratoma formation observed following transplantation of ~1×105 ESCs (8). Nevertheless, because of their potential advantages, ESC therapy is an active field of investigation. Prior to clinical use of these cells, researchers must therefore develop techniques that can pre-differentiate these cells reliably to lessen the likelihood of teratoma formation.
Another potential cell source is mesenchymal stem cells (MSCs), which are adult stem cells purified from whole bone marrow following in vitro expansion. One important characteristic of MSCs is that they are potentially “immunoprivileged”--they cannot stimulate T-cells proliferation because they do not express HLA class II antigens, B7 costimulatory molecules, or CD40 (9). These immunoprivileged properties, the ability to home in on the heart following infarct, and anti-inflammatory benefits (10) make these cells attractive candidates for allogenic transplantation. In 2009, Osiris Therapeutics announced the completion of the Phase I Osiris Prochymal study (11). This study looked at the safety of allogeneic MSC transplantation in patients with acute MI and found that over the two-year study, 47.4% of placebo patients had cardiac arrhythmia compared to only 11.8% of Prochymal patients (p=0.006). The patients who received MSCs also had a higher LVEF at 2 years (12).
Bone marrow mononuclear cells have been used in both animal models and human clinical trials, and this work has stirred both controversy and excitement. Being free from formation of arrhythmias or teratomas, these cells came into the spotlight of the field of cardiac regenerative medicine after initial reports of transdifferentiation following transplantation in mice in 2001 (13), results which have not been reproduced by later studies (14–16). Although transdifferentiation remains controversial, significant neomyogenesis is unlikely to be a major contributor given that <1% of the bone marrow cells survive 8 weeks following transplantation (16). Transplantation of autologous bone marrow stem cells has also been tested in pigs, and transplantation of cells expressing pro-angiogenic factors such as vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF) has been shown to increase cardiac contractility and perfusion, suggesting that some benefit may be due to paracrine effects (17). Translation to human clinical trials of cardiac bone marrow cell transplant post-infarct has thus far shown mixed results. Two large trials found that bone marrow cell transfusion within 6 days after myocardial infarction had no effect on left ventricular function at six months (18, 19). By contrast, the Bone Marrow Transfer to Enhance ST-Elevation Infarct Regeneration (BOOST) trial found that left ventricular ejection function (LVEF) did improve with bone marrow cell infusion at 6 months (20), though this improvement was not present at the 18-month follow-up (21). The Reinfusion of Enriched Progenitor Cells and Infarct Remodeling in Acute Myocardial Infarction (REPAIR-AMI) trial showed increased LVEF in the bone-marrow cell infusion group vs. placebo (5.5±7.3% vs. 3.0±6.5%; P=0.01) at four months, and reported greatest improvement in those with poorest baseline LVEF (22). These same patients later had a reduced risk for repeat MI or death. A meta-analysis of clinical trials through 2007 found that cell therapy in an acute MI setting increased LVEF, and reduced risk for death and rehospitalization from heart failure (23). However, results from these studies suggest that if bone marrow mononuclear stem cells could provide functional benefit and reduce mortality, they do so by accelerating recovery in the acute stages post-MI. Early cell engraftment may be a key indicator of functional outcome, necessitating sensitive noninvasive imaging of stem cell therapies for clinical use.
The contradictory results from many pre-clinical and clinical studies employing diverse patient populations, delivery methods, and cell types highlight the need for more basic research into mechanisms of stem cell repair. Functional benefit could be due to factors as diverse as paracrine effects, progenitor cell mobilization, or neovascularization (24, 25). Though cell therapies are singularly useful, it may also be important to reduce cell injury through other mechanisms, such as reducing reperfusion injury post-ischemia by mediating chemokine activity (26) or by preconditioning (27). Molecular imaging that provides information about cell behavior in vivo can elucidate the mechanisms of cell therapy and possibly settle, or clarify, the contradictory reports of functional outcomes. Given the diverse mechanisms that underlie functional effects of cardiac stem cell therapies, the urgent need to improve, validate, and evaluate current techniques of tracking stem cells must be met.
MOLECULAR IMAGING
Physical labeling of cells requires the use of an intracellular probe that is detectable externally using imaging technology. Genetic labeling requires a reporter gene to be transiently or stably integrated into the cellular chromosomes. Following labeling of the cells (either physical or genetic), imaging can then be performed using currently available detectors such as positron emission tomography (PET), single photon emission computed tomography (SPECT), magnetic resonance imaging (MRI), and optical charged coupled device (CCD). The present review will outline the basic principles, applications to cell therapy, benefits, and drawbacks of four major imaging modalities: radionuclide imaging, magnetic resonance imaging, bioluminescence imaging, and fluorescence imaging. A schematic of these modalities can be seen in Figure 1. While the ideal imaging modality should be highly sensitive in detecting stem cell viability, nontoxic, longitudinally valid, and highly specific, no single modality currently encompasses all of these features. Our review will take a detailed look at the methods and applications of the 4 major imaging modalities to assess their relative benefits and drawbacks.
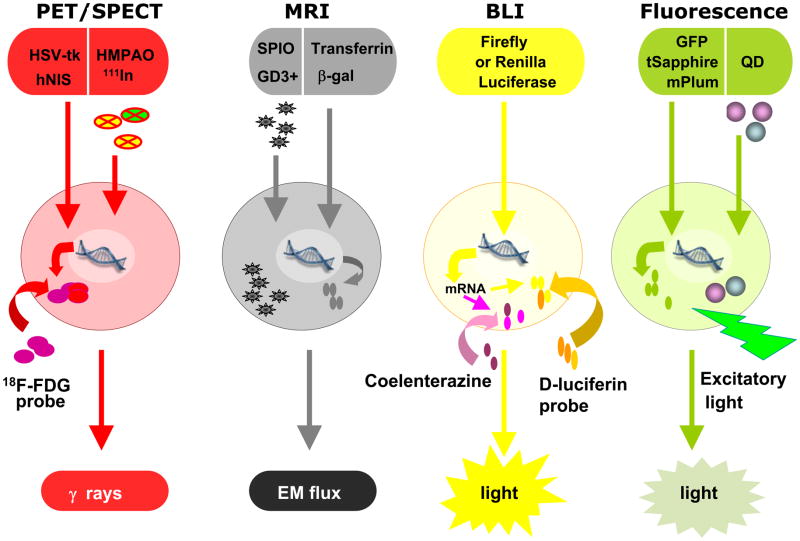
Figure 1. Schematic diagram of key imaging modalities, labeling methods, and probes.
PET = Positron Emission Tomography. SPECT = Single Photon Emission Computed Tomography. HSV-tk = herpes simplex virus thymidinekinase. hNIS = human sodium iodide symporter. MRI: Magnetic Resonance Imaging. EM = Electromagnetic flux. GFP = Green Fluorescent Protein. QD = Quantum Dot.
RADIONUCLIDE IMAGING
Both PET and SPECT are highly sensitive tools for investigating in vivo biodistribution of cells. PET scanners can record 511-keV γ photons that are emitted from positron-emitting isotopes of elements such as carbon, oxygen, nitrogen, and fluorine. SPECT scanners use scintillation crystals to detect lower energy γ photons emitted from radioisotope tracers. PET has a sensitivity of 10−11–10−12 mole/L, whereas SPECT has lower sensitivity, around 10−10–10−11mol/L (28). Radionuclide imaging can be used in two ways: direct labeling and reporter gene labeling. Direct labeling requires isotopes such as 18O, 11C, 13N, and 18F, and genetic labeling can be performed with reporter genes such as herpes simplex virus thymidinekinase (HSV-tk) (29) or human sodium iodide symporter (hNIS) (30).
Direct radioisotope labeling of stem cells has been used in both animal models and human studies. For example, [In-111]oxyquinolone (oxine) (111In), with a half-life of almost 3 days, has been used clinically for over 20 years as a white blood cell scan to assess infection such as osteomyelitis (31). For cardiac stem cell therapy, SPECT imaging has been used to monitor the trafficking of 111In labeled mesenchymal stem cells in a porcine myocardial infarction model (32). More recently, PET imaging has been used to monitor the homing of 2-[F-18]-fluoro-2-deoxy-D-glucose ([18F]-FDG) labeled bone marrow mononuclear cells (33). In this study, three patient groups received bone marrow mononuclear cells with varying results. Using unselected bone marrow cells and intravenous delivery, no cells were visualized in the heart. Using unselected cells and intracoronary delivery, only about 1.3–2.6% were seen in the infarct area. On the other hand, using CD 34+ enriched bone marrow cells and intracoronary infusion, 14–39% of these cells homed in on infarcted myocardium, specifically in the ischemic border zone. These studies demonstrate the clinical feasibility and utility of PET and SPECT imaging technologies to track and monitor stem cells used for cardiac therapy.
Preclinical studies of PET or SPECT reporter genes have been used to track stem cell survival longitudinally. In order to do this, cells are either transiently or stably transduced with a reporter gene (e.g., HSV-tk or hNIS) in vitro prior to transplantation. This technique has been used to track MSCs stably transduced with HSV-tk (via lentiviral vector) (34) or MSCs transiently transduced with HSV-tk (via adenoviral vector) (35). In both cases, the PET reporter probe (9-(4-[fluorine 18]-fluoro-3-hydroxymethylbutyl)-guanine([18F]-FHBG) was used to determine the degree of uptake by transplanted MSCs expressing HSV-tk, which indirectly reflect their survival following quantitative image analysis as shown in Figure 2. The PET reporter probe [18F]-FHBG has been approved by the FDA as an Investigational New Drug for imaging to track HSV-tk reporter gene expression in humans (36). In 2009, a clinical study demonstrated for the first time that the PET reporter gene and PET reporter probe approach can be used to track cell fate in human patients (37). In this study, a patient with grade-IV glioblastoma multiforme was enrolled in a FDA-approved clinical trial of adoptive cellular gene immunotherapy. The patient’s cytolytic T cells were isolated and electroporated with a plasmid encoding interleukin 13 zetakine gene (which targets T-cells to glioblastoma tumors) and HSV-tk. After 5 weeks of cell infusion, [18F]-FHBG accumulation was detected at the tumor site, demonstrating the feasibility and safety of the PET reporter gene approach to track cell fate in humans. The major advantage of reporter genes over direct labeling is that the signal directly reflects cell viability, because the readout depends on reporter gene expression and the interaction of the reporter gene product (i.e., HSV-TK protein) and the reporter probe. In contrast, because direct labeling depends on the decay of the radioisotope, positive results do not necessarily equate cell viability, but only denote probe presence.
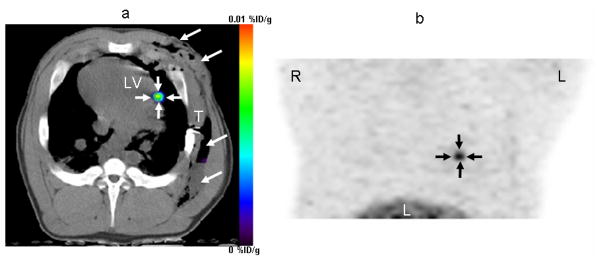
Figure 2. PET Image of MSCs in porcine heart.
MSCs were transduced with adenovirus containing the CMV promoter driving HSV-tk reporter gene in vitro followed by transplantation into the porcine myocardium through a left thoracotomy. Cells can then be visualized following [18F]-FHBG injection, seen in this reconstructed image of the left ventricle taken 4 hours after intravenous administration of the PET reporter probe. Arrows show localization of cells at injection site in the heart. Permission from (35).
Drawbacks of Radionuclide Imaging
Radionuclide imaging is highly sensitive and clinically applicable. Its current probes are biostable, cleared from blood via hepatic metabolism or renal excretion, and the radiation exposure is acceptable. Both [18F]-FDG and [111In]-oxine have been used in the clinic for various cardiac and non-cardiac applications over the past few decades. Direct labeling with radioisotopes has drawbacks such as leakage of tracer to non-target tissues, impairment of cell viability, and unsuitability for long-term use. As most PET tracers have short half-lives (e.g., 20 minutes for 11C and 110 minutes for 18F), other methods must be used for longitudinal tracking (38). Genetic labeling provides more valuable information regarding cell fate compared to direct labeling but is hampered by the need for introducing foreign genetic materials into the cells.
MAGNETIC RESONANCE IMAGING
Magnetic resonance images are created when the magnetic dipoles of hydrogen nuclei align in a pulse of a magnetic field and then return to baseline, a change detected as electromagnetic (EM) flux (28). This flux can vary depending on tissue characteristics such as blood flow or nucleus density. MRI can measure simultaneously molecular and anatomical data, and is therefore useful for real-time cell-survival tracking along with such efficacy measures as cardiac contractility. Though MRI has a high spatial resolution (micrometers), the sensitivity of MRI is low, in the micromolar range with gadolinium chelates and in the millimolar range with iodine-based contrast agents (39). This is because the percentage of dipoles that align correctly in the magnetic pulse is very low, so large amounts of contrast must be used (28). MRI is less sensitive than PET, SPECT, bioluminescence, and fluorescence, but also presents considerable advantages in spatial resolution. Novel molecular agents are being developed to increase the sensitivity of this imaging modality. MRI of cardiac stem cells can also be performed via two labeling methods: physical labeling (e.g., gadolinium chelates, iron oxide particles) or genetic labeling (e.g., β-gal, transferrin receptor).
Physical labeling requires that cells be labeled with detectable contrast agents such as lanthanide gadolinium or super paramagnetic iron oxide (SPIO) particles in vitro prior to injection. Gadolinium chelates localize in the cytoplasm via electroporation and decrease the T1 relaxivity. Gadolinium labels have been used to track migration of transplanted stem cells to a lesion following stroke in rat models (40). But the large amount of gadolinium chelate required to detect a signal, as well as concerns regarding the effect on cellular proliferation, have made other direct-label agents more preferable options (41). Micrometer-sized SPIO (about 1 micrometer in diameter) particles are the most sensitive particles for MR detection. This is because they contain thousands of iron particles on their surface, which then alter the magnetic field experienced by protons near these particles, allowing detection at low nanomolar, or even picomolar, concentrations (39, 42). Serial imaging of SPIO labeled MSCs injected into porcine myocardium post-MI has allowed visualization of cells for up to three weeks (43). While SPIO particles can be imaged much longer than radionuclide tracers used for PET, concerns over toxicity must also be addressed. In light of this, recent studies have confirmed that in vitro labeling of hematopoietic stem cells and MSCs with ferumoxides-protamine sulfate complexes such as Feridex did not affect differentiation capacity or expression of CD34, CD31, CD20, and other key surface markers (44). Further work is needed to determine optimal concentrations for labeling, imaging parameters, correlation with MR signal and cell count, and possible toxic effects of iron labels on cells.
MR reporter genes rely on the same detection principles as direct labels. T1 lengthening paramagneticlanthanides can be detected in tissues that uptake gadolinium, and T2 shortening iron accumulation or SPIO can be detected in iron-binding enzymes. Iron-binding enzymes make good candidates for MR reporter genes (45). A modified gadolinium-containing sugar substrate called (1-(2-(β-galactopyranosyloxy)propyl)-4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecane)gadolinium(III) can be used to increase contrast. When the galactopyranose of this substrate is cleaved by β-gal, one water coordination site is open, so T1 relaxivity increases, which allows for MR detection of β-gal activity (46). Though gadolinium chelates are clinically applicable, iron binding proteins are better candidates for MR reporter genes because of the low detection threshold of iron particles and SPIO. Transferrin-bound iron enters cells via the transferrin receptor (TR). Excess iron binds to and accumulates in the ferritin protein, making it a useful reporter gene (45). Ferritin overexpression has allowed for MR detection of modified cells without administration of a contrast agent (47). Detection can also be enhanced via an engineered TR that lacks regions regulated by iron levels and mRNA-destabilizing motifs (48). This overexpresses the receptor and results in over 500% increase in iron binding of cells (49), allowing for very low detection thresholds. MR reporter genes are better indicators of cell proliferation and viability than physical labels, and can take advantage of an already widely used imaging modality and clinically-approved probes.
Drawbacks of Magnetic Resonance Imaging
One obstacle to optimization of MRI studies remains the low sensitivity. In studies of small and large animals, the lowest number of cells that could be detected was ~100,000 (50). Lower detection thresholds are possible, and single-cell detection has been demonstrated with a 1.63 micron ultra-small SPIO (51); however, further validation is still needed. In addition, magnetic labels can also leak to nearly cells, resulting in measurement errors. Another problem with MRI is that physical labeling with SPIO does not distinguish viable from non-viable cells, and cannot provide information about cell proliferation, since the initial number of iron particles used to label the parent cell can remain after cell death (52). While gadolinium contrast agents have been shown to impair cell proliferative capacity, SPIO particles have not been as toxic to stem cells. The therapeutic benefit following transplantation of SPIO-labeled versus unlabeled cells has been shown to be nearly identical (53). Another problem is that MRI is not widely used in patients with implantable devices such as pacemakers or implantable cardioverter defibrillators because of the potential interference between the device and the magnetic field (54, 55). Finally, while genetic modification to express MR reporter genes is highly useful and sensitive to both cell proliferation and viability, hurdles involving detection sensitivity and the effects of modification on cell characteristics have yet to be resolved.
BIOLUMINESCENCE IMAGING
Bioluminescence imaging (BLI) requires incorporation of a reporter gene such as firefly luciferase (from Photinus pyralis). Photons are emitted when the optical probe, d-luciferin, administered intraperitoneally or intravenously, is oxidized by the firefly luciferase enzyme, which is encoded by the Fluc gene (28). Unlike fluorescence imaging, BLI requires no excitatory light source. The amount of background autoflourescence in vivo is low, so images are highly sensitive. Other types of bioluminescence reporter gene and reporter probe (e.g., renilla luciferase from sea pansy (Renilla reniformis) and coelenterazine) are also available (56).
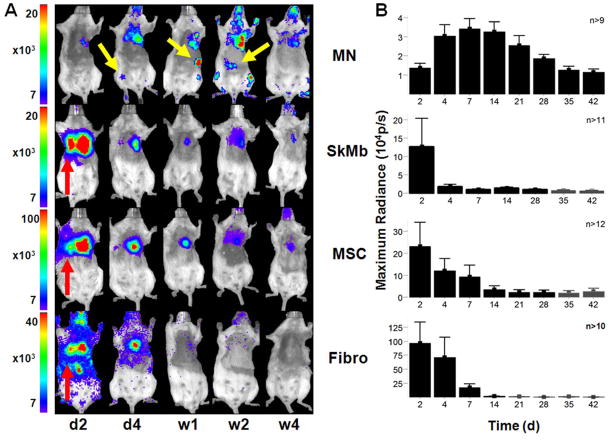
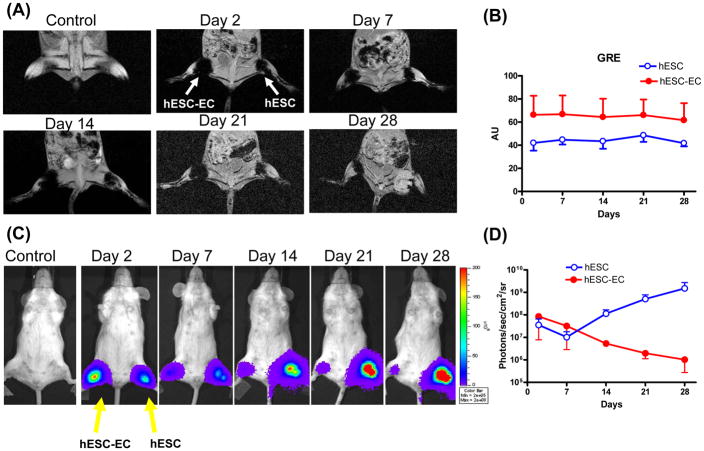
Cao et al. demonstrated the utility of BLI by tracking survival and proliferation of mouse ESCs following cardiac injection in rats (57). BLI has been used to study the efficacy of different immunosuppressive drugs in preventing immunogenic rejection of human ESCs (58). BLI has also been used to compare the survival differences of bone marrow mononuclear cells, MSCs, and SKMs in the ischemic myocardium (Figure 3) (16). BLI is a highly sensitive method for tracking cell survival, especially compared to physical labels used in MR, or to short-lived PET radionuclide tracers. A head-to-head comparison of BLI and MRI using human ESCs in immunodeficient mouse hindlimb models found that MR images showed stable and similar signals in both undifferentiated ESCs and differentiated endothelial cells for 4 weeks, whereas BLI showed divergent survival profiles for the two groups (Figure 4) (52). The study concluded that use of the reporter gene imaging (via Fluc) is preferred for tracking cell viability, and MRI (via SPIO) is better for anatomic detection of cell location. In addition to tracking cell survival, BLI can also be used to track differentiation by inserting the Fluc reporter gene downstream of tissue-specific promoters. This has been done to observe in vitro differentiation of PC12 and F11 cells into neurons (59), a principle that could be applied to visualization of expression of any desired gene in vivo (60).
Figure 3. Stem cell survival kinetics using BLI.
(A) BLI can be used to visualize Fluc-expressing cells. When the injected d-luciferin probe is oxidized by the Fluc enzyme, the emitted photons can be detected by a CCD camera such as the Xenogen In Vivo Imaging System (IVIS) system. Photon emission data are then overlaid on a black and white photograph of the animals. This image shows Fluc-expressing bone marrow mononuclear cells (MN), MSCs, skeletal myoblasts (SkM), and fibroblasts following transplantation of 5×105 cells into the infarcted mouse hearts. Images were acquired 20–25 minutes after d-luciferin injection. (B) Quantification of BLI results shows acute donor cell death during the 6 week period. Permission from (16).
Figure 4. Direct comparison of reporter gene imaging (genetic labeling) vs. iron particle imaging (physical labeling) for tracking stem cells.
(A) Human ESCs were cultured under normal conditions or on gelatin/fibronectin coated plates to induce endothelial cell differentiation. These pre-differentiated human ESC-derived endothelial cells (hESC-ECs) and undifferentiated ESCs were SPIO-labeled (with Feridex) and 1×106 cells were then injected into mouse hindlimbs. MR images of one representative animal show the cells at days 2, 7, 14, 21 and 28. (B) MR does not show survival differences between the two groups, as the signal is steady throughout all imaging timepoints, with a higher signal in the hESC-EC group through day 28. (C) These same ESCs were transduced with the human ubiquitin promoter driving firefly luciferase (Fluc) and enhanced green fluorescence protein (eGFP). These cells were then cultured as in (A) prior to transplantation into the hindlimb of a mouse. (D) BLI showed divergent survival profiles for the two groups, with proliferation of ESC and acute donor cell death of pre-differentiated hESC-ECs. This study demonstrated that MRI provided detailed information on the anatomical location of cells, but not on cell viability. Reporter gene imaging is a better indicator of cell viability and proliferation. Permission from (52).
Drawbacks of Bioluminescence Imaging
Though BLI is a highly-sensitive and versatile imaging tool, it has several disadvantages. First, light transmission through an opaque animal is dependent on tissue type and depth. In addition, photon scatter and signal loss are nonlinear as a function of depth (61). Second, current BLI produces only 2-D images and there is no human equivalent (28). A final problem is one shared by all tracking methods involving genetic manipulation. Reporter gene expression such as Fluc can decrease over time due to epigenetic silencing, especially when a viral promoter (e.g., cytomegalovirus promoter) is used (62).
FLUORESCENCE IMAGING
In order to acquire a fluorescent image, excitatory visible light is aimed at the subject; the resultant shifts in wavelength are then recorded. Fluorescent reporter proteins are well-established, versatile tools for tracking gene expression and localization. In the case of green fluorescent protein (GFP) derived from the jellyfish Aequorea victoria, excitatory violet light leads to GFP emission of green (509 nm) light (28). GFP has been used to histologically verify the presence of transduced bone marrow mononuclear cells following transplantation into myocardium (16). To date, there are many Aequorea-derived fluorescent proteins with point mutations that lead to different fluorescent properties (63). These range in excitation wavelength from far-red mPlum (590 nm excitation) to T-sapphire (399 nm excitation), with many other classes of fluorescent proteins within this range. The emission peaks of such proteins are all in the 500–650 nm range (64). Though most fluorescent proteins scatter at shallow tissue depths, deep tissue penetration is possible with fluorescence techniques such as reflectance fluorescence imaging or fluorescence mediated molecular tomography (FMT). The advent of FMT permits multiple “source-detector” pairs of points to be illuminated, and then recorded prior to reconstruction into an image that provides detailed tomographic information at up to 1 mm in depth (65).
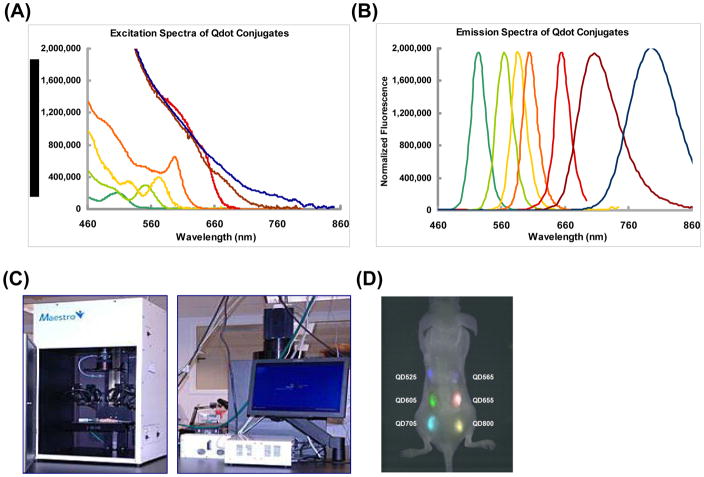
Fluorescent imaging has also been performed with physical labeling using semiconductor nanocrystals, or quantum dots (QDs). These can be uniquely synthesized to certain excitation and emission wavelengths, which allow for construction of QDs for a single study with many non-overlapping emission peaks. While traditional fluorescent probes have been limited because of cellular autofluorescence, the photostability of QDs allows them to be distinguished from autofluorescing cells because they can be imaged over long periods of time (66). QDs ranging from 525 to 800 nm emission have been used to label mouse ESCs, with greatest fluorescence observed with QD800 (67). Labeling did not affect cell differentiation ability. Multiplex imaging is possible as QDs with similar excitation wavelengths and different emission wavelengths can be distinguished in vivo using the same detection camera (Figure 5).
Figure 5. Imaging of cell fate using quantum dots.
(A) Excitation spectra of selected QDs. (B) Emission spectra of same selected QDs. For both the excitation and emission spectra, the colors representing specific QDs as are follow: dark green = QD 525; green = QD 565; yellow = QD 585; orange = QD 605; red = QD 655; brown = QD 705; blue = QD 800 nm. (C) Maestro Imaging System used to detect QDs in vivo. (D) Representative image shows 1×106 ESCs labeled with different QDs and transplanted into the back of an immunodeficient mouse in six different locations. Multiplex in vivo imaging allows for different QDs to be imaged with the same excitatory wavelength as their emission wavelengths do not overlap. Permission from (67).
Drawbacks of Fluorescence Imaging
In vivo fluorescence imaging has a relatively high background due to significant autofluorescence, and is also limited by scattered and shallow tissue penetration (68). Signal loss can occur due to the fact that hemoglobin, deoxyhemoglobin, water, and lipids absorb visible light readily (69). NIR fluorochromes (700–1000 nm) maximize detectable target signal relative to background autofluorescence because hemoglobin does not as readily absorb near-infrared light as it does visible light (28). Another drawback of fluorescence imaging based on physical labeling (e.g., with QDs) is the potential for leakage to neighboring cells, a problem shared by SPIO-based MRI. Leakage to nearby cells or to macrophages following cell death could lead to inaccurate measurements and false-positives, since signal would no longer correlate with cell viability. Use of QDs must be further investigated, as they are a new and exciting class of fluorescent label. While detrimental effects on proliferation have not been observed, injection of large numbers of QDs (5×109) into Xenopus blastomeres can lead to late-stage embryological abnormalities (70).
DISCUSSION
While regeneration of cardiac tissue with stem cell therapy is promising, its progress will depend on further incorporation of clinical research with novel molecular imaging technologies. Each imaging modality discussed here presents both benefits and drawbacks. Identifying the specific needs for each patient can allow for selection of the modality that provides the most benefit (Table 1).
Table 1.
| Imaging Modality | Labeling Method | Label/Probe | Clinical Translation | Advantages | Disadvantages |
|---|---|---|---|---|---|
| PET/SPECT | Direct | 18F-FDG HMPAO 111In | Direct | High sensitivity | Radioactivity exposure, radioisotope decay, leakage to non-target cells |
| Reproter gene | HSV-1-TK hNIS | ||||
| MRI | Direct | Gadolinium SPIO | Direct | Simultaneous molecular and anatomical data | Low sensitivity, signal may not represent viable cells |
| Reporter gene | β-gal transferrin | ||||
| Bioluminescence | Reporter gene | Firefly luciferase, renilla luciferase | No human equivalent | Long-term serial tracking in single animal, low background, high specificity | Two-dimensional, non-linear signal loss with depth, epigenetic silencing attenuates signal |
| Fluorescence | Direct | Quantum dot | No human equivalent | No substrate, can be used in vitro or in vivo | Low signal-to-noise ratio, scatter, shallow penetration, epigenetic silencing |
| Reporter gene | Range from far-Red to UV-excitable green |
As outlined previously, the ideal imaging modality would bring high sensitivity, high resolution and low toxicity. While each of the four modalities discussed has distinct sets of advantages, closer examination of the individual features and applications will be necessary to choose the most effective imaging methods for any given clinical studies of cell therapy.
Direct radionuclide labeling allows for detection of small numbers of cells and has been used clinically to track cardiac stem cell therapies (33, 71) with a sensitivity of around 10−11 mol/L for both PET and SPECT. The spatial resolution of clinical PET and SPECT scanners is around 4–8 mm (28).While the short half-lives of isotope-based tracers is limiting, genetic labeling with PET-detectable genes and probes overcomes this lack of longitudinal applicability and is a promising new area of research. The clinical approval of [18F]-FHBG as a tracer and subsequent use to track HSV-tk expression open the possibility of using PET imaging in a way that is both highly sensitive and potentially useful for long-term tracking of cell therapies (37). Since reporter gene expression is a reflection of cell viability, the use of PET reporter genes presents a promising imaging system. However, genetic manipulation of cells prior to transplantation may have potentially negative effects on cell characteristics due to introduction of foreign genetic material and further validation studies will be needed.
Physical MR labels allow for acquisition of detailed spatial information regarding cell location, but SPIO-based MRI is limited by lower detection sensitivity and inability to distinguish viable from nonviable cells. The relatively low sensitivity of MR, in the 10−3 to 10−5 mol/L range with current clinical scanners, makes it a less ideal candidate for tracking of stem cell therapies. However, MR does have a higher spatial resolution than PET or SPECT, around 25–100μm. Feridex-labeled neural stem cells have been tracked with clinical MR scanners following transplantation in a patient with brain trauma (72) within a short incubation period (and no genetic manipulation). While a key benefit of MR lies in its high spatial resolution, its use for detailed and long-term tracking cell therapy is limited because of the low sensitivity.
A key consideration in deciding between choosing MR versus PET/SPECT is whether cell location or cell number is more important for a particular patient group. If cell location is important, the high spatial resolution of MR is a favorable factor to consider. However, cell viability and cell number can be more accurately tracked with direct radionuclide labeling (short term) or reporter gene labeling (long term).
Optical imaging such as bioluminescence and fluorescence is highly sensitive (estimates of BLI sensitivity posit values in the femtomolar range) and provides valuable information via small animal models. The spatial resolution of optical bioluminescence is in the 3–5 mm range, while fluorescent imaging sensitivity is around 2–3 mm (28). While highly sensitive, both of these optical imaging modalities have rather low spatial resolutions and limited tissue penetrance, thus hindering their clinical translatability at present.
In summary, while stem cell therapies provide hope for treatment of cardiac disease, a full understanding of these therapies requires detailed mechanistic visualization via molecular imaging. At the same time, imaging modalities must be tailored to answer specific scientific questions. Further joint efforts are therefore needed from stem cell biologists and imaging experts to develop, validate, and accelerate impressive progress already made using current imaging tools, which have illuminated different facets of stem cell biology in vivo for the first time.
References
- 1.Lloyd-Jones D, Adams R, Carnethon M, et al. Heart disease and stroke statistics--2009 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009 January 27;119(3):480–6. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008 Feb 21;451(7181):937–42. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 3.Borchardt TBT. Cardiovascular regeneration in non-mammalian model systems: What are the differences between newts and man? Thromb Haemost. 2007;98(2):311–8. [PubMed] [Google Scholar]
- 4.Gorlach A. Vascular remodelling processes: cells, signals and their integration. Thromb Haemost. 2007 Nov;98(5):919–21. [PubMed] [Google Scholar]
- 5.Menasche P, Alfieri O, Janssens S, et al. The Myoblast Autologous Grafting in Ischemic Cardiomyopathy (MAGIC) Trial: first randomized placebo-controlled study of myoblast transplantation. Circulation. 2008 March 4;117(9):1189–200. doi: 10.1161/CIRCULATIONAHA.107.734103. [DOI] [PubMed] [Google Scholar]
- 6.Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127(6):1101–4. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 7.Cao F, Wagner RA, Wilson KD, et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2008;3(10):e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AS, Tang C, Cao F, et al. Effects of cell number on teratoma formation by human embryonic stem cells. Cell Cycle. 2009 Aug 24;8(16) doi: 10.4161/cc.8.16.9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tse WT, Pendleton JD, Beyer WM, et al. Suppression of allogeneic T-cell proliferation by human marrow stromal cells: implications in transplantation. Transplantation. 2003 Feb 15;75(3):389–97. doi: 10.1097/01.TP.0000045055.63901.A9. [DOI] [PubMed] [Google Scholar]
- 10.Barbash IM, Chouraqui P, Baron J, et al. Systemic delivery of bone marrow-derived mesenchymal stem cells to the infarcted myocardium: feasibility, cell migration, and body distribution. Circulation. 2003 August 19;108(7):863–8. doi: 10.1161/01.CIR.0000084828.50310.6A. [DOI] [PubMed] [Google Scholar]
- 11.Taupin P. OTI-010 Osiris Therapeutics/JCR Pharmaceuticals. Curr Opin Investig Drugs. 2006 May;7(5):473–81. [PubMed] [Google Scholar]
- 12.Osiris T. Osiris reports positive two-year data on stem cell treatment for acute myocardial infarction: Prochymal shows lasting clinical benefit in heart attack patients. 2009 [cited; Available from: http://investor.osiris.com/releasedetail.cfm?ReleaseID=364825]
- 13.Orlic D, Kajstura J, Chimenti S, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001 Apr 5;410(6829):701–5. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 14.Balsam LB, Wagers AJ, Christensen JL, et al. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428(6983):668–73. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- 15.Murry CE, Soonpaa MH, Reinecke H, et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428(6983):664–8. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- 16.van der Bogt KE, Sheikh AY, Schrepfer S, et al. Comparison of different adult stem cell types for treatment of myocardial ischemia. Circulation. 2008 Sep 30;118(14 Suppl):S121–9. doi: 10.1161/CIRCULATIONAHA.107.759480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs S, Baffour R, Zhou YF, et al. Transendocardial delivery of autologous bone marrow enhances collateral perfusion and regional function in pigs with chronic experimental myocardial ischemia. J Am Coll Cardiol. 2001 May;37(6):1726–32. doi: 10.1016/s0735-1097(01)01200-1. [DOI] [PubMed] [Google Scholar]
- 18.Janssens S, Dubois C, Bogaert J, et al. Autologous bone marrow-derived stem-cell transfer in patients with ST-segment elevation myocardial infarction: double-blind, randomised controlled trial. The Lancet. 2006 Jan 20;367(9505):113–21. doi: 10.1016/S0140-6736(05)67861-0. [DOI] [PubMed] [Google Scholar]
- 19.Lunde K, Solheim S, Aakhus S, et al. Intracoronary injection of mononuclear bone marrow cells in acute myocardial infarction. N Engl J Med. 2006 September 21;355(12):1199–209. doi: 10.1056/NEJMoa055706. [DOI] [PubMed] [Google Scholar]
- 20.Wollert KC, Meyer GP, Lotz J, et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: the BOOST randomised controlled clinical trial. The Lancet. 2004 Jul 16;364(9429):141–8. doi: 10.1016/S0140-6736(04)16626-9. [DOI] [PubMed] [Google Scholar]
- 21.Meyer GP, Wollert KC, Lotz J, et al. Intracoronary bone marrow cell transfer after myocardial infarction: eighteen months’ follow-up data from the randomized, controlled BOOST (BOne marrOw transfer to enhance ST-elevation infarct regeneration) Trial. Circulation. 2006 March 14;113(10):1287–94. doi: 10.1161/CIRCULATIONAHA.105.575118. [DOI] [PubMed] [Google Scholar]
- 22.Schachinger V, Erbs S, Elsasser A, et al. Intracoronary bone marrow-derived progenitor cells in acute myocardial infarction. N Engl J Med. 2006 September 21;355(12):1210–21. doi: 10.1056/NEJMoa060186. [DOI] [PubMed] [Google Scholar]
- 23.Lipinski MJ, Biondi-Zoccai GGL, Abbate A, et al. Impact of intracoronary cell therapy on left ventricular function in the setting of acute myocardial infarction: a collaborative systematic review and meta-analysis of controlled clinical trials. Journal of the American College of Cardiology. 2007;50(18):1761–7. doi: 10.1016/j.jacc.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 24.Rosenzweig A. Cardiac cell therapy -- mixed results from mixed cells. N Engl J Med. 2006 September 21;355(12):1274–7. doi: 10.1056/NEJMe068172. [DOI] [PubMed] [Google Scholar]
- 25.Gorlach A. The response to cardiovascular injury: A field of emerging complexity. Thromb Haemost. 2007;98(2):259–61. [Google Scholar]
- 26.Steffens S, Montecucco F, Mach F. The inflammatory response as a target to reduce myocardial ischaemia and reperfusion injury. Thromb Haemost. 2009 Aug;102(2):240–7. doi: 10.1160/TH08-12-0837. [DOI] [PubMed] [Google Scholar]
- 27.Hausenloy DJ. Signalling pathways in ischaemic postconditioning. Thromb Haemost. 2009 Apr;101(4):626–34. [PubMed] [Google Scholar]
- 28.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003 Mar 1;17(5):545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 29.Wu JC, Chen IY, Sundaresan G, et al. Molecular imaging of cardiac cell transplantation in living animals using optical bioluminescence and positron emission tomography. Circulation. 2003 September 16;108(11):1302–5. doi: 10.1161/01.CIR.0000091252.20010.6E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terrovitis J, Kwok KF, Lautamäki R, et al. Ectopic expression of the sodium-iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. Journal of the American College of Cardiology. 2008;52(20):1652–60. doi: 10.1016/j.jacc.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters AM, Saverymuttu SH. The value of indium-labeled leucocytes in clinical practice. Blood Reviews. 1987;1(1):65–76. doi: 10.1016/0268-960x(87)90021-x. [DOI] [PubMed] [Google Scholar]
- 32.Chin BB, Nakamoto Y, Bulte JW, et al. 111In oxine labelled mesenchymal stem cell SPECT after intravenous administration in myocardial infarction. Nucl Med Commun. 2003 Nov;24(11):1149–54. doi: 10.1097/00006231-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann M, Wollert KC, Meyer GP, et al. Monitoring of bone marrow cell homing into the infarcted human myocardium. Circulation. 2005 May 3;111(17):2198–202. doi: 10.1161/01.CIR.0000163546.27639.AA. [DOI] [PubMed] [Google Scholar]
- 34.Gyöngyösi MBJ, Marian T, Tron L, Petnehazy Ö, Petrasi Z, Hemetsberger R, Rodriguez J, Font G, Pavo I, jr, Kertesz I, Balkay L, Pavo N, Posa A, Emri M, Galuska L, Kraitchman DL, Wojta J, Huber K, Glogar D. Serial non-invasive in vivo positron emmission tomographyc (PET) tracking of percutaneously intramyocardially injected autologous porcine mesenchymal stem cells modified for transgene reporter gene expression. Circulation: Cardiovascular Imaging. 2008;1:94–3. doi: 10.1161/CIRCIMAGING.108.797449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willmann JK, Paulmurugan R, Rodriguez-Porcel M, et al. Imaging gene expression in human mesenchymal stem cells: from small to large animals. Radiology. 2009 July 1;252(1):117–27. doi: 10.1148/radiol.2513081616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaghoubi SS, Couto MA, Chen CC, et al. Preclinical safety evaluation of 18F-FHBG: a PET reporter probe for imaging herpes simplex virus type 1 thymidine kinase (HSV1-tk) or mutant HSV1-sr39tk’s expression. J Nucl Med. 2006 Apr;47(4):706–15. [PubMed] [Google Scholar]
- 37.Yaghoubi SS, Jensen MC, Satyamurthy N, et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Prac Oncol. 2009;6(1):53–8. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humm JL, Rosenfeld A, Del Guerra A. From PET detectors to PET scanners. Eur J Nucl Med Mol Imaging. 2003 Nov;30(11):1574–97. doi: 10.1007/s00259-003-1266-2. [DOI] [PubMed] [Google Scholar]
- 39.Sosnovik DE. Molecular imaging in cardiovascular magnetic resonance imaging: current perspective and future potential. Top Magn Reson Imaging. 2008 Feb;19(1):59–68. doi: 10.1097/RMR.0b013e318176c57b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modo M, Mellodew K, Cash D, et al. Mapping transplanted stem cell migration after a stroke: a serial, in vivo magnetic resonance imaging study. NeuroImage. 2004;21(1):311–7. doi: 10.1016/j.neuroimage.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 41.Budde MD, Frank JA. Magnetic tagging of therapeutic cells for MRI. J Nucl Med. 2009 February 1;50(2):171–4. doi: 10.2967/jnumed.108.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morawski AM, Winter PM, Crowder KC, et al. Targeted nanoparticles for quantitative imaging of sparse molecular epitopes with MRI. Magnetic Resonance in Medicine. 2004;51(3):480–6. doi: 10.1002/mrm.20010. [DOI] [PubMed] [Google Scholar]
- 43.Kraitchman DL, Heldman AW, Atalar E, et al. In vivo magnetic resonance imaging of mesenchymal stem cells in myocardial infarction. Circulation. 2003 May 13;107(18):2290–3. doi: 10.1161/01.CIR.0000070931.62772.4E. [DOI] [PubMed] [Google Scholar]
- 44.Arbab AS, Yocum GT, Rad AM, et al. Labeling of cells with ferumoxides-protamine sulfate complexes does not inhibit function or differentiation capacity of hematopoietic or mesenchymal stem cells. NMR in Biomedicine. 2005;18(8):553–9. doi: 10.1002/nbm.991. [DOI] [PubMed] [Google Scholar]
- 45.Gilad AA, Winnard PT, Jr, van Zijl PC, et al. Developing MR reporter genes: promises and pitfalls. NMR Biomed. 2007 May;20(3):275–90. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- 46.Louie AY, Huber MM, Ahrens ET, et al. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotech. 2000;18(3):321–5. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 47.Cohen B, Dafni H, Meir G, et al. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005 Feb;7(2):109–17. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basilion JP, Kennedy MC, Beinert H, et al. Overexpression of iron-responsive element-binding protein and its analytical characterization as the RNA-binding form, devoid of an iron-sulfur cluster. Archives of Biochemistry and Biophysics. 1994;311(2):517–22. doi: 10.1006/abbi.1994.1270. [DOI] [PubMed] [Google Scholar]
- 49.Weissleder R, Moore A, Mahmood U, et al. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6(3):351–4. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 50.Beeres SLMA, Bengel FM, Bartunek J, et al. Role of imaging in cardiac stem cell therapy. Journal of the American College of Cardiology. 2007;49(11):1137–48. doi: 10.1016/j.jacc.2006.10.072. [DOI] [PubMed] [Google Scholar]
- 51.Shapiro EM, Sharer K, Skrtic S, et al. In vivo detection of single cells by MRI. Magnetic Resonance in Medicine. 2006;55(2):242–9. doi: 10.1002/mrm.20718. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Suzuki Y, Huang M, et al. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem Cells. 2008;26(4):864–73. doi: 10.1634/stemcells.2007-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Au K-W, Liao S-Y, Lee Y-K, et al. Effects of iron oxide nanoparticles on cardiac differentiation of embryonic stem cells. Biochemical and Biophysical Research Communications. 2009;379(4):898–903. doi: 10.1016/j.bbrc.2008.12.160. [DOI] [PubMed] [Google Scholar]
- 54.Sierra M, Machado C. Magnetic resonance imaging in patients with implantable cardiac devices. Rev Cardiovasc Med. 2008 Fall;9(4):232–8. [PubMed] [Google Scholar]
- 55.Zhang SJ, Wu JC. Comparison of imaging techniques for tracking cardiac stem cell therapy. The Journal of Nuclear Medicine. 2007;48(12):3. doi: 10.2967/jnumed.107.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):377–82. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao F, Lin S, Xie X, et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006 February 21;113(7):1005–14. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swijnenburg R-J, Schrepfer S, Cao F, et al. In vivo imaging of embryonic stem cells reveals patterns of survival and immune rejection following transplantation. Stem Cells and Development. 2008;17(6):1023–30. doi: 10.1089/scd.2008.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang D, Kang J, Jeong J, et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. European Journal of Nuclear Medicine and Molecular Imaging. 2008;35(1):135–45. doi: 10.1007/s00259-007-0561-8. [DOI] [PubMed] [Google Scholar]
- 60.Gruber PJ, Li Z, Li H, et al. In vivo imaging of mlc2v-luciferase, a cardiac-specific reporter gene expression in mice1. Academic Radiology. 2004;11(9):1022–8. doi: 10.1016/j.acra.2004.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ntziachristos V, Ripoll J, Wang LV, et al. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotech. 2005;23(3):313–20. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 62.Krishnan M, Park JM, Cao F, et al. Effects of epigenetic modulation on reporter gene expression: implications for stem cell imaging. FASEB J. 2005 October 24;:05–4551fje. doi: 10.1096/fj.05-4551fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang JH, Chung J-K. Molecular-genetic imaging based on reporter gene expression. J Nucl Med. 2008 June 1;49(Suppl_2):164S–79. doi: 10.2967/jnumed.107.045955. [DOI] [PubMed] [Google Scholar]
- 64.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Meth. 2005;2(12):905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 65.Graves EE, Ripoll J, Weissleder R, et al. A submillimeter resolution fluorescence molecular imaging system for small animal imaging. Medical Physics. 2003;30(5):901–11. doi: 10.1118/1.1568977. [DOI] [PubMed] [Google Scholar]
- 66.Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005 January 28;307(5709):538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin S, Xie X, Patel M, et al. Quantum dot imaging for embryonic stem cells. BMC Biotechnology. 2007;7(1):67. doi: 10.1186/1472-6750-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sutton E, Henning T, Pichler B, et al. Cell tracking with optical imaging. European Radiology. 2008;18(10):2021–32. doi: 10.1007/s00330-008-0984-z. [DOI] [PubMed] [Google Scholar]
- 69.Tung C. Fluorescent peptide probes for in vivo diagnostic imaging. Peptide Science. 2004;76(5):391–403. doi: 10.1002/bip.20139. [DOI] [PubMed] [Google Scholar]
- 70.Dubertret B, Skourides P, Norris DJ, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002 November 29;298(5599):1759–62. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 71.Schachinger V, Aicher A, Dobert N, et al. Pilot trial on determinants of progenitor cell recruitment to the infarcted human myocardium. Circulation. 2008 Sep 30;118(14):1425–32. doi: 10.1161/CIRCULATIONAHA.108.777102. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J, Zhou L, XingWu F. Tracking Neural Stem Cells in Patients with Brain Trauma. N Engl J Med. 2006 November 30;355(22):2376–8. doi: 10.1056/NEJMc055304. [DOI] [PubMed] [Google Scholar]