Abstract
MicroRNAs are known to regulate gene function in many tissues and organs, but their expression and function, if any, in tooth development are elusive. We sought to identify them by microRNA screening analyses and reveal their overall roles by inactivating Dicer1 in the dental epithelium and mesenchyme. Discrete sets of microRNAs are expressed in molars compared with incisors as well as epithelium compared with mesenchyme. Conditional knockout (cKO) of Dicer1 (mature microRNAs) in the dental epithelium of the Pitx2-Cre mouse results in multiple and branched enamel-free incisors and cuspless molars, and change in incisor patterning and in incisor and molar size and shape. Analyses of differentiating dental epithelial markers reveal a defect in ameloblast differentiation. Conversely, the cervical loop (stem cell niche) is expanded in Dicer1 cKO. These results demonstrate that tooth development is tightly controlled by microRNAs and that specific microRNAs regulate tooth epithelial stem cell differentiation.
Keywords: Dicer1, dental stem cells, craniofacial and tooth microRNAs, epithelial differentiation
Introduction
The mouse incisor grows continuously throughout the life of the mouse. Evidence suggests that there are tooth epithelial stem cells located in the cervical loop region at the posterior end of the incisor (Harada et al., 1999; Wang et al., 2007; Klein et al., 2008). These stem cells give rise to transit-amplifying (T-A) cells that insert into the inner enamel epithelium (Smith, 1998; Gritli-Linde et al., 2002). Transit-amplifying cells will undergo differentiation processes as they move anteriorly (Appendix Fig. 1). The different cell types are present during development and morphogenesis in the order of posterior to anterior and make the mouse incisor a good model system for the study of epithelial cell differentiation.
MicroRNAs are a new family of small, non-coding RNAs that regulate gene function post-transcriptionally (Bartel, 2004; He and Hannon, 2004). It has been predicted that more than a third of the protein-coding genes are under the control of microRNAs (Lewis et al., 2005). MicroRNAs are small RNAs with stem-loop structures called pri-miRNA; these are converted to pre-miRNA through the cleavage activity of the Drosha enzyme. Drosha crops the flanking regions of pri-miRNA to liberate the 60- to 70-nt pre-miRNA (Shalgi et al., 2007). The pre-miRNA is exported to the cytoplasm by Exportin-5. The pre-miRNA is processed into 18-22 nucleotide miR duplexes by Dicer and, in humans, its partner TRBP. One strand of the duplex is degraded, and the other strand accumulates as a mature miRNA. Animal miRNAs are imperfectly paired to the 3′UTR of target mRNA and inhibit protein production. These miRs can also bind to other regions of the mRNA to degrade the target mRNA or inhibit protein synthesis. More and more developmental and physiological processes have been found to rely on microRNAs, but their functions during tooth development are not known (Chen et al., 2004; Poy et al., 2004; Wienholds and Plasterk, 2005; Zhao et al., 2007; Wang et al., 2008).
In this study, microRNAs were deleted in the oral epithelium and dental mesenchyme in Pitx2-Cre and Wnt-1 Cre mice, respectively. These mice should reveal the function of microRNAs during tooth development.
Materials & Methods
Animals
All animals were housed in the Program of Animal Resources of the Institute of Biosciences and Technology, and were handled in accordance with the principles and procedure of the Guide for the Care and Use of Laboratory Animals. All experimental procedures were approved by the Texas A&M Health Science Center, Institutional Animal Care and Use Committee. Mice carrying floxed Dicer1 alleles (Dicer1flox/flox) were mated to mice carrying a Pitx2-Cre (Liu et al., 2003) and the Wnt1-Cre transgenic line (Danielian et al., 1998) to generate tooth epithelial and mesenchyme conditional Dicer1 KO. Embryos were collected at various timepoints, with the day of observation of a vaginal plug considered to be embryonic day (E) 0.5.
MicroRNA Microarray
Incisor and molar tooth germs were dissected from P0 mice with the use of a dissection microscope. To separate epithelium and mesenchyme, we treated the tooth germs with Dispase II and Collagenase I (Worthington, Lakewood, NJ, USA)) for 30 min at 37°C. This procedure separated the epithelium from the mesenchyme and allowed for specific RNA extraction of the 2 tissue types. Total RNAs including microRNA were prepared with the use of a miRNeasy Mini Kit from Qiagen (Valencia, CA, USA). LC Sciences (Houston, TX, USA) performed the microRNA microarray analyses.
Histology
Mouse embryos or heads were dissected in phosphate-buffered saline (PBS). For adult mice, heads were first decalcified in 8% EDTA/PBS solution. Embryos or decalcified heads were fixed with 4% paraformaldehyde-PBS solution for 4 hrs or overnight. We used standard hematoxylin and eosin to examine tissue morphology. For trichrome staining, samples were first stained with Azocarmine for 1 hr, and then stained with Orange G and Aniline Blue for 2 hrs.
Immunohistochemistry
Tissue was prepared by 4% formaldehyde fixation of whole embryos or heads, which were paraffin-wax-embedded and sectioned at a thickness of 7 µm. The antigens were retrieved by autoclaving in Tris-HCl buffer (pH 9.0) for 5 min. Primary antibodies against the following proteins were used: Amelogenin (Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:300), Ameloblastin (Santa Cruz, 1:300), Dspp (Santa Cruz, 1:300), Ki67 (Abcam, Cambridge, UK, 1:500), lunatic-fringe (Santa Cruz, 1:150). Normal IgG was used as a negative control. The IHC experiments were repeated at least twice, with the wild-type mice serving as positive controls. All sections (wild-type and mutant) were place on one slide and processed together under the same conditions. Primary antibodies were diluted in TBS/0.1% Triton X-100/5% goat serum/1% BSA, incubated overnight at 4°C, and detected with a biotinylated goat anti-rabbit IgG conjugate (1:200; Vector Labs, Burlingame, CA, USA), avidin-biotin complex formation (Vector Labs), and an AEC Staining Kit (Sigma, St. Louis, MO, USA).
Results
Differential Expression of microRNAs in Incisors vs. Molars and Dental Epithelial vs. Mesenchyme Tissues
To identify microRNAs expressed during late-stage tooth development, we isolated P0 mouse incisor and molar total RNAs. MicroRNA microarrays revealed discrete sets of microRNAs expressed in molars and incisors at this stage (Appendix Table). The incisor was further separated into epithelium and mesenchyme, and microRNA microarrays found differentially expressed microRNAs between these 2 compartments (Table). Furthermore, specific microRNAs were identified in the cervical loop region of developing incisors [microRNA and transcription factor target (mechanism) database at crantoomir.org].
Table.
MicroRNA Expression Profile of P0 Incisor Epithelium and Mesenchyme*
| microRNAs Highly Expressed in P0 Epithelium |
microRNAs Highly Expressed in P0 Mesenchyme |
||||
|---|---|---|---|---|---|
| miRNA | Signal Intensity | Frequency | miRNA | Signal Intensity | Frequency |
| mmu-miR-709 | 18,459 | 10.51% | mmu-miR-709 | 35,250 | 8.43% |
| mmu-miR-26a | 10,130 | 5.77% | mmu-let-7a | 30,518 | 7.30% |
| mmu-let-7a | 9,903 | 5.64% | mmu-let-7f | 28,725 | 6.87% |
| mmu-miR-23b | 8,692 | 4.95% | mmu-let-7c | 25,462 | 6.09% |
| mmu-let-7c | 8,502 | 4.84% | mmu-let-7d | 22,079 | 5.28% |
| mmu-let-7f | 7,409 | 4.22% | mmu-miR-26a | 19,400 | 4.64% |
| mmu-miR-1894-3p | 6,700 | 3.81% | mmu-miR-199a-3p | 18,567 | 4.44% |
| mmu-let-7b | 6,619 | 3.77% | mmu-let-7b | 17,570 | 4.20% |
| mmu-let-7d | 6,307 | 3.59% | mmu-miR-705 | 11,445 | 2.74% |
| mmu-miR-200c | 5,774 | 3.29% | mmu-miR-762 | 11,332 | 2.71% |
| mmu-miR-23a | 5,557 | 3.16% | mmu-miR-23b | 10,711 | 2.56% |
| mmu-miR-125b-5p | 5,482 | 3.12% | mmu-miR-125b-5p | 10,494 | 2.51% |
| mmu-miR-205 | 4,868 | 2.77% | mmu-let-7i | 9,600 | 2.29% |
| mmu-miR-24 | 4,709 | 2.68% | mmu-miR-214 | 9,312 | 2.23% |
Bold: more than three-fold difference in frequency compared with other cell compartments.
The highest-expressed microRNAs in P0 dental epithelium compared with P0 dental mesenchyme. Frequency: the percentage of individual microRNA signal intensity among total microRNA signal intensity. Craniofacial-specific microRNAs have been cloned and transgenic mice made. For more information, visit our database at crantoomir.org.
Knockout of Epithelial Mature microRNAs Resulted in Branched and Multiple Incisors Lacking Enamel
Our microRNA screening results led us to investigate the overall function of microRNAs in tooth development. Dicer1, an RNase III endonuclease essential for maturation of microRNAs, was conditionally knocked out in tooth epithelial cells with the Pitx2-Cre. Pitx2 is expressed in the oral ectoderm as early as E10.5 and is restricted to the oral and dental epithelial cell lineages (Mucchielli et al., 1997; Hjalt et al., 2000; Liu et al., 2003). Analyses of the Pitx2-Cre/Dicer1f/f (cKO) mice revealed severely malformed incisors upon eruption. The one-month-old wild-type mouse incisor became acinaciform because of the deposition of enamel at the labial side, resulting in an incisor with a yellow-brown color (Fig. 1a). In contrast, the Pitx2-Cre/Dicer1 cKO incisor was straight, relatively thin, and white in color (Fig. 1b). Interestingly, there were multiple incisors in the lower and upper jaws (Fig. 1b). Incisors were excised from the lower jaw, and we observed a branched incisor (denoted by asterisks, Fig. 1b). This was a completely new anomaly, and to understand this branching process, we sectioned through decalcified heads of two-week-old Pitx2-Cre/Dicer1 cKO mice. Surprisingly, we found that one incisor branched from another incisor due to the formation of a connecting alternative cervical loop or new stem cell niche (Appendix Figs. 2a, 2b).
Figure 1.
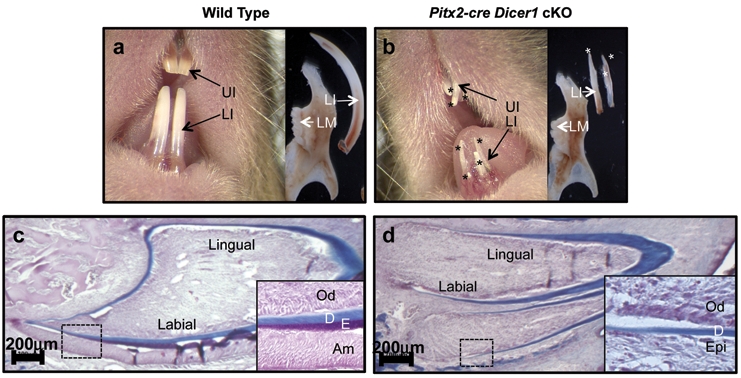
Absence of enamel and formation of supernumerary incisors in Pitx2-Cre/Dicer1 cKO. (a) Two yellow-brown lower incisors (LI) and 2 yellow-brown upper incisors (UI) in one-month-old wild-type (WT) mice. (b) Supernumerary white incisors (asterisk) in one-month-old Pitx2-Cre/Dicer1 cKO mice. (c,d) Trichrome staining of two-week-old WT and Pitx2-Cre/Dicer1 cKO incisor. A complete absence of enamel (E, staining for red) in Dicer1 cKO incisor. Dentin (D, staining for blue) is present at both sides of the incisor in Dicer1 cKO, the same as in WT. Note that in Dicer1 cKO, odontoblast (Od) differentiation is relatively unaffected compared with ameloblasts (Am). UI, upper incisors; LI, lower incisors; LM, lower molars; Od, odontoblasts; Am, ameloblasts; D, dentin; E, enamel; Epi, epithelium.
The loss of labial-lingual asymmetry and white color indicated an enamel formation defect in the Pitx2-Cre/Dicer1 cKO incisor. To demonstrate an enamel defect, we used the trichrome staining method, which stains dentin and enamel to blue and red, respectively. In the two-week-old Pitx2-Cre/Dicer1 cKO mice, we found dentin formation on both sides, similar to wild-type; however, enamel was absent (Fig. 1d). To confirm the lack of enamel in the Pitx2-Cre/Dicer1 cKO mice, we took microradiographs of three-month-old wild-type and Pitx2-Cre/Dicer1 cKO incisors using a Faxitron X-ray system. Consistent with trichrome staining results, the Pitx2-Cre/Dicer1 cKO incisor completely lacked enamel (Appendix Figs. 3g, 3h).
MicroRNAs Control Molar Cusp and Palate Formation
The Pitx2-Cre/Dicer1 cKO molars were small, abnormal, and lacking cusps compared with wild-type molars (Appendix Fig. 3f). Sections through E16.5 and P0 Pitx2-Cre/Dicer1 cKO heads revealed abnormal molar morphology with a lack of cusp formation (Appendix Figs. 3b, 3d). Analysis of these data indicated that microRNAs control molar size and cusp formation. Analysis of these data, taken together, revealed an important role for microRNAs in tooth size, shape, and patterning.
To understand the role of the neural-crest-derived dental mesenchyme microRNAs in tooth development, we crossed the Wnt1-Cre mouse with the Dicer1f/f mouse. This mouse is embryonic-lethal, and at E17.5 it has an extremely severe craniofacial phenotype devoid of many craniofacial structures and a complete loss of tooth morphogenesis (Appendix Fig. 4a). Sections through the oral cavity revealed severe malformations, with a lack of a defined tongue, oral cavity, and mandible (Appendix Fig. 4c).
Interestingly, some Pitx2-Cre/Dicer1 cKO mice had a cleft palate, demonstrating a role for microRNAs in palate fusion processes (Appendix Fig. 4f).
MicroRNAs Regulate Dental Epithelial Cell Differentiation
The lack of enamel suggested an epithelial (ameloblast) differentiation defect in the Pitx2-Cre/Dicer1 cKO incisor. We sectioned through E14.5 and P0 wild-type and Pitx2-Cre/Dicer1 cKO incisors to reveal an ameloblast differentiation defect. At the E14.5 early stage of incisor development, the wild-type epithelial cells were becoming polarized and ordered; however, in the Dicer 1 cKO, the epithelial cells were misshapen and poorly polarized (Fig. 2b). Secretory ameloblast cells are tall and polarized, as shown on the labial side (bottom of each slide) of P0 wild-type incisors (Figs. 2c, 2e, 2g). However, only short and poorly polarized ameloblast cells were found on the labial side of the Pitx2-Cre/Dicer1 cKO incisor (Figs. 2d, 2f, 2h). Conversely, the cervical loops (stem cell niche) were significantly expanded in the Pitx2-Cre/Dicer1 cKO incisor. Pitx2-Cre is not expressed in the mesenchyme; however, odontoblast differentiation appears to be delayed compared with that in wild-type mice. Interestingly, dentin was present in the Pitx2-Cre/Dicer1 cKO incisor, although dentin formation was affected (Appendix Fig. 3h). The correct formation of dentin requires reciprocal interactions between dental epithelium and mesenchyme.
Figure 2.
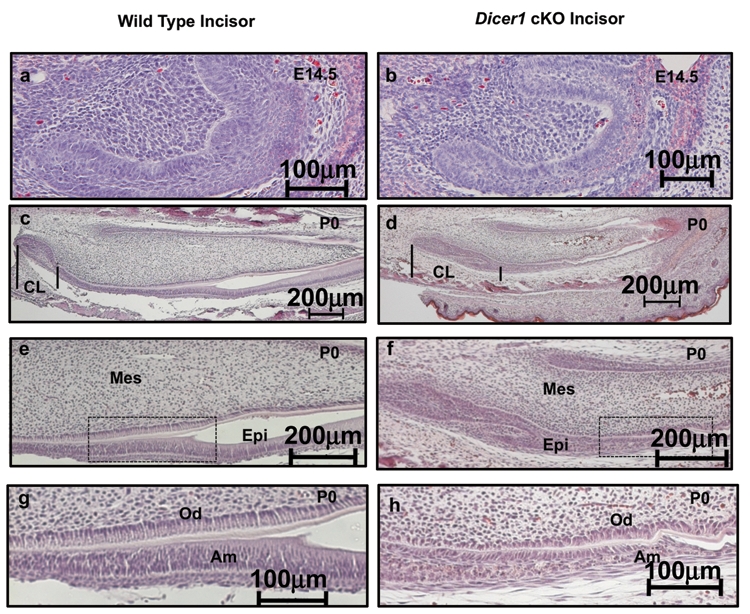
Expanded cervical loop region of incisors and repressed ameloblast differentiation in Pitx2-Cre/Dicer1 cKO. (a,b) Sagittal sections of E14.5 lower incisors, demonstrating smaller incisors in the Pitx2-Cre/Dicer1 cKO mice. (c,d) At P0, the expanded cervical loops and reduced epithelial cell differentiation were clearly observed in the mutant. Note that the cervical loop regions (CL, where tooth epithelial stem cells are located) of Pitx2-Cre/Dicer1 cKO are expanded compared with those in WT mice (lines bracket the CL). (e,g) In the WT lower incisor, tall and polarized ameloblast cells are present on the labial side. (f,h) In the Dicer1 cKO lower incisor, ameloblast cells are short and poorly polarized. (e,f) Higher magnification of (c,d). (g,h) Higher magnification of (e,f) boxed region. CL, cervical loop; Mes, mesenchyme; Epi, epithelium; Od, odontoblasts; Am, ameloblasts.
Analyses of Ameloblast Differentiation Markers
For further investigation of the ameloblast differentiation process in the Pitx2-Cre/Dicer1 cKO mouse, markers of different stages were used to determine the stages of ameloblast differentiation. Amelogenin is secreted by secretory-stage ameloblasts, which constitute 80-90% of total enamel protein, and is expressed transiently in odontoblasts (Paine et al., 2003; Hu et al., 2007). Another major enamel protein is ameloblastin, which constitutes roughly 5% of enamel protein (Fukumoto et al., 2004). Both amelogenin and ameloblastin were strongly expressed at the anterior end of the labial side of the P0 wild-type incisor (Figs. 3a, 3c). In contrast, amelogenin and ameloblastin expression was dramatically decreased in the Pitx2-Cre/Dicer1 cKO incisor (Figs. 3b, 3d). DNA microarray and real-time PCR analyses confirmed the dramatic decrease of amelogenin and ameloblastin transcripts in the Pitx2-Cre/Dicer1 cKO incisor. Dentin sialophosphoprotein (DSPP) was expressed predominantly by odontoblasts and transiently by pre-secretory-stage ameloblasts, while secretory-stage ameloblasts did not express DSPP (Fig. 3e) (Fleischmannova et al., 2008). In the Pitx2-Cre/Dicer1 cKO incisor, DSPP was clearly expressed in the mesenchyme (Fig. 3f). In the epithelium, only a few cells were observed expressing DSPP (Fig. 3f). Similar to P0, the P4 incisors revealed a dramatic decrease of amelogenin and ameloblastin (Appendix Figs. 5b, 5d), implying that loss of microRNAs led to an ameloblast differentiation defect rather than to a delay in developmental fate.
Figure 3.
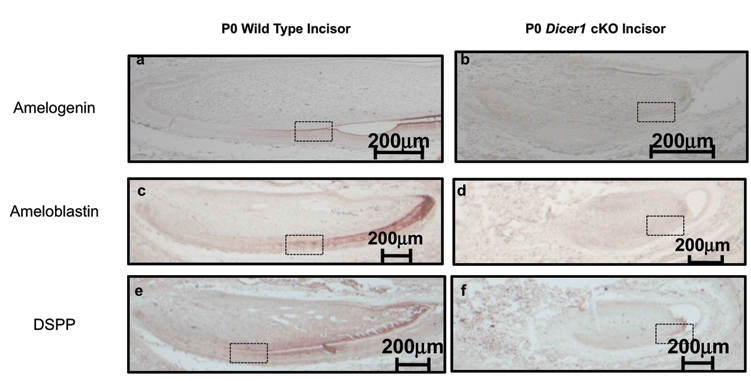
Ameloblast differentiation is repressed in the Pitx2-Cre/Dicer1 cKO. (a-f) Ameloblast differentiation markers observed by immunohistochemistry of newborn (P0) wild-type and Pitx2-Cre/Dicer1 cKO lower incisors. (a-d) Secretory-stage ameloblast marker: Amelogenin and Ameloblastin. Note the dramatic decrease of amelogenin and ameloblastin in Pitx2-Cre/Dicer1 cKO compared with WT. (e,f) Pre-secretory-stage ameloblast marker: DSPP. DSPP was expressed in odontoblasts; however, it was expressed only in pre-secretory-stage ameloblasts. In Pitx2-Cre/Dicer1 cKO, DSPP was clearly expressed in odontoblasts. There are also a few cells in the epithelium compartment that express DSPP (arrows). Higher-magnification photos are shown in Appendix Fig. 6.
When dental epithelial cells become pre-secretory-stage ameloblasts, they are post-mitotic cells (Appendix Fig. 1). The Ki-67 protein is a cellular marker for proliferation and is present in all proliferative cells, but not post-mitotic cells. In wild-type P0 incisors, Ki-67 was strongly expressed at the posterior region, but was gradually lost at the anterior end of the labial side (Appendix Fig. 6g). In the Pitx2-Cre/Dicer1 cKO P0 incisor, we see a similar pattern of Ki-67 expression (Appendix Fig. 6h). Analysis of these data suggests that cells proliferate without miRNAs. Lack of enamel is probably due to repression of cell differentiation, but not due to a loss of cell proliferation. Because the stem cell niche (cervical loop) is expanded in the Pitx2-Cre/Dicer1 cKO incisor, we asked whether there were more undifferentiated cells or stem cells in the cKO incisor than in the wild-type. Lunatic-fringe (Lfng) is a marker for undifferentiated tooth epithelial cells (Harada and Ohshima, 2004). In the wild-type P0 incisors, Lfng staining identified a few cells in the cervical loop region (Appendix Figs. 6i, 6k). However, Lfng was dramatically up-regulated in P0 Pitx2-Cre/DicerI cKO incisors. The expanded cervical loop and anterior regions revealed increased Lfng expression (Appendix Figs. 6j, 6l). Thus, the lack of microRNAs appeared to repress tooth epithelial cell differentiation.
We used genome-wide DNA microarrays to identify genes differentially expressed in Pitx2-Cre/Dicer1 cKO compared with wild-type incisors. These arrays revealed approximately 350 genes consistently up-regulated in repeat experiments. The BMP inhibitors Noggin and Follistatin were increased, and real-time PCR confirmed the up-regulation of these genes in the mutant mice (data not shown). Ectopic expression of Noggin and Follistatin has been shown to inhibit ameloblast differentiation. Analysis of these data suggests that microRNAs target BMP inhibitors like Noggin and Follistatin in normal tooth development, to allow BMP signals to induce ameloblast differentiation.
Discussion
Previous studies have shown that murine Dicer 1 is required for normal embryonic development and stem cell maintenance (Bernstein et al., 2003). Dicer1-deficient ES cells formed emybroid body-like structures, but failed to differentiate in multiple assays (Kanellopoulou et al., 2005). Furthermore, this differentiation defect was rescued in Dicer1-reconstructed clones. Analysis of these data, taken together with our results, suggests that microRNAs could be a major regulator of stem cell proliferation and differentiation during tooth development.
By conditionally knocking out the microRNAs in the dental epithelium, the cervical loop regions were expanded with proliferating progenitor cells that did not fully differentiate in vivo. These proliferative progenitor cells gave rise to extra incisors, including branched incisors. The branched incisor phenotype appeared to originate from an expansion of the lingual cervical loop stem cells.
Extra incisors were formed in the neonate mouse. Four-week-old mice had more incisors (6 lower incisors, 4 upper incisors) than two-week-old mice (4 lower incisors, 3 upper incisors), indicating that this is a continual process regulated by microRNAs. Because the murine molars do not contain a stem cell niche since they do not grow continuously, they have defects in enamel formation due to the lack of ameloblast differentiation. Furthermore, due to the lack of differentiation, the progenitor cells continued to proliferate and were capable of forming multiple erupted incisors after birth. MicroRNA screens have shown that small specific sets of microRNAs are expressed in the incisor vs. molar and epithelium vs. mesenchyme. Furthermore, the cervical loop region contains specific sets of microRNAs that regulate cell differentiation. We have identified a code of microRNAs required for tooth patterning, size, and shape, and we are currently screening for microRNA expression at earlier stages.
Supplementary Material
Acknowledgments
We thank Drs. Tord A. Hjalt (University of Lund, Sweden) and Robert Schwartz (TAMHSC-IBT) for reagents and technical advice, and members of the Amendt laboratory for helpful discussions.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
Support for this research was provided from grant DE 13941 from the National Institute of Dental and Craniofacial Research to Brad A. Amendt.
References
- Bartel DP. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. (2003). Dicer is essential for mouse development. Nat Genet 35:215-217 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. (2004). MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83-86 [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8:1323-1326 [DOI] [PubMed] [Google Scholar]
- Fleischmannova J, Matalova E, Tucker AS, Sharpe PT. (2008). Mouse models of tooth abnormalities. Eur J Oral Sci 116:1-10 [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol 167:973-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP. (2002). Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development 129:5323-5337 [DOI] [PubMed] [Google Scholar]
- Harada H, Ohshima H. (2004). New perspectives on tooth development and the dental stem cell niche. Arch Histol Cytol 67:1-11 [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. (1999). Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol 147:105-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522-531 [DOI] [PubMed] [Google Scholar]
- Hjalt TA, Semina EV, Amendt BA, Murray JC. (2000). The Pitx2 protein in mouse development. Dev Dyn 218:195-200 [DOI] [PubMed] [Google Scholar]
- Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. (2007). Enamel formation and amelogenesis imperfecta. Cells Tissues Organs 186:78-85 [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, et al. (2008). An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135:377-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15-20 [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Lu MF, Martin JF. (2003). Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development 130:6375-6385 [DOI] [PubMed] [Google Scholar]
- Mucchielli ML, Mitsiadis TA, Raffo S, Brunet JF, Proust JP, Goridis C. (1997). Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev Biol 189:275-284 [DOI] [PubMed] [Google Scholar]
- Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. (2003). A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem 278:19447-19452 [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, MacDonald PE, et al. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432:226-230 [DOI] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y. (2007). Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. (1998). Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med 9:128-161 [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, et al. (2007). An integrated gene regulatory network controls stem cell proliferation in teeth. Plos Biol 5:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Keys DN, Au-Young JK, Chen C. (2008). MicroRNAs in embryonic stem cells. J Cell Physiol 218:251-255 [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. (2005). MicroRNA function in animal development. FEBS Lett 579:5911-5922 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129:303-317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


