Abstract
Accurately corresponding a population of human cortical surfaces provides important shape information for the diagnosis of many brain diseases. This problem is very challenging due to the highly convoluted nature of cortical surfaces. Pairwise methods using a fixed template may not handle well the case when a target cortical surface is substantially different from the template. In this paper, we develop a new method to organize the population of cortical surfaces into pairs with high shape similarity and only correspond such similar pairs to achieve a higher accuracy. In particular, we use the geometric information to identify co-located gyri and sulci for defining a new measure of shape similarity. We conduct experiments on 40 instances of the cortical surface, resulting in an improved performance over several existing shape-correspondence methods.
1 Introduction
The cerebral cortex plays a key role in human brain functions such as memory, attention, perceptual awareness, thought, language, and consciousness. Different cortical folding or cortical thickness have been correlated with various brain diseases, including schizophrenia, Alzheimer’s disease, depression, and multiple sclerosis. For identifying the disease-effected cortical regions using neuroimages, it is important to bring all individual cortical surfaces into the common space for comparison (between normal and abnormal groups). To achieve this, one way is to identify the correspondences between different cortical surfaces. Then, the corresponding cortical thickness (or folding features) can be compared at different parts of the brain cortex, thus facilitating the detection of the statistically significant regions that are related to the specific diseases.
Multiple 3D shape correspondence has been studied by many researchers [1, 2, 4, 9]. Some of them correspond each shape instance in the population to a fixed template [1, 2] in a pairwise way, which may produce large errors when a target shape instance is substantially different from the template. Other methods consider the entire population simultaneously [4, 9, 11, 12]. Such groupwise methods usually require complex optimization schemes with high computing complexity and may not be guaranteed to produce the desirable optimal solutions. Specific brain mapping models and algorithms have also been developed [5, 6, 14] to identify corresponded landmarks on cortical surfaces. Most of them are based on geometric and anatomic features and their performances are highly dependent on certain pre-processing steps, such as the accurate extraction of the gyri and sulci.
Recently, Munsell et al. [10] suggested a shape organization approach for improving shape correspondence. The basic idea is to organize the shape population into a rooted tree, where each node represents a shape instance and a parent-child pair represents two similar shape instances. Shape correspondence starts from the root and propagates to its children, ending at the leaf nodes. Particularly, in [10], a minimum spanning tree (MST) is constructed, where the tree-edge weight describes the pairwise shape dissimilarity. However, the correspondence error accumulates during the propagation in this approach. While such an accumulation error is not obvious when corresponding simple 2D shape instances [10], it becomes serious in corresponding complex 3D shape instances, such as cortical surfaces, as revealed by the experiments in Section 4. Similar approach of shape organization was also used for the interactive navigation of an image database [8].
In this paper, we develop a new method of shape organization for multiple cortical surface correspondence. As in [10], it organizes the whole shape population into a tree. However, we not only require each parent-child pair to describe similar shape instances, but also control the height of the tree to reduce the accumulation error in the propagation. In this paper, we use the Freesurfer software (http://surfer.nmr.mgh.harvard.edu) to co-register a pair of cortical surfaces [7], with which we define the pairwise shape similarity and build the correspondence between a parent-child pair for the propagation. In the experiments, we compare the performance of the proposed method to the pairwise method using a fixed template, the groupwise method developed in [11, 12], and the MST-based shape-organization method developed in [10].
2 Problem Description
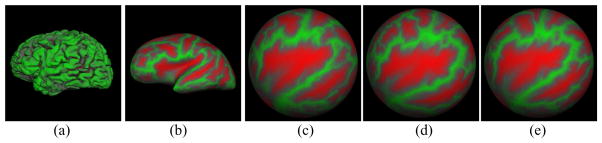
Each cortical surface consists of two hemispheres, which are usually corresponded independently. We represent each hemisphere, or subject, S by (i) point cloud extracted from MR images representing the pial surface, (ii) triangle mesh constructed from the point cloud, as shown in Fig. 1(a), and (iii) spherical representation S̃, as shown in Fig. 1(c), which is the spherical mapping of the triangle mesh. These can be obtained from a given MR image using the method of [3] available in the Freesurfer software. The goal of correspondence is to identify N corresponded landmarks across a population of subjects S1, S2, …, SM.
Fig. 1.
An illustration of the left hemisphere of cortical surfaces and their co-registration. (a) The pial surface of a subject. (b) The inflated version of the pial surface (a). (c) The spherical mapping, S̃, of (a). (d) Spherical mapping of a second subject. (e) Deformed version of (d) after co-registration with (c).
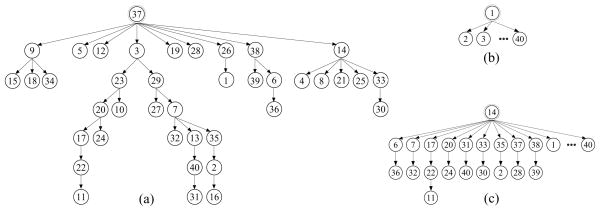
As mentioned above, we use shape organization to facilitate shape correspondence in this paper. In general, it consists of the following steps. First, the given M subjects are organized into a tree, e.g., the ones shown in Fig. 2, where each node represent a subject. Second, we take the subject represented by the root as the template and sample a set of N landmarks on the template. Third, for each child of the root, we treat the subject represented by this node as the target and match the target to the template by identifying N corresponded landmarks on the target. Fourth, each target (with corresponded landmarks) in the previous step is then treated as the new template to match its own child subjects in the tree by identifying the N corresponded landmarks. This pairwise matching process is repeated until propagated to the subjects represented by the leaf nodes, which leads to the final corresponded landmarks across all the subjects.
Fig. 2.
The shape-organization trees constructed from LONI LPBA40 dataset using (a) the MST-based method in [10], (b) the pairwise method with a fixed template, and (c) the proposed method. In (b) and (c), the ellipsis represents all the unlisted subjects, which are directly linked to the root as its children.
Clearly, the structure of the constructed tree is important to the performance of the resulting shape correspondence. If we select one subject as the root and set all the other subjects as the children of the root, as shown in Fig. 2(b), the above method is reduced to the widely used pairwise shape correspondence method with a fixed template. The method may lead to large errors when there exist subjects that are substantially different from the template and cannot be accurately corresponded to the template using a pairwise matching method. In [10], an MST is constructed such that each parent-child pair represents very similar subjects. This way, we only need to match very similar subjects, which is relatively simple and can be achieved in high accuracy. However, the constructed MST may have a larger height, as shown in Fig. 2(a), and the correspondence error may accumulate in the propagation and finally affect the consistency of landmarks as revealed in the later experiments.
3 Proposed Method
In this section, we describe a new strategy for constructing the shape-organization tree and apply it for cortical surface correspondence. We focus on addressing the following two problems: (i) measuring the shape dissimilarity between a pair of subjects and (ii) constructing the tree to organize a set of subjects.
3.1 Shape Dissimilarity using Registration
We use the Freesurfer software to obtain the continuous co-registration between a pair of subjects. Specifically, this software implements the method described in [7]. In registering the subjects Si and Sj, this method takes as input their spherical representations S̃i and S̃j and delivers an output spherical representation , which is a deformed version of S̃j on the sphere and is co-registered with S̃i. For example, the spherical representation shown in Fig. 1(e) is the deformed version of the subject shown in Fig. 1(d) after co-registration with the subject shown in Fig. 1(c). With this co-registration, we can uniformly sample K co-registered points on both spherical representations S̃i and and define the dissimilarity between these two subjects by
| (1) |
where ci(k) = 0 if the curvature value at the k-th sampled point on pial surface i is negative and ci(k) = 1 otherwise. This dissimilarity measure reflects whether the co-registered points from these two subjects show consistent presence of gyri and sulci. Considering the numerical sensitivity involved in computing curvature values, we do not use the exact curvature values in defining this dissimilarity measure.
3.2 Shape Organization using a Low-Height Tree
In contrast to the MST used in [10], we combine two preferences in constructing the shape-organization tree: (i) the parent-child pair represents similar subjects and (ii) the constructed tree has a low height, which reduces the propagation length and therefore, reduces the accumulation error. We first construct a fully connected graph G, with each node representing a subject and the edge weight between two nodes being the dissimilarity of the subjects represented by these two nodes. We then prune the edges in G sequentially in the descending order of their weights, until one more pruning will make the graph disconnected. From the pruned graph G′, we construct the tree T using the following steps:
Find the node with the largest number of incident edges in G′ and add it to the initially empty T as the root.
Let VT and V̄T be the sets of nodes that are in T and not in T respectively.
If V̄T ≠ ∅, for each node in V̄T with edge links to nodes in VT in G′, we add this node to T as a child to its linked node in VT with the smallest edge weight. Note that, in this step, we check every node in V̄T for possible additions to T without updating VT and V̄T. This way, the height of T will only increase by one after all possible node additions in this step.
Update VT and V̄T and go back to Step 3 until V̄T = ∅.
An example of a tree constructed using this method is shown in Fig. 2(c), which will be discussed in more detail in the later experiments.
As for the pairwise subject correspondence in the propagation, we use the Freesurfer software to continuously co-register the template and the target subjects. This registration maps the N landmarks on the template to N points on the target and we simply take them as the corresponded landmarks.
4 Experiments
For experiments, we use the LONI Probabilistic Brain Atlas (LPBA40) dataset, which consists of 40 3D MR images of the brain [13]. On each image, 56 anatomic structures have been identified by labeling all the relevant voxels. We extract the cortical surface from these MR images using Freesurfer for all 40 cases and then take the left hemisphere of the extracted cortical surface as the test subjects. For the pairwise shape dissimilarity (1), we uniformly sample the angular-spherical coordinates, together with two poles, to construct K = 4, 952 co-registered points on each subject.
To evaluate the performance, we use anatomic-structure labels provided with this dataset: we check whether the corresponded landmarks across the population show consistent labels. First, for each identified landmark on each subject, we find its label by searching for the closest labeled voxel in the original image. Second, for each set of corresponded landmarks across the population, e.g., the first landmarks on all 40 subjects, we perform a majority voting to find the label shared by the largest number of subjects. We use this majority label as the true label for these 40 landmarks. This way, for the j-th landmark on the subject i, we have its label and true label . If , we find on image i the closest voxel to such that this voxel has a label . We calculate the Euclidean distance between and this closest voxel as the error for landmark . If , we simply set . We then define the average error on the subject as , the average error for the set of j-th landmarks on all subjects as , and use these errors to evaluate the correspondence performance.
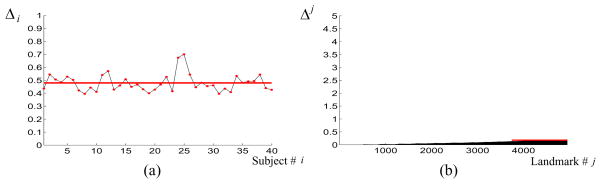
Figure 2(c) is the low-height tree constructed in our experiment using the proposed method, where subject 14 is chosen as the root and we uniformly sample the angular-spherical coordinates, together with two poles, to construct N = 4, 952 landmarks on this subject to start the propagation. We also define total average error as . Figure 3 shows the the Δ average errors Δi for all 40 subjects and Δj for all 4, 952 landmarks, with the horizontal lines being the total average error Δ = 0.479.
Fig. 3.
Average of the error in terms of (a) each subject and (b) each set of corresponded landmarks, for the correspondence obtained by the proposed method.
To further evaluate the proposed method, we conduct a quantitative comparison with three other methods: (M1) a pairwise method with a fixed template, (M2) the groupwise method developed in [11, 12], and (M3) the MST-based shape-organization method developed in [10]. For M1, we choose a subject as the fixed template and then correspond all the other 39 subjects to this template, as illustrated in Fig. 2(b). By selecting different templates, the resulting total average error Δ ranges from 0.509 to 0.684. For the proposed method, the total average Δ = 0.479 (the horizontal lines in Fig. 3) is better than the result from M1 using the best template (subject 29). Note that, in practice, we do not have ground-truth labels and we may not be able to find this best template.
For M2, we downloaded the source code from http://www.ia.unc.edu/dev/tutorials/InstallLib/index.htm and also identify N = 4, 952 landmarks on each subject, by including the sulcal depth as an attribute in the similarity metric defined in this code. As shown in Table 1, its resulting total average error Δ = 0.791 is also higher than the proposed method. M2 is a groupwise method and by reading all subjects at once, it takes 11GB of memory in our experiments, while the proposed method takes no more than 2GB of memory.
Table 1.
Average errors produced by the proposed method and three comparison methods. The variances and standard deviations are calculated over Δi, i = 1, 2, …, M and Δj, j = 1, 2, …, N respectively. The numbers for M1 come from the best template (subject 29) that leads to smallest total average error Δ.
| In terms of Subject | In terms of Landmark | |||||||
|---|---|---|---|---|---|---|---|---|
| M1(best) | M2 | M3 | Proposed | M1(best) | M2 | M3 | Proposed | |
| Total Average (Δ) | 0.509 | 0.791 | 0.532 | 0.479 | 0.509 | 0.791 | 0.532 | 0.479 |
| Variance | 0.006 | 0.073 | 0.010 | 0.004 | 0.333 | 0.717 | 0.352 | 0.266 |
| Standard Deviation | 0.082 | 0.270 | 0.100 | 0.067 | 0.577 | 0.846 | 0.594 | 0.515 |
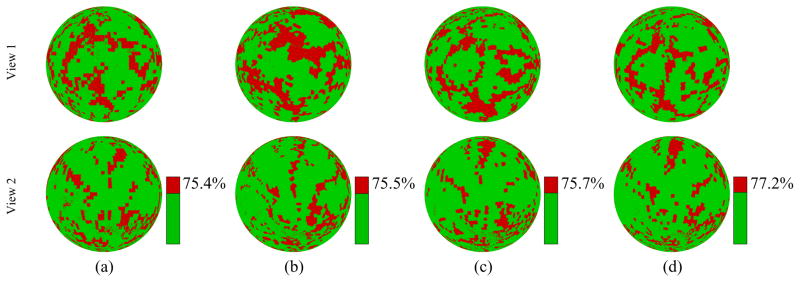
For M3, we use the same dissimilarity measure in Section 3.1 to construct an MST as shown in Fig. 2(a) and also use uniformly sampled N = 4, 952 landmarks on the root subject (subject 37) to start the propagation. From Table 1, its resulting total average error Δ = 0.532 is also higher than the proposed method. This is mainly caused by the accumulation error during the propagation. Figure 4 visualizes the error for all 4, 952 landmarks on subject 18, by using each of the three comparison methods and the proposed method: red area indicates and green area indicates . This is shown on the spherical representation of the subject for clarity. These experiments show that the corresponded landmarks identified by the proposed method show better consistency in terms of the true labels than the other three comparison methods.
Fig. 4.
A visualization of the error on the spherical mapping of one subject (subject 18): red area indicates and green area indicates . The top and bottom rows show the views that are the same as and diametrically opposite to the one used in Fig. 1(c), respectively. (a–d) the results from M1, M2, M3, and the proposed method, with green area accounting for 75.4%, 75.5%, 75.7% and 77.2% of the sphere, respectively.
5 Conclusion
We have developed a new method for corresponding highly convoluted 3D cortical surfaces. A new shape similarity measure between a pair of cortical surfaces was developed by using Freesurfer co-registration results. The cortical surfaces are then organized into a low-height tree where parent-child pairs represent similar subjects. Multiple shape correspondence was obtained by propagating the pairwise correspondence from the root to the leaves in the constructed tree. Experiments on 40 LONI LPBA40 images showed that the proposed method produces a better performance than three other existing shape-correspondence methods. The shape dissimilarity measure based on the Freesurfer registration is computationally expensive. In the future, we plan to develop more efficient algorithms to measure such pairwise dissimilarity for shape organization.
Acknowledgments
This work was funded, in part, by AFOSR FA9550-07-1-0250, NSF IIS0951754, NIH EB006733, EB008374, EB009634, and MH088520. Experiments were conducted on a shared memory computer funded by NSF CNS0708391. We would like to thank Ipek Oguz for help with the code of M2.
Contributor Information
Pahal Dalal, Email: dalalpk@email.sc.edu.
Feng Shi, Email: fengshi@med.unc.edu.
Dinggang Shen, Email: dgshen@med.unc.edu.
Song Wang, Email: songwang@cec.sc.edu.
References
- 1.Chui H, Rangarajan A. A new algorithm for non-rigid point matching. CVPR; 2000. pp. 44–51. [Google Scholar]
- 2.Dalal P, Munsell B, Wang S, Tang J, Oliver K, Ninomiya H, Zhou X, Fujita H. A fast 3D correspondence method for statistical shape modeling. CVPR; 2007. [Google Scholar]
- 3.Dale A, Fischl B, Sereno M. Cortical surface-based analysis I: Segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 4.Davies R, Twining C, Cootes T, Waterton J, Taylor C. A minimum description length approach to statistical shape modeling. IEEE TMI. 2002;21(5):525–537. doi: 10.1109/TMI.2002.1009388. [DOI] [PubMed] [Google Scholar]
- 5.Essen DCV. Surface-based approaches to spatial localization and registration in primate cerebral cortex. NeuroImage. 2004;23:S97–S107. doi: 10.1016/j.neuroimage.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Faugeras O, et al. Variational, geometric, and statistical methods for modeling brain anatomy and function. NeuroImage. 2004;23:S46–S55. doi: 10.1016/j.neuroimage.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Fischl B, Sereno M, Dale A. Cortical surface-based analysis II: Inflation, flat-tening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 8.Joshi S, Horn JV, Toga A. Interactive exploration of neuroanatomical meta-spaces. Frontiers in Neuroinformatics. 2009;3 doi: 10.3389/neuro.11.038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotcheff A, Taylor C. Automatic construction of eigen-shape models by genetic algorithm. IPMI; 1997. pp. 1–14. [Google Scholar]
- 10.Munsell B, Temlyakov A, Wang S. Fast multiple shape correspondence by pre-organizing shape instances. CVPR; 2009. pp. 840–847. [Google Scholar]
- 11.Oguz I, Cates J, Fletcher T, Whitaker R, Cool D, Aylward S, Styner M. Cortical correspondence using entropy-based particle systems and local features. ISBI; 2008. pp. 1637–1640. [Google Scholar]
- 12.Oguz I, Niethammer M, Cates J, Whitaker R, Fletcher T, Vachet C, Styner M. Cortical correspondence with probabilistic fiber connectivity. IPMI; 2009. pp. 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shattuck D, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr K, Poldrack R, Bilder R, Toga A. Construction of a 3D probabilistic atlas of human cortical structures. NeuroImage. 2007;39(3):1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosun D, Rettmann M, Han X, Tao X, Xu C, Resnick S, Pham D, Prince J. Cortical surface segmentation and mapping. NeuroImage. 2004;23:S108–S118. doi: 10.1016/j.neuroimage.2004.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]