Abstract
A magnetoresistive biosensing platform based on a single magnetic tunnel junction (MTJ) scanning probe and DNA microarrays labeled with magnetic particles has been developed to provide an inexpensive, sensitive and reliable detection of DNA. The biosensing platform was demonstrated on a DNA microarray assay for quantifying bacteria capable of degrading methyl tertiary-butyl ether (MTBE), where concentrations as low as 10 pM were detectable. Synthetic probe bacterial DNA was immobilized on a microarray glass slide surface, hybridized with the 48 base pair long biotinylated target DNA and subsequently incubated with streptavidin-coated 2.8 μm diameter magnetic particles. The biosensing platform then makes use of a micron-sized MTJ sensor that was raster scanned across a 3 mm by 5 mm glass slide area to capture the stray magnetic field from the tagged DNA and extract two dimensional magnetic field images of the microarray. The magnetic field output is then averaged over each 100 μm diameter DNA array spot to extract the magnetic spot intensity, analogous to the fluorescence spot intensity used in conventional optical scanners. The magnetic scanning result is compared with results from a commercial laser scanner and particle coverage optical counting to demonstrate the dynamic range and linear sensitivity of the biosensing platform as a potentially inexpensive, sensitive and portable alternative for DNA microarray detection for field applications.
Keywords: Magnetic Tunnel Junction, DNA Microarray, Scanometric, Magnetic Particles, Methyl Tertiary Butyl Ether
1. Introduction
Sequence-selective DNA detection has become an increasingly important tool used in understanding molecular biology and unraveling the genetic basis of disease. By employing DNA microarrays in a highly multiplex and parallel format, the arrays and its accompanying imaging platform enable the high throughput biological detection required in areas such as medical diagnostics (Clarke et al. 2001; Heller 2002), drug discovery (Chin and Kong 2002) and environmental monitoring (Loy et al. 2002). DNA and protein microarrays represent two of the best examples of how microfabrication technology enables hybridization and detection to be carried out in microminiaturized, highly-parallel formats.
The gold standard in DNA microarray technology is the fluorescence based solid-phase assay format. Although hampered by the need for sophisticated fluorescence microscopes/scanners as well as strongly environment-dependent quantum yields of the fluorescent tags, no other scheme for readout is likely to supersede fluorescence detection for standard use in centralized bulk laboratory facilities. However, the current instrumentation has limitations in both flexibility and portability, two important factors for the assay and sensing platform to be deployed in field applications.
Other assay formats have been developed based on either label-free methodologies (Anderson et al. 2008; Piscevic et al. 1995) or using other types of labels such as gold nanoparticles (Reichert et al. 2000; Taton et al. 2000), quantum dots (Gerion et al. 2003) and magnetic particles (Baselt et al. 1998). Although the label-free approach is attractive for its simple operating protocol that eliminates undesirable effects such as steric impediments and instabilities of the labels, the signal detection mechanism is more complicated. Since both target and probe are of the same nature, and often both contribute to the signal, incremental changes due to binding or hybridization events are extremely difficult to sense. On the other hand, magnetic labels have many advantageous characteristics such as robustness, non-toxicity and stable properties over time. The ability to manipulate these particles with on-chip or external magnetic fields (Graham et al. 2005; Wirix-Speetjens et al. 2007), together with the absence of magnetic background in most biological materials, make magnetic particles labeling an extremely promising approach.
Biosensors using highly sensitive magnetic sensor technology are among the most sensitive and amenable to miniaturization. Biosensor chips based on magnetic sensor arrays have been proposed to create easy-to-use portable lab-on-a-chip devices that are sensitive, versatile and easily integrated with standard silicon integrated-circuit technology. In a typical magnetic array chip, underneath each magnetically-labeled DNA spot sits a magnetoresistive (MR) sensor, using either giant magnetoresistive (GMR), spin valves or tunnel junction sensor designs (Ferreira et al. 2003; Megens and Prins 2005; Rife et al. 2003; Shen et al. 2008). Here, the number of sensors and DNA spots are equal; an array format containing 103 DNA spots will thus require 103 sensors for a complete analysis. This increases the cost and complexity of the biochip and introduces many technical challenges in designing the biochip for efficient multiplexing. In addition, a good passivation layer between the sensor and the biological solutions is required to ensure sensor integrity and prevent spurious signals due to contamination of the sensor surface, while stringent washing processes are needed in order to reuse the expensive sensor substrate (Schotter et al. 2004). Moreover, errors due to sensor offset drift occurring during the hybridization or washing process complicates the discrimination of true signals even when reference sensors are present on the array (Graham et al. 2004; Xu et al. 2008).
In this paper we describe a different biosensing platform that combines the advantages of stable magnetic labels and highly sensitive MR sensor in a scanning probe format similar to that of a hard disk drive. The biosensing platform is comprised of a reusable magnetic “reader” unit and low-cost disposable assay substrates printed with DNA probes and labeled with magnetic tags. The reader consists of a single mechanically-scanned MR sensor and associated readout electronics, while the passive, disposable substrate retains the use of the standard glass microscope slide used in conventional fluorescence based assays. By using a single micron-sized sensor to scan across the whole glass slide, large assay areas can be imaged with high spatial resolution. Moreover, the same sensor can be applied to different assays by changing just the disposable substrate without the need to expose the sensor to any biochemical or washing solutions. This platform aims to demonstrate the potential of using small sensitive sensors in a scanning format resembling a hard disk drive to develop a portable biosensing platform for on site environmental monitoring.
In this approach, a true magnetic measurement of the DNA array is captured, free from sensor offset errors since the same sensor images both the magnetically-labeled spot and the label-free background. This sensing platform is used to quantify Methylibium petroleiphilum PM1 bacteria, an organism that is naturally present at aquifers contaminated with methyl tert-butyl ether (MTBE) and have been linked to the biodegradation of MTBE (Hristova et al. 2003).
2. Materials and Methods
2.1 Oligonucleotide probe design
Linear DNA oligoprobes were designed based on the Methylibium petroleiphilum PM1 16S rDNA gene sequence. Table 1 shows the single stranded DNA sequences for both the commercially synthesized 22-mer oligonulecotide probe and 48-mer complementary target. All DNA sequences were purchased from Integrated DNA Technologies, (IDT, IL). The probes were amino modified at the 5' end to enable covalent immobilization of probes onto a solid support. The targets used for fluorescent labeling were tagged with a cyanine 3 (Cy3) fluorophore, while targets for magnetic labeling had a biotinylated end which serves as the interaction point with the streptavidin coated magnetic particles. A C-C mismatch was inserted into the middle of the sequence at two locations to create a 2 base pair mismatch target.
Table 1.
PM1 bacterial 16S rDNA sequences used in microarray experiments. The sequence in the target that is complementary to the probe is in boldface. Mismatched base pairs are underlined, while the 5' end of targets used for magnetic labeling and fluorescent labeling is in italic.
| Oligonucleotide | Sequence |
|---|---|
| Probe: | 5' – NH2 – ACA CGA GCT GAC GAC GGC CAT G – 3' |
| Perfect Match Target (Magnetic): | 3'-TTGTAGAGTGC TGT GCT CGA CTG CTG CCG GTA C GTCGTGGACACAAGA–biotin-5' |
| Perfect Match Target (Fluorescent): | 3'-TTGTAGAGTGC TGT GCT CGA CTG CTG CCG GTA C GTCGTGGACACAAGA–Cy3-5' |
| 2 basepairs Mismatch Target | 3'-TTGTAGAGTGC TGT GCT CCA CTG CTC CCG GTA C GTCGTGCACACAAGA–biotin-5' |
2.2 Surface functionalization and spotting
Expoxysilane glass slides, Nexterion® E (Schott, NY) were used as the base substrate for DNA microarrays. The epoxysilane coating serves as a uniform surface for biomolecule immobilization via the covalent interaction between epoxide end groups in the coating and nucleophilic groups on the DNA probe, as illustrated in Fig. 1a. These glass slides were spotted with a periodic array of ~100 μm diameter spots using a Lucidea Microarrayer (GE, Piscataway, NJ). All probes were spotted at 20 mM concentration in 1× Nexterion™ spotting buffer solution. The spotted arrays were left in a humid chamber (70% humidity) overnight and washed the next day. The washing procedure includes four washes using 6× SSPE and 0.01% Tween. These arrays were subsequently dried in nitrogen gas for later use.
Fig. 1.
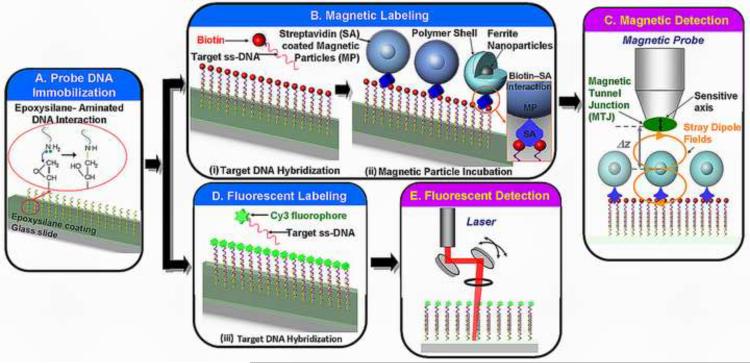
Bioassay protocol for magnetic and fluorescent labeled DNA microarrays. (A) The oligonucleotide probe was immobilized on epoxysilane glass slides. The microarrays were subjected to labeling with either (B) magnetic particles that involved a two-step process of: (i) hybridization with biotinylated target DNA and (ii) incubation with streptavidin coated magnetic particles, or (D) Cy3 fluorophore conjugated target DNA. The DNA duplex structure was then scanned using (C) a magnetic tunnel junction (MTJ) probe close to the magnetic particles for the magnetically labeled microarray or (D) the commercial laser scanner for the fluorescently labeled microarrays.
2.3 Oligonucleotide hybridization
The complete sequence of hybridization and incubation steps to produce both magnetically and fluorescently labeled arrays is summarized in Fig. 1a–b. A prehybridization blocking step (1% bovine serum albumin (BSA), 1× Denhardt solution, 1× saline sodium citrate (SSC) and 0.1% sodium dodecyl sulfate (SDS)) was performed to reduce undesirable non-specific binding of target DNA on the slides. After a washing step using 1× SSC and drying by dry nitrogen gas, hybridization was allowed to occur between the 5' end biotinylated target and the probe DNA in 20 μl volumes of the hybridization buffer on the glass slide. This hybridization step was carried out in an oven at 42 °C with 70% humidity for ~8 hours. 10 μl of the suspended target at varying concentrations was denatured by heating at 95°C for 3 min in a heat block. A quick spin in a microcentrifuge was performed to re-suspend the oligonucleotide probes in 20 μl of the hybridization buffer (DIG Easy Hyb buffer, Roche, Switzerland). After the end of the hybridization step, three consecutive rinsing steps using 1× SSC, 0.5× SSC and 0.1× SSC were performed to remove the remaining non-hybridized DNA. The glass slide was then dried in nitrogen. The whole process ends at this step for the fluorescently labeled microarrays, while the magnetically labeled microarrays require an additional incubation step with the magnetic particles.
2.4 Magnetic particle incubation
The magnetic labels used in the bioassay were streptavidin-coated 2.8 μm diameter paramagnetic particles (Dynal M280, Invitrogen, CA). These particles are superparamagnetic and contain ~12% Fe2O3 magnetic material. Particles suspended in a phosphate buffered saline (1× PBS and 0.1% SDS) were incubated with the hybridized biotinylated target DNA on the glass for one hour at ambient temperature to allow for the bio-specific interactions between biotin and streptavidin to occur. A series of washing steps were performed in 1× phosphate buffered saline (PBS) to remove any unbound magnetic particles from the glass slide.
2.5 Fluorescent scanning
Fluorescently-labeled microarray slides were scanned with an Axon laser scanner Genepix 4000B (Molecular Devices, CA) using a 532 nm laser excitation source for the Cy3 fluorophore tag. The fluorescence signals were quantified using the GenePix Pro6 microarray image analysis software (Molecular Devices, CA). A grid of individual circles defining the location of each spot on the array was superimposed on the image to designate each fluorescent spot to be quantified. The mean signal intensity was determined for each spot. In addition, the mean signal intensity of the local background area surrounding the spots was also extracted.
2.6 Optical imaging or counting
To provide a reference for the analysis of magnetically-labeled microarrays, a MATLAB (Mathworks) particle counting algorithm was developed to count the number of particles visible in optical images of each DNA spot. After thresholding to create a binary (black and white) image, the optical images were analyzed to extract the percentage of the spot covered with magnetic particles by segmenting, measuring and counting objects.
2.7 Magnetic tunnel junction (MTJ) sensor
The magnetoresistive sensor used in the magnetic scanning setup is a current perpendicular to plane tunnel junction sensor (Gallagher et al. 1997) manufactured by MicroMagnetics Inc (STJ-030). The tunnel junction is elliptically shaped with a dimension of 2 μm by 4 μm. The device has a high magnetoresistive ratio (ΔR/R ~ 20%), a zero-field resistance of ~1.3 kΩ, and a measured sensitivity of 0.6 %/mT over the operating field range of ±100 mT. The sensor has a magnesium oxide (MgO) tunnel barrier layer sandwiched between two ferromagnetic layers. The magnetization of one of the ferromagnetic layer is pinned along a fixed axis, while the other can freely rotate in response to external field. When imaging magnetically-labeled microarrays, stray fields from the magnetic labels rotate the magnetization of the free layer which in turn changes the sensor resistance.
2.8 Magnetic scanning and detection
The glass microarray slide was seated on top of a XY translation stage (Prior Scientific Inc.), with the magnetic sensor mounted on a probe and positioned via a piezoelectric stage (Nanocube, PI L.P.) for precise control of the sensor-sample distance (Δz), as shown in Fig. 1c. Because magnetic field intensity is a strong function of this distance, each individual scan area was limited to less than 3 mm × 5 mm to minimize the effect of height variations due to bowing or warpage of the glass slide surface. First-order leveling of the tip/tilt of the slide relative to the scanning system was performed using optical measurements.
During the magnetic image collection, an external DC magnetic field (Bdc) of 7.8 mT was applied to magnetize the paramagnetic beads on the microarray slide. The MTJ sensor is (to first order) insensitive to the field in this axis due to shape anisotropy, hence preventing the applied field from saturating the sensor. A constant current source of 0.1 mA was applied to bias the MTJ element and a bridge configuration with subsequent amplification and filtering using a preamplifier (Stanford Research SR560) was used to extract the magnetic signature. Synchronized stage motion and data acquisition was achieved through LabView (National Instruments) software-based PC control, while post-acquisition image processing was done in MATLAB using custom-built algorithms.
Using a 1500 μm/s scan speed in the x-direction and a y-step resolution of 2 μm, an area of 4200 μm by 2800 μm was imaged with the sensor. To improve the signal-to-noise ratio, a total of N = 4 scans were made across each x-axis scan line, and the magnetic images captured were formed by averaging the four measurements. In the first series of scans, the MTJ sensor was positioned away from the array and an initial background scan was collected to measure any spatial variations in the DC magnetic field. Subsequently, in the second series of scans, the sensor was moved to a height of ~20 μm away from the surface of the microarray. The background due to any misalignment or variation in the DC field detected in the first scan was low pass filtered and subtracted from the second scan to capture the data solely due to the magnetic contribution from the particles.
3. Results and Discussion
3.1 Imaging of magnetically labeled DNA microarrays
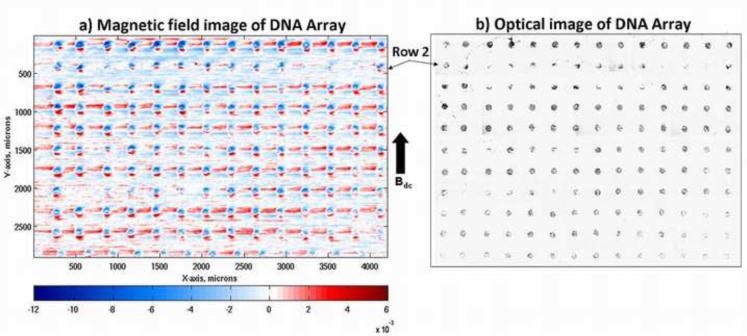
After washing and drying, the sample was scanned with the magnetic scanning setup described above. Optical micrographs of a 11 × 14 array of magnetically-labeled DNA spots along with magnetic field intensity images of these spots are shown in Fig. 2a and 2b, respectively. Each of these 100 μm diameter DNA spots appears as a single magnetic dipole aligned with respect to the applied external field, Bdc. The large sensor-to-sample distance blurs the field signature from each individual bead, resulting in an ensemble image of the field arising from all the beads on each spot. The two images were taken at the same location on the slide, where the sparsely populated DNA spots in the second row of the optical image correspond to the weak magnetic spots in the second row of the magnetic image. These images show a good correlation between the densities of the magnetic particles and the strength of the representative dipole of each DNA spot, and illustrate a high contrast of the particle coverage between complementary binding at the DNA spots and non-complementary DNA binding at the non-printed areas on the glass.
Fig. 2.
(a) A two-dimensional magnetic map of a magnetically labeled DNA microarray obtained with an applied bias magnetic field, Bdc of 7.8 mT and a scan speed of 1.5 mm/s. Each circular dipole indicates a 100 μm diameter DNA spot, with the high background noise coming from the nonspecifically bound particles, and (b) Optical image of the same magnetically labeled DNA microarray showing the good correlation between the spot intensities in both magnetic and optical images.
Quantification of the measured magnetic field from each spot was carried out via image processing techniques analogous to those used in the analysis of fluorescent microarrays. After gridding and segmenting each image, the spots were analyzed by extracting from the foreground data a quantitative measure of the magnetic field from the particles bound within the spot. This measure can then be correlated to the concentration of target DNA. Often, during the incubation process, the magnetic particles were positioned randomly within the 100 μm spot depending upon a number of factors such as particle weight, accessibility of the biotinylated end of the target to streptavidin on the particle, and steric hindrance. Hence, the distribution of particles within each spot was not uniform and clusters of beads appeared as multiple localized peaks/valleys in the magnetic image, reflecting the bead distribution captured in the optical image. A parameter, Bave, that considers the average field across a 100 μm DNA spot, was used as a measurand for spot-to-spot comparison.
The upper limit of the dynamic range of the assay is defined by the maximum number of 2.8 μm beads that can be packed into a 100 μm DNA spot while the noise floor of the instrument determines the lower limit. The background of the magnetic signal can be partially attributed to the presence of non-specifically bound beads distributed randomly on the glass surface. At the current sensor-bead spacing of ~20 μm, the noise limited resolution is approximately 27 beads, corresponding to an average field of 800 nT within the 100 μm diameter spot.
3.2 Fluorescent versus magnetic labeled DNA microarray
In order to examine the quantitative potential of using magnetic labeling and the magnetic scanning setup, we systematically studied the signal dependence on varying concentrations of target DNA using the assay protocol described above. Glass slides were printed with six replicate arrays consisting of 240 spots within each array. Each replicate array was assayed with the same amount (0.5 mg) of magnetic particles, but was exposed to a different target DNA concentration during the hybridization step.
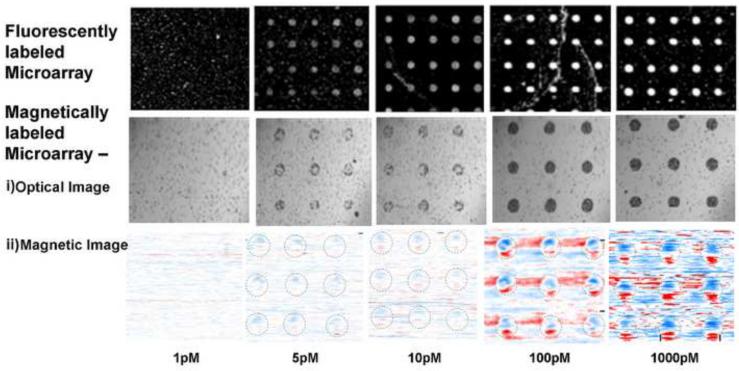
Fig. 3 shows a summary of the magnetic images extracted from two different glass slides, each hybridized with the same range of target DNA concentrations but assayed separately with fluorescent and magnetic probes. The fluorescent response signal and the density of magnetic particles increased with the target concentration in the range of 1 pM to 1 nM. The magnetic images plotted using the same color scale also indicate that the magnetic signal exhibits a positive correlation with concentration. The magnetic signal for a 5 pM concentration is visible at a SNR of ~1.5 while the signal for 100 pM has a higher SNR of ~4.5.
Fig. 3.
Microarray quantification of the PM1 bacterial 16S rDNA using fluorescent and magnetic labels over five different concentration zones. The (i) optical and (ii) magnetic images were taken of the magnetically labeled arrays. For the magnetic images, the data are plotted with the same color intensity scale to show the contrast of the DNA spots increasing with the density of particles over the dynamic range.
3.3 Dynamic Range and Limits of Detection
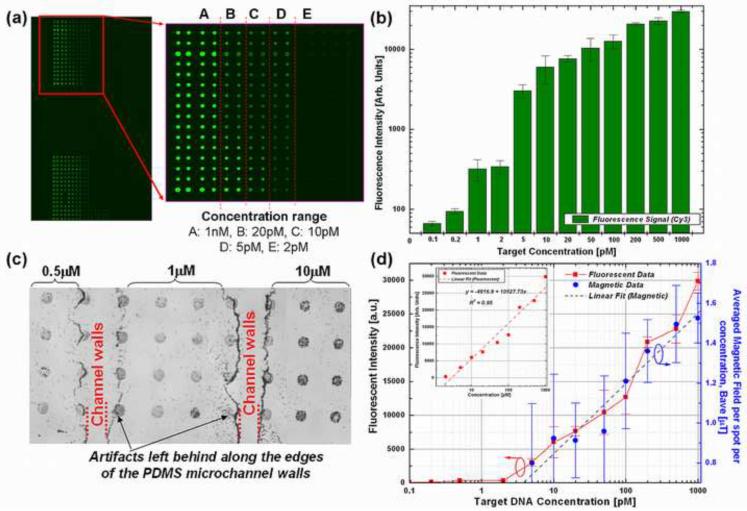
Parallel hybridization experiments were conducted to determine the detection limit and dynamic range of this magnetic scanning platform and eliminate any slide-to-slide variation in the fluorescent or magnetic signal strength due to differences in washing procedure. Each of these slides were printed with the same concentration of probe DNA but hybridized with multiple target DNA concentrations. In order for parallel hybridization of multiple samples to take place within a millimeter-scale region on the slide, the pre-hybridization blocking and hybridization processes were performed using microfluidic techniques. Microchannels molded into poly-dimethylsiloxane (PDMS) were reversibly bonded to the glass slide and removed once the hybridization process was completed. Each microchannel carried a 5 μl volume of a specific concentration of target DNA, covering an array of >30 spots, as illustrated in Fig. 4a and c. A more stringent flagging of the DNA spots disregarded spots located near or at the channel edge (see Fig. 4c) distorted by the channel imprints, thus reducing the number of spots used in the analysis of each concentration to ~20.
Fig. 4.
(a) Fluorescent micrographs of the fluorescently-labeled microarray slide containing arrays of 100 μm diameter DNA probe spots after hybridization with varying concentrations of Cy3 target DNA, (b) fluorescent intensity vs. different Cy3 target concentration (varying from 1 pM to 10 nM) at different regions on the same glass slide using microfluidic channel hybridization. Volume of target DNA used per microchannel is 5 μl. (c) Optical micrograph of the magnetically labeled microarrays containing the same 100 μm spots after hybridization with varying target concentrations, (d) Experimental data from fluorescently labeled and magnetically labeled DNA microarrays scanned using a fluorescent scanner and the prototype magnetic scanning microscope setup (respectively) plotted along two different axes, showing the same linear response to target DNA concentration. The signal and error bars represent the average and standard deviation based on fluorescent intensity measurement from 20 spots per concentration zone.
For the fluorescently labeled microarrays, a linear trend in the target DNA concentration against fluorescent intensity is observable in Fig. 4b. Increasing the concentration of ss-target DNA alone enhanced the signal of hybridization very sharply, where the fluorescence intensity reached a maximum at a concentration of 1000 pM. The saturation is due in part to the settings of the PMT gain (350) during imaging. The spot intensities showed a linear relationship (R2 = 0.95) with target DNA over the dynamic range of interest, as shown in Fig. 4c. The background fluorescence for these measurements are at a level of 30–50 AU, while the signal from a 1 pM target DNA can be clearly visualized with an SNR of ~5.
Figure 4d shows the experimental results extracted from the fluorescent assay laser scanning readout and magnetic scanning plotted together along two different axes. Each data point shown in the plots is the averaged fluorescent data and the Bave data from 20 spots in the same concentration range. Both show a linear response to the target DNA concentration demonstrating that magnetic scanning is a feasible alternative to fluorescent scanning.
The lower limit of the dynamic range however is defined by the detection limit of the scanning system and MTJ sensor. The detection limit is defined as the lowest number of beads discernable from the background noise of the image. With the 20 μm sensor-bead spacing employed here, the limit is ~30 particles, corresponding to a target concentration of ~5 pM.
The larger scatter in the magnetic data as compared to the fluorescent data is due to the high binding variation resulting from the use of micron-sized magnetic particles, as well as the manual rinsing and drying processes in the bioassay protocol. These larger magnetic particles suffer from larger drag effects during the rinsing steps as compared to the much smaller fluorescent particles. By using smaller magnetic labels, steric hindrance at the solid glass slide surface can be reduced to improve the binding efficiency of the bioassay, while the reduced drag minimizes assay variability due to sample preparation.
In addition, using magnetic forces in the final rinsing steps, either through magnetic gradient fields generated by on-chip current lines or an external magnetic field ensure that a more repeatable bioconjugation step can be obtained.
Unlike optical-scanners where the lens captures far-field fluorescence, the near-field nature of magnetic bead detection renders this mechanically-scanned biosensing platform subject to a set of different challenges in establishing and controlling the sensor-to-sample distance. The most important limitation of the MTJ sensor is the large distance of ~18 μm between the elliptical tunnel junction and the polished edge of the silicon probe. Since the magnetic field has a cubic dependence on the sensor-to-sample spacing (1/Δz3), this distance greatly affects the detection limit of the system. Reducing this distance would allow smaller magnetic particles to be employed, which would in turn help to reduce steric hindrance effects and enable a broader dynamic range.
At the lower detection limit, as the concentration of the particles decreases, the effects of other noise sources becomes more prominent. These noise sources come from the total distance variability, Δz that can arise from both stage motion and the slide flatness. As the sensor-sample distance decreases, scanning stage control becomes even more critical. At this point, the scanning is performed in an open loop format, resulting in a relatively large out of plane displacement of ~1 μm. To reduce this variation, sensors can be used to maintain a constant sensor-sample distance via a closed-loop feedback control. The sensor can come in the form of a reflectance probe to detect the surface of the glass slide or by making use of the existing MTJ sensor to detect the nickel thin film patterns deposited on the same glass slide on which the DNA hybridization will take place. The sensitivity of this system, which has yet to be optimized, points toward a potential method for detecting oligonucleotide targets at femtomolar concentrations.
3.4 Specificity
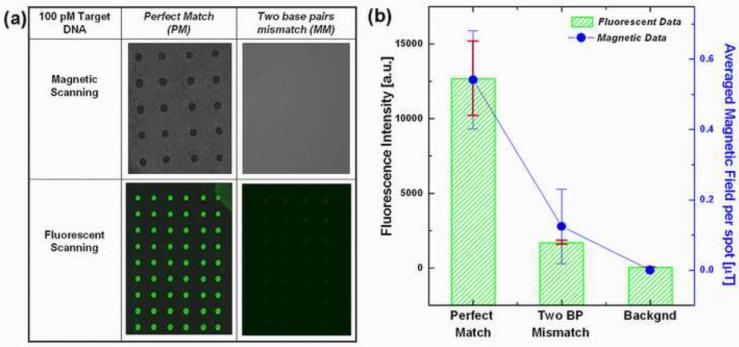
Differentiating a target DNA sequence from its congeneric sequence having only a few mismatches or identifying a mutation with single nucleotide polymorphism for genotyping represent the most stringent selectivity and specificity metrics for field-use assays and biosensing platforms. To demonstrate the specificity of this platform, a parallel comparison experiment was performed on the same glass slide under the same hybridization, incubation and detection procedures. In this set of experiments, the same 100 pM concentration of two different targets: a perfectly-matched complementary target (PM) and a two base pair mismatched (MM) target were used. The mismatch target was designed to represent an example of the 16s rDNA gene from a phylogenetically related strain to M. petroleiphilum PM1. We observed more than seven-fold smaller fluorescence intensity in the mismatched targets (compared to perfectly matched probe sequence) for the fluorescently labeled microarrays, as shown in Fig. 5.
Fig. 5.
Sequence selectivity of both the fluorescent and magnetic labeled assay, where 100 pM of target DNA with complementary and two base pairs mismatch sequences were hybridized with probe DNA spotted on the glass slide. (a) Table of optical micrographs showing the magnetically and fluorescently labeled DNA microarrays having different label concentrations for a perfect match and two base pair mismatch targets, (b) Comparison of the fluorescent and magnetic field intensities for the perfect and mismatched targets. Points represent the average fluorescence intensities from more than 60 replicate test probe spots, from two separate glass slides under two separate labeling experiments. Error bars represent the standard deviation at each point.
The optical images from the magnetically labeled microarray also displayed a similar contrast, where the number of magnetic particles bound to DNA probe spots hybridized with the mismatch targets showed fewer than five particles per spot, which is below the present detection limit of the magnetic scanning system. The background level in fluorescence measurements results from low-level fluorescence of the unprinted surface of the slide, while the background in the magnetic data results from MTJ sensor offset. In Fig. 5b, the sensor offset is subtracted from the magnetic field data resulting in zero average background level in these measurements. Both the magnetic and fluorescent results demonstrated the specificity of the bioassay in its capability to discriminate a non-target DNA with a mismatch of only two base pairs in the sequence.
4. Conclusion
The magnetic scanometric platform with integrated highly sensitive magnetic tunnel junction sensor was developed to detect magnetically labeled DNA spots in a standard microarray format. A bioassay protocol was designed and tailored to allow DNA hybridization reactions and fluorescent dye or magnetic particles conjugation to take place on microarray glass slides in a conventional as well as PDMS microchannel format. These large arrays of 100 μm DNA spots labeled with either 2.8 μm diameter Dynal particles or Cy3 fluorescent dye were used to compare between the magnetic and the fluorescent scanning platform. Scanning the micron size MTJ sensor across the array allows both a large scanning area (> 1 cm2) and high spatial resolution (~ 1 μm). Measurements demonstrated on detecting Methylibium petroleiphilum PM1 bacterial DNA yield a detection limit of ~30 particles in a 100 μm DNA spot with high signal-to-noise ratio, three decades of dynamic range and the limit of detection estimated to be 10 pM. Although the detection limit of the current magnetic scanning system is higher as compared to the fluorescent standard, the use of smaller magnetic particles coupled with a closer sensor to sample spacing promise to dramatically increase the sensitivity of the detection. The selectivity of the bioassay and the use of a single sensor in a large area scanning format opens opportunities in the development of a fundamental technology for a low-cost high throughput portable disk-drive based bioassay system.
Acknowledgement
This work was supported by the National Science Foundation under Grant No. ECCS 0601383. Partial support has been provided by grant number 5 P42 ES004699 from the National Institute of Environmental Health Sciences (NIEHS), NIH and the contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH. The authors would like to thank J. Pfeiff from the Arraycore Facility at UC Davis for the DNA microarray printing as well as Micromagnetics Inc. for valuable discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson EP, Daniels JS, Yu H, Karhanek M, Lee TH, Davis RW, Pourmand N. Sens. Actuator B-Chem. 2008;129(1):79–86. doi: 10.1016/j.snb.2007.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselt DR, Lee GU, Natesan M, Metzger SW, Sheehan PE, Colton RJ. Biosensors and Bioelectronics. 1998;13(7–8):731–739. doi: 10.1016/s0956-5663(98)00037-2. [DOI] [PubMed] [Google Scholar]
- Chin KV, Kong ANT. Pharm. Res. 2002;19(12):1773–1778. doi: 10.1023/a:1021425004264. [DOI] [PubMed] [Google Scholar]
- Clarke PA, Poele RT, Wooster R, Workman P. Biochem. Pharmacol. 2001;62(10):1311–1336. doi: 10.1016/s0006-2952(01)00785-7. [DOI] [PubMed] [Google Scholar]
- Ferreira HA, Graham DL, Freitas PP, Cabral JMS. Journal of Applied Physics. 2003;93(10):7281–7286. [Google Scholar]
- Gallagher WJ, Parkin SSP, Lu Y, Bian XP, Marley A, Roche KP, Altman RA, Rishton SA, Jahnes C, Shaw TM, Xiao G. Journal of Applied Physics. 1997;81(8):3741–3746. [Google Scholar]
- Gerion D, Chen FQ, Kannan B, Fu AH, Parak WJ, Chen DJ, Majumdar A, Alivisatos AP. Anal. Chem. 2003;75(18):4766–4772. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- Graham DL, Ferreira HA, Feliciano N, Freitas PP, Clarke LA, Amaral MD. Sensors and Actuators B-Chemical. 2005;107(2):936–944. [Google Scholar]
- Graham DL, Ferreira HA, Freitas PP. Trends in Biotechnology. 2004;22(9):455–462. doi: 10.1016/j.tibtech.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Heller MJ. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- Hristova K, Gebreyesus B, Mackay D, Scow KA. Appl. Environ. Microbiol. 2003;69(5):2616–2623. doi: 10.1128/AEM.69.5.2616-2623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M. Appl. Environ. Microbiol. 2002;68(10)):5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens M, Prins M. Journal of Magnetism and Magnetic Materials. 2005;293:702–708. [Google Scholar]
- Piscevic D, Lawall R, Veith M, Liley M, Okahata Y, Knoll W. Applied Surface Science. 1995;90(4):425–436. [Google Scholar]
- Reichert J, Csaki A, Kohler JM, Fritzsche W. Anal. Chem. 2000;72(24):6025–6029. doi: 10.1021/ac000567y. [DOI] [PubMed] [Google Scholar]
- Rife JC, Miller MM, Sheehan PE, Tamanaha CR, Tondra M, Whitman LJ. Sensors and Actuators A - Physical. 2003;107(3):209–218. [Google Scholar]
- Schotter J, Kamp PB, Becker A, Puhler A, Reiss G, Bruckl H. Biosensors and Bioelectronics. 2004;19(10):1149–1156. doi: 10.1016/j.bios.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Shen WF, Schrag BD, Carter MJ, Xie J, Xu CJ, Sun SH, Xiao G. Journal of Applied Physics. 2008;103(7) [Google Scholar]
- Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289(5485):1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- Wirix-Speetjens R, Reekmans G, De Palma R, Liu C, Laureyn W, Borghs G. Sensors and Actuators B: Chemical. 2007;128(1):1–4. [Google Scholar]
- Xu L, Yu H, Han SJ, Osterfeld S, White RL, Pourmand N, Wang SX. IEEE Transactions on Magnetics. 2008;44(11):3989–3991. doi: 10.1109/TMAG.2008.2002795. [DOI] [PMC free article] [PubMed] [Google Scholar]