Abstract
Three heterozygous missense mutations in the motor domain of myosin 1c (Myo1c), which mediates adaptation in the inner ear, are associated with bilateral sensorineural hearing loss in humans. With transient kinetic analyses, steady-state ATPase and motility assays, and homology modeling, we studied the interaction of these mutants with nucleotide and actin using a truncated construct, Myo1c1IQ-SAH, which includes an artificial lever arm. Results indicate that mutation R156W, near switch 1, affects the nucleotide-binding pocket and the calcium binding by disrupting switch 1 movement. Mutation V252A, in the K helix of the upper 50 kDa domain, showed reduced actin affinity consistent with disruption of communication between the actin- and nucleotide-binding sites. T380M, in a Myo1c-specific insert in the HO linker, displayed aberrant changes in most kinetic parameters and uncoupling of the ATPase from motility. These data allow for an interpretation of how these mutations might affect adaptation.
Keywords: Myo1c, Myosin, Mutation, Hearing, Adaptation
Introduction
The prevailing model for mechanotransduction is one in which bundles of actin-filled stereocilia on the specialized hair cells of the inner ear deflect in unison in response to sound (cochlea) or head movement (vestibule), opening and closing transduction channels (for a recent review see [1]). Extracellular filaments known as tip links connect the tips of stereocilia to the adjacent taller stereocilia and are involved in gating of the transduction channels [2, 3]. Adaptation permits hair cells to remain sensitive to new stimuli by decreasing the response to a maintained stimulus [4]. In the case of fast adaptation, Ca2+ enters the transduction channels and closes them on the order of a few milliseconds in both cochlear and vestibular hair cells, although the mechanisms might be distinct. Slow adaptation also closes the transduction channels, but on a much slower timescale and through a mechanism distinct from that of fast adaptation [5]. In the model for slow adaptation that has prevailed for many years, an adaptation motor complex consisting of multiple myosin molecules associated with the transduction channels resets the resting tension of the gating spring [6]. During an excitatory stimulus, tension is initially high, opening the channels. The motor complex then slips down the actin cytoskeleton reducing tip link tension and allowing the channels to close (slipping adaptation). During a negative stimulus, tension is initially low and the channels close. The motor complex then ascends the cytoskeleton restoring the resting tension and reopening the channels (climbing adaptation).
The class I myosin, Myo1c, is located near the insertion point for the upper tip link into the stereociliary membrane, an electron-dense region known as the upper tip link density (UTLD) [1, 7, 8]. Based in part on studies in which adaptation was measured in utricular cells isolated from mice expressing Myo1c sensitive to inhibition by an ADP analog [9], Myo1c is thought to be the adaptation motor. Recently, new evidence indicates that the transduction channels are only present in electron-dense regions close to the insertion points of the lower tip link density (LTLD) [10], requiring some refinement of the existing model for adaptation. A new notion is that the entry of Ca2+ through a transduction channel affects the adaptation motor complex associated with the next tip link down in the same stereocilium [1].
Kinetic, structural, and single-molecule mechanical studies show that Myo1c is kinetically slow and exhibits a strain-sensing ADP-release mechanism, which allows it to adapt to mechanical load [11, 12]. Studies of the ATP-driven interaction with actin of a truncated Myo1c consisting of the motor domain and first calmodulin-binding domain (Myo1c1IQ) show that Myo1c has unusual calcium dependence [13]. Calcium binding to calmodulin associated with the first IQ domain has little effect on the steady-state ATPase or motor activity, but alters specific steps of the ATPase cycle. ATP hydrolysis is inhibited sevenfold by calcium while ADP release from acto-Myo1c is accelerated by tenfold. These two changes together would reduce the lifetime of the actin-attached states and increase the lifetime of the actin-detached state without altering the overall cycle time. In combination these properties appear to be ideal to modulate the activity of Myo1c in response to a calcium transient in the inner ear.
Unlike studies with myosins VI, VII, and VX, where the availability of mutants has allowed substantial insight into the roles of these myosins in the inner ear [14, 15], until recently no mutations in Myo1c have been associated with deafness. Mutational screening of 450 patients with bilateral sensorineural hearing loss has recently identified six heterozygous missense mutations in Myo1c in a study to evaluate Myo1c as a candidate deafness gene [16]. Three of these mutations, R156W, V252A, and T380M, reside in the motor domain of Myo1c, which contains the nucleotide- and actin-binding sites (Table 1). To investigate the effect of these mutations on the interaction of Myo1c with both actin and nucleotide, we expressed wild-type and mutant constructs consisting of the Myo1c motor domain, the first calmodulin-binding domain, and an artificial lever arm consisting of a single α-helix (Myo1c1IQ-SAH), purified these proteins and studied them using steady- and transient-state kinetics as well as in vitro motility assays. Understanding how the biochemical properties of Myo1c are affected by specific mutations allows us to predict their effects on adaptation.
Table 1.
Myo1c mutations associated with hearing loss
| Mutation | Location of mutation in motor domain | Phenotype of patientsa |
|---|---|---|
| Myo1c R156W | Immediately preceding switch 1 | Bilateral sensorineural hearing loss |
| Myo1c V252A | In helix K in the upper 50 kDa domain | Moderate bilateral sensorineural hearing loss |
| Myo1c T380M | In helix O of transducer near switch 2 | Progressive bilateral sensorineural hearing loss |
aData from Zadro et al. 2009 [16]
Materials and methods
Preparation of constructs and expression in insect cells
For protein expression in insect cells, we prepared PCR products encoding wild-type or mutant Myo1c motor domain and the first calmodulin-binding (IQ) domain (amino acids 1–725) followed by a SAH domain from myosin-X (amino acids 805–843) and both myc and FLAG tags using cDNA encoding the entire open reading frame of mouse Myo1c and myosin X (both kind gifts of T. Friedman, National Institute of Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD, USA); we refer to this construct as Myo1c1IQ-SAH. After treatment with the appropriate restriction enzymes, the purified PCR products were ligated into the pFastBacDUAL transfer vector (Gibco BRL, Gaithersburg, MD, USA) downstream of the polyhedrin promoter; the p10 promoter cloning site contained the gene coding for calmodulin [17]. The plasmids were transformed into DH5α cells and selected by antibiotic resistance. Colonies were grown, and the isolated DNA was tested for the presence of inserts by restriction analysis. The sequences of plasmids containing Myo1c inserts were confirmed with automatic sequencing using internal and vector-specific oligonucleotides. The recombinant donor plasmid was transformed into DH10Bac E. coli cells (Invitrogen, Carlsbad, CA, USA) for transposition into bacmid. Recombinant bacmid DNA was isolated by potassium acetate precipitation as described in the Bac-to-Bac Baculovirus Expression Systems Instruction Manual supplied by Invitrogen. Virus was produced by transfecting the recombinant bacmid DNA into Spondoptera frugiperda 9 (Sf9) insect cells with Cellfectin reagent (Invitrogen) then allowing the cells to grow for 3 days. Subsequently, amplified virus was used to infect Sf9 cells in suspension. Infection was allowed to proceed for 4 days, after which time cells were harvested by centrifugation. Cell pellets were either used immediately for protein isolation or frozen in liquid N2 and stored at −80°C for future use.
Protein purification
To isolate proteins expressed in insect cells, cell pellets were homogenized in 10 mM Tris, pH 7.5, 0.2 M NaCl, 4 mM MgCl2, 2 mM ATP, and protease inhibitors, then centrifuged at 183,000 g for 50 min. The supernatant was applied to an anti-FLAG column, and after washing the column, the expressed proteins were eluted with a step gradient of FLAG peptide. Fractions containing protein were identified by SDS-polyacrylamide gel electrophoresis, pooled and dialyzed against 10 mM Tris, pH 7.5, 50 mM KCl, and 1 mM DTT. The yield exceeded 2 mg purified protein/L insect cells with the pooled fraction being 2–3 ml at ~0.5 mg/ml. Proteins were either used immediately or stored at –80°C for future use.
ATPase activity
The steady-state actin-activated Mg2+-ATPase activity of wild-type and mutant Myo1c1IQ-SAH was measured using a colorimetric assay [18] in 10 mM Tris, pH 7.5, 50 mM KCl, and 1 mM MgCl2 as well as CaCl2 and EGTA to effect pCa 4 and 8.9.
Motility assays
The ability of the purified myosins to translocate actin filaments in vitro was determined as previously described [19]. Briefly, flow chambers were made by attaching nitrocellulose-coated coverslips face down to a glass microscope slide with double-sided sticky tape. Myosin was mixed with 0.05 mg/ml F-actin and 10 mM ATP for 20 min and then spun at 24,500 g and 4°C for 20 min; the supernatant was collected. The flow chambers were coated with ~12 μl of 0.05 mg/ml myc antibody (Invitrogen) in PBS for 15 min and then 75 μl of 5 mg/ml bovine serum albumin (BSA) in motility buffer [25 mM imidazole (pH 7.5), 1 mM EGTA, 25 mM KCl, 4 mM MgCl2] for 10 min to block nonspecific sites. Thirty microliters myosin supernatant was applied to the flow chamber for 15 min and then washed with motility buffer containing BSA for 10 min. Thirty microliters of ~0.14 μM rabbit skeletal muscle F-actin labeled with rhodamine phalloidin (Molecular Probes, Eugene, OR, USA) was applied to the flow chamber for 5 min. The chambers were washed with 75 μl motility buffer plus BSA before adding 50 μl motility buffer containing 0.5% methylcellulose, 3 mg/ml glucose, 20 mM DTT, 0.1 mg/ml glucose oxidase, 0.05 mg/ml catalase, and 2 mM ATP. Slides were examined at room temperature with a Nikon fluorescence microscope using a 100× Plan Fluor 1.3 oil immersion lens and an EF-4 G-1B filter cube for rhodamine (546/10 nm excitation band, 565 nm dichroic reflector, and 590 nm long-pass barrier filter) (Nikon, Melville, NY, USA). Images were captured with a Dage MTI CCD-300T-RC camera and recorded digitally using a FlashBus MV PCI bus frame grabber (Integral Technologies, Indianapolis, IN, USA). The speed of the filaments was determined using Track Points in MetaMorph bioimaging software (version 6.3r2, Universal Imaging, Downingtown, PA, USA).
Transient enzyme kinetics
Rapid kinetic measurements were taken at 20°C with a standard Hi-Tech Scientific SF-61 DX2 stopped-flow system fitted with a 75 W Xe-Hg lamp and monochromator for wavelength selection. Intrinsic tryptophan fluorescence was excited at 295 nm and emission monitored through a WG320 cutoff filter, while pyrene fluorescence was excited at 365 nm and emission monitored through a KV 389 nm cutoff filter. The stopped-flow transients were fitted to one or two exponentials as described previously by nonlinear least squares curve fitting using the Kinetic Studio software (TgK Scientific). All experiments were carried out at 20°C in 20 mM MOPS buffer, pH 7.0 containing 100 mM KCl and 5 mM MgCl2. In addition the buffer contained either 1 mM EGTA for measurements without calcium or 1.1 mM EGTA and 1.2 mM CaCl2 for measurements carried out in the presence of calcium.
Data analysis
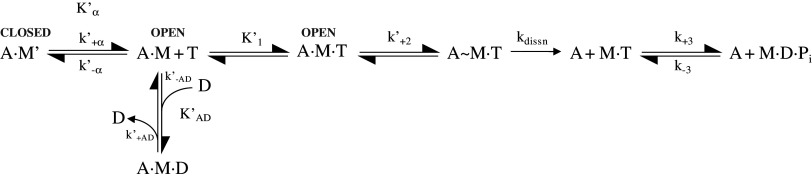
Data were analyzed as previously described [19]. In the absence of actin, the interaction of ATP and ADP with Myo1c was interpreted in terms of the seven-step mechanism for the myosin ATPase cycle (Scheme 1) in which ATP binding to myosin occurs in two steps (a binding step and a protein conformational change) followed by reversible ATP hydrolysis [20, 21]. Pi release and ADP release then occur sequentially with each dissociation event preceded by a protein isomerization. Our previous work established that ATP binding is accompanied by an increase in tryptophan fluorescence and that there is no measurable fluorescence change associated with ADP binding [13]. We assume that the fluorescence change associated with ATP binding is therefore coupled to the hydrolysis step because Myo1c has the conserved tryptophan at the end of the relay loop that is known to signal the protein conformational change associated with switch 2 closure and the hydrolysis step in other myosins [22, 23].
Scheme 1.

M myosin, T ATP, D ADP, Pi inorganic phosphate, asterisks indicate a quench in fluorescence
The results of the kinetic interaction of actin-Myo1c with nucleotide were interpreted in terms of the model in Scheme 2, which we described previously for Myo1b [24] and Myo1c [11, 13]. The model assumes that acto-Myo1c exists in two conformations, A.M and A.M′, in the absence of nucleotide and that the interconversion between the two conformations is defined by the equilibrium constant, Kα, where K′α = k′+α/k′−α. A.M′ represents the complex with the nucleotide pocket in the “closed” conformation, which must isomerize into the “open-pocket” form, A.M, before nucleotide binding/release can occur. This isomerization is presumably coupled to a swing of the converter/IQ regions of the myosin motor domain [11, 25]. The ATP-induced dissociation of actin-Myo1c is therefore biphasic [11, 13]. The fast phase represents ATP binding to A.M, and the observed rate constant is hyperbolically dependent upon ATP concentration with kobs,fast = K′1k+2[ATP]/(1 + K′1[ATP]). The slow phase has a maximum observed rate constant of k′+α and the ratio of the two amplitudes (ampfast/ampslow) defines [A.M.]/[A.M′] = K′α.
Scheme 2.
A actin, M myosin, A·M actomyosin complexes, T ATP, D ADP, Pi inorganic phosphate, rate constants denoted with ′ were determined in the presence of actin
Two similar complexes are also assumed to exist in the presence of ADP, A.M.D and A.M′.D, with an open and closed nucleotide pocket, respectively. In the case of Myo1c, the binding and release of ADP from A.M.D and A.M′.D cannot be distinguished because the rate constants governing ATP binding are slower than ADP release. The apparent ADP affinity for A.M. (Kapp) was estimated by measuring the ADP inhibition of the ATP-induced dissociation of acto-Myo1c. In the case of Myo1c, ADP is in rapid equilibrium with A.M and A.M′ on the time scale of the slow phase of the reaction. Thus, the kobs of the slow phase is reduced with a hyperbolic dependence upon ADP concentration (independent of whether the ADP is premixed with the protein or the ATP) and Kapp = (K′AD/(1 + 1/K′α), where K′AD is the affinity of A.M for ADP.
Other
Rabbit skeletal muscle actin was purified as previously described [26].
Results
Constructs used in biochemical study
In previous studies, we determined that use of a truncated form of Myo1c consisting of the motor domain and first IQ domain (Myo1c1IQ) facilitates our studies because its expression level exceeds that of full-length Myo1c, and it exhibits kinetics resembling that of native full-length Myo1c [13, 19]. Although useful for steady-state and transient-state kinetic analyses, this 1-IQ construct is not useful for motility assays because its shortened lever arm results in slow motility. To overcome this problem, we investigated the practicality of attaching a SAH domain found in myosin X [27] as an artificial lever arm much the same way that α-actinin has been used [28]. In fact, a new report shows that substitutions of IQ domains with SAH domains can be made in myosin V [29]. We originally placed the SAH domain following IQ 2 based on sequence homology between Myo1c and myosin X and found improvement in protein expression, but little, if any, improvement in motility [19] over full-length Myo1c (30 nm/s) [30]. We show here that placement of the SAH domain immediately after the first IQ domain results in a molecule that both expresses at high levels (~2 mg/L insect cells) and is able to move actin filaments at high rates (0.55 ± 0.17 μm/s). For these reasons, we expressed wild-type Myo1c and the hearing mutants as Myo1c1IQ-SAH constructs.
Steady-state ATPase activity
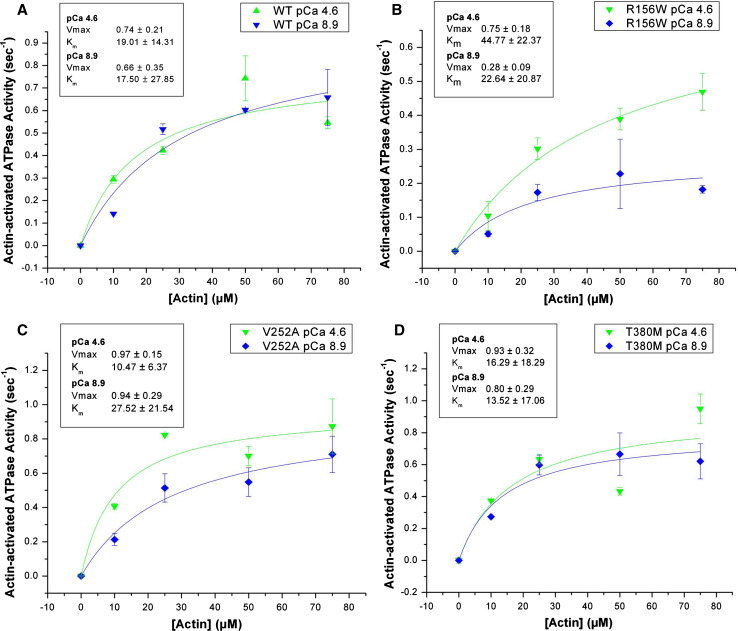
The basal ATPase of wild-type Myo1c is very low (≪0.1 s−1) and relatively insensitive to calcium. Similar results were observed for Myo1c1IQ-SAH-WT and the hearing mutants. The steady-state actin-activated Mg2+-ATPase activity of Myo1c1IQ-SAH-WT and the hearing mutants is shown in Fig. 1 and Table 2. The ATPase activity of Myo1c1IQ-SAH-WT was very similar to the construct without the SAH domain and exhibited little calcium sensitivity at pCa 4.6 vs. 8.9 (Vmax 0.74 ± 0.21 s−1 at pCa 4.6; 0.66 ± 0.35 s−1 at pCa 8.9) with Km values just below 20 μM. Each of the three mutants was activated with increasing amounts of actin and at pCa 4.6, the Vmax of the mutants was not dramatically different from that of Myo1c1IQ-SAH-WT. However, at pCa 8.9, the Vmax of Myo1c1IQ-SAH-R167 W was reduced by ~60% (Vmax 0.28 ± 0.09 s−1 at pCa 8.9 vs. 0.75 ± 0.18 s−1 at pCa 4.6). Myo1c1IQ-SAH-V252A exhibited a reduced Km of 10.47 ± 6.37 μM at pCa 4.6.
Fig. 1a–d.
Actin-activated Mg2+-ATPase activity. The Mg2+-ATPase activity of a Myo1c1IQ-SAH-WT, b Myo1c1IQ-SAH-R156W, c Myo1c1IQ-SAH-V252A, and d Myo1c1IQSAH-T380M was measured at 37°C and actin concentrations of 0–75 μM in the presence (pCa 4.6) and absence (pCa 8.9) of calcium
Table 2.
Analysis of actin-activated Mg2+-ATPase activity of Myo1c1IQ-SAH-WT and hearing mutants measured at 37°C
| pCa | Km, mean ± SD (μM) | Vmax, mean ± SD (s−1) | |
|---|---|---|---|
| Myo1c1IQ-SAH-WT | 4.6 | 19.01 ± 14.31 | 0.74 ± 0.21 |
| 8.9 | 17.50 ± 27.85 | 0.66 ± 0.35 | |
| Myo1c1IQ-SAH-R156W | 4.6 | 44.77 ± 22.37 | 0.75 ± 0.18 |
| 8.9 | 22.64 ± 20.87 | 0.28 ± 0.09 | |
| Myo1c1IQ-SAH-V252A | 4.6 | 10.47 ± 6.37 | 0.97 ± 0.15 |
| 8.9 | 27.52 ± 21.54 | 0.94 ± 0.29 | |
| Myo1c1IQ-SAH-T380M | 4.6 | 16.29 ± 18.29 | 0.93 ± 0.32 |
| 8.9 | 13.52 ± 17.06 | 0.80 ± 0.29 |
Translocation of actin
The ability of the constructs to translocate fluorescently labeled actin filaments was tested at pCa 8.9 with antibody-assisted motility assays (Table 3). Myo1c1IQ-SAH-WT translocated actin filaments at a rate of 0.55 ± 0.17 μm s−1. A 20% reduction in velocity was observed for Myo1c1IQ-SAH-R156W (0.45 ± 0.17 μm s−1). In the case of Myo1c1IQ-SAH-V252A, filaments bound to the myosin-coated surface in the absence of ATP, but no filaments were observed bound to the surface in the presence of ATP. These results are different from those for Myo1c1IQ-SAH-T380M, where filaments bound but were not translocated in ATP.
Table 3.
Translocation of rhodamine-labeled actin filaments in vitro by Myo1c1IQ-SAH-WT and hearing mutants in the absence of calcium
| Velocity of actin (number of filaments) | |
|---|---|
| Myo1c1IQ-SAH-WT | 0.55 ± 0.17 μm s−1 (n = 61) |
| Myo1c1IQ-SAH-R156W | 0.45 ± 0.11 μm s−1 (n = 34) |
| Myo1c1IQ-SAH-V252A | Filaments bound −ATP but were released from the surface +ATP |
| Myo1c1IQ-SAH-T380M | Immobile (n > 2,000) |
Motility was only measured at pCa 8.9 because at pCa 4.6 actin filaments on coverslips coated with antibody and Myo1c1IQ-SAH-WT fragmented upon the addition of ATP. The reason is unclear and cannot be simply attributed to higher motor activity in calcium vs. EGTA because the ATPase assays show little difference in activity ± calcium. Fragmentation of actin in sliding actin filament assays can occur at high myosin concentrations [31], but this also does not appear to be the cause because fragmentation only occurred in the presence of calcium. The possibility of the presence of Ca2+-activated actin-severing proteins in the actin preparation was also ruled out by actin cosedimentation assays showing that the critical concentration of actin was not affected by increasing the calcium concentration (data not shown). That fragmentation was ATP-dependent suggests that calcium has a direct effect on the motor activity of Myo1c1IQ-SAH-WT.
Transient kinetic analyses
A detailed kinetic analysis of the events in the ATPase cycles of Myo1c1IQ-SAH and acto-Myo1c1IQ-SAH was carried out essentially as previously described [13, 19]. The myosin ATPase analysis uses the intrinsic tryptophan signal from myosin to measure the apparent second order rate constant of ATP binding (assigned to K1k+2 in Scheme 1) and the maximum observed rate constant of ATP binding (assigned to k+3+k−3). The amplitude of the fluorescence change (~6%) for Myo1c1IQ-SAH-WT was similar to that previously measured for Myo1c1IQ-WT [13, 19]. For Myo1c1IQ-SAH-WT, the value of k+3+k−3 is 43 s−1 in the absence of calcium; this is reduced to 9 s−1 in the presence of calcium. The value in the absence of calcium is very similar to that found for Myo1c1IQ-WT (44.3 ± 1.4 s−1); however, the value in the presence of calcium is 30% higher than measured for Myo1c1IQ-WT (6.3 ± 0.1 s−1). The affinity of ADP for Myo1c1IQ-SAH-WT in the absence of calcium was measured by competition with ATP binding (K6K7 = 1.9 μM), and the rate constant for ADP release (k+6 = 15 s−1) was measured by displacement of ADP from myosin by an excess of ATP. Since in the case of Myo1c1IQ-WT these values were 3.3 μM and 6 s−1, respectively, it appears that events occurring at the ADP pocket are sensitive to the presence of the SAH domain. In the presence of calcium, these measurements are unreliable because the rate constants for the maximum value for both ATP binding and ADP release are similar.
The acto-myosin kinetics were studied using the pyrene label on actin to report the association and dissociation of Myo1c with actin and how this is influenced by ATP and ADP. Like Myo1c1IQ-WT, Myo1c1IQ-SAH-WT showed two phases of the ATP-induced dissociation of the complex. The maximum observed rate constants for the ATP-induced dissociation of actin from the complex for the fast phase (k′+2) and the slow phase (k′+α) as well as the ratio of the amplitudes of the two phases (K′α) are all very similar in the absence of calcium. When calcium is present, k′+2 is reduced twofold for Myo1c1IQ-WT and almost threefold for Myo1c1IQ-SAH-WT, while k′+α (2.7–4.0 s−1) and K′α (0.115–0.24) are both increased 1.5- to 2-fold in the presence of the SAH domain. The affinity of ADP for the A.M complex (K′AD) was threefold weaker in the absence of calcium (4.4 vs. 1.6 μM) when SAH was present and twofold weaker (34 vs. 17 μM) in the presence of calcium. This is consistent with ADP release (k′+AD) being accelerated tenfold for Myo1c1IQ-WT and ca. eightfold for Myo1c1IQ-SAH-WT on addition of calcium. Overall the data are consistent with our earlier conclusion that the presence of calcium on the calmodulin of the first IQ domain influences events at the ATP hydrolysis site and the ADP binding pocket. The presence of the SAH domain can modulate this interaction to a modest extent.
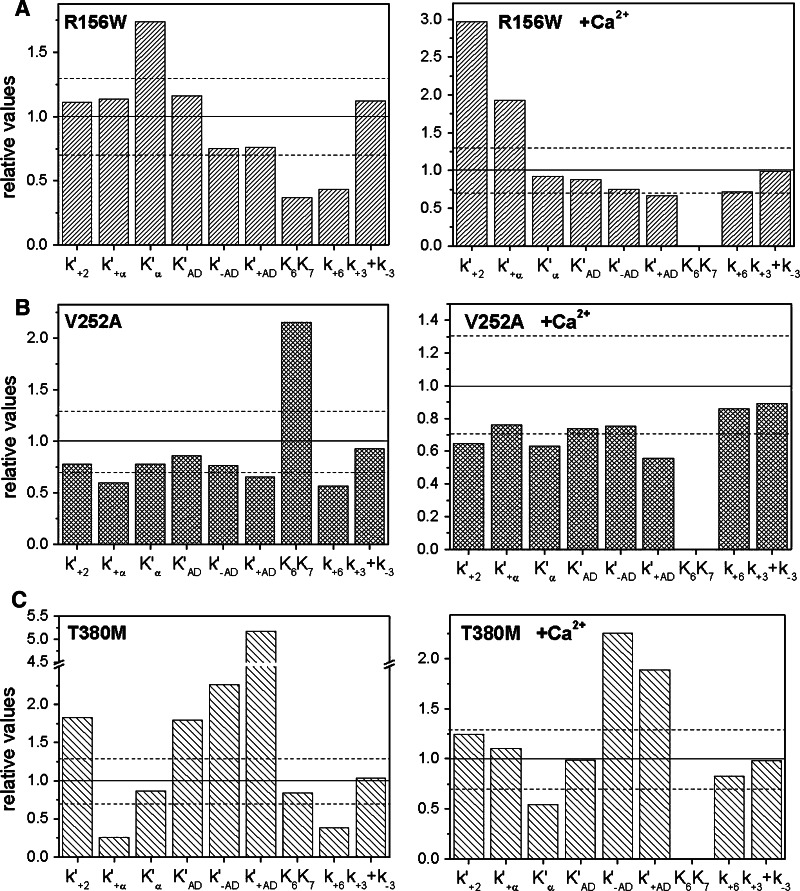
The three point mutations in the motor domain had different effects on the ATPase cycle. The data are summarized in Fig. 2 and the values are given in Table 4. In the absence of calcium most of the measured values for Myo1c1IQ-SAH-R156W were within 30% of the values for Myo1c1IQ-SAH-WT. The exceptions were the affinity for ADP (K6K7) and the rate constant of ADP release from myosin (k+6) in the absence of actin, which had 2- to 2.5-fold smaller values for Myo1c1IQ-SAH-R156W than seen for Myo1c1IQ-SAH-WT, and the value of K′α was decreased by 50% compared to Myo1c1IQ-SAH-WT. In calcium again most values were similar to those of Myo1c1IQ-SAH-WT except that the value of k′+2 was threefold larger for Myo1c1IQ-SAH-R156W than Myo1c1IQ-SAH-WT and the rate constant of the isomerization step (k′+α) increased nearly twofold. This construct showed ~30% loss in tryptophan fluorescence compared to Myo1c1IQ-SAH-WT; loss in amplitude is not unexpected since this construct contains an extra tryptophan.
Fig. 2a–c.
Transient kinetic parameters determined by stopped-flow spectroscopy for hearing mutants. Displayed are values for a Myo1c1IQ-SAH-R156W, b Myo1c1IQ-SAH-V252A, and c Myo1c1IQ-SAH-T380M, relative to those obtained for Myo1c1IQ-SAH-WT. A value of 1.0, indicated by the solid horizontal line, indicates that the two values are equal. The dashed lines represent 30% deviation from 1.0. Left EGTA, right calcium
Table 4.
Transient kinetics analyses of Myo1c1IQ-SAH and hearing mutants
| Parameter | Units | Myo1c1IQ-WT | SAH-WT | SAH-R156W | SAH-V252A | SAH-T380M | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| −Ca2+ | +Ca2+ | −Ca2+ | +Ca2+ | −Ca2+ | +Ca2+ | −Ca2+ | +Ca2+ | −Ca2+ | +Ca2+ | ||
| K′1k′+2 a | μM−1s−1 | 0.081 | 0.012 | 0.0195 | 0.0108 | 0.0555 | 0.026 | 0.023 | 0.015 | 0.123 | 0.0038 |
| K′1 | μM−1 | 1.97 | 0.602* | 0.376 | 0.780 | 0.968 | 0.645 | 0.576 | 1.69* | 1.30* | 1.34* |
| k′+2 | s−1 | 41.2 | 19.6 | 51.7 | 13.8 | 57.4 | 41.0 | 40.5 | 8.9 | 94.4 | 17.2 |
| k′+α | s−1 | 2.0 | 2.7 | 2.59 | 3.99 | 2.95 | 7.7 | 1.55 | 3.03 | 0.67 | 4.41 |
| K′α | 0.11 | 0.115 | 0.11 | 0.238 | 0.2 | 0.22 | 0.09 | 0.15 | 0.1 | 0.13 | |
| K′aAD | μM | 1.6 | 17.2 | 4.4 | 34.0 | 5.1 | 30.0 | 3.8 | 25.0 | 7.9 | 33.5 |
| k′−AD | μM−1s−1 | 1.2 | (1.2) | 1.33 | (1.33) | 1.0 | (1.0) | 1.02 | (1.0) | 3.0 | (3.0) |
| k′+AD a | s−1 | 1.9 | 20.6 | 5.8 | 45 | 4.4 | 30 | 3.8 | 25 | 30.0 | 85 |
| K6K7 | μM | 3.3* | – | 1.9 | – | 0.7 | – | 4.1 | – | 1.6* | – |
| k+6 | s−1 | 6 | 5–20 | ~15 | ~7 | 6–7 | ~5 | ~8.5 | ~6.0 | ~5.8 | ~5.8 |
| k+3+k−3 | s−1 | 44.3 | 6.3 | 42.9 | 9.3 | 48.2 | 9.2 | 40.0 | 8.3 ± 0.4 |
44.4, fast 5.4, slow |
9.09 |
Parentheses indicate values that are assumed to be independent of calcium. Asterisks indicate errors in the range 30–50%; all other errors are <20%
aCalculated. K′AD = Kapp × (1 + 1/K′α), k′+AD = K′AD × k′−AD
The values of most of the kinetic parameters of the V252A mutant, both in the absence and presence of calcium, were similar or slightly reduced in value compared to those of WT (i.e., 20–50% smaller) with one significant exception: in the absence of calcium the affinity of ADP (K6K7) was twofold weaker for Myo1c1IQ-SAH-V252A than for Myo1c1IQ-SAH-WT.
The T380M mutation showed the most significant and widespread changes in kinetic behavior of the three mutants tested. The value of k′+2 in the absence of calcium was 1.7-fold faster and k′+α was reduced to 0.2 of the value for Myo1c1IQ-SAH-WT; however, no differences in these parameters from Myo1c1IQ-SAH-WT were seen when calcium was present. The ADP affinity for A.M (K′AD) was weaker and both the ADP-binding rate constant (k′-AD) and the ADP release from A.M (k′+AD) were accelerated twofold and fivefold, respectively. The ADP release from myosin on its own (k+6) for T380M was 50% of the value seen with Myo1c1IQ-SAH-WT. Notable is the presence of an additional slow phase (kmax = 4 s−1, Aslow = 2.3%) in the protein fluorescence transients of ATP binding to Myo1c1IQ-SAH-T380M. The origin of this extra phase is unknown, but it was reproducible in kmax and amplitude. The extra phase in the transient was not present in calcium. In the presence of calcium, the K′AD was unaltered, but the rate constants for both ADP binding and ADP release for A.M were accelerated twofold as compared to SAH-WT values.
Discussion
The studies show that attachment of an artificial lever arm comprised of the SAH domain from myosin X immediately following the first IQ domain of Myo1c (at L720) has relatively modest effects on the kinetic properties of Myo1c, but substantially increases its ability to translocate actin filaments in vitro. The construct therefore has a lever arm that is ~2.5 times the length of the 1IQ construct (slightly less if allowance is made for the fulcrum of the lever being within the motor domain). The motility of Myo1c1IQ-SAH is ca. fivefold faster than the velocity of Myo1c1IQ and suggests that Myo1c1IQ with its very short lever arm may have more interaction with the surface, which interferes with its movement. The SAH domain used here comprises 38 amino acids (5.7 nm α-helix in length), which equates to ~1.5 IQ motifs. The SAH domain is expected to be remote from the motor domain and is unlikely to affect the motor domain directly, but the interface between the SAH domain and the calmodulin associated with the first IQ domain may be important. The use of an SAH domain here is consistent with recent studies showing that the SAH domain from Dictyostelium MyoM can replace IQ domains in myosin V to retain processivity and step size [29]. The exact positioning of the SAH domain in Myo1c after the first IQ domain was not critical with similar motility measured for constructs in which the SAH domain was placed at L720, K723, or L725; or when placed after L720 and two glycine molecules (data not shown). That the insertion of two glycine residues between the IQ and the SAH domains did not affect motility is surprising as these were expected to form a flexible joint between the two rigid domains thus reducing step size and motility. In the motility assays, the actin filaments move against no force so changes in the stiffness of the lever arm might occur, although these are not detectable with this method.
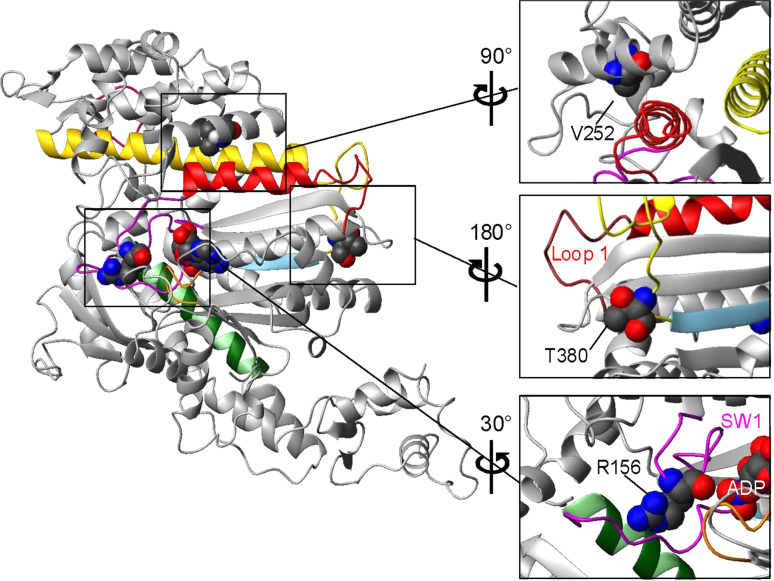
The different classes of myosins share sequence homology in the motor domain, which contains the nucleotide- and actin-binding sites. Although there is no solved crystal structure of Myo1c, the similarity among all myosin motor domains allows us to gain insight into its structure based on that of other myosins, in particular MyoE, a class I myosin from Dictyostelium, and currently the only solved structure of a class I myosin. Myo1c and MyoE share 46% sequence identity, greater than the 36% identity between Myo1c and Dictyostelium myosin II (PDB: 1G8X), which was used in homology modeling of Myo1c by Zadro and colleagues [16]. Figure 3 shows the motor domain structure predicted for Myo1c generated by homology modeling (Swiss-Model; [32]) using MyoE, which has ADP and vanadate bound to the active site and is in the pre-power stroke conformation (PDB 1LKX) [33]. The locations of the three mutations associated with bilateral sensorineural hearing loss [16] are shown in Fig. 3.
Fig. 3.
Homology model of the crystal structure of the Myo1c motor domain showing the positions of three residues that are associated with hearing disorders when mutated. The regions around each of the three residues are enlarged and rotated for the best view (right). The model was created with Swiss-Model [32] using the solved crystal structure of Dictyotelium MyoE (ADP.vanadate structure) as a template (PDB 1LKX-A). Color-coded structural elements: magenta switch 1 and switch 2, orange P-loop, green SW-2 helix, sky blue β-stand 5, red helix G and loop 1, yellow helix O and HO-linker, dark pink myopathy loop. Bound nucleotide (ADP) and point mutations are shown in space-fill mode
Mutation R156W is in a highly conserved amino acid positioned immediately adjacent to switch 1 (residues 157–162 in Myo1c), which has a consensus sequence NXNSSR and is one of three loops together with switch 2 and the P-loop that surround bound nucleotide in myosins [34]. In the best-studied myosins, closure of switch 1 onto ATP is required before ATP-induced dissociation of actin occurs through coupling the movement of switch 1 with the opening of the large 50 kDa cleft that divides the actin-binding regions of myosins [35]. Mutations in switch 1 can severely affect the ability of switch 1 to function normally [36–38]. There are no published reports of mutations at the position of the conserved R156 in Myo1c; however, the same R to W mutation has been identified in human cardiac myosin II (R237W) in a patient with familial dilated cardiomyopathy suggesting a possible association of his mutation with disease [39]. The mutation from the charged residue, arginine, to the hydrophobic residue, tryptophan, is quite dramatic. Thus, apart from any direct local effect of the side chain itself, the mutation could disrupt the structure and movement of switch 1.
We observed a threefold faster value for k′+2, the maximum rate constant of the ATP-induced dissociation of actin from myosin, in the presence of calcium in R156W compared to wild type, which is compatible with the mutation affecting the ATP-induced movement of switch 1 and actin dissociation. In contrast, in the absence of calcium, R156W produces only a small reduction in k′+2 (~25%). This suggests that disruption to the switch-1 region affects the actin-binding site and decreases the calcium sensitivity of the ATP-induced dissociation in Myo1c. The only other large changes seen in R156W were an increase in K′α, which is linked to a conformational change in the lever arm that affects the nucleotide-binding pocket, and a weaker affinity for ADP in the absence of actin. Both of these effects are seen only in the absence of calcium. Our studies also demonstrate that this mutation results in a 60% reduction in the Vmax of the actin-activated Mg2+-ATPase activity and a 20% reduction in motility in EGTA; however, in Ca2+ the ATPase level remains high while the motility cannot be measured. These results suggest that this mutation affects the communication between the nucleotide pocket and the presence of calcium on calmodulin.
The equivalent of R156 in scallop myosin II, residue R236, makes a complex salt bridge with two glutamic acids, E177 in the P-loop (E106 in Myo1c) and E675 (D595 in Myo1c) in the upper 50 kDa domain [40]. Although this salt bridge is not seen in the MyoE crystal structure or in our homology model of Myo1c, the presence of the E and D residues at the relevant sites in Myo1c suggests that formation of the same salt bridge is possible in Myo1c.
Our homology model shows residue R156 forming a weak hydrogen bond with the backbone peptide amine group of V392. V392 follows E391 of switch 2, which makes a well-characterized salt bridge to residue R162 of switch 1 when both switch 1 and switch 2 close onto ATP [41]. Mutation of R156 to W may disrupt these interactions with V392 and the bulkier tryptophan may disrupt entry or exit of nucleotide from the nucleotide pocket, consistent with our finding that ADP release is slower in R156W than in wild type. Our data also show changes in the calcium sensitivity of this mutation (Table 5) and suggest that this mutation could ultimately affect the communication between calcium binding to calmodulin and the nucleotide pocket by disruption of communication between switch 1 and the switch-2 relay helix.
Table 5.
The lifetime of the detached M.ATP state compared to the lifetime of the post power stroke A.M.D and A.M states
| −Ca2+ | +Ca2+ | |||||
|---|---|---|---|---|---|---|
| Lifetime of M.ATP (tdet) (s) | Lifetime of A.M.D + A.M (tatt) (s) | Ratio | Lifetime of M.ATP (tdet) (s) | Lifetime of A.M.D + A.M (tatt) (s) | Ratio | |
| WT | 0.023 | 0.55 | 24 | 0.16 | 0.10 | 0.63 |
| SAH-WT | 0.023 | 0.19 | 8.2 | 0.11 | 0.10 | 0.88 |
| R156W | 0.021 | 0.24 | 12 | 0.11 | 0.06 | 0.53 |
| V252A | 0.025 | 0.29 | 11.5 | 0.12 | 0.15 | 1.3 |
| T380M | 0.044 | 0.44 | 1.95 | 0.11 | 0.07 | 0.64 |
Mutation V252A is in the middle of an α-helix (named K) in the upper 50 kDa domain on the same side of the molecule as loop 1. In our homology model, V252 shows hydrophobic interactions with F149 on helix G, the helix that links switch 1 with surface loop 1. We have recently shown that loop 1 modulates ADP release in both Myo1b and Myo1c [19, 42]. The location of V252A in the upper 50 kDa domain close to loop 1 and switch 1 suggests that loss of hydrophobic contacts as a consequence of the mutation could disrupt coupling between the actin- and nucleotide-binding sites.
There are relatively few changes in the biochemical kinetic properties of V252A. The only large change is an enhancement in the affinity of ADP for myosin when actin and calcium are absent. The observation that K6K7 (= KD for ADP) is twofold higher in the V252A mutant while the K′AD value shows little change means that the thermodynamic coupling constant (K′AD/KD) decreases from 2 for wild type to 1 in the case of V252A. This in turn requires an equivalent change in the affinity of actin for myosin, K′A/K′DA, in the presence and absence of ADP.
In steady-state assays, we see little change in the Vmax of the ATPase, but the Km for actin is smaller for V252A in the absence of calcium, consistent with a disruption of the communication between the actin- and nucleotide-binding sites. The lack of filaments binding to immobilized Myo1c1IQ-SAH-V252A in the motility assay also suggests a weaker affinity for actin at steady state or a lower duty ratio for this construct.
Mutation T380M is the final amino acid in a six-amino-acid-long insertion (residues 375–380 with sequence SWRSTT) found in the mammalian Myo1c isoforms, which is at the end of the HO linker in the transducer region of the motor domain. The HO linker provides a connection between switch 2 and strand 5 of the β-sheet on one side and the long helix O and the actin-binding myopathy loop on the other side of the linker. The role of this linker and the Myo1c-specific insert may be in coupling the actin- and nucleotide-binding sites. The central β-sheet, which is referred to as the transducer region, is thought to have a role in the power stroke, which couples actin binding and Pi release.
Changes observed for the T380M mutant are far more diverse than those seen with the other two mutants. For the transient kinetic parameters measured at low calcium, only K′α, K6K7, and k+3+k−3 were similar to wild type. There were fewer changes in the presence of calcium, emphasizing how large the effects of calcium are on Myo1c. In contrast, there is little change in the steady-state ATPase activity of T380M vs. wild type; however, T380M did not translocate actin filaments in vitro. These results are consistent with disruptions to the communication pathways in the motor domain that uncouple enzymatic activity from motility. Mutations in other myosins also uncouple ATPase activity from motility; however, these mutations are mainly in switch 2 or the SH-1/SH-2 region [43–47].
According to current models of how stereocilia respond to sound and movement, Myo1c generates enough tension in the tip links such that the transduction channels partially open, setting them near the optimal point to respond to small movements of the stereocilia [1]. Myo1c also senses changes in tension and resets the tension in the tip links on the time scale of slow adaptation (>10 ms) either by slipping down the stereocilia in response to a positive stimulus or climbing up the core bundle of actin filaments in response to a negative stimulus. Given this highly specialized function, it may not be surprising that subtle changes to the properties of Myo1c could result in sensorineural hearing loss. Small changes in properties may be less critical to the role(s) of Myo1c in other cell types in the same way that some mutations in β-myosin affect the heart more than slow twitch muscle fibers [48]. However, surprisingly, some of the identified Myo1c mutants have large defects such as T380M′s inability to translocate actin filaments. Rather than transporting cargo long distances, it is likely that Myo1c acts largely as a tension sensor in the cytoskeleton and responds to changes in load and local changes in calcium concentration. This is consistent with a recent study showing that membrane tension increases in cells over-expressing Myo1c [49].
Changes in Myo1c activity as a consequence of mutation can be expected to have an effect on both the level of tension set by the motor (the stall force) and slow adaptation; either effect could result in hearing loss. A decrease in the velocity of Myo1c as exhibited by mutation R156W in low Ca2+ will decrease the rate of climbing adaptation. Mutant V252A also displayed aberrant Ca2+ sensitivity, which is expected to affect its speed during climbing adaptation. In addition to aberrant Ca2+ sensitivity, the motility studies presented here appear to indicate that the affinity of the V252A mutant for actin is weaker than that of wild type. This might reduce the steady-state tension. The mutation, T380M, causes uncoupling of the ATPase activity from motility resulting in a nonfunctional motor. Adaptation would be compromised in patients expressing the T380M mutation because the ability to reset the tip-link tension during climbing adaptation would be compromised.
We have previously argued that the calcium sensitivity exhibited by Myo1c may be important in slow adaptation [11, 13]. The model we proposed is that, as a consequence of the calcium transient and increase in tip-link tension, Ca2+ binds to the calmodulin of the first IQ motif and accelerates ADP release and ATP-induced detachment of Myo1c from actin. At the same time, the Ca2+-calmodulin slows down the rate constant of the ATP-hydrolysis step, thereby increasing the lifetime of the detached M.ATP state. Together, this would reduce the duty ratio of the motor allowing it to slip thereby reducing tip-link tension and closing the channels. A change in calcium sensitivity due to mutation may affect adaptation leading to the observed phenotypes.
Table 5 summarizes changes in the calcium sensitivity of the lifetime of the more important complexes. The value of 1/(k+3+k−3) defines the lifetime of the M.ATP detached state, tdet, and the effect of calcium on this value is similar for all of the mutations studied (20–22 ms in the absence of calcium and ~110 ms in the presence of calcium). The net lifetime of the force-holding A.M.D and A.M states, tatt, is controlled by both k′+2 and k′+AD, where tatt = (k′+2 + k′+AD)/k′+2 × k′+AD and, in the absence of calcium, has a value of 550 ms for the WT construct and 190–290 ms for all of the SAH constructs except T380M, which is much shorter at 44 ms. When calcium is present, these values decrease to 150 ms for V252A, 100 ms for WT and the SAH-WT, and 60 ms for R156W. For T380M the value of tatt increases from 44 to 70 ms.
The parameter of interest here is then the ratio of the lifetime of the detached M.ATP complex and the force-holding states (A.M.D + A.M). These values, to be treated cautiously because they are derived from several measurements with accumulated errors, are summarized in Table 5. In the presence of calcium, the ratio is reduced tenfold or more to a value of 0.5–1.3 for all constructs reflecting the lower duty ratio expected with the two lifetimes now being comparable. In the absence of calcium, the ratio is 24 for WT and 8–12 for the other constructs (except T380M) reflecting the 10- to 20-fold longer lifetime of the A.M.D state compared to M.ATP. For T380M, the lifetime of the attached state is greatly reduced because of a large value of k′+AD. This may explain why this construct does not support motility.
Acknowledgments
We thank Dr. Marieke Bloemink (University of Kent) for invaluable help with the structural models and critical discussion of the manuscript. This work was supported by National Institutes of Health Grant R01 DC08793 and DC08793-01S1 to L.M.C. and Wellcome Trust Grant 085309 to M.A.G.
Abbreviations
- IQ domain
A calmodulin-binding domain with a consensus sequence usually containing isoleucine (I) and glutamine (Q)
- LCBD
Light-chain-binding domain
- LTLD
Lower tip link density
- RT
Room temperature
- SAH
Single α-helix
- UTLD
Upper tip link density
References
- 1.Gillespie PG, Müller U. Mechanotransduction by hair cells: models, molecules, and mechanics. Cell. 2009;139:33–44. doi: 10.1016/j.cell.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickles JO, Comis SD, Osborne MP. Cross-links between stereocilia in the guinea pig organ of Corti, and their possible relation to sensory transduction. Hear Res. 1984;15:103–112. doi: 10.1016/0378-5955(84)90041-8. [DOI] [PubMed] [Google Scholar]
- 3.Hudspeth AJ. Hair-bundle mechanics and a model for mechanoelectrical transduction by hair cells. Soc Gen Physiol Ser. 1992;47:357–370. [PubMed] [Google Scholar]
- 4.LeMasurier M, Gillespie PG. Hair-cell mechanotransduction and cochlear amplification. Neuron. 2005;48:403–415. doi: 10.1016/j.neuron.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie PG, Cyr JL. Myosin-1c, the hair cell′s adaptation motor. Annu Rev Physiol. 2004;66:521–545. doi: 10.1146/annurev.physiol.66.032102.112842. [DOI] [PubMed] [Google Scholar]
- 6.Howard J, Hudspeth AJ. Mechanical relaxation of the hair bundle mediates adaptation in mechanoelectrical transduction by the bullfrog′s saccular hair cell. Proc Natl Acad Sci USA. 1987;84:3064–3068. doi: 10.1073/pnas.84.9.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia JA, Yee AG, Gillespie PG, Corey DP. Localization of myosin-Ibeta near both ends of tip links in frog saccular hair cells. J Neurosci. 1998;18:8637–8647. doi: 10.1523/JNEUROSCI.18-21-08637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steyger PS, Gillespie PG, Baird RA. Myosin I is located at tip link anchors in vestibular hair bundles. J Neurosci. 1998;18:4603–4615. doi: 10.1523/JNEUROSCI.18-12-04603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt JR, Gillespie SKH, Provance DW, Shah K, Shokat KM, Corey DP, Mercer JA, Gillespie PG. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell. 2002;108:371–381. doi: 10.1016/S0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 10.Beurg M, Fettiplace R, Nam J-H, Ricci AJ. Localization of inner hair cell mechanotransducer channels using high speed imaging. Nat Neurosci. 2009;12:553–558. doi: 10.1038/nn.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batters C, Arthur CP, Lin A, Porter J, Geeves MA, Milligan RA, Molloy JE, Coluccio LM. Myo1c is designed for the adaptation response in the inner ear. EMBO J. 2004;23:1433–1440. doi: 10.1038/sj.emboj.7600169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batters C, Wallace MI, Coluccio LM, Molloy JE. A model of stereocilia adaptation based on single molecule mechanical studies of myosin I. Phil Trans R Soc B. 2004;359:1895–1905. doi: 10.1098/rstb.2004.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamek N, Coluccio LM, Geeves MA. Calcium sensitivity of the cross-bridge cycle of Myo1c, the adaptation motor of the inner ear. Proc Natl Acad Sci USA. 2008;105:5710–5715. doi: 10.1073/pnas.0710520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman TB, Sellers JR, Avraham KB. Unconventional myosins and the genetics of hearing loss. Am J Med Genet. 1999;89:147–157. doi: 10.1002/(SICI)1096-8628(19990924)89:3<147::AID-AJMG5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- 16.Zadro C, Alemanno MS, Bellacchio E, Ficarella R, Donaudy F, Melchionda S, Zalante L, Rabionet R, Hilgert N, Estivill X, Van Camp G, Gasparini P, Carella M. Are Myo1C and Myo1F associated with hearing loss? Biochim Biophys Acta. 2009;1792:27–32. doi: 10.1016/j.bbadis.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Perreault-Micale C, Shushan AD, Coluccio LM. Truncation of a mammalian myosin I results in loss of Ca2+-sensitive motility. J Biol Chem. 2000;275:21618–21623. doi: 10.1074/jbc.M000363200. [DOI] [PubMed] [Google Scholar]
- 18.Pollard TD (1982) Myosin purification and characterization. In: Wilson L (ed) Methods in Cell Biology, vol 24. The cytoskeleton, pt. A. Academic Press, New York pp 333–371 [DOI] [PubMed]
- 19.Adamek N, Lieto-Trivedi A, Geeves MA, Coluccio LM. Modification of loop 1 affects the nucleotide-binding properties of Myo1c, the adaptation motor in the inner ear. Biochemistry. 2010;49:958–971. doi: 10.1021/bi901803j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagshaw CR, Trentham DR. The characterization of myosin-product complexes and of product-release steps during the magnesium ion-dependent adenosine triphosphatase reaction. Biochem J. 1974;141:331–349. doi: 10.1042/bj1410331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trentham DR, Eccleston JF, Bagshaw CR. Kinetic analysis of ATPase mehanisms. Q Rev Biophys. 1976;9:217–281. doi: 10.1017/S0033583500002419. [DOI] [PubMed] [Google Scholar]
- 22.Malnasi-Csizmadia A, Pearson DS, Kovacs M, Woolley RJ, Geeves MA, Bagshaw CR. Kinetic resolution of a conformational transition and the ATP hydrolysis step using relaxation methods with a Dictyostelium myosin II mutant containing a single tryptophan residue. Biochemistry. 2001;40:12727–12737. doi: 10.1021/bi010963q. [DOI] [PubMed] [Google Scholar]
- 23.Malnasi-Csizmadia A, Woolley RJ, Bagshaw CR. Resolution of conformational states of Dictyostelium myosin II motor domain using tryptophan (W501) mutants: implications for the open-closed transition identified by crystallography. Biochemistry. 2000;39:16135–16146. doi: 10.1021/bi001125j. [DOI] [PubMed] [Google Scholar]
- 24.Geeves MA, Perreault-Micale C, Coluccio LM. Kinetic analyses of a truncated mammalian myosin I suggest a novel isomerization event preceding nucleotide binding. J Biol Chem. 2000;275:21624–21630. doi: 10.1074/jbc.M000342200. [DOI] [PubMed] [Google Scholar]
- 25.Coluccio LM, Geeves MA. Transient kinetic analysis of the 130 kDa myosin I (myr 1 gene product) from rat liver: a myosin I designed for maintenance of tension? J Biol Chem. 1999;274:21575–21580. doi: 10.1074/jbc.274.31.21575. [DOI] [PubMed] [Google Scholar]
- 26.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 27.Knight PJ, Thirumurugan K, Xu Y, Kalverda AP, Stafford WFI, Sellers JR, Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 28.Anson M, Geeves MA, Kurzawa SE, Manstein DJ. Myosin motors with artificial lever arms. EMBO J. 1996;15:6069–6074. [PMC free article] [PubMed] [Google Scholar]
- 29.Baboolal TG, Sakamoto T, Forgacs E, White HD, Jackson SM, Takagi Y, Farrow RE, Molloy JE, Knight PJ, Sellers JR, Peckham M. The SAH domain extends the functional length of the myosin lever. Proc Natl Acad Sci U S A. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu T, Sata M, Ikebe M. Functional expression of mammalian myosin I beta: analysis of its motor activity. Biochemistry. 1996;35:513–522. doi: 10.1021/bi952053c. [DOI] [PubMed] [Google Scholar]
- 31.Toyoshima YY, Kron SJ, McNally EM, Niebling KR, Toyoshima C, Spudich JA. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 32.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kollmar M, Dürrwang U, Kliche W, Manstein DJ, Kull FJ. Crystal structure of the motor domain of a class-I myosin. EMBO J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 35.Kintses B, Gyimesi M, Pearson DS, Geeves MA, Zeng W, Bagshaw CR, Malnasi-Csizmadia A. Reversible movement of switch 1 loop of myosin determines actin interaction. EMBO J. 2007;26:265–274. doi: 10.1038/sj.emboj.7601482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XD, Rhodes TE, Ikebe R, Kambara T, White HD, Ikebe M. Effects of mutations in the gamma-phosphate binding site of myosin on its motor function. J Biol Chem. 1998;273:27404–27411. doi: 10.1074/jbc.273.42.27404. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki N, Sutoh K. Structure-mutation analysis of the ATPase site of Dictyostelium discoideum myosin II. Adv Biophys. 1998;35:1–24. [PubMed] [Google Scholar]
- 38.Shimada T, Sasaki N, Ohkura R, Sutoh K. Alanine scanning mutagenesis of the switch I region in the ATPase site of Dictyostelium discoideum myosin II. Biochemistry. 1997;36:14037–14043. doi: 10.1021/bi971837i. [DOI] [PubMed] [Google Scholar]
- 39.Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, Nauman D, Burgess D, Partain J, Litt M. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. 2008;1:21–26. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risal D, Gourinath S, Himmel DM, Szent-Gyorgyi AG, Cohen C. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci USA. 2004;101:8930–8935. doi: 10.1073/pnas.0403002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furch M, Fujita-Becker S, Geeves MA, Holmes KC, Manstein DJ. Role of the salt-bridge between switch-1 and switch-2 of Dictyosteliumd myosin. J Mol Biol. 1999;290:797–809. doi: 10.1006/jmbi.1999.2921. [DOI] [PubMed] [Google Scholar]
- 42.Clark R, Ansari MA, Dash S, Geeves MA, Coluccio LM. Loop 1 of transducer region in mammalian class I myosin, Myo1b, modulates actin affinity, ATPase activity, and nucleotide access. J Biol Chem. 2005;280:30935–30942. doi: 10.1074/jbc.M504698200. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki N, Ohkura R, Sutoh K. Dictyostelium myosin II mutations that uncouple the converter swing and ATP hydrolysis cycle. Biochemistry. 2003;42:90–95. doi: 10.1021/bi026051l. [DOI] [PubMed] [Google Scholar]
- 44.Murphy CT, Rock RS, Spudich JA. A myosin II mutation uncouples ATPase activity from motility and shortens step size. Nat Cell Biol. 2001;3:311–315. doi: 10.1038/35060110. [DOI] [PubMed] [Google Scholar]
- 45.Ruppel KM, Spudich JA. Myosin motor function: structural and mutagenic approaches. Curr Opin Cell Biol. 1995;7:89–93. doi: 10.1016/0955-0674(95)80049-2. [DOI] [PubMed] [Google Scholar]
- 46.Ruppel KM, Spudich JA. Structure-function studies of the myosin motor domain: importance of the 50 kDa cleft. Mol Biol Cell. 1996;7:1123–1136. doi: 10.1091/mbc.7.7.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yengo CM, Ananthanarayanan SK, Brosey CA, Mao S, Tyska MJ. Human deafness mutation E385D disrupts the mechanochemical coupling and subcellular targeting of myosin-1a. Biophys J. 2008;94:L5–L7. doi: 10.1529/biophysj.107.122689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/S0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 49.Nambiar R, McConnell RE, Tyska MJ. Control of cell membrane tension by myosin-I. Proc Natl Acad Sci USA. 2009;106:11972–11977. doi: 10.1073/pnas.0901641106. [DOI] [PMC free article] [PubMed] [Google Scholar]