Abstract
Flaviviruses enter their host cells by receptor-mediated endocytosis, a well-orchestrated process of receptor recognition, penetration and uncoating. Recent findings on these early steps in the life cycle of flaviviruses are the focus of this review.
Keywords: FLAVIVIRUS, CELL ENTRY, RECEPTOR, ENDOCYTOSIS, ENVELOPE PROTEIN, PH ENVIRONMENT, STRUCTURE
The Flaviviridae are a family of small, enveloped RNA viruses that is comprised of three genera: Flavivirus, Hepacivirus, and Pestivirus. More than 70 viruses have been classified in the genus Flavivirus [1]. The genus includes various noteworthy mosquito- and tick-borne human pathogens, such as Yellow Fever virus (YFV), dengue virus (DENV), Tick-borne Encephalitis Virus (TBEV), and West Nile virus (WNV), that are subclassified into several antigenic complexes and phylogenetic groups [2]. Flaviviruses are typically associated with mild systemic disease, but can also cause severe symptoms such as hemorrhagic fever, encephalitis or death. Many of these viruses are resurgent, are spreading to new environments and are responsible for substantial morbidity and mortality around the globe. Flaviviruses are primarily transmitted through arthropod vectors. Humans and other mammals are not known to commonly develop infectious-level viremias and thus are probably incidental hosts with the exceptions of YFV, DENV and TBEV that are sufficiently well-adapted to a mammalian host.
1. The flavivirus lifecycle
The basic stages of the flavivirus life cycle include attachment to the cell surface, internalization into the host cell, transfer of the viral RNA genome into the cytoplasm, translation of the viral proteins, replication of the genomic RNA, assembly and maturation of the virions, and ultimately the release of progeny viruses from the cell.
Flaviviruses are lipid-enveloped viruses (Fig. 1A). After recognition and attachment to specific receptor molecules on the surface of the cell, the virus is internalized into the host cell by clathrin-dependent endocytosis. Uncoating is induced by the low pH environment of the endosomes, where the viral proteins enter into a fusion-active state and initiate the merging of the viral envelope with the endosomal membrane, thereby releasing the viral RNA genome into the cytoplasm.
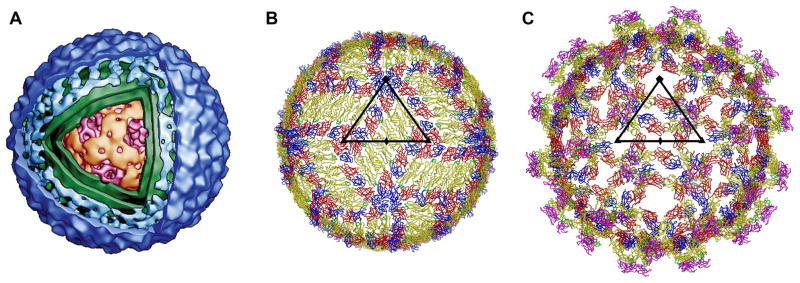
Fig. 1.
The flavivirion structure. (A) 3D rendering of a cryoEM density map of DENV (kindly provided by Wei Zhang, Institute of Molecular Virology, University of Minnesota, USA). The cut-out window allows the view inside the virion at the nucleocapsid core (magenta/orange) composed of the C protein and the RNA genome. The core is surrounded by a host-derived lipid envelope (green). The outer icosahedral shell of the virus (blue) is formed by two membrane-anchored glycoproteins, E and M. (B) Prefusion arrangement of the E glycoprotein on the surface of the mature flavivirion, viewed down an icosahedral twofold axis. Domains DI, DII, and DIII of each E monomer are colored red, yellow, and blue, respectively. The fusion loop is shown in green. A total of 180 E monomers are associated in 30 rafts of three, nearly parallel E homodimers that form a distinct herringbone pattern. One icosahedral asymmetric unit (ASU), the smallest unit from which the particle can be generated by applying icosahedral symmetry, is outlined by a black triangle. The positions of the neighboring icosahedral five-, three- and twofold symmetry axes are marked with symbols (pentagons, triangles, and ovals, respectively). Each ASU contains the equivalent of three E monomers. (C) Arrangement of the E glycoproteins and pr peptides on the surface of an immature, fusion-incompetent flavivirion, viewed down an icosahedral twofold axis. Domains DI, DII, and DIII of each E monomer and the fusion loop are colored as in (A). The pr peptide is shown in magenta. One ASU is outlined by a black triangle and the positions of the symmetry axes are indicated. The E proteins are icosahedrally arranged in 60 trimeric spikes, composed of prM/E heterodimers. The pr peptide of the prM protein covers the fusion peptide in DII of E (on the tip of each spike) to prevent premature fusion.
Flaviviruses have a single-stranded, positive-sense, ~11-kb RNA genome with a single open reading frame that is directly translated into a polyprotein precursor [1]. The polyprotein is subsequently glycosylated by cellular glycosyltransferases and cleaved by a combination of viral and host proteases to release three structural (C, prM, and E) and seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5). NS3 and NS5 are the best-characterized non-structural proteins that assemble with several other viral and host proteins to form the replication complex. NS3 combines helicase/NTPase, serine protease, and RNA triphosphatase activity [3, 4]. The functional domains of NS5 include S-adenosyl methyltransferase and RNA-dependent RNA polymerase activities [5]. The replication complex is responsible for the synthesis of a negative-sense RNA that acts as the template for the synthesis of the viral genomic progeny RNA. The viral structural proteins are the capsid (C) protein that associates with the RNA genome to form the nucleocapsid core, the envelope (E) protein that functions in receptor binding, membrane fusion, and viral assembly, and the pre-membrane (prM) protein that facilitates the folding and trafficking of the E protein during virus particle biogenesis [6, 7] (Fig. 1).
The assembly of virions occurs in the endoplasmic reticulum when the newly synthezised viral proteins and nucleic acid combine to form immature, fusion-incompetent virus particles [1] (Fig. 1C). The budding into the endoplasmic reticulum is also responsible for the gain of the host-derived lipid envelope. After assembly, immature virions undergo a protease- and pH-dependent maturation during their transit from the endoplasmic reticulum through the trans-Golgi network to the cell surface [8–11]. During maturation, the E proteins go through a significant positional reorganization while the prM protein is cleaved by the cellular subtilisin-like endoproteinase furin [9]. The proteolytic cleavage releases the glycosylated, N-terminal pr fragment from the small, membrane-anchored M portion that will remain virion-associated. The pr peptide is subsequently shed from the virion during the exocytotic release of virus progeny [11]. The removal of the previously covalently attached pr peptides from the virions primes the viral particles in their mature, metastable structural state (Fig 1B) ready for the low-pH triggered fusion events during the upcoming cell entry [12, 13].
2. The structure of the flavivirus particle
Mature, fusion-competent flavivirions are roughly spherical with a diameter of about 500 Å (Figs. 1A and 1B). Their nucleocapsid core, formed by the RNA genome and multiple copies of the highly basic, largely α-helical C protein [14], is surrounded by a host-derived lipid bilayer and a smooth, spikeless outer glycoprotein shell [1]. The outer protein shell has icosahedral symmetry and is composed of 180 copies, each, of the membrane-anchored M and E glycoproteins.
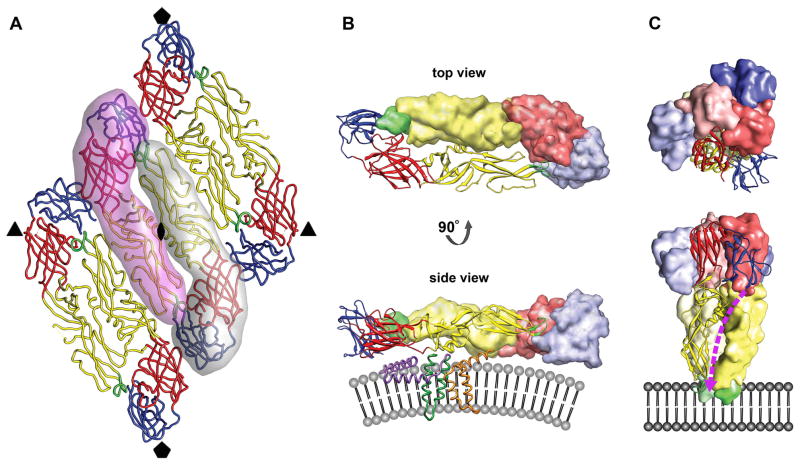
The E protein has an important role in the replication and maturation process of the virus, being involved in receptor binding, membrane fusion and virus assembly. On the viral surface, sets of three E head-to-tail homodimers are tightly associated into 30 rafts that form a herringbone pattern [15] (Figs. 1B and 2A). Each E monomer is structurally divided into a two-helix transmembrane region and an elongated ectodomain that stretches out along the viral lipid envelope (Fig. 2B). An α-helical stem region connecting the C-terminus of the E ectodomain with its transmembrane anchor lies essentially flat against the lipid bilayer underneath the ectodomain, half-buried in the outer lipid leaflet, and might alleviate electrostatic repulsion forces between the E ectodomain and the membrane [15, 16] (Fig. 2B). The ectodomain has three distinct, structurally defined domains (DI to DIII) that are joined by flexible hinges [17, 18] (Fig. 2). The eight-stranded β-barrel of the central domain DI is flanked by DII and DIII and participates in the conformational changes induced by endosomal acidification during cell entry [19, 20]. A four-stranded hinge connects DI and the finger-like oligomerization domain DII, that is formed by two large, highly β-structured loops connecting adjacent strands of the DI β-barrel. DII contains a hydrophobic fusion loop at its tip hidden at the dimer interface in a hydrophobic pocket that is supplied by DI and DIII of the second E monomer of the homodimer (Fig. 2B). This fusion loop is indispensable for virus-cell membrane fusion [21]. The immunoglobulin-like C-terminal domain DIII is connected to DI by a single polypeptide linker and undergoes a major positional rearrangement during cell entry [17, 19, 20] (Figs. 2B and 2C). DIII projects slightly above the general viral surface [15] and is the proposed receptor binding domain [2, 22–24]. The three E monomers per icosahedral asymmetric unit (Figs. 1B and 2A) are situated in distinctly different chemical environments that influence their availability for antibody or receptor binding. Buried under the E protein layer is the M protein, also anchored in the viral envelope by two antiparallel transmembrane helices [16] (Fig. 2B). The small, predominantly α-helical M protein is the product of proteolytic processing of the prM protein by the cellular protease furin during virus maturation. Neither M nor E interact directly with the nucleocapsid core in the mature virion.
Fig. 2.
The oligomeric structure of the E glycoprotein. (A) Raft organization of three E homodimers as observed for mature flavivirions. The relative positions of the neighboring icosahedral symmetry axes are indicated by symbols. The elongated E ectodomain has three distinct, structurally defined domains that are joined by flexible hinges. DI, DII, and DIII are colored red, yellow, and blue, respectively. The fusion loop is shown in green. Two E monomers forming the central homodimer at the icosahedral twofold axis are emphasized by shadows. (B) The prefusion head-to-tail homodimer arrangement of the E glycoprotein shown from the top (looking towards the center of the virus) and from the side after a 90° rotation around its long axis. One monomer is rendered as a ribbon diagram, showing its three domains largely adopting β-sheet folds. The other monomer is represented as a surface shaded volume. The fusion loop of one E protein is buried in a hydrophobic pocket at the DI–DIII interface of the other E molecule. The side view shows that each E monomer is structurally divided into the ectodomain that elongates parallel to the viral lipid envelope and a two-helix transmembrane anchor (dark green). The α-helical stem region (purple), half-buried in the outer lipid leaflet, connects the C-terminus of the E ectodomain with the transmembrane helices. The M protein (orange) also contains two antiparallel transmembrane helices and is essentially buried under the E protein layer. The stem-anchor region is only shown for the E monomer depicted as a ribbon diagram. Similarly, only one M protein is illustrated. (C) The postfusion homotrimer arrangement of the E glycoprotein shown from the top and from the side after a 90° rotation. The E monomers are oriented parallel to one another with their fusion peptides exposed on one end of the trimer. DIII of E has undergone a major rotational displacement to the side of DI and towards DII. The stem region (dashed purple line) is predicted to bind in a hydrophobic groove that extends along the interface of two neighboring DIIs of the E trimer towards the fusion loop. The irreversible fold-back conversion of the E protein into the post-fusion hairpin-like structure and zipping up of the stem brings the E transmembrane helices (connected via the stem to DIII of the ectodomain) in juxtaposition with the membrane-embedded fusion loop at the tip of DII.
The E proteins on the surface of immature, newly synthesized flavivirus particles are icosahedrally arranged in 60 trimeric spikes composed of prM/E heterodimers [25, 26] (Fig. 1C). The pr peptide components [27] of the prM proteins cover the fusion peptides in DII of the E glycoproteins to prevent premature fusion [11, 26, 28]. These immature particles are fusion-incompetent and non-infectious [29]. However, they can be rendered infectious by entering Fc receptor-bearing cells as virus-antibody complexes [30]. The maturation of these virions seems to belatedly occur in the endosomal environment, although the mechanism for shedding the pr peptide under these circumstances is unclear.
Whereas the proteolytic processing of prM is a necessary step in the virus life cycle [8–10], studies have shown that complete maturation is not required for infectivity or fusion activity [29]. Partially matured particles that are heterogeneous in residual prM content and in the arrangement of E proteins on the virion, are present in flavivirus populations [31–34]. These can be infectious, presumably by attaching to cells via mature surface portions and subsequently completing maturation in the endosomal compartment.
3. The early stages of the flavivirus life cycle
The early stages in the life cycle of flaviviruses involve entry into susceptible cells by receptor-mediated endocytosis with the ultimate goal to release the RNA genome from the protective envelope and protein shell into the cytoplasm. Receptor recognition, internalization and uncoating involve the formation and release of numerous specific molecular interactions and large-scale re-organization of the structural proteins and their domains on the viral surface. In flaviviruses, these seemingly simple, but not well understood mechanisms are mediated by the membrane-anchored E protein that combines both receptor binding and fusogenic activity.
3.1 Receptor recognition and attachment
The entry of flaviviruses into their target cells is mediated by the interaction of the E glycoprotein with cellular surface receptor molecules. Receptor recognition and attachment is likely to be a process in which multiple cellular receptor molecules are used in combination or consecutively to gain entry. Receptor usage is both cell-type and virus-specific and contributes to host range, tissue tropism and viral pathogenicity.
After vector inoculation of flaviviruses, the initial round of viral replication is considered to be primarily supported in dendritic cells (DC) of the skin [1, 32, 35–38]. Infected DCs migrate to draining lymph nodes, and a second round of replication occurs in lymphoid tissues leading to the viremic stage, when virus enters the circulation and spreads to internal organs through unknown mechanisms. Monocytes and macrophages are thought to be the major target cells in DENV infection and replication, but studies have further suggested a wide variety of cells as potential targets: B lymphocytes, T lymphocytes, hepatocytes, endothelial cells, epithelial cells, and fibroblasts [37, 39–45]. For WNV, neurons or macrophages are proposed to be the physiologically relevant target cell types [46, 47]. Various putative attachment factors and/or receptor elements have been implicated for flaviviruses, most of which are poorly characterized and are of unclear physiological relevance.
Highly sulfated, negatively charged glycosaminoglycans, such as heparan sulfate, can be utilized by several flaviviruses as low-affinity attachment factors that concentrate the virus on the cell surface, mainly mediated by DIII of E [48–55]. Similar to observations made for other Flaviviridae, low-density lipoprotein receptor may play a role as an attachment receptor of non-heparan sulfate adapted JEV strains in mammalian cells, and might be responsible for the neurovirulence of the virus [56]. Another cell surface carbohydrate determinant identified for DENV interaction with mammalian cells is the terminal disaccharide of a glycosphingolipid, neolactotetraosylceramide [57].
Putative primary protein receptors for flaviviruses are DC-SIGN (Dendritic Cell-Specific Intercellular adhesion molecule-3 (ICAM3)-Grabbing Non-integrin) (or CD209) on monocyte-derived dendritic cells and DC-SIGNR on microvascular endothelial cells [32, 37, 58, 59]. These tetrameric C-type lectins bind mannose-rich N-linked glycans with high affinity and have been shown to mediate DENV and WNV infection as attachment factors, presumably requiring the existence of additional entry receptor molecules for virus internalization. For DENV, cryoEM studies on the virus-receptor complex showed that DC-SIGN preferentially utilizes carbohydrate moieties at Asn67 in DII, a site unique to DENV [60]. Another Ca2+-dependent lectin, the mannose receptor, that carries a ligand specificity different from DC-SIGN and is constitutively internalized by clathrin-mediated endocytosis, has been shown to bind DENV, JEV and TBEV and is proposed to be an internalization receptor of DENV in human macrophages [61]. The glycosylation pattern (the number, distribution, and structure of carbohydrate moieties) on the viral surface varies among different flaviviruses or even among different strains of the same virus and influences cell/tissue tropism, infectivity and infection efficiency [38, 58, 62–67].
The presence of a classic RGD motif in DIII of the YFV-17D envelope protein and similar motifs in other flaviviruses suggested a mechanism for interaction with integrins [68]. However, DC infection with the YFV-17D vaccine strain does not depend on the RGD motif, nor on DC-SIGN or on various integrins [38]. For WNV and JEV, αvβ 3 integrin has been documented as a functional receptor in permissive mammalian cells apparently mediated by DIII and likely in a non-RGD-dependent manner [23]. However, the cellular receptors on neurons or macrophages remain unknown. Similarly, it has been suggested that RGD-dependent integrin binding is not essential for entry of Murray Valley encephalitis virus and that multiple and/or alternative receptors may be involved in its cell entry [69]. Recent studies have reported that WNV entry into embryonic mouse fibroblasts and hamster melanoma cells is independent of αvβ3 integrin, suggesting alternative receptor molecules for different cell types or strain differences [70]. Another cell adhesion molecule, the 37/67-kDa high-affinity laminin receptor has been described as a DENV-1 specific binding protein on human liver cells [71]. However, DENV serotypes 2 and 3 have also been shown to interact with this receptor protein extracted from porcine kidney cells [72]. These contrasting results suggest that differential receptor usage by flaviviruses might depend on cell-specific posttranslational modifications of the receptor molecule itself and not only on viral factors [73]. In mosquito cells, a closely related laminin-binding protein may be involved in DENV-3/4 infection [73].
Hsp90 and Hsp70, heat shock proteins associated with membrane microdomains (or lipid rafts), participate in DENV entry as a receptor complex in human neuroblastoma and monocytic cell lines as well as in human monocytes/macrophages [74], but do not play a role for internalization into liver cells [75]. A related stress protein, the glucose-regulated protein GRP78 or BiP, involved in the unfolded protein response, has been identified as a minor receptor element for DENV-2 internalization into human liver cells [75, 76]. Hsp70 might serve as the putative receptor of JEV in mouse neuroblastoma cells [77]. Two heat shock related proteins (gp45 and a 74-kD protein) were indicated as part of the DENV receptor complex in mosquito cells [78], while others hypothesize that heat shock cognate protein 70 (HSC70) mediates JEV entry into mosquito cells as a penetration receptor [79]. Apart from the above, several proteins of varying molecular mass and of unknown identity have been described as putative flavivirus receptors in different cell lines and cell types.
Alternatively to direct attachment of the E glycoprotein to a receptor, flaviviruses can infect monocytes and macrophages in an antibody-dependent manner by binding as virus-antibody complexes to immunoglobulin Fcγ receptors that recognize the constant region of IgG molecules [30, 80, 81].
3.2 Internalization and fusion
Flaviviruses enter their target cell by receptor-mediated, clathrin-dependent endocytosis [82, 83]. The receptor-bound virus is internalized via pre-formed clathrin-coated pits that bud into the cytosol and deliver their cargo to early endosomes. Single particle-tracking in living cells showed that these early endosomes mature to late endosomes, in which membrane fusion seem to take place predominantly [82]. However, the requirement for endocytic trafficking, specifically the maturation stage of the endosome at which virus fusion occurs, appears to vary between different flaviviruses and/or virus strains [82–84], possibly dependent on the pH threshold for fusion of the particular virus.
Upon exposure of the virion to mildly acidic pH in endocytic vesicles a series of molecular events leading to membrane fusion is evoked that have currently been detailed or inferred using biochemical and structural approaches (Fig. 3). The E proteins on the viral surface rearrange from a pre-fusion homodimeric array (Figs. 1B, 2A and 2B) to fusion-active homotrimers. Large-scale conformational changes in the E domain arrangement, made possible by the flexible hinges between the three E domains, bring the viral and the target membrane in close proximity and facilitate the fusion of the two bilayers [12]. The formation of a fusion pore results in the release of the viral RNA genome into the cytoplasm for initiation of replication and translation (Fig. 3).
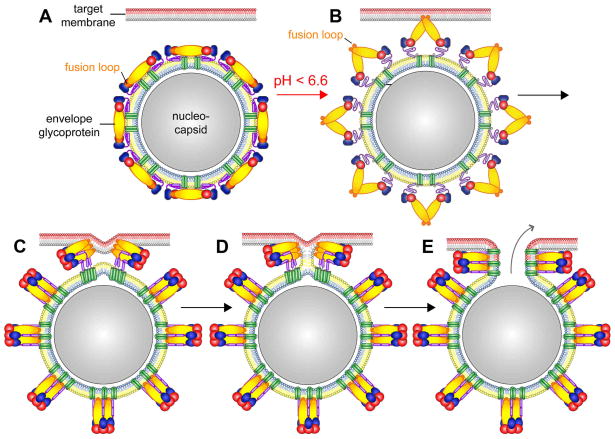
Fig. 3.
Schematic of the proposed steps of the flavivirus membrane fusion process, modified from the original figure drawn by Stiasny et al. (2009) [18]. Domains DI, DII, and DIII of E are colored red, yellow, and blue, respectively. The fusion loop at the distal end of DII is indicated in orange. The stem region is shown in purple and the transmembrane helices in green. The outer/inner leaflets of the viral lipid envelope and the target membrane are colored yellow/blue and gray/red, respectively. (A) Pre-fusion E homodimers on the virus surface. (B) Low endosomal pH weakens inter- and intramolecular E contacts and induces the outward extension of the stem region, followed or accompanied by the dissociation of the E dimers into monomers. The outward projection of the E monomers allows the interaction of the fusion loop with the endosomal target membrane. (C) E trimer formation, back-folding of DIII, and ‘‘zipping-up’’ of the stem. (D) Hemifusion intermediate in which only the outer leaflets of viral and target membranes have mixed. (E) Formation of the post-fusion E trimer and opening of the fusion pore allows the release of the C protein-associated RNA genome (nucleocapsid) into the cytoplasm.
Fusion of flaviviruses with liposomes occurs on a time scale of a few seconds, is strictly dependent on mildly acidic pH (pH threshold ~6.6) and does not require a protein or carbohydrate receptor in the target lipid membrane [29, 85, 86]. Exposure of the virus alone to low pH results in inactivation of its fusion activity. The lipid composition of the target membrane influences fusion efficiency and pH threshold [13, 29, 87, 88]. Studies have suggested that flaviviruses utilizes cholesterol-rich lipid rafts (membrane microdomains enriched with cholesterol und glycosphingolipids) during initial stages of internalization presumably by interacting with raft-associated proteins for attachment or signaling [70, 74]. However, while WNV fusion activity is strongly promoted by the presence of cholesterol in the target membrane, the presence of lipid rafts might not be essential for fusion [29].
The molecular mechanisms prompting viral fusion are not well understood, but histidine residues have been hypothesized to function as sensors of pH that trigger conformational changes in viral fusion proteins. The binding of protons is to lower the energetic barrier separating the pre-fusion from the post-fusion conformation or to provide activation energy [12]. All E domains contain histidine residues that could potentially function as pH sensors [18], either by specifically or cumulatively increasing the surface net charges upon acidification. Studies of TBEV subviral particles identified a conserved histidine at the interface between DI and DIII of E as the critical pH sensor required for the initiation of viral membrane fusion [89], but mutational studies on WNV do not support a requirement for any single histidine of E and suggest that non-histidine residues of the E protein might be involved in initiating the pH-dependent exposure of the hydrophobic fusion loop [90].
The flavivirus E glycoprotein with its three domains, each largely adopting β-sheet folds, has been assigned as a class II fusion protein [91] (Fig 2). Triggered by low pH, the E proteins undergo extensive conformational and positional transformations that drive membrane fusion, expose the fusion loop to interact with the target membrane, and establish a close positioning of viral and target membranes leading to fusion pore formation [13] (Fig. 3). The pre-fusion conformation of E at neutral pH (Figs. 2A and 2B) and the structure of the E ectodomain at low pH, after fusion completion (Fig. 2C), is known for several flaviviruses [18]. The post-fusion structure is a trimeric arrangement of E, with the E monomers parallel to one another and with their long axis oriented perpendicular to the plane of the membrane [13] (Fig. 2C). The fusion peptides are exposed on one end of the trimer, suitable for penetrating into the outer leaflet of the associated lipid bilayer. Hydrophobic grooves, in which the stem regions are predicted to bind, are generated by the trimerization and extend along the interface of two neighboring DIIs towards the fusion loop. Compared to the pre-fusion dimer (Fig. 2B), DIII of E has undergone a major rotational displacement to the side of DI and towards DII (Fig. 2C). The fold-back of DIII along the sides of the trimer brings the transmembrane domains in the viral envelope spatially close to the fusion peptide at the tip of DII that is embedded in the target membrane (Fig. 3). At the same time, the membranes are locally deformed and destabilized.
The oligomerization state of the post-fusion E structure suggests the presence of a pre-fusion trimeric arrangement of E. The formation of such fusogenic E trimers has to be preceded by the protonation-dependent disruption of the raft arrangement on the surface of the virion, and the dissociation of E dimers into monomers [92] (Fig. 3B). The resultant exposure of the fusion loop on the tip of the outward projected DII allows its insertion in the outer leaflet of the target membrane, thereby anchoring the E protein simultaneously in viral and target membrane, followed by the reassociation of E molecules into trimers [13] (Figs. 3B and 3C). Trimerization has been shown to be essential for fusion activity of synthetic fusion peptides derived from TBEV [93]. The large-scale reorganization of E molecules is aided by the release and extension of the E stem region away from the viral membrane [94] supporting the idea of formation of an extended “pre-hairpin” intermediate [12] before its collapse into a fold-back conformation (Fig. 3). The release of the E stem region from the viral lipid envelope is a pre-requisite for the conversion into the post-fusion E trimer, in which the stem is stretched out along the newly generated groove between associated DII domains (Fig. 2C). The irreversible fold-back conversion of the E protein into the post-fusion hairpin-like structure requires the rotational repositioning of DIII and zipping up of the stem bringing the E transmembrane helices in the viral membrane in juxtaposition with the target membrane-anchored fusion loop [19, 20] (Fig. 3). The fusion process progresses via a hemifusion intermediate to the formation of the post-fusion E trimer and opening of the fusion pore [95]. The free energy liberated by the rearrangement of the viral surface proteins from a metastable homodimer to a more stable homotrimer formation is likely used to overcome the kinetic barrier hindering fusion of the two membrane bilayers [12, 13]. The number of E trimers necessary for the cooperative formation of the fusion pore is still a matter of debate [13, 95, 96].
4. Concluding remarks
Significant advances towards understanding flavivirus cell entry have been made and a series of conformational events can be inferred from available structures and biochemical data. However, further studies will be necessary to dissect the kinetics of the virus-cell fusion pathway and to understand the order of the large, conformational changes at a molecular level. The information obtained from structure can provide insight for the development of preventive and therapeutic means against the spread and effect of pathogenic flaviviruses, allowing the design and targeting of antiviral agents or vaccines.
Acknowledgments
We thank Sheryl L. Kelly for her help in the preparation of the manuscript. We are grateful to Wei Zhang and Karin Stiasny for providing the originals of Figs. 1A and 3. We apologize to all authors whose original work could not be quoted because of limitation in the allowed number of references. This work was supported by NIH grant 1 R01 AI76331 to M.G.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- 2.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 3.Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Assenberg R, Mastrangelo E, Walter TS, Verma A, Milani M, Owens RJ, Stuart DI, Grimes JM, Mancini EJ. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol. 2009;83:12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson AD. Chapter 2. New insights into flavivirus nonstructural protein 5. Adv Virus Res. 2009;74:41–101. doi: 10.1016/S0065-3527(09)74002-3. [DOI] [PubMed] [Google Scholar]
- 6.Konishi E, Mason PW. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J Virol. 1993;67:1672–1675. doi: 10.1128/jvi.67.3.1672-1675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allison SL, Stadler K, Mandl CW, Kunz C, Heinz FX. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72:1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- 9.Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J GenVirol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 11.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 12.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stiasny K, Heinz FX. Flavivirus membrane fusion. J GenVirol. 2006;87:2755–2766. doi: 10.1099/vir.0.82210-0. [DOI] [PubMed] [Google Scholar]
- 14.Dokland T, Walsh M, Mackenzie JM, Khromykh AA, Ee KH, Wang S. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 18.Stiasny K, Fritz R, Pangerl K, Heinz FX. Molecular mechanisms of flavivirus membrane fusion. Amino Acids. 2009 doi: 10.1007/s00726-00009-00370-00724. [DOI] [PubMed] [Google Scholar]
- 19.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 20.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J Gen Virol. 2005;86:405–412. doi: 10.1099/vir.0.80411-0. [DOI] [PubMed] [Google Scholar]
- 23.Lee JW, Chu JJ, Ng ML. Quantifying the specific binding between West Nile virus envelope domain III protein and the cellular receptor αVβ3 integrin. J Biol Chem. 2006;281:1352–1360. doi: 10.1074/jbc.M506614200. [DOI] [PubMed] [Google Scholar]
- 24.Huerta V, Chinea G, Fleitas N, Sarria M, Sanchez J, Toledo P, Padron G. Characterization of the interaction of domain III of the envelope protein of dengue virus with putative receptors from CHO cells. Virus Res. 2008;137:225–234. doi: 10.1016/j.virusres.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Wengler G. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J Virol. 1989;63:2521–2526. doi: 10.1128/jvi.63.6.2521-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 28.Heinz FX, Stiasny K, Puschner-Auer G, Holzmann H, Allison SL, Mandl CW, Kunz C. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 29.Moesker B, Rodenhuis-Zybert IA, Meijerhof T, Wilschut J, Smit JM. Characterization of the functional requirements of West Nile virus membrane fusion. J GenVirol. 2010;91:389–393. doi: 10.1099/vir.0.015255-0. [DOI] [PubMed] [Google Scholar]
- 30.Rodenhuis-Zybert IA, van der Schaar HM, Silva Voorham JM, Ende-Metselaar H, Lei HY, Wilschut J, Smit JM. Immature dengue virus: a veiled pathogen? PLoS Pathog. 2010;6:e1000718. doi: 10.1371/journal.ppat.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randolph VB, Winkler G, Stollar V. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology. 1990;174:450–458. doi: 10.1016/0042-6822(90)90099-d. [DOI] [PubMed] [Google Scholar]
- 32.Davis CW, Nguyen HY, Hanna SL, Sanchez MD, Doms RW, Pierson TC. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J Virol. 2006;80:1290–1301. doi: 10.1128/JVI.80.3.1290-1301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zybert IA, Ende-Metselaar H, Wilschut J, Smit JM. Functional importance of dengue virus maturation: infectious properties of immature virions. J Gen Virol. 2008;89:3047–3051. doi: 10.1099/vir.0.2008/002535-0. [DOI] [PubMed] [Google Scholar]
- 34.Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 2009;28:3269–3276. doi: 10.1038/emboj.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston LJ, Halliday GM, King NJ. Phenotypic changes in Langerhans' cells after infection with arboviruses: a role in the immune response to epidermally acquired viral infection? J Virol. 1996;70:4761–4766. doi: 10.1128/jvi.70.7.4761-4766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marovich M, Grouard-Vogel G, Louder M, Eller M, Sun W, Wu SJ, Putvatana R, Murphy G, Tassaneetrithep B, Burgess T, Birx D, Hayes C, Schlesinger-Frankel S, Mascola J. Human dendritic cells as targets of dengue virus infection. J Investig Dermatol Symp Proc. 2001;6:219–224. doi: 10.1046/j.0022-202x.2001.00037.x. [DOI] [PubMed] [Google Scholar]
- 37.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, Steinman RM, Schlesinger S, Marovich MA. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barba-Spaeth G, Longman RS, Albert ML, Rice CM. Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J Exp Med. 2005;202:1179–1184. doi: 10.1084/jem.20051352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mentor NA, Kurane I. Dengue virus infection of human T lymphocytes. Acta Virol. 1997;41:175–176. [PubMed] [Google Scholar]
- 40.Bielefeldt-Ohmann H. Analysis of antibody-independent binding of dengue viruses and dengue virus envelope protein to human myelomonocytic cells and B lymphocytes. Virus Res. 1998;57:63–79. doi: 10.1016/s0168-1702(98)00087-2. [DOI] [PubMed] [Google Scholar]
- 41.King AD, Nisalak A, Kalayanrooj S, Myint KS, Pattanapanyasat K, Nimmannitya S, Innis BL. B cells are the principal circulating mononuclear cells infected by dengue virus. Southeast Asian J Trop Med Public Health. 1999;30:718–728. [PubMed] [Google Scholar]
- 42.Moreno-Altamirano MM, Sanchez-Garcia FJ, Munoz ML. Non Fc receptor-mediated infection of human macrophages by dengue virus serotype 2. J GenVirol. 2002;83:1123–1130. doi: 10.1099/0022-1317-83-5-1123. [DOI] [PubMed] [Google Scholar]
- 43.Wei HY, Jiang LF, Fang DY, Guo HY. Dengue virus type 2 infects human endothelial cells through binding of the viral envelope glycoprotein to cell surface polypeptides. J GenVirol. 2003;84:3095–3098. doi: 10.1099/vir.0.19308-0. [DOI] [PubMed] [Google Scholar]
- 44.Suksanpaisan L, Cabrera-Hernandez A, Smith DR. Infection of human primary hepatocytes with dengue virus serotype 2. J Med Virol. 2007;79:300–307. doi: 10.1002/jmv.20798. [DOI] [PubMed] [Google Scholar]
- 45.Kou Z, Quinn M, Chen H, Rodrigo WW, Rose RC, Schlesinger JJ, Jin X. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J MedVirol. 2008;80:134–146. doi: 10.1002/jmv.21051. [DOI] [PubMed] [Google Scholar]
- 46.Ceccaldi PE, Lucas M, Despres P. New insights on the neuropathology of West Nile virus. FEMS Microbiol Lett. 2004;233:1–6. doi: 10.1016/j.femsle.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 47.Rios M, Zhang MJ, Grinev A, Srinivasan K, Daniel S, Wood O, Hewlett IK, Dayton AI. Monocytes-macrophages are a potential target in human infection with West Nile virus through blood transfusion. Transfusion. 2006;46:659–667. doi: 10.1111/j.1537-2995.2006.00769.x. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 49.Hilgard P, Stockert R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology. 2000;32:1069–1077. doi: 10.1053/jhep.2000.18713. [DOI] [PubMed] [Google Scholar]
- 50.Thullier P, Demangel C, Bedouelle H, Megret F, Jouan A, Deubel V, Mazie JC, Lafaye P. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J Gen Virol. 2001;82:1885–1892. doi: 10.1099/0022-1317-82-8-1885. [DOI] [PubMed] [Google Scholar]
- 51.Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RW, Zarski JP, Drouet E. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology. 2002;292:162–168. doi: 10.1006/viro.2001.1232. [DOI] [PubMed] [Google Scholar]
- 52.Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virology. 2003;308:92–100. doi: 10.1016/s0042-6822(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 53.Lee E, Lobigs M. E protein domain III determinants of yellow fever virus 17D vaccine strain enhance binding to glycosaminoglycans, impede virus spread, and attenuate virulence. J Virol. 2008;82:6024–6033. doi: 10.1128/JVI.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen HL, Her SY, Huang KC, Cheng HT, Wu CW, Wu SC, Cheng JW. Identification of a heparin binding peptide from the Japanese encephalitis virus envelope protein. Biopolymers. 2010;94:331–338. doi: 10.1002/bip.21371. [DOI] [PubMed] [Google Scholar]
- 55.Kozlovskaya LI, Osolodkin DI, Shevtsova AS, Romanova LI, Rogova YV, Dzhivanian TI, Lyapustin VN, Pivanova GP, Gmyl AP, Palyulin VA, Karganova GG. GAG-binding variants of tick-borne encephalitis virus. Virology. 2010;398:262–272. doi: 10.1016/j.virol.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Chien YJ, Chen WJ, Hsu WL, Chiou SS. Bovine lactoferrin inhibits Japanese encephalitis virus by binding to heparan sulfate and receptor for low density lipoprotein. Virology. 2008;379:143–151. doi: 10.1016/j.virol.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 57.Aoki C, Hidari KI, Itonori S, Yamada A, Takahashi N, Kasama T, Hasebe F, Islam MA, Hatano K, Matsuoka K, Taki T, Guo CT, Takahashi T, Sakano Y, Suzuki T, Miyamoto D, Sugita M, Terunuma D, Morita K, Suzuki Y. Identification and characterization of carbohydrate molecules in mammalian cells recognized by dengue virus type 2. J Biochem. 2006;139:607–614. doi: 10.1093/jb/mvj067. [DOI] [PubMed] [Google Scholar]
- 58.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lozach PY, Burleigh L, Staropoli I, Navarro-Sanchez E, Harriague J, Virelizier JL, Rey FA, Despres P, Arenzana-Seisdedos F, Amara A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J Biol Chem. 2005;280:23698–23708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- 60.Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 61.Miller JL, de Wet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, Gordon S. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis CW, Mattei LM, Nguyen HY, Ansarah-Sobrinho C, Doms RW, Pierson TC. The location of asparagine-linked glycans on West Nile virions controls their interactions with CD209 (dendritic cell-specific ICAM-3 grabbing nonintegrin) J Biol Chem. 2006;281:37183–37194. doi: 10.1074/jbc.M605429200. [DOI] [PubMed] [Google Scholar]
- 64.Bryant JE, Calvert AE, Mesesan K, Crabtree MB, Volpe KE, Silengo S, Kinney RM, Huang CY, Miller BR, Roehrig JT. Glycosylation of the dengue 2 virus E protein at N67 is critical for virus growth in vitro but not for growth in intrathoracically inoculated Aedes aegypti mosquitoes. Virology. 2007;366:415–423. doi: 10.1016/j.virol.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Mondotte JA, Lozach PY, Amara A, Gamarnik AV. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J Virol. 2007;81:7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martina BE, Koraka P, van den DP, Rimmelzwaan GF, Haagmans BL, Osterhaus AD. DC-SIGN enhances infection of cells with glycosylated West Nile virus in vitro and virus replication in human dendritic cells induces production of IFN-α and TNF-α. Virus Res. 2008;135:64–71. doi: 10.1016/j.virusres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 67.Murata R, Eshita Y, Maeda A, Maeda J, Akita S, Tanaka T, Yoshii K, Kariwa H, Umemura T, Takashima I. Glycosylation of the West Nile Virus envelope protein increases in vivo and in vitro viral multiplication in birds. Am J Trop Med Hyg. 2010;82:696–704. doi: 10.4269/ajtmh.2010.09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Most RG, Corver J, Strauss JH. Mutagenesis of the RGD motif in the yellow fever virus 17D envelope protein. Virology. 1999;265:83–95. doi: 10.1006/viro.1999.0026. [DOI] [PubMed] [Google Scholar]
- 69.Hurrelbrink RJ, McMinn PC. Attenuation of Murray Valley encephalitis virus by site-directed mutagenesis of the hinge and putative receptor-binding regions of the envelope protein. J Virol. 2001;75:7692–7702. doi: 10.1128/JVI.75.16.7692-7702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Medigeshi GR, Hirsch AJ, Streblow DN, Nikolich-Zugich J, Nelson JA. West Nile virus entry requires cholesterol-rich membrane microdomains and is independent of αvβ3 integrin. J Virol. 2008;82:5212–5219. doi: 10.1128/JVI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thepparit C, Smith DR. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J Virol. 2004;78:12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tio PH, Jong WW, Cardosa MJ. Two dimensional VOPBA reveals laminin receptor (LAMR1) interaction with dengue virus serotypes 1, 2 and 3. Virol J. 2005;2:25. doi: 10.1186/1743-422X-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakoonwatanyoo P, Boonsanay V, Smith DR. Growth and production of the dengue virus in C6/36 cells and identification of a laminin-binding protein as a candidate serotype 3 and 4 receptor protein. Intervirology. 2006;49:161–172. doi: 10.1159/000089377. [DOI] [PubMed] [Google Scholar]
- 74.Reyes-Del Valle J, Chavez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol. 2005;79:4557–4567. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cabrera-Hernandez A, Thepparit C, Suksanpaisan L, Smith DR. Dengue virus entry into liver (HepG2) cells is independent of hsp90 and hsp70. J Med Virol. 2007;79:386–392. doi: 10.1002/jmv.20786. [DOI] [PubMed] [Google Scholar]
- 76.Jindadamrongwech S, Thepparit C, Smith DR. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch Virol. 2004;149:915–927. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- 77.Das S, Laxminarayana SV, Chandra N, Ravi V, Desai A. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology. 2009;385:47–57. doi: 10.1016/j.virol.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 78.Salas-Benito J, Reyes-Del Valle J, Salas-Benito M, Ceballos-Olvera I, Mosso C, Del Angel RM. Evidence that the 45-kD glycoprotein, part of a putative dengue virus receptor complex in the mosquito cell line C6/36, is a heat-shock related protein. Am J Trop Med Hyg. 2007;77:283–290. [PubMed] [Google Scholar]
- 79.Ren J, Ding T, Zhang W, Song J, Ma W. Does Japanese encephalitis virus share the same cellular receptor with other mosquito-borne flaviviruses on the C6/36 mosquito cells? Virol J. 2007;4:83. doi: 10.1186/1743-422X-4-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Halstead SB, O'Rourke EJ. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J Exp Med. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daughaday CC, Brandt WE, McCown JM, Russell PK. Evidence for two mechanisms of dengue virus infection of adherent human monocytes: trypsin-sensitive virus receptors and trypsin-resistant immune complex receptors. Infect Immun. 1981;32:469–473. doi: 10.1128/iai.32.2.469-473.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Schaar HM, Rust MJ, Chen C, Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Acosta EG, Castilla V, Damonte EB. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J Gen Virol. 2008;89:474–484. doi: 10.1099/vir.0.83357-0. [DOI] [PubMed] [Google Scholar]
- 84.Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gollins SW, Porterfield JS. pH-dependent fusion between the flavivirus West Nile and liposomal model membranes. J Gen Virol. 1986;67(Pt 1):157–166. doi: 10.1099/0022-1317-67-1-157. [DOI] [PubMed] [Google Scholar]
- 86.Corver J, Ortiz A, Allison SL, Schalich J, Heinz FX, Wilschut J. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology. 2000;269:37–46. doi: 10.1006/viro.1999.0172. [DOI] [PubMed] [Google Scholar]
- 87.Koschinski A, Wengler G, Repp H. The membrane proteins of flaviviruses form ion-permeable pores in the target membrane after fusion: identification of the pores and analysis of their possible role in virus infection. J Gen Virol. 2003;84:1711–1721. doi: 10.1099/vir.0.19062-0. [DOI] [PubMed] [Google Scholar]
- 88.Stiasny K, Heinz FX. Effect of membrane curvature-modifying lipids on membrane fusion by tick-borne encephalitis virus. J Virol. 2004;78:8536–8542. doi: 10.1128/JVI.78.16.8536-8542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. 2008;183:353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelson S, Poddar S, Lin TY, Pierson TC. Protonation of individual histidine residues is not required for the pH-dependent entry of West Nile virus: evaluation of the "histidine switch" hypothesis. J Virol. 2009;83:12631–12635. doi: 10.1128/JVI.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 92.Stiasny K, Kossl C, Lepault J, Rey FA, Heinz FX. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 2007;3:e20. doi: 10.1371/journal.ppat.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan J, Lai CB, Scott WR, Straus SK. Synthetic fusion peptides of tick-borne encephalitis virus as models for membrane fusion. Biochemistry. 2010;49:287–296. doi: 10.1021/bi9017895. [DOI] [PubMed] [Google Scholar]
- 94.Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 2009;5:e1000672. doi: 10.1371/journal.ppat.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harrison SC. The pH sensor for flavivirus membrane fusion. J Cell Biol. 2008;183:177–179. doi: 10.1083/jcb.200809175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]