Abstract
In plants, Ca2+, phosphatidylinositol phosphates (PtdInsPs) and inositol phosphates are major components of intracellular signaling. Several kinds of proteins and enzymes, such as calmodulin (CaM), protein kinase, protein phosphatase, and the Ca2+ channel, mediate the signaling. Two new Ca2+-binding proteins were identified from Arabidopsis thaliana and named PCaP1 and PCaP2 [plasma membrane (PM)-associated Ca2+(cation)-binding protein 1 and 2]. PCaP1 has an intrinsically disordered region in the central and C-terminal parts. The PCaP1 gene is expressed in most tissues and the PCaP2 gene is expressed predominantly in root hairs and pollen tubes. We recently demonstrated that these proteins are N-myristoylated, stably anchored in the PM, and are bound with phosphatidylinositol phosphates, especially PtdInsP2s. Here we propose a model for the switching mechanism of Ca2+-signaling mediated by PtdInsPs. Ca2+ forms a complex with CaM (Ca2+-CaM) when there is an increase in the cytosol free Ca2+. The binding of PCaPs with Ca2+-CaM causes PCaPs to release PtdInsPs. Until the release of PtdInsPs, the signaling is kept in the resting state.
Key words: calcium signal, calmodulin, inositol phosphate, intrinsically disordered protein, myristoylation, phosphatidylinositol phosphate, plasma membrane
Introduction
The versatile Ca2+ signaling in cells derives from the functioning of several Ca2+-binding proteins, such as calmodulin (CaM), calreticulin, calnexin and annexin, which are localized in their specific organelles. These Ca2+-binding proteins transfer signals through change in Ca2+ concentration by interacting with partner proteins. Two Ca2+-binding proteins recently identified in Arabidopsis thaliana, AtPCaP1 and AtPCaP2 (hereafter referred to as PCaP1 and PCaP2), have been found to have unique characteristics: they are N-myristoylated and associated with the PM stably,1–3 and bind with the Ca2+-CaM complex and phosphatidylinositol phosphates (PtdInsPs). In this report, we suggest that PCaP1 and PCaP2 are involved in the crosstalk between CaM and PtdInsPs signaling.
Structural Characteristics and Association with the PM
The N-myristoylation of PCaP1 and PCaP2 and their stable association with the PM have been demonstrated by in vitro myristoylation assay and single amino acid substitution.2,3 Both proteins have a putative N-myristoylation consensus sequence, Met-Gly-X-X-X-Ser-Lys.4 Several lines of evidence show that PCaP1 can stably bind with the PM. For example PCaP1 is not released after treatment with NaCl, urea or NaCO3.2
PCaP1 and PCaP2 are rich in glutamate, lysine, proline and valine residues (PEVK-rich domain). It should be noted that PCaP1 has an intrinsically disordered region in the central and C-terminal parts.5 The intrinsically disordered (ID) protein is defined to possess a relatively long sequence of more than 50 amino acid residues that is intrinsically disordered or has no folded structure. Most ID proteins are rich in glutamate, lysine, proline, serine or glutamine residues.6 A PCaP orthologue in radish (RVCaB) has also been demonstrated to be an intrinsically disordered protein.7 These PCaPs might have an irregular tertiary structure instead of a typical globular one. Indeed, they migrated abnormally in SDS-polyacrylamide gel electrophoresis. Recombinant PCaP1 and PCaP2, with a size of 36 and 43 kDa, respectively, have been detected although their calculated molecular weights are 24,584 and 18,549.1–3
ID proteins or ID regions have a flexible structure and can bind with ligands or partner proteins without structural hindrance. For this reason, RVCaB can bind with a large number of Ca.2–7 Recently it has been proposed that ID regions confer hub proteins with the ability to interact with multiple proteins in interaction networks.6 Association of ID protein with a partner protein causes a transition to highly ordered structure.8 This structural change might be essential for the function of ID proteins including PCaPs.
Interaction with PtdInsPs and Ca2+/CaM
The property of PCaPs most important for its physiological role is the ability to bind with PtdInsPs. PCaPs were shown to interact with PtdInsPs using PIP Strips™ and PIP Array™ sheet.2,3 Various proteins exhibit their physiological roles by associating with PtdInsPs.9 However, PCaP1 and PCaP2 are distinct from these proteins in their primary sequences. Both PCaP1 and PCaP2 were found to bind with PtdIns(3,4)P2, PtdIns(3,5) P2, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in an in vitro assay. These PtdInsPs except for PtdIns(3,4,5)P3 exist in plant cells.10 Therefore, PtdInsP2s are good candidates of ligands. The N-terminal part of 25 amino acid residues is essential for PCaP1 to interact with PtdInsPs, because the mutant PCaP1 lacking the part completely had no PtdInsP-binding ability.2 PCaP2 has a N-terminal sequence similar to that of PCaP1 (18 out of 25 residues are identical) and is thought to interact specifically with PtdInsPs at its N-terminal region.
A Ca2+-CaM complex, but not a free CaM, is another ligand, and its binding to PCaPs causes dissociation of PtdInsPs from PCaPs.2,3 This reaction can mediate the signal of Ca2+ to PtdInsPs in living cells. The site or sequence of PCaPs binding with the Ca2+-CaM complex is unclear at present, although a certain N-myristoyl protein interact with the Ca2+-CaM complex at its N-terminal domain.11
PCaP2 associated with microtubules is also called MAP18.12 Microtubules may be involved in the tip growth of root hairs and pollen tubes.13 In our experiments using PCaP2 fused with GFP, however, the green fluorescence of GFP was clearly detected only on the PM.3 There is a possibility that the GFP portion of the fusion protein disturbs the interaction between microtubules and PCaP2. Although the interaction of MAP18 (PCaP2) with microtubules needs to be discussed, here we focus on the interaction of PCaP2 with CaM and PtdInsPs.
Hypothetical Role of PCaPs: A Cross Talk between Ca2+ and PtdInsPs
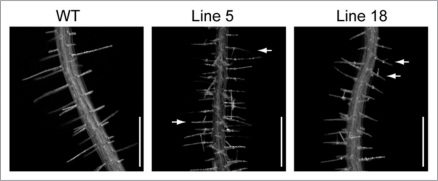
PCaP2 is expressed predominantly in root hairs and elongating pollen tubes, which are categorized as tip growth cells.3 PtdInsPs have been reported to be involved in polarized cell growth of these cells.13–15 We introduced a PCaP2-GFP construct with its own promoter into the wild type of A. thaliana. Therefore, the transgenic plants are mild overexpression lines. Accumulation of the protein was confirmed by detection of GFP fluorescence on the PM. Most lines showed normal growth of roots with straight root hairs. However, branched root hairs were observed in several lines, which showed relatively strong fluorescence of GFP (Fig. 1), indicating involvement of PCaP2 in root hair growth. This is the only morphological alteration observed in the mild overexpression lines. This point needs to be confirmed using other overexpression mutants with the root-hair specific promoter.
Figure 1.
Morphological changes in mild overexpression lines of PCaP2. A PCaP2-GFP construct with its own promoter was introduced into the wild type of A. thaliana. Morphological changes were observed in root hairs of several mutant lines. Roots of 9-day-old seedlings were photographed. Branched root hairs, which are indicated by arrows, were observed in most mutant lines. Bar = 0.5 mm.
In plant cells, PtdInsPs constitute a minor fraction of the total membrane lipids.16,17 The amount of PCaP1 protein is assumed to be balanced with the amount of PtdInsP2 protein in the PM. PCaP1 protein, but not PCaP2, can be detected in the membrane fractions prepared from shoots and roots by immunoblotting.1 The absence of PCaP2 is due to its low content and restricted expression, because the protein is expressed in root hairs, pollen tubes, and root epidermal cells. Therefore, PCaP2 may play a role in these specific cells. The amount of PCaP1, PCaP2 and PtdInsP2 in the PM remains to be quantified.
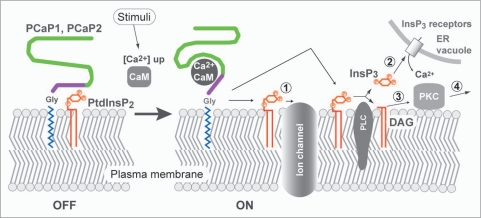
We propose a model for the function of PCaPs as a molecular switch of the Ca2+ signaling mediated by PtdInsPs and CaM. At resting cytosolic [Ca2+], PCaPs bind to phosphate moieties on the inositol ring of PtdInsPs in the PM. Indeed, Ca2+-signaling plays a key role in the polarized growth of root hairs.18 When the cytosolic [Ca2+] is elevated, Ca2+ forms a Ca2+-CaM complex that promotes dissociation of PtdInsPs from PCaPs. The free PtdInsP2 can interact with particular ion channels such as K+ channel and regulates their gating19–21 (Fig. 2, step 1). Inositol(1,4,5)P3 (InsP3) and diacylglycerol (DAG) are produced by hydrolysis of PtdIns(4,5)P2 by phospholipase C (PLC). Consequently the release of Ca2+ from ER and vacuole are enhanced by the activation of Ca2+ channels (step 2). DAG can function as a signal mediator and activates protein kinase C (PKC) together with Ca2+ (step 3). The activated PKC phosphorylates specific target molecules.
Figure 2.
Schematic representation of the proposed role of PCaP1 and PCaP2 in the PM. During the resting state, PCaPs hold PtdlnsPs on the membrane surface. Increased cytosolic [Ca2+] leads formation of a Ca2+-CaM complex. Interaction with Ca2+CaM complex stimulates release of PtdlnsP2 from PCaPs. the free PtdlnsP2 interacts with particular ion channels and regulates its function (step 1). In another case, Ptdins(4,5)P2 is hydrolyzed by PLC and a product InsP3 enhances the release of ca2+ from ER and vacuole by activation of Ca2+ channels (InsP3 receptors) (step 2). Remaining DAG activates PKC together with increased [Ca2+] (step 3). The activated PKC modifies target molecules (step 4).
PCaPs have been shown to interact with ligands, PtdInsPs and Ca2+-CaM, and the binding with Ca2+-CaM subsequently affects its affinity for PtdInsPs. However, the down stream of PtdInsP signaling remains to be examined. Furthermore we should examine the following possibilities. (1) PCaPs are distributed unevenly or are clustered in the PM and function in a spatio-temporal dependent manner. (2) PCaPs can interact with particular partner proteins in addition to CaM to exhibit their biochemical function as novel hub proteins. PCaPs are located at a cross-talk point between PtdInsPs and Ca2+/CaM. The Arabidopsis genome harbors seven CaMs and about 50 CaM-like genes.22 Examination of the interaction of each CaM with PCaP1 and PCaP2, and phenotypic analysis of the knockout and overexpression of the PCaP genes might help elucidate the signaling pathway.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11825
References
- 1.Ide Y, Nagasaki N, Tomioka R, Suito M, Kamiya T, Maeshima M. Molecular properties of a novel, hydrophilic cation-binding protein associated with the plasma membrane. J Exp Bot. 2007;58:1173–1183. doi: 10.1093/jxb/erl284. [DOI] [PubMed] [Google Scholar]
- 2.Nagasaki N, Tomioka R, Maeshima M. A hydrophilic cation-binding protein of Arabidopsis thaliana AtPCaP1 is localized to plasma membrane via N-myristoylation and interacts with calmodulin and phosphatidylinositol phosphates, PtdIns(3,4,5)P3 and PtdIns(3,5)P2. FEBS J. 2008;275:2267–2282. doi: 10.1111/j.1742-4658.2008.06379.x. [DOI] [PubMed] [Google Scholar]
- 3.Kato M, Nagasaki-Takeuchi N, Ide Y, Maeshima M. An Arabidopsis hydrophilic Ca2+-binding protein with a PEVK-rich domain, PCaP2, is associated with the plasma membrane and interacts with calmodulin and phosphatidylinositol phosphates. Plant Cell Physiol. 2010;51:366–379. doi: 10.1093/pcp/pcq003. [DOI] [PubMed] [Google Scholar]
- 4.Rosenhouse-Dantsker A, Logothetis DE. Molecular characteristics of phosphoinositide binding. Pflugers Arch—Eur J Physiol. 2007;455:45–53. doi: 10.1007/s00424-007-0291-6. [DOI] [PubMed] [Google Scholar]
- 5.Nagasaki-Takeuchi N, Miyano M, Maeshima M. A plasma membrane-associated protein of Arabidopsis thaliana AtPCaP1 binds copper ions and changes its higher order structure. J Biochem. 2008;144:487–497. doi: 10.1093/jb/mvn092. [DOI] [PubMed] [Google Scholar]
- 6.Ishijima J, Nagasaki N, Maeshima M, Miyano M. RVCaB, a calcium-binding protein in radish vacuoles, is predominantly an unstructured protein with a polyproline type II helix. J Biochem. 2007;142:201–211. doi: 10.1093/jb/mvm130. [DOI] [PubMed] [Google Scholar]
- 7.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Kleerekoper QK, Putkey JA. PEP-19, an intrinsically disordered regulator of calmodulin signaling. J Biol Chem. 2009;284:7455–7464. doi: 10.1074/jbc.M808067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaadige MR, Ayer DE. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J Biol Chem. 2006;281:28831–28836. doi: 10.1074/jbc.M605624200. [DOI] [PubMed] [Google Scholar]
- 10.Meijer HJG, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- 11.Matsubara M, Nakatsu T, Kato H, Taniguchi H. Crystal structure of a myristoylated CAP-23/NAP22 N-terminal domain complexed with Ca2+/calmodulin. EMBO J. 2004;23:712–718. doi: 10.1038/sj.emboj.7600093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Zhu L, Liu B, Wang L, Jin L, Zhao Q, et al. Arabidopsis MICROTUBLE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell. 2007;19:877–889. doi: 10.1105/tpc.106.048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusano H, Testerink C, Vermeer JEM, Tsuge T, Shimada H, Oka A, et al. The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell. 2008;20:367–380. doi: 10.1105/tpc.107.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helling D, Possart A, Cottier S, Klahre U, Kost B. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell. 2006;18:3519–3534. doi: 10.1105/tpc.106.047373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Bak G, Choi Y, Chuang WI, Cho HT, Lee Y. Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol. 2008;147:624–635. doi: 10.1104/pp.108.117341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF. Inositol signaling and plant growth. Trends Plant Sci. 2000;5:252–258. doi: 10.1016/s1360-1385(00)01652-6. [DOI] [PubMed] [Google Scholar]
- 17.Jung JY, Kim YW, Kwak JM, Hwang JU, Young J, Schroeder JI, et al. Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell. 2002;14:2399–2414. doi: 10.1105/tpc.004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monshausen GB, Messerli MA, Gilroy S. Imaging of the yellow cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 2008;147:1690–1698. doi: 10.1104/pp.108.123638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma X, Shor O, Diminshtein S, Yu L, Im YJ, Perera I, et al. Phosphatidylinositol (4,5) bisphosphate inhibits K+-efflux channel activity in NT1 tobacco cultured cells. Plant Physiol. 2009;149:1127–1140. doi: 10.1104/pp.108.129007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh BC, Hille B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu Rev Biophys. 2008;37:175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clapham DE. Cacium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 22.MaCormack E, Tsai YC, Braam J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005;10:383–389. doi: 10.1016/j.tplants.2005.07.001. [DOI] [PubMed] [Google Scholar]