Abstract
Deposition of core histones into chromatin in vivo transpires through an active, ATP-dependent mechanism catalyzed by molecular motor proteins, such as CHD1.
The organization of chromatin affects all aspects of nuclear DNA metabolism in eukaryotes. H3.3 is an evolutionarily conserved histone variant and a key substrate for replication-independent chromatin assembly. Elimination of maternal chromatin remodelling factor CHD1 in Drosophila embryos abolishes incorporation of H3.3 into the male pronucleus, renders the paternal genome unable to participate in zygotic mitoses and leads to the development of haploid embryos. Furthermore, CHD1 but not ISWI interacts with HIRA in cytoplasmic extracts. Our findings establish CHD1 as a major factor in replacement histone metabolism in the nucleus and reveal a critical role for CHD1 in the earliest developmental instances of genome-scale, replication-independent nucleosome assembly. Furthermore, our results point to the general requirement of ATP-utilizing motor proteins for histone deposition in vivo.
Histone-DNA interactions constantly change during various processes of DNA metabolism. Recent studies have highlighted the importance of histone variants, such as H3.3, CENP-A or H2A.Z, in chromatin dynamics (1, 2). Incorporation of replacement histones into chromatin occurs throughout the cell cycle, whereas nucleosomes containing canonical histones are assembled exclusively during DNA replication. A thorough understanding of the replication-independent mechanisms of chromatin assembly, however, is lacking.
In vitro, chromatin assembly requires the action of histone chaperones and ATP-utilizing factors (3). Histone chaperones may specialize for certain histone variants. For example, H3.3 associates with a complex containing HIRA, whereas canonical H3 is in a complex with CAF-1 (4). The molecular motors known to assemble nucleosomes are ACF, CHRAC and RSF, which contain the Snf2 family member ISWI as the catalytic subunit (5-7), and CHD1, which belongs to the CHD subfamily of Snf2-like ATPases (8). These factors have not been shown to mediate deposition of histones in vivo. We previously demonstrated that CHD1 together with the chaperone NAP-1 assembles nucleosome arrays from DNA and histones in vitro (9). Here we investigated the role of CHD1 in chromatin assembly in vivo in Drosophila.
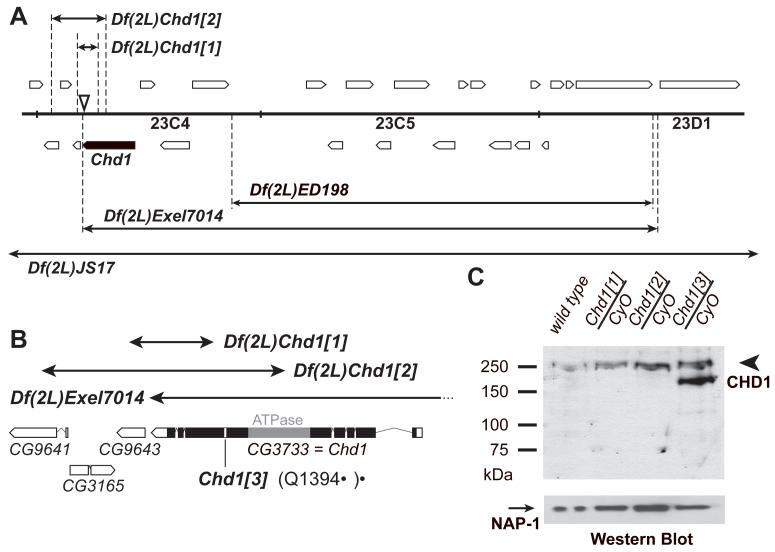
We generated Chd1 alleles by P-element-mediated mutagenesis (Fig. 1A). Two excisions, Df(2L)Chd1[1] and Df(2L)Chd1[2], deleted fragments of the Chd1 gene and fragments of unrelated adjacent genes. Heterozygous combinations, however, of Chd1[1] or Chd1[2] with Df(2L)Exel7014 affect both copies of only the Chd1 gene (Fig. 1B). We also identified a single point mutation that results in premature translation termination of Chd1 (Q1394*) in a previously described lethal allele, l(2)23Cd[A7-4] (FlyBase.org). Hence, l(2)23Cd[A7-4] was renamed Chd1[3].
Fig. 1.
Characterization of Chd1 mutant alleles. (A) Genomic structure of the Chd1 locus. Df(2L)JS17 and Df(2L)Exel7014 uncover Chd1. Open boxes, predicted genes. Closed box, Chd1. Arrows, chromosome deficiencies. Dashed lines, deficiency breakpoints. Triangle, P{EPgy2}EY07345 insertion that was used for excisions. (B) The Chd1[1] and Chd1[2] excisions delete 296 and 958 amino acids, respectively, from the C-terminus of CHD1. Chd1[3] has a nonsense mutation resulting in a stop at Q1394. The distal breakpoint of Df(2L)Exel7014 is located immediately downstream of the Chd1 3′-UTR. Open boxes, predicted genes. Closed box, Chd1 coding sequence. Gray box, Chd1 ATPase domain. (C) Western blot of heterozygous mutant embryos. Truncated CHD1 polypeptides are not detected in heterozygous Chd1[1] or Chd1[2] embryos. Heterozygous Chd1[3] embryos express a truncated (residues 1–1394) CHD1 polypeptide. Arrowhead, wild-type CHD1 (250 kDa). Arrow, NAP-1 (loading control). CyO, second-chromosome balancer.
Western analysis of embryos from heterozygous Chd1[3] flies revealed the presence of a truncated polypeptide besides full-length CHD1 (Fig. 1C). No truncated polypeptides were detected in heterozygous Chd1[1] or Chd1[2] embryos. Therefore, the corresponding deficiencies result in null mutations of Chd1. Crosses of heterozygous Chd1 mutant alleles with Df(2L)JS17/CyO or Df(2L)Exel7014/CyO produced sub-viable adult homozygous mutant progeny (Fig. S1). Both males and females were sterile. Homozygous null females mated to wild-type males laid fertilized eggs that died before hatching. Therefore, maternal CHD1 is essential for embryonic development.
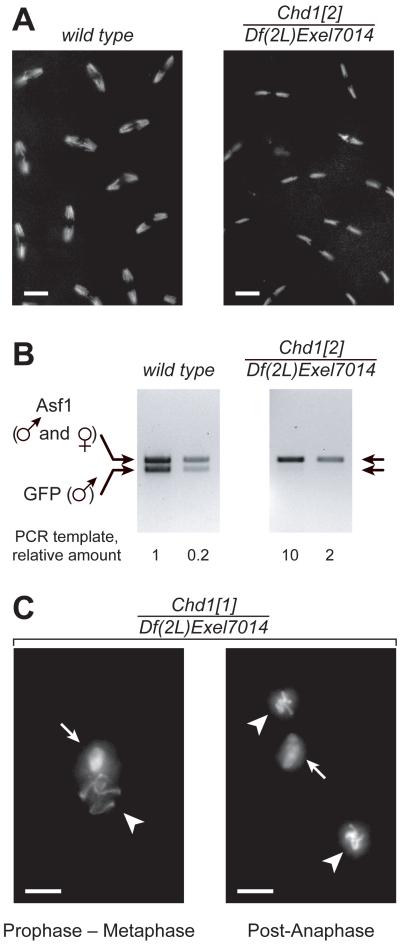
When we examined the chromosome structure of 0–4 hr old embryos laid by Chd1 null females, we observed that during syncytial mitoses (cycles 3–13) the nuclei appeared to be abnormally small. The observed numbers of anaphase chromosomes suggested that they were haploid (Fig. 2A). To confirm this observation, we mated wild-type or Chd1 null females with males that carried a GFP transgene. Embryonic DNA was amplified with primers detecting male-specific GFP and a reference gene, Asf1. In wild-type embryos, both primer pairs produced PCR products, whereas only the Asf1 fragment was amplified in the mutants (Fig. 2B). Thus, Chd1 embryos develop with haploid, maternally derived chromosome content.
Fig. 2.
Embryos from homozygous Chd1 mutant females are haploid. (A) Propidium iodide (PI) staining reveals the haploid chromosome content in Chd1 null embryos (right). Cycle 10 embryos are shown. (B) Propagation of only the maternal genome is detected by PCR in embryos from Chd1 females that have been mated with males carrying a GFP transgene. Primers for GFP recognize the paternal DNA, primers for Asf1 amplify sequences from both male and female genomes. (C) The absence of maternal CHD1 results in the inability of one pronucleus (arrows) to enter the first mitosis. The other pronucleus (arrowheads) continues with divisions (left, prophase – metaphase; right, post-anaphase). Labeling above the panels refers to genotypes of mothers. Scale bars, 10 μm.
To investigate the causes of haploidy in mutant animals, we compared distributions of various developmental stages in samples of wild-type and Chd1 null embryos (Table S1). The lack of maternal CHD1 dramatically changed this distribution. Most notably, at 0–4 hr after egg deposition, the majority of Chd1 embryos (56%) remained at a very early stage of development in contrast to the wild type (24%; Table S1).
In Drosophila eggs, meiosis gives rise to four haploid nuclei. Upon fertilization, one is selected as a female pronucleus, whereas the remaining three form the polar body. After breakdown of the sperm nuclear envelope, the compacted sperm chromatin is decondensed, and sperm-specific protamines are replaced with maternal histones. The male and female pronuclei juxtapose in the middle of the embryo and undergo one round of separate haploid mitoses. The resulting products fuse with their counterparts to give two diploid nuclei (10). In the majority of Chd1 embryos, we observed partial decondensation of the sperm chromatin and normal apposition of parental pronuclei. Then, however, one pronucleus underwent mitosis, whereas the other one did not (Fig. 2C). Considering the subsequent loss of paternal DNA (Fig. 2B), we conclude that mitotic progression of the male pronucleus is hindered in Chd1 embryos.
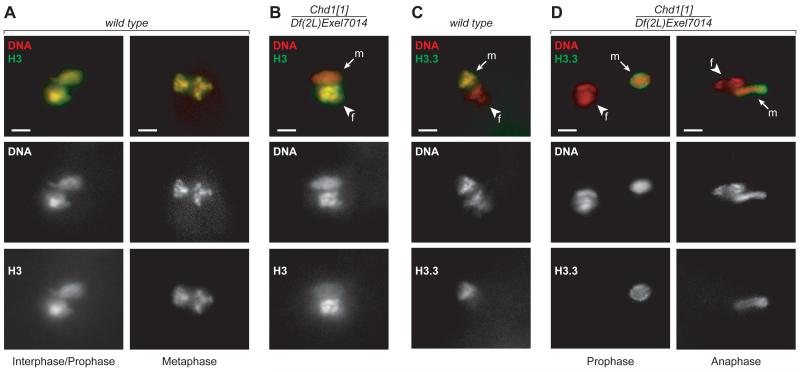
Since CHD1 can assemble nucleosomes in vitro, we asked whether the absence of CHD1 affects histone incorporation into the male pronucleus. Embryos from wild-type or Chd1 null females were stained with an antibody against histone H3. In wild-type embryos, we observed uniform staining in both parental pronuclei (Fig. 3A). In contrast, in Chd1 null embryos only the female chromatin was brightly stained. The male pronucleus contained considerably less histone H3 (Fig. 3B). These observations indicate that CHD1 is necessary for nucleosome assembly during sperm decondensation.
Fig. 3.
CHD1 is required for incorporation of histones into decondensing sperm chromatin. (A) Histone H3 co-localizes with DNA in both parental pronuclei of wild-type embryos (left, interphase/prophase; right, metaphase). (B) In Chd1 mutant embryos, the male pronucleus fails to undergo mitosis and accumulates abnormally little H3. (C) H3.3-FLAG is incorporated into chromosomes of the male pronucleus in wild-type embryos. Panels show the first metaphase. (D) In Chd1 mutant eggs, H3.3-FLAG accumulates in the periphery of the male pronucleus. The female pronucleus proceeds with mitosis (left, prophase; right, anaphase). Arrows, male pronuclei (m). Arrowheads, female pronuclei (f). Labeling above the panels refers to genotype of mothers. Red, PI; green, H3 (A and B) or H3.3-FLAG (C and D). Scale bars, 10 μm.
Sperm DNA does not replicate during decondensation, and histones are deposited by replication-independent assembly mechanisms, which involve the variant histone H3.3 but not canonical H3 (11). It has been shown in Drosophila and mice that H3.3 is specifically present in the male pronucleus (12, 13). We analyzed the distribution of H3.3 in embryos derived from Chd1 null females that carry an H3.3-FLAG transgene. In wild-type embryos, we observed co-localization of the H3.3-FLAG signal with male pronuclear DNA during migration and apposition. No H3.3-FLAG was detectable in the maternal pronucleus (Fig. 3C). In Chd1 null embryos, the male pronucleus showed altered H3.3-FLAG staining. The signal did not co-localize with the DNA but remained constrained to the nuclear periphery in a “sac”-shaped pattern (Fig. 3D).
These findings suggest that in the earliest phases of Drosophila development CHD1 is essential for the incorporation of H3.3 and normal assembly of paternal chromatin. In contrast, CHD1 does not appear to affect the organization of maternal chromatin. We conclude that CHD1 is required for replication-independent nucleosome assembly in the decondensing male pronucleus but is dispensable for replication-coupled incorporation of H3.
It was shown recently that the sèsame (ssm) mutation of Drosophila histone chaperone HIRA caused the development of haploid embryos and abolished H3.3 deposition into the male pronucleus. Chd1 and ssm mutants, however, differ profoundly in the manifestation of this phenotype. In ssm embryos, H3.3 is absent from the male pronucleus. In contrast, in Chd1 null embryos, H3.3 delivery to the male pronucleus appears to be unaffected. Thus, our observations allow to mechanistically discern the roles of CHD1 and HIRA. Whereas HIRA is essential for histone delivery to the sites of nucleosome assembly, CHD1 directly facilitates histone deposition (Fig. S3). Our findings are consistent with observations in vitro that histone chaperones either do not assemble nucleosomes or assemble them at a greatly reduced rate in the absence of ATP-utilizing factors. Our data provide evidence that histone deposition in vivo also transpires through an ATP-dependent mechanism.
CHD1 has been implicated in transcription elongation-related chromatin remodeling (14). We demonstrate that CHD1 functions in nucleosome assembly in the early Drosophila embryo, which is transcriptionally silent. The biological role of CHD1, therefore, is not confined to transcription-related processes. The S. pombe homolog of CHD1, Hrp1, has been shown to function in loading of the centromere-specific H3 variant CENP-A (15). Similar to H3.3, incorporation of CENP-A into chromatin is not restricted to S-phase. Therefore, CHD1 may have a general role in replication-independent nucleosome assembly.
Sperm decondensation involves not only histone incorporation but also eviction of protamines. To examine if CHD1 has a role in this process, we analyzed the fate of protamine B (Mst35Bb) in Chd1 null embryos. Although we detected GFP-tagged Mst35Bb in the sperm head immediately upon fertilization, we did not observe Mst35Bb-GFP signal in the male pronucleus (Fig. S2). Thus, like HIRA (16), CHD1 is dispensable for protamine removal. We have shown that the male pronucleus in Chd1 null embryos contains very low amounts of histones (Fig. 3), yet the DNA is not packaged with protamines. It remains an open question whether other DNA-protein complexes exist in the male pronucleus.
Drosophila eggs contain stores of both known chromatin assembly factors CHD1 and ISWI (Fig. S4A) and (17). Nevertheless, ISWI is unable to substitute for CHD1 in the deposition of H3.3. To examine whether CHD1 and ISWI differ in their ability to interact with the H3.3 chaperone HIRA, we performed co-immunoprecipitation experiments with extracts from embryos expressing FLAG-HIRA. CHD1 signal was readily detected in anti-FLAG immunoprecipitates, whereas ISWI did not co-immunoprecipitate with HIRA (Fig. S4B). Thus, a fraction of CHD1, but not ISWI, physically associates with HIRA. This property of CHD1 may account for its unique function in the H3.3 deposition process.
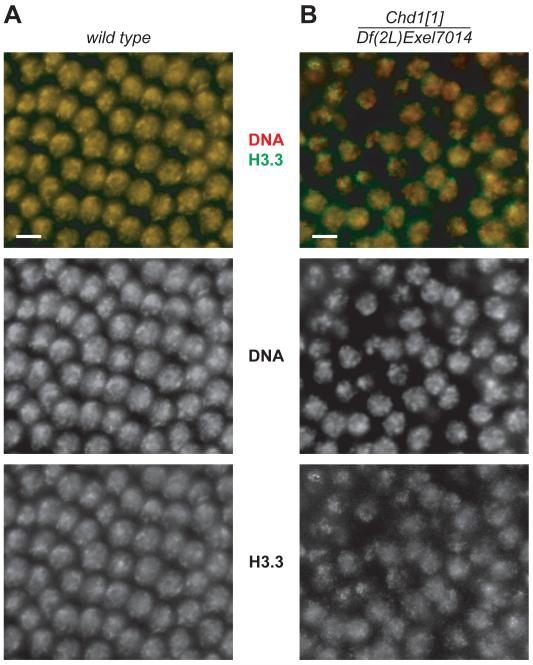
A subpopulation of Chd1 mutant haploid embryos survives beyond apposition stage (Table S1). Therefore, we asked whether H3.3 deposition is altered in Chd1 mutant embryos during later developmental stages. In wild-type nuclei, the H3.3-FLAG signal originating from the male pronucleus becomes undetectable after 2–3 divisions. Most maternal H3.3 remains distributed diffusely throughout the syncytium. After cycle 11 (roughly correlating with the onset of zygotic transcription) H3.3-FLAG is re-distributed into the nuclei, where it co-localizes with the DNA (Fig. 4A). In contrast, incorporation of H3.3 into Chd1 mutant nuclei was impaired. H3.3-FLAG produced a speckled staining with numerous bright dots that poorly overlapped with the maxima of DNA staining (Fig. 4B). It is important to note that in the ssm (HIRA) mutant, H3.3 incorporation defects in tissues or developmental stages other than the apposition stage were not observed. This result is consistent with the idea that misincorporation of H3.3 in Chd1 embryos is a direct effect of CHD1 deletion rather than a consequence of haploid development. We also conclude that CHD1 functions in H3.3 deposition during later stages of embryonic development, possibly in a HIRA-independent fashion.
Fig. 4.
Deposition of H3.3 into chromatin during syncytial blastoderm is compromised in Chd1 mutants. (A) H3.3-FLAG overlaps with DNA in syncytial nuclei of a cycle 14 wild-type embryo. (B) H3.3-FLAG co-localizes poorly with DNA in Chd1 mutant embryos. Labeling above the panels refers to genotypes of mothers. Red, PI; green, H3.3-FLAG. Scale bars, 10 μm.
This study provides evidence that ATP-dependent mechanisms are utilized for histone deposition during chromatin assembly in vivo. Thus, molecular motor proteins, such as CHD1, function not only in remodeling of existing nucleosomes but also in de novo nucleosome assembly from DNA and histones. Finally, our work identifies CHD1 as a specific factor in the assembly of nucleosomes that contain variant histone H3.3.
Supplementary Material
Acknowledgments
We thank M. Goralik-Schramel for technical assistance. We thank B. Loppin and R. Renkawitz-Pohl for fly lines and R. Perry for antibodies. We thank K. Beirit, B. Birshtein, V. Elagin, T. Juven-Gershon, M.-C. Keogh, P. Loidl, S. Morettini, M. Scharff, D. Sharp and A. Skoultchi for critical reading of the manuscript. We thank A. Tokareva for discussions and advice. This work was supported by grants from the National Institutes of Health to D.V.F. (GM74233) and J.T.K. (GM58272), the Austrian Science Foundation (Y275-B12) and Tyrolean Science Foundation to A.L., and the Swiss National Science Foundation to V.P. D.V.F is a Scholar of the Sidney Kimmel Foundation for Cancer Research.
Footnotes
(Materials and Methods are available as supporting material in Science Online).
Deposition of core histones into chromatin in vivo transpires through an active, ATP-dependent mechanism catalyzed by molecular motor proteins, such as CHD1.
References
- 1.Jin J, et al. Trends Bioch. Sci. 2005;30:680. [Google Scholar]
- 2.Henikoff S, Ahmad K. Ann. Rev. Cell Dev. Biol. 2005;21:133. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 3.Fyodorov DV, Kadonaga JT. Cell. 2001;106:523. doi: 10.1016/s0092-8674(01)00478-0. [DOI] [PubMed] [Google Scholar]
- 4.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116:51. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 5.Ito T, et al. Genes Dev. 1999;13:1529. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varga-Weisz PD, et al. Nature. 1997;388:598. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 7.Loyola A, et al. Mol. Cell. Biol. 2003;23:6759. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Proc. Natl. Acad. Sci. USA. 1997;94:11472. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lusser A, Urwin DL, Kadonaga JT. Nat. Struct. Mol. Biol. 2005;12:160. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 10.Callaini G, Riparbelli MG. Dev. Biol. 1996;176:199. doi: 10.1006/dbio.1996.0127. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad K, Henikoff S. Mol. Cell. 2002;9:1191. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 12.Loppin B, et al. Nature. 2005;437:1386. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 13.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Intl. J. Dev. Biol. 2006;50:455. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 14.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Genes Dev. 2004;18:2437. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 15.Walfridsson J, et al. Nucl. Acids Res. 2005;33:2868. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raja SJ, Renkawitz-Pohl R. Mol. Cell. Biol. 2005;25:6165. doi: 10.1128/MCB.25.14.6165-6177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deuring R, et al. Mol. Cell. 2000;5:355. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.