Abstract
Microfluidic devices offer unparalleled capability for digital microfluidic automation of sample processing and complex assay protocols in medical diagnostic and research applications. In our own work, monolithic membrane valves have enabled the creation of two platforms that precisely manipulate discrete, nanoliter-scale volumes of sample. The digital microfluidic Automaton uses two-dimensional microvalve arrays to combinatorially process nanoliter-scale sample volumes. This programmable system enables rapid integration of diverse assay protocols using a universal processing architecture. Microfabricated emulsion generator array (MEGA) devices integrate actively controlled 3-microvalve pumps to enable on-demand generation of uniform droplets for statistical encapsulation of microbeads and cells. A MEGA device containing 96 channels confers the capability of generating up to 3.4 × 106 nanoliter-volume droplets per hour for ultrahigh-throughput detection of rare mutations in a vast background of normal genotypes. These novel digital microfluidic platforms offer significant enhancements in throughput, sensitivity, and programmability for automated sample processing and analysis.
1. Introduction
The development of microfluidic sample processing and microvalve technology offers significant opportunities for the miniaturization and large scale integration of automated laboratory systems. Integrated microvalve control enables precise metering of nanoliter scale sample volumes through networks of microchannels.1–4 Functions including on-chip pumping, reagent mixing, and droplet generation have been utilized to automate a wide range of biomolecular assays. The structure of the normally-closed, monolithic membrane valves developed by our group is illustrated in Figure 1. These monolithic membrane valves have been used to automate a wide range of applications from SNP-based DNA computing5 to nanoliter-scale Sanger DNA sequencing.6 Here we report on recent advances in the development of two digital microfluidic platforms enabled by microvalve technology that achieve massively parallel biomarker analysis, and an unprecedented level of programmability for assay automation.
Figure 1.
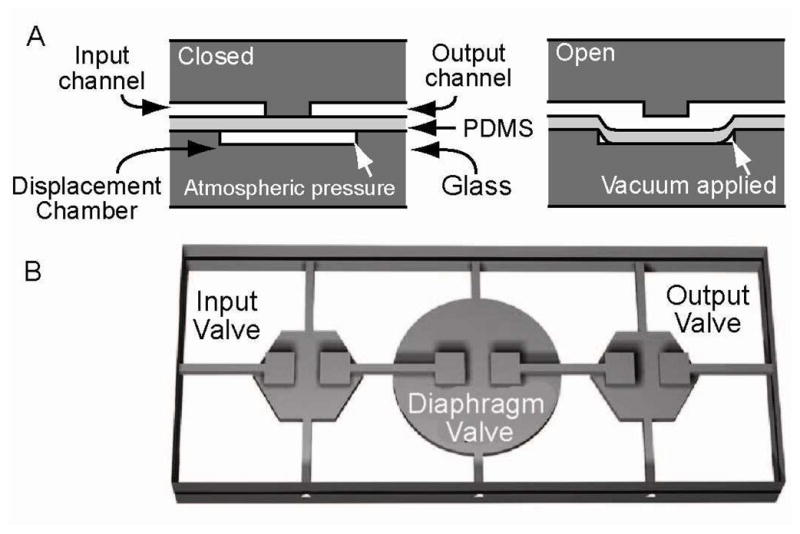
Cross sectional view through a monolithic membrane valve. The microvalves are composed of a featureless PDMS membrane sandwiched between a discontinuous fluidic channel and a pneumatic displacement chamber. (A) The valves are normally closed with the PDMS membrane resting on the valve seat. Application of a vacuum to the displacement chamber through a control channel pulls the PDMS membrane away from the discontinuity, allowing fluid to fill the chamber and/or flow across the discontinuity in the fluid channel. (B) Three independently actuated microvalves in series form an integrated micropump for transport of samples and reagents.
2. Microfluidic Automaton
A digital microfluidic Automaton has been developed, based on 2-dimensional microvalve array technology, for sample processing and analysis (Figure 2). Digital transfer of fluids between microvalves enables precise and rapid metering of nanoliter scale sample volumes through programmable valve networks within the array.7 The basic program for the transfer of fluids between microvalves in a rectilinear array begins with a single open microvalve filled with fluid. An adjacent microvalve is opened, drawing fluid from the first valve. The first valve is then closed with an applied pneumatic pressure, forcing the remainder of the fluid into the second valve. A 120 nL bolus of fluid is transferred between the microvalves under optimal conditions. Programs for reagent routing, mixing, rinsing, serial dilution, storage/retrieval and many other operations have been developed. High level device programming enables rapid automation of diverse assay protocols on a common chip format. Previous programmable digital microfluidic platforms have been demonstrated using electrowetting arrays,8–10 however these systems often suffer from significant imprecision in droplet splitting operations, thus limiting quantitative control.11
Figure 2.
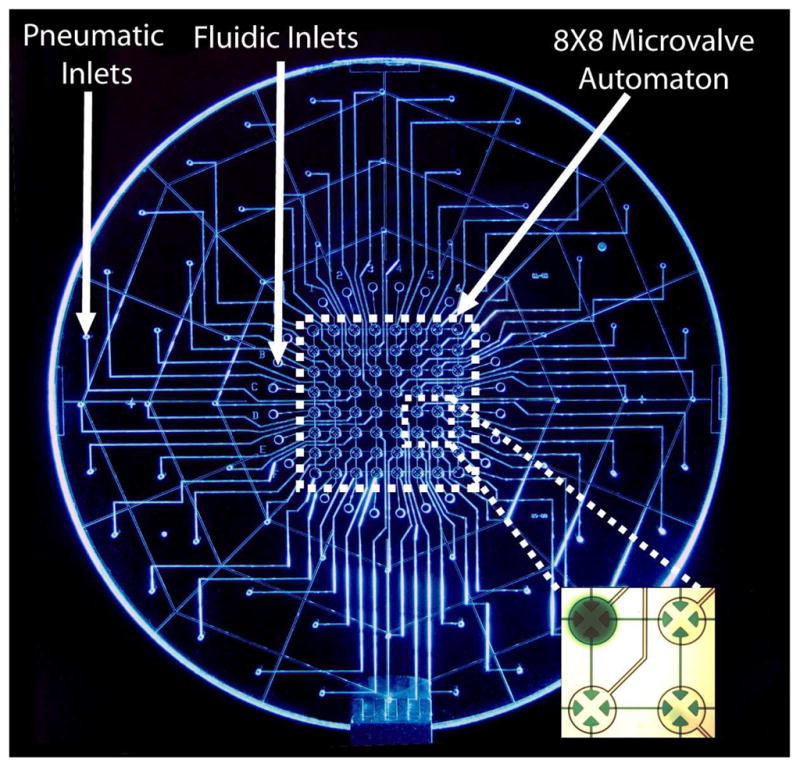
Photograph of the digital microfluidic Automaton. The 4-way microvalves control fluid flow through a rectilinear grid of discontinuous fluidic channels. The inset shows a close up portion of the array with a single microvalve storing dye in an automated program. The individual microvalves in this array serve as 120 nL reaction chambers as well as fluidic control mechanisms.
The pneumatically actuated microvalve array is composed of a 3-layer glass PDMS (polydimethylsiloxane) hybrid structure,1 and incorporates a rectilinear network of fluidic channels. Vacuum and pressure are supplied to the microvalves through drilled inputs on the pneumatic layer via computer-actuated solenoid valves. Samples and reagents can be loaded from drilled fluidic reservoirs to any subset of the microvalves by a programmed actuation sequence. The channels in the pneumatic layer are isotropically etched to a depth of 70 microns whereas the fluidic features are etched to a depth of 30 microns to reduce dead volumes between the microvalves.
Programmable microfluidic systems for assay automation typically require active mixing mechanisms due to the laminar flow profile of fluids within microchannels.12 Rapid reagent mixing is achieved with the Automaton by cyclically transferring the contents of two or more valves within a loop. For instance, an 8-step, 640 ms subroutine is iterated to cycle the contents of two microvalves through a 4-valve loop. Dilution of fluorescein standards with buffer and analysis by fluorescence microscopy indicate this mixing is complete in less than 5 seconds (Figure 3a). The digital fluidic transfers used in these programs result in mixing operations that are more than five times faster than traditional microfluidic mixing loops.13
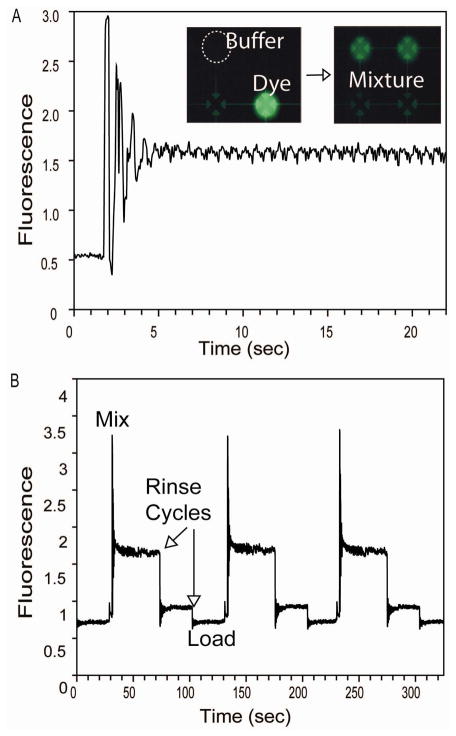
Figure 3.
(A) Confocal fluorescence data and epifluorescent images from a reagent mixing/dilution program. Fluorescein and buffer were loaded into separate valves in a 4-valve mixing circuit. Cyclic transfer of the volumes within the circuit results in rapid reagent mixing. As the fluorescein is diluted with buffer, the data follow a dampened oscillatory curve during the initial mixing phase and rapidly (<5 sec) reach equilibrium at an intermediate intensity value. (B) Three consecutive mixing operations with intermediate washing steps. These data demonstrate both the reproducibility of mixing operations and the efficiency of washing steps.
Figure 3b shows data from three consecutive fluorescein dilution operations with intermediate valve rinsing steps. This serial processing program results in highly reproducible mixing proportions for nanoliter-scale sample volumes. Furthermore, rinsing programs enable reuse of valve networks for different operations at different time points in a program. The output from a mixing circuit can be stored for future retrieval, or it can be used as the input for downstream processing operations. Iteration of the mixing program enables rapid generation of precise serial dilutions with adjustable dilution factors. Since the dead volumes of an individual microvalve can be used as a carryover fraction, as little as 14 nL can be precisely metered and mixed using the current design, and sub-nanoliter volumes could be similarly controlled by reducing the etch depth of fluidic features.
More complex operations involving larger reagent sets can be achieved by using larger microvalve circuits for mixing. Figure 4 shows video frames of a program in which all possible combinations of a set of four reagents are prepared. The program is initialized by loading a subset of the four input reagents to the mixing loop. The same program can then be used to mix the reagents regardless of the size of the loaded subset. The scale of these operations can be increased by simply including more microvalves in the mixing circuit.
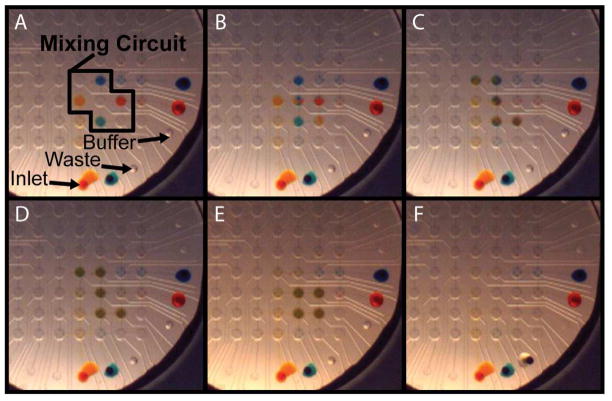
Figure 4.
Combinatorial mixing program. (A) Any subset of four unique input samples can be loaded into a 7-valve mixing circuit. Reagents are partially mixed during the first cycle of recirculation (B, C) and completely mixed in subsequent cycles (D, E). Mixing circuit microvalves are cleared by a washing program (F).
Several of the basic operations described above were combined to develop a rapid, quantitative assay for H2O2, a biomarker of oxidative stress (Figure 5). Elevated serum H2O2 is an indicator for oxidative cellular damage in conditions such as chronic allopathic neuropathy, the leading cause of kidney transplant failure.14 In this program, a H2O2 standard was serially diluted, stored, mixed with horseradish peroxidase and aminophenyl fluorescein, and analyzed using laser induced fluorescence to produce an on-chip calibration. Following introduction of sample, this fully-automated program achieves a sub-micromolar limit of detection with a 14 minute runtime. Samples of H2O2 (5.0 μM) prepared at the normal human serum concentration are quantified with a high level of precision and less that 4% error. Furthermore, this program can be extended to a wide range of metabolic analytes without modification.
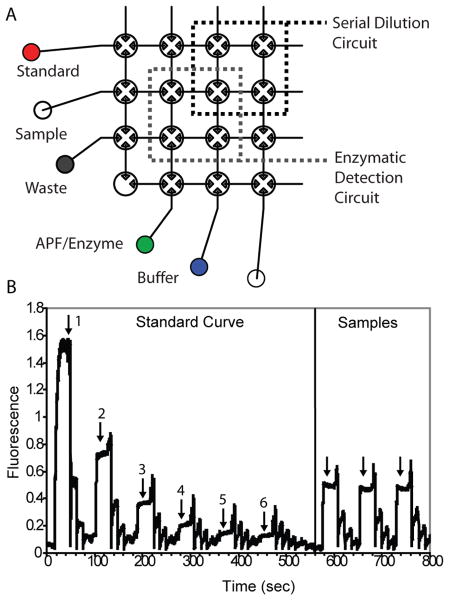
Figure 5.
(A) Details of a fully-automated assay for H2O2, a biomarker for oxidative stress. A H2O2 standard is diluted with buffer in the serial dilution circuit. A portion of the dilution is stored, and the remainder is transferred to the enzymatic detection circuit where it is mixed with aminophenyl fluorescein and horseradish peroxidase. During this phase, the aminophenyl fluorescein is oxidized to form a highly fluorescent product. After rinsing the enzymatic detection circuit, the stored H2O2 dilution is further diluted and the program is repeated to generate a standard curve. Finally, 120 nL samples are loaded and mixed with detection reagents for quantification. (B) Confocal fluorescence data acquired from the detection circuit during the performance of the assay. Arrows indicate the points in the assay where mixing is complete and data are collected.
We have recently enhanced the capabilities of this platform by developing procedures for inhomogeneous immunoassays. Magnetic microspheres coated with a capture antibody are loaded into specific microvalves in the array by trapping them in an external magnetic field (Figure 6a). An individual capture microvalve is held continuously open while adjacent microvalves are actuated to transfer the antibody coated microbeads from an inlet. External application of an 1/8th inch diameter cylindrical neodymium magnet to the pneumatic wafer traps the magnetic microbeads only in the selected capture microvalve. The amount of beads loaded to the capture microvalve is adjusted based on the actuation rate and total program runtime.
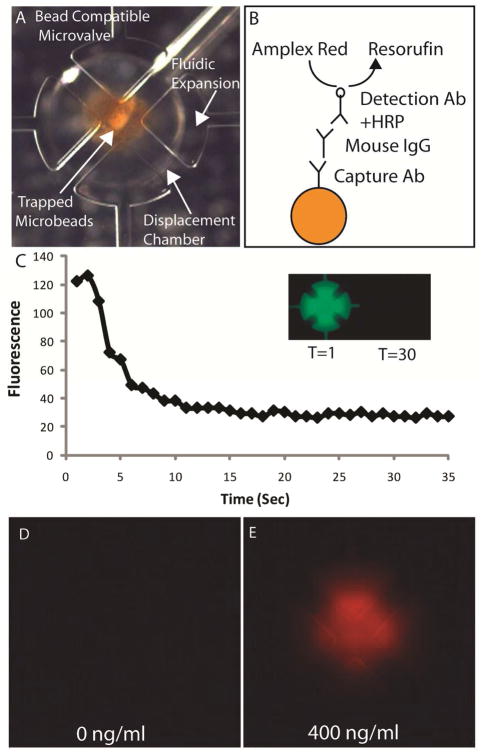
Figure 6.
(A) Trapping microvalve loaded with approximately 120 ng of magnetic microbeads coated with antimouse IgG. The bead-compatible digital microfluidic Automaton uses microvalves with larger fluidic expansions to prevent the beads from becoming trapped as they are transferred through the array. (B) Schematic of the immune complex formed in the immunoassay for mouse IgG. (C) Fluorescence profile obtained while rinsing 10 uM fluorescein dye from the trapping microvalve. The same program was used to remove unbound analyte and detection antibody after their corresponding incubations. Epifluorescence images of trapping valves after performing the assay with a negative control (D) and 400 ng/ml mouse IgG sample (E).
To demonstrate a fluorescence ELISA assay, we utilized magnetic microbeads coated with Fc-specific goat anti-mouse IgG (Bangs Labs, BM550). Figure 6b illustrates the immune complex formed to detect mouse IgG. Prior to starting the assay, all microvalves and channels were treated with Superblock (Thermo Scientific). Approximately 150 ng of beads were transferred to each capture microvalve as described above. Capture microvalves were held continuously opened for the duration of the assay. A total of 3 uL mouse IgG (Trevgen 4360-MC-100) in Superblock was transferred through the capture microvalve in a continuous pumping mode to a designated waste outlet. A similar program was used to rinse unbound analyte from the capture microvalves. We found that opened capture microvalves could be effectively rinsed in less than 30 seconds using TBST (Figure 6c). Afer loading 0.8 μg/ml HRP-conjugated anti-mouse IgG (Santa Cruz Biotechnology, SC-2371) and rinsing as described above, capture microvalves were filled with amplex red detection reagent (Invitrogen, A22188). After a 5 minute incubation, the capture microvalves were imaged using epifluorescence microscopy. Figure 6d and 6e shows the increase in fluorescence signal observed for a mouse IgG positive control after the 5 minute incubation. These results indicate successful capture and detection of a protein target using the digital microfluidic Automaton without additional surface modification steps.
We have shown the utility of our digital microfluidic Automaton for the performance of diverse bioassay protocols on a common chip format. With the current 64-bit processor, 1.84 × 1019 unique states can be defined for each step of a program. This, in combination with the capabilities described above, demonstrates an unprecedented level of programmability for the automation of microfluidic bioassays. The scaling of the current device is only limited by the one-to-one correspondence between off-chip pneumatic solenoid actuators to microvalves in the array. Up to 96 solenoid valves could easily be controlled by a single USB input/output card (National Instruments, USB-6509). However, we have previously shown that binary demultiplexing pneumatic circuits connected to pneumatic latching valves enable the control of 2N-1 microfluidic valves using N off-chip controllers.15 Although the longer actuation times required for the demultiplexed latching valves (> 120 ms) would reduce the speed of the digital microfluidic Automaton, further design optimization of these structures may enable more rapid sample processing capabilities.
3. Microfluidic Emulsion Generator Array
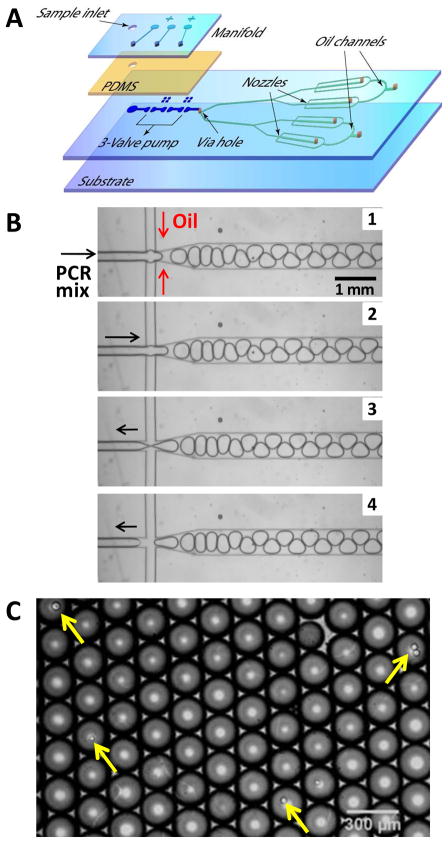
Single cell analysis is imperative to understanding cellular heterogeneity which underlies mechanisms in complex biological systems and diseases, such as cancer. Monolithic membrane valve technology has enabled the development of a microfluidic platform for high-performance single cell genetic analysis. As illustrated in Figure 7a, we have devised a microfluidic emulsion generator array device (MEGA) with an integrated micropump that operates four parallel cross-injectors for aqueous droplet formation.16 The micropump consists of a serial array of three microvalves connected through a via hole to a microfabricated network enclosed between two thermally bonded glass wafers.17 The microfluidic network is composed of symmetrically bifurcated microchannels to form four parallel crosses or droplet nozzles at the junctions. This system is used to partition individual cells within the droplets for high throughput genetic screening applications.
Figure 7.
(A) Exploded view of the four-layer, 4-channel MEGA device with an integrated micropump driving four parallel nozzles for droplet generation. (B) Image sequence of a cycle of droplet formation at a frequency of 5.6 Hz. PCR mix is pumped through the cross-injector and pinched by the oil flow infused from side channels (Images 1 and 2). The aqueous solution is pulled back at the end of each pumping cycle, which causes the release of the droplets (Images 3 and 4). As a result, the droplet formation is synchronized with the pump actuation, and the droplet size is determined by the flow rate of pumped fluid. (C) Optical micrograph of highly uniform droplets generated by our method containing a predictable stochastic distribution of primer-functionalized agarose beads (~34 μm, indicated by arrows). For this experiment, the average bead concentration was 0.1 beads per 3 nL droplet.
On-chip pumping confers the capability of on-demand generation of droplets with well controlled frequency and droplet size. The on-chip pump drives an aqueous PCR mix into the cross-injector where it is pinched by the oil flow infused from side channels (Figure 7b, images 1 and 2). Due to the pulsatile nature of valve actuation, the aqueous solution is pulled back at the end of one pumping stroke, causing the release of the droplets (images 3 and 4). As a result, the frequency of droplet formation is controlled by the pump actuation frequency. In addition, the droplet size can be independently tuned by varying the actuation pressure to change the flow rate of pumped fluid. In contrast, other passive microfluidic droplet techniques, such as flow focusing, rely on the interfacial instability to form droplets which involves complicated interplay between many parameters including channel geometry, flow rates of two phases, and surfactant.18–20 We found that the pulsatile pumping allows effective transport of large microbeads against gravity sedimentation to ensure the predictable stochastic encapsulation of single beads/cells into uniform nanoliter droplets (Figure 7c). The size deviation of 3 nL droplets was found to be 3.8%, comparable to that reported by using passive microdroplet array generators (1.3 nL droplets, CV 3.7%).20 These results demonstrate that our technique presents a robust platform for actively controlled generation of uniform nanoliter droplets, which is crucial to achieve quantitative single cell genetic measurement.
Our microfluidic technique is readily scalable, as the 4-channel array defines a basic unit for multiplexed MEGA devices. To achieve a higher density, we have designed a novel ring micropump composed of three pairs of coaxial ring-shaped valve seats connected by offset channels, as well as corresponding circular displacement trenches. This compact micropump permits the high density integration of 96 T-shaped droplet generation channels on a 4-inch wafer (Figure 8). A custom-made assembly module is used to deliver reagents and collect droplets from the 96-channel MEGA. The 96-channel MEGA system has a maximum droplet production rate of 3.4 × 106 droplets per hour, which greatly improves the detection limit and processing time for detection of low-frequency events.
Figure 8.
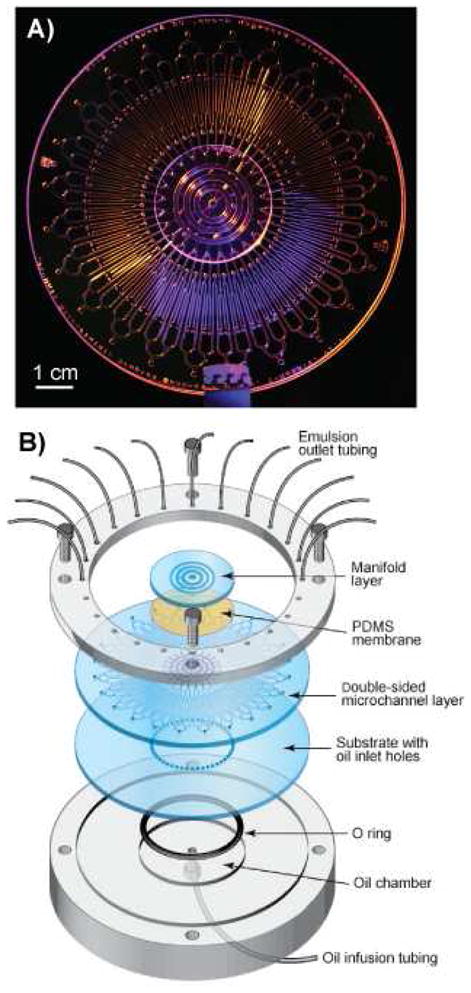
(A) Photograph of a 96-channel MEGA device. (B) Exploded view of the chip with world-to-chip interfacing manifolds used for pneumatic actuation and oil flow.
We have utilized the 96-channel MEGA to perform high-throughput bead-based multiplex PCR assays for single cell genotyping. Multiplex single cell genetic analysis (SCGA) enables the detection and quantification of both normal and mutant cells by targeting genes specific to individual cell types. In this approach, beads and cells are diluted in the PCR mix and partitioned into individual uniform reaction droplets dispersed in a carrier oil phase using a MEGA device. Statistical distribution of single beads and cells leads to a fraction of droplets containing both one bead and one or more cells. Beads are functionalized with forward primers targeting both cell types, and the PCR mix contains reverse primers each labelled with a unique fluorescent dye. Thousands of such droplets, generated by MEGA chips within minutes, are collected in standard PCR tubes and thermally cycled in parallel. Post-PCR beads are recovered from the emulsion and rapidly analyzed by flow cytometry for multi-color fluorescent digital counting of each single cell detection event. Beads coexisting with only one cell type will carry one type of dye-labelled double-stranded amplicons, while beads compartmentalized with different types of target cells will be labelled with two dyes.
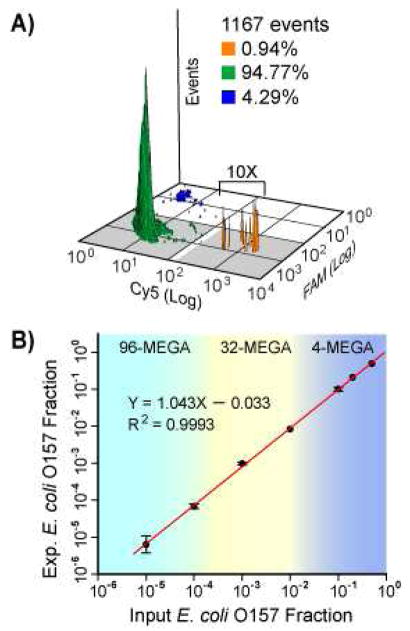
We have applied the MEGA devices for low-frequency detection of pathogenic E. coli O157 bacteria in a background of non-pathogenic E. coli K12 cells (Figure 9). Quantitative detection of low-frequency O157 cells can be achieved at an average cell concentration up to 100 E. coli cells per 2.5 nL droplet, which greatly increases the analysis throughput and hence the detection sensitivity, without excessively extending the droplet production time (Figure 9a). This result indicates that the use of nanoliter droplets provides sufficient reagents for efficient and specific multiplex PCR amplification, and confers tolerance to PCR inhibition, as the inhibitors released from cells are significantly diluted in the large droplets. This performance would be very challenging using picoliter droplets. Compared to other droplet-format microdevices for PCR assays, such as electro-wetting-on-dielectric (EWOD) chip21 and SlipChi,22 our method offers the ability of automated mixing and single bead/cell encapsulation with much higher throughput. A detection limit of 1/105 has been achieved by screening ~106 cells with only 30 minutes of droplet generation when using the 96-channel MEGA (Figure 9b). The entire procedure, including PCR thermal cycling, post-PCR cleanup, and flow cytometry takes approximately 4 hours and compares favourably to standard PCR-based detection assays while providing much better sensitivity. Our quantitative digital format outperforms previously reported microsystems which detect only one bacterial strain by PCR with detection limits ranging from a few to 104 bacterial cells.25 Since our multiplex SCGA approach digitally detects single cells, the detection limit can be further improved to 1/106 or lower by extending droplet generation time to analyze more cells. Such sensitivity makes the technique a promising candidate for many applications, such as food safety, where microbial pathogen detection needs to meet a zero tolerance policy for many foods.26 These results also indicate the feasibility of the MEGA platform for other applications, for instance, the analysis of cancer development and progression, circulating tumor cells, and stem cell differentiation, where high-throughput genetic variation analysis of mammalian cells at the single cell level may facilitate a deeper understanding of the biological mechanisms involved.
Figure 9.
(A) Two-dimensional flow cytometry results of fluorescent amplicon beads showing detection of pathogenic E. coli O157 in a background of harmless E. coli K12 at a ratio of 10−4. (B) Differently multiplexed MEGA devices enable quantitative detection of O157 frequency over five orders of magnitude with a detection limit of 1/105.
4. Summary
Advances in monolithic membrane valve technology have enabled the development of robust platforms for digital microfluidic assay automation. The programmability, speed, throughput, and low sample volume requirements conferred by these systems offer significant advantages compared to conventional benchtop robotic laboratory automation systems and specialized microfluidic processing platforms. The digital transfer of fluids between microvalves in the microfluidic Automaton platform enables the implementation of diverse serial and combinatorial sample processing operations on a common chip format. The programmability of this platform can be exploited to replace the specialized microfluidic circuits used in conventional lab-on-a chip devices. Similarly, the use of microvalve pumps in the MEGA platform enables high throughput droplet generation with programmable droplet volumes and formation rates. This programmability has been instrumental for high-throughput screening of single cells for low frequency genetic variations. These novel, digital sample processing technologies represent significant advances in the field of microfluidic laboratory automation.
Acknowledgments
Financial support for this work was provided by grant U54ES016115 from the U.S. National Institute for Environmental Health Sciences (NIEHS) through the trans-NIH Genes, Environment and Health Initiative. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institute of Environmental Health Sciences or the National Institutes of Health. Additional funding was provided by Samsung Corporation. Device fabrication was performed in the UC Berkeley Microlab and Center and Biomolecular Nanotechnology Center (BNC). R.A.M. has a financial interest in IntegenX, Inc. that is commercially developing aspects of the technologies presented here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grover WH, Skelley AM, Liu CN, Lagally ET, Mathies RA. Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices. Sens Actuators B. 2003;89:315–323. [Google Scholar]
- 2.Thorsen T, Sebastian J, Maerkl SJ, Quake SR. Microfluidic large scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 3.Oh KW, Ahn CH. A Review of Microvalves. J Micromech Microeng. 2006;16:R13–39. [Google Scholar]
- 4.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. A fully integrated genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grover WH, Mathies RA. An integrated microfluidic processor for single nucleotide polymorphism based DNA computing. Lab Chip. 2005;5:1033–1040. doi: 10.1039/b505840f. [DOI] [PubMed] [Google Scholar]
- 6.Blazej RG, Kumaresan P, Mathies RA. Microfabricated bioprocessor for integrated nanoliter-scale Sanger DNA sequencing. Proc Natl Acad Sci U S A. 2006;103:7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen EC, Bhat BP, Mathies RA. A digital microfluidic platform for the automation of quantitative biomolecular assays. Lab Chip. 2010;10:685–691. doi: 10.1039/b920124f. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan V, Pamula VK, Fair RB. Droplet-based microfluidic lab-on-a-chip for glucose detection. Anal Chim Acta. 2004;507:145–150. [Google Scholar]
- 9.Miller EM, Wheeler AR. A digital microfluidic approach to homogeneous enzyme assays. Anal Chem. 2008;80:1614–1619. doi: 10.1021/ac702269d. [DOI] [PubMed] [Google Scholar]
- 10.Teh SY, Lin R, Hung LH, Lee AP. Droplet microfluidics. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 11.Urbanski JP, Thies W, Rhodes C, Amarasinghe S, Thorsen T. Digital microfluidics using soft lithography. Lab Chip. 2006;6:96–104. doi: 10.1039/b510127a. [DOI] [PubMed] [Google Scholar]
- 12.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 13.Paegel BM, Grover WH, Skelley AM, Mathies RA, Joyce GF. Microfluidic serial dilution circuit. Anal Chem. 2006;78:7552. doi: 10.1021/ac0608265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djamali A, Sadowski EA, Muehrer RJ, Reese S, Smavatkul C, Vidyasagar A, Fain SB, Lipscomb RC, Hullett DH, Samaniego-Picota M, Grist TM, Becker BN. BOLD-MRI assessment of intrarenal oxygenation and oxidative stress in patients with chronic kidney allograft dysfunction. Am J Physiol. 2007;292:F513–F522. doi: 10.1152/ajprenal.00222.2006. [DOI] [PubMed] [Google Scholar]
- 15.Grover WH, Ivester RHC, Jensen EC, Mathies RA. Development and multiplexed control of latching pneumatic valves using microfluidic logical structures. Lab Chip. 2006;6:623–631. doi: 10.1039/b518362f. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Y, Novak R, Shuga J, Smith MT, Mathies RA. High-performance single cell genetic analysis using microfluidic emulsion generator arrays. Anal Chem. 2010;82:3183–3190. doi: 10.1021/ac902683t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumaresan P, Yang CJ, Cronier SA, Blazej RG, Mathies RA. High-throughput single copy DNA amplification and cell analysis in engineered nanoliter droplets. Anal Chem. 2008;80:3522–3529. doi: 10.1021/ac800327d. [DOI] [PubMed] [Google Scholar]
- 18.Song H, Chen DL, Ismagilov RF. Reactions in droplets in microflulidic channels. Angew Chem Int Ed Engl. 2006;45:7336–56. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christopher GF, Anna SL. Microfluidic methods for generating continuous droplet streams. J Phys D: Appl Phys. 2007;40:R319–R336. [Google Scholar]
- 20.Nisisako T, Torii T. Microfluidic large-scale integration on a chip for mass production of monodisperse droplets and particles. Lab Chip. 2008;8:287–293. doi: 10.1039/b713141k. [DOI] [PubMed] [Google Scholar]
- 21.Chang YH, Lee GB, Huang FC, Chen YY, Lin JY. Integrated polymerase chain reaction chips utilizing digital microfluidics. Biomed Microdevices. 2006;8:215–225. doi: 10.1007/s10544-006-8171-y. [DOI] [PubMed] [Google Scholar]
- 22.Shen F, Du W, Davydova EK, Karymov MA, Pandey J, Ismagilov RF. Nanoliter multiplex PCR arrays on a SlipChip. 2008;80:3522–3529. doi: 10.1021/ac1007249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, Sensabaugh GF, Riley LW, Mathies RA. Integrated portable genetic analysis microsystem for pathogen/infectious disease detection. Anal Chem. 2004;76:3162–3170. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 24.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 25.Cady NC, Stelick S, Kunnavakkam MV, Batt CA. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens Actuators, B: Chem. 2005;107:332–341. [Google Scholar]
- 26.Batt CA. Materials science: Food pathogen detection. Science. 2007;316:1579–1580. doi: 10.1126/science.1140729. [DOI] [PubMed] [Google Scholar]