Abstract
The disintegrin and metalloproteinase Adam10 has been implicated in the regulation of key signaling pathways that determine skin morphogenesis and homeostasis. To address the in vivo relevance of Adam10 in the epidermis, we have selectively disrupted Adam10 during skin morphogenesis and in adult skin. K14-Cre driven epidermal Adam10 deletion leads to perinatal lethality, barrier impairment and absence of sebaceous glands. A reduction of spinous layers, not associated with differences in either proliferation or apoptosis, indicates that loss of Adam10 triggers a premature differentiation of spinous keratinocytes. The few surviving K14-Adam10-deleted mice and mice in which Adam10 was deleted postnatally showed loss of hair, malformed vibrissae, epidermal hyperproliferation, cyst formation, thymic atrophy and upregulation of the cytokine thymic stromal lymphopoetin (TSLP), thus indicating non cell-autonomous multi-organ disease resulting from a compromised barrier. Together, these phenotypes closely resemble skin specific Notch pathway loss-of-function phenotypes. Notch processing is indeed strongly reduced resulting in decreased levels of Notch intracellular domain fragment and functional Notch signaling. The data identify Adam10 as the major Site-2 processing enzyme for Notch in the epidermis in vivo, and thus as a central regulator of skin development and maintenance.
Keywords: Adam10, Epidermis, Mouse, Notch, Shedding, Skin
INTRODUCTION
The epidermis is a stratifying epithelium that protects the organism from dehydration, mechanical trauma and microbial invasion. The epidermis and its appendages are separated by a basement membrane from the underlying mesenchymally derived dermis (Koster and Roop, 2004). A tightly controlled program orchestrates the morphogenesis of a single-layered ectoderm into a multi-layered stratified epithelium (consisting of the viable basal, spinous and granular layers and the dead cornified layer). Each of these layers is characterized by a specific expression repertoire of keratins and other intracellular and cell surface-associated proteins (Koster and Roop, 2007). Different populations of stem cells guarantee life-long renewal to maintain and restore the interfollicular epidermis (IFE), hair follicles and sebaceous glands (Blanpain and Fuchs, 2009; Oshima et al., 2001; Taylor et al., 2000; Watt et al., 2008). Partial failure of epidermal morphogenesis or disturbed maintenance of the epidermis results in a variety of human skin disorders (Okuyama et al., 2008; Thelu et al., 2002).
Loss- and gain-of-function studies of either Notch receptors, key regulators of Notch signaling and Notch-dependent target genes (Lin and Kopan, 2003; Lin et al., 2000; Nicolas et al., 2003; Pan et al., 2004; Rangarajan et al., 2001; Uyttendaele et al., 2004; Yamamoto et al., 2003) have revealed a crucial role for Notch signaling in the proper stratification and development of the epidermis and its appendages (Moriyama et al., 2008; Vauclair et al., 2005). Keratinocytes require Notch signaling to downregulate basal genes and induce spinous layer markers, thereby switching from proliferation to differentiation (Blanpain and Fuchs, 2009; Blanpain et al., 2006; Rangarajan et al., 2001). Ablation of Notch signaling during skin embryogenesis invoked early postnatal death mainly due to a disturbed epidermal barrier (Demehri et al., 2008). Postnatal knockout of Notch-1 led to a hyperproliferative epidermis, hair loss and epidermal cyst formation (Blanpain et al., 2006; Nicolas et al., 2003). Loss of epidermal Notch-1 or Notch-1/Notch2 disturbed skin homeostasis and resulted in inflammatory skin disease (Demehri et al., 2009; Dumortier et al., 2010). Mice with impaired Notch-1 signaling developed skin tumors, indicating a tumor suppressor function for Notch-1 in the skin (Nicolas et al., 2003; Proweller et al., 2006).
Notch signaling is initiated by ligand-receptor interactions between neighboring cells. The γ-secretase complex generates the cytoplasmic domain of Notch (NICD), which translocates to the nucleus to activate transcription (Mumm and Kopan, 2000). This is preceded by a disintegrin and metalloproteases (ADAMs)-dependent cleavage within the Notch extracellular-juxtamembrane region. Although in vitro data suggested a role for both Adam10 and Adam17 in the regulation of Notch shedding, it is unknown whether both are required to regulate Notch signaling in vivo in all tissues or whether different tissues require different ADAMs (Bozkulak and Weinmaster, 2009; Brou et al., 2000; Mumm et al., 2000). Our studies using complete Adam10 knockout mice (Hartmann et al., 2002), suggested a role for this protease in the cleavage of Notch1 during mouse embryogenesis.
To understand the in vivo physiological function of Adam10 in the epidermis and to address whether Adam10 serves as a key in vivo regulator of the different Notch signaling pathways that control skin morphogenesis and homeostasis, we decided to inactivate epidermal Adam10 during skin morphogenesis and under steady-state conditions in the adult epidermis. Epidermal deletion of Adam10 led to a precocious epidermal differentiation and to hyperproliferation when Adam10 was deleted in adult epidermis. These phenotypes were directly linked to defective Notch processing and signaling. Our data conclusively demonstrate that Adam10 is a key regulator of Notch signaling in the skin not only during morphogenesis but also in adult skin.
MATERIALS AND METHODS
Generation of Adam10 cKO and inducible cKO mice
The Adam10flox/flox mice were generated as described (Jorissen et al., 2010) and crossed with keratin 14 (K14) Cre mice (Dassule et al., 2000). To obtain conditional knockout mice (cKO), homozygous Adam10 floxed mice and mice heterozygous for the floxed Adam10 allele and the K14 transgene were bred. Adam10flox/flox, Adam10flox/+ or K14-Cre; Adam10flox/+ were used as controls. Adam10flox/flox/K5tTA+/–/tet-Cre+/– were obtained by breeding Adam10flox/+/K5tTA+/–/tet-Cre+/– (Diamond et al., 2000) with Adam10flox/flox mice. Deletion was induced by replacement of doxycycline-containing chow for normal chow. Littermates of Adam10flox/flox/K5tTA+/–/tet-Cre+/– mice without doxycyline chow, as well as Adam10flox/flox/K5tTA+/–/tet-Cre+/– with doxycyline chow were used as controls.
Western blot analysis
Epidermal lysates from newborn mice were prepared as described previously (Tunggal et al., 2005). Protein was loaded on 10% SDS-PAGE gels and transferred to PVDF membranes (Roth) to perform western blot analysis. The following antibodies were used: B42.1 polyclonal Adam10 serum (a gift from W. Annaert, University of Leuven, The Netherlands), Notch1 rat mAb (Okochi et al., 2006), Notch rabbit polyclonal antibody (ab18925, Abcam), keratin 10 polyclonal (PRB-159P, covance), anti-Fl-335 against GAPDH (Santa Cruz) and anti-actin (Sigma-Aldrich). Blots were developed using the ECL Detection System (Amersham).
Preparation, histology and ultrastructural analysis
Back skin samples of newborn mice were fixed in glutaraldehyde (6% in PBS) and embedded in araldite according to routine procedures. For light microscopy, 1 μm semi-thin sections were stained with Toluidine Blue. For electron microscopy tissue was postfixed in OsO4 for 2 hours, and processed in araldite according to routine procedures. Ultrathin sections were observed with a Zeiss EM 900 microscope.
Immunohistochemical analysis
Samples were fixed in 4% buffered formalin. After overnight postfixation, dehydratation and paraffin embedding 5 μm sections were stained with antibodies against Adam10 (RnD Systems AB946), keratin 14, keratin 10, loricrin and keratin 15 (1:500, all Covance), Ki-67 (1:100; Abcam), Scd1 (1:200, Santa Cruz), Giemsa Stain (Sigma-Aldrich) and Fluorometric TUNEL System (Promega). Cryostat sections (7 μm) were stained with 0.15 mg/ml Nile Red for 2 minutes and counterstained with DAPI (Gareus et al., 2007). Wholemounts of mouse tail epidermis, preparation and immunofluorescence were performed as described previously (Braun et al., 2003). A laser scanning microscope (Olympus Fluoview FV1000) was used for image acquisition.
Serology
Serum thymic stromal lymphopoietin (TSLP) levels were measured according to the manufacturer's instructions in the Quantikine mouse TSLP ELISA Kit (R&D Systems).
Primary keratinocytes, growth curve and colony-forming assays
Primary keratinocytes were isolated from the skin of newborn mice and passaged until P3 as described previously (Hennings et al., 1980). For growth curve analysis 5000 cells were plated in triplicate in a 96-well plate and analyzed with CellTiter 96 Proliferation Assay (Promega). For colony-forming assays 4000 cells were plated in triplicate in a six-well plate and cultured for 2-3 weeks in the presence of feeders. Cells were fixed with 1% PFA for 15 minutes and stained for 1 hour with 0.05% Crystal Violet in PBS.
Water loss and barrier function assays
The weight of newborn mice at 37°C was monitored every hour. Hematocrit was measured by centrifuging blood in HEPES-coated capillary tubes. Transepidermal water loss (TEWL) was measured using a TEWAmeter (Courage and Khazaka). Toluidine Blue dye staining was carried out as described previously (Hardman et al., 1998).
Quantitative RT-PCR
RNA was extracted from epidermal splits or from E17.5 embryos using the NucleoSpin RNAII Kit (Macherey Nagel, Germany). RNA (2 μg) was reverse transcribed using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Germany). Hes5, Hey1, Hey2, Acer1, Adam8, Alox12e and Scd1 expression were determined by real-time PCR analysis of 0.5 μl cDNA on a 7900HT Fast Real-time PCR System (Applied Biosystems, USA) in 10 μl reaction volume. The expression levels of the genes were shown as % of GAPDH expression using ΔCt for calculation. For the calculation of the t-test, ΔΔCt values were calculated using the mean Ctr value.
Microarray analysis
Total RNA from epidermal splits were extracted (Hasler et al., 2009) and hybridized to an Affymetrix Mouse Gene 1.0 ST array. Data were normalized using RMA (AGCC, Affymetrix) and signals that were not stronger than the 95th percentile of all intron probes on the array were excluded from further analysis. For the Gene Ontology analysis and the cluster only genes with a fold change greater than +2 or less than –2; rank sum difference ≥16 were included. Gene ontology analysis was performed as previously published (Tavazoie et al., 1999) and the hierarchical cluster was generated based on regulated genes using the correlation as the similarity measure and UPGMA (unweighted average) as the clustering method (Spotfire Decision Site 9.1.1). Data sets have been submitted to GEO [Gene Expression Omnibus (www.ncbi.nlm.gov/geo), Accession Number GSE25480].
Statistical analysis
Statistical significance was performed as unpaired Student's t-test using Microsoft Excel software. Error bars indicate the mean±s.d. of the mean. *P<0.05, **P<0.005, ***P<0.0005.
RESULTS
Generation of mice lacking Adam10 in the epidermis
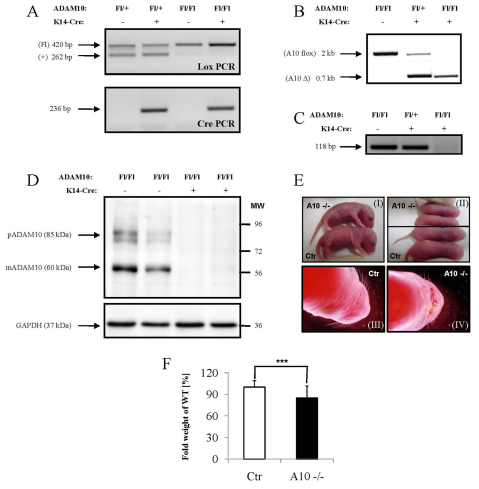
To examine the contribution of Adam10 to the morphogenesis and function of stratified epithelia we employed the Cre/loxP recombination system and crossed Adam10 floxed mice (Jorissen et al., 2010) with mice that express Cre under the control of the keratin 14 (K14) promoter to drive expression in the basal layer of the epidermis and its appendages, such as hair follicles and sebaceous glands (Hafner et al., 2004). PCR analysis on tail DNA identified the expected fragments (Fig. 1A). Genomic deletion upon Cre expression was close to 100% (Fig. 1B). RT-PCR analysis further confirmed efficient deletion of exon 2 (Fig. 1C) (see Fig. S1C in the supplementary material). Western blot analysis showed an almost complete loss of Adam10 from the epidermis (Fig. 1D). Thus, K14-driven Cre efficiently ablates Adam10 during epidermal morphogenesis.
Fig. 1.
Epidermal deletion of Adam10 (A10). (A) PCR analysis on genomic DNA showing the different genotypes of the mice. (B) PCR analysis on genomic DNA from split epidermis showing the efficiency of deletion of the floxed region in the presence of the keratin 14-driven Cre recombinase. (C) RT-PCR analysis on mRNA from split epidermis showing the loss of targeted Adam10 transcripts. (D) Western blot analysis on epidermal lysates using antibodies against the C-terminal domain of Adam10. pAdam10, precursor of Adam10; mAdam10, mature form of Adam10. (E) Macroscopic appearance of control mice (I, III) and mice with an epidermal deletion of Adam10 (II, IV). (F) Weight analysis of newborn mice (<24 hours). The average weight of control mice (Ctr) among the litter was set as 100%. n=84 for control mice, n=34 for A10–/– mice. ***P<0.0005. Results are mean±s.e.m.
Adam10 deletion in basal cells of the epidermis leads to perinatal death and defects in the inside-out barrier of newborn mice
Over 95% of the Adam10flox/flox K14Cre (Adam10epi–/–) mice died during the first 24 hours of life while heterozygous and homozygous floxed Adam10 mice without Cre transgene and the Adam10Fl/+ hemizygous Cre-positive mice were viable and fertile without any obvious phenotype (from here on referred to as control mice). The different genotypes were born in the expected Mendelian ratio. Only 1 out of the 153 (0.6%) homozygous Adam10 floxed null mutant mice survived longer than 3 weeks, whereas eight (3.9%) reached an age between P9 and P18, after which they had to be sacrificed in order to avoid suffering.
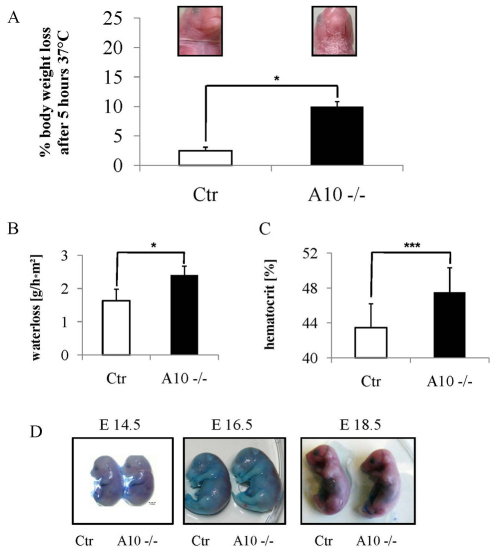
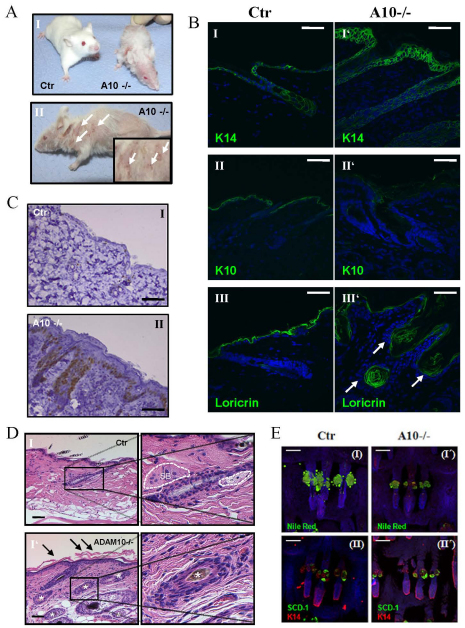
Macroscopically, loss of Adam10 during murine epidermal morphogenesis resulted in a shiny, reddish and translucent skin, and short aberrantly formed vibrissae (Fig. 1E). A few hours after birth Adam10epi–/– mice but not controls showed scaling of the upper ventral side. These macroscopic defects suggested a perturbed skin barrier function. Weight analysis revealed a small but significant weight reduction in newborn (Fig. 1F) but not in E17.5 Adam10epi–/– mice (see Fig. S1A in the supplementary material). This weight difference, in addition to a smaller overall body size (e.g. Fig. 1E, parts I and II), became more prominent in the small number of surviving mice (Fig. 4A,B). These mice showed a much more dramatic loss of weight when kept for 5 hours in a dry environment after separation from their mothers (Fig. 2A), indicating water loss. Transepidermal water loss was indeed significantly increased in Adam10epi–/– mice compared with controls (Fig. 2B), accompanied by an increased hematocrit (Fig. 2C), further demonstrating loss of fluids.
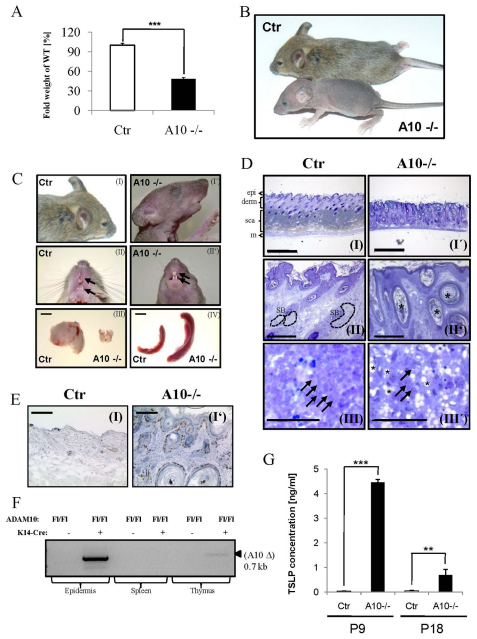
Fig. 4.
Postnatal deficiency of Adam10 in the epidermis leads to hair loss, hyperplasia, epidermal cyst formation, loss of sebaceous glands and multi-organ disease. (A) Weight analysis shows that deletion of Adam10 leads to severe growth retardation and difficulty to thrive (n=7 for cKO and control). Results are mean±s.e.m. ***P<0.0005. (B) Macroscopic appearance of P18 control and mutant mice. (C) Adam10 deficiency leads to thymic atrophy, splenomegaly and lymphadenopathy (data not shown), as well as malformation of vibrissae and teeth architecture and signs of hyperkeratinization. Scale bars: 2 mm for III and IV. (D) Histological analysis of P18 back skin sections show epidermal cyst formation (indicated by black asterisks) and absence of sebaceous glands (SB; surrounded by broken lines) from hair follicle in Adam10 mutant (I′, II′) in contrast to control mice (I, II). Epi, epidermis; derm, dermis; sca, subcutaneous adipose fat tissue; m, muscle. The atrophic thymus of Adam10 mutant mice is characterized by abnormal thymic epithelial cells exhibiting large vacuole structures [black asterisks (III, III′)], representative thymocytes are indicated by black arrows. Scale bars: 500 μm for I, I′; 200 μm for II, II′; 50 μm for III, III′. (E) Ki67 staining showed massive hyperproliferation in the epidermis of Adam10-deficient mice (I′) in contrast to control littermates (I). (F) PCR-analysis on genomic DNA from diverse tissues showed successful deletion of Adam10 in epidermis and thymus at P18, while other tissues were unaffected. (G) ELISA measurement revealed strongly elevated levels of keratinocyte-derived chemokine TSLP in plasma of Adam10 cKO mice. Analysis was carried out using plasma from n=3 mice for each genotype; **P<0.005, ***P<0.0005. Results are mean±s.e.m.
Fig. 2.
Barrier defect assays showing disturbance of outside-in barrier in Adam10-deficient mice. (A) Adam10 mutant mice (kept at 37°C) show pronounced water loss with subsequent death between 5 and 7 hours. Insets show the ventral side of control and mutant mice incubated at 37°C for 5 hours. (B) Transepidermal water loss (TEWL) measurements of control and Adam10-deficient mice; n=4 for each genotype. (C) Hematocrit analysis of blood from control (n=16) and Adam10-deficient mice (n=13). Results are mean±s.e.m. *P<0.05, ***P<0.0005. (D) Epidermal loss of Adam10 does not affect the outside-in barrier: Toluidene Blue penetration assay of E14.5, E16.5 and E18.5 shows no time-course dependent difference of barrier formation between control and Adam10-deficient mice.
To examine whether the barrier defect resulted from impaired function of the stratum corneum, Toluidine Blue exclusion assays were performed (Fig. 2D). As expected, at E16.5 the barrier was only partially functional in the back skin of control and Adam10epi–/– mice, as judged by penetration of Toluidine Blue; whereas at E18.5 Toluidine Blue was excluded both in control and Adam10epi–/– mice, thus suggesting that in the absence of Adam10 the stratum corneum barrier developed relatively normally. Overall, the data showed a perturbed water barrier function upon loss of Adam10, probably explaining why the majority of mice die within the first day of birth.
Reduction of spinous layers due to premature differentiation of keratinocytes in Adam10-deficient epidermis
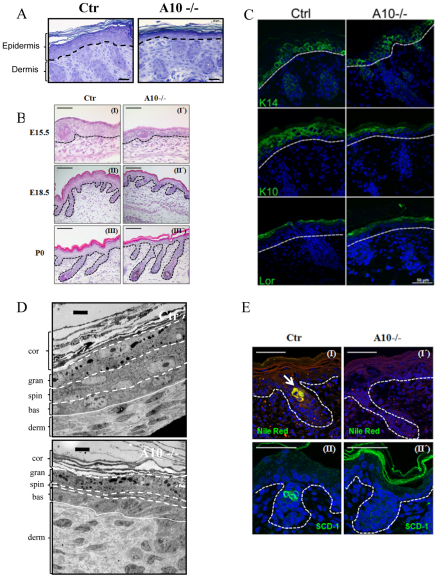
To analyze how epidermal Adam10 deletion results in impaired barrier formation, we examined the skin architecture of newborn mice. Semi-thin sections of back skin revealed a strong reduction in epidermal thickness in Adam10epi–/– mice compared with control mice (Fig. 3A) (see Fig. S1B in the supplementary material). This decrease was already evident at E15.5 and persisted throughout later developmental stages (Fig. 3B), even though hair follicle formation was not obviously disturbed. Nevertheless, all four layers of the interfollicular epidermis (IFE) could be distinguished based on their morphological characteristics (Fig. 3A,B). To examine more directly whether loss of Adam10 alters epidermal differentiation, different markers identifying the different epidermal layers were used (Fig. 3C). Staining for keratin 14 as a marker for the basal layer was unaltered in control and Adam10epi–/– mice. Similarly, no obvious difference was observed for loricirin, a marker for the stratum granulosum. However, staining for keratin 10 (K10), which marks the stratum spinosum and stratum granulosum, revealed a reduction in suprabasal layers positively stained for K10, indicating that the reduction in epidermal thickness is due to a decrease in the number of spinous layers (Fig. 3C, parts III and III′) (see Fig. S1C in the supplementary material). Ultrastructural analysis of back skin sections (Fig. 3D) confirmed the observation that the typical character of all epidermal layers was sustained with the difference that only one layer of spinous cells could be observed in the mutant mice in contrast to more spinous layers in the control mice. These results show that Adam10 does not regulate epidermal differentiation itself but instead regulates the timing of the spinous to granular layer transition.
Fig. 3.
Histological and ultrastructural analysis of newborn Adam10-deficient and control mice. (A) Toluidene Blue staining of back skin sections from newborn back skin. Scale bars: 20 μm. A representative example of n=6 mice for each genotype is shown. Broken lines indicate basement membrane layer. (B) Hematoxylin and Eosin staining of back skin sections show regular hair follicle formation in Adam10-deficient mice. Scale bars: 50 μm for I and I′; 100 μm for II, III, II′ and III′. A representative example of n=3 mice for each genotype is shown. Broken lines indicate basement membrane layer. (C) Immunofluorescence analysis of paraffin sections from newborn back skin as detected with the following markers: keratin 14, stratum basale; keratin 10, stratum spinosum; loricrin, stratum granulosoum. Broken white line indicates basement membrane. Scale bars: 20 μm. (D) Ultrastructural analysis revealed strongly reduced thickness, albeit regular morphological appearance, of the spinous layer in Adam10 mutant mice. bas, stratum basale; spin, stratum spinosum; gran, stratum granulosum; cor, stratum corneum; derm, dermis. Scale bars: 1 μm. Broken and unbroken lines indicate the boundaries of the epidermal layers. (E) Cryosections of newborn mice were stained with Nile Red (I,I′) or Scd1 antibodies (II,II′), and counterstained with DAPI. Polar lipids are stained red, non-polar lipids are stained green (I,I′). Representative pictures of sections from three different mice from each genotype are shown. A sebaceous gland in Ctr mice is indicated by white arrow. Broken lines indicate epidermal/dermal junction. Scale bars: 100 μm.
Loss of Adam10 did not obviously affect the ultrastructure and number of cell adhesion structures, such as hemidesmosomes, desmosomes and corneodesmosomes (data not shown). We also asked whether alterations in the balance between proliferation and apoptosis might explain the reduction in spinous layers. However, Ki67 and TUNEL staining did not identify a difference in the number of either proliferating or apoptotic cells (see Fig. S1D,E in the supplementary material).
To examine whether production of lipids, major components of the epidermal barrier, was altered we conducted Nile Red staining of back skin (Fig. 3EI,I′). Whereas control newborn mice showed positive Nile Red staining in a few cells at the position of developing sebaceous glands in the hair follicle (indicated by white arrow), no positive cells could be detected within hair follicles in Adam10epi–/– mice. This reduced lipid production could either be due to a reduced production by the few sebocytes that already have developed in newborns or by the absence of properly differentiated cells. We therefore stained for Scd1 (Miyazaki et al., 2001), a marker for mature sebocytes, but, in contrast to controls, could not detect any stained cells (Fig. 3E, parts II and II′), indicating the absence of mature sebaceous gland cells. This was further confirmed by real time PCR analysis for Scd1 on epidermal splits (see Fig. S3C in the supplementary material). Moreover, well-known genes of lipid-metabolism, e.g. alkaline ceramidase 1 (Acer1) and epidermal arachidonate 12-lipoxygenase (Alox12e) crucial for barrier formation (Johnson et al., 1999; Mao and Obeid, 2008), were significantly downregulated in Adam10-deficient epidermal splits (see Fig. S3 in the supplementary material). This coincided with a small reduction in total free lipid deposition in the stratum corneum of Adam10-deficient epidermis as judged by Nile Red staining (Fig. 3E). Together, these results strongly point to an accelerated and prematurely timed differentiation program of suprabasal cells in the Adam10 mutant mice during embryonic development in combination with either a lack or strong reduction in the formation of sebaceous glands. This then may result in improper stratum corneum formation and lipid production and deposition, and thus to altered skin barrier function.
Adam10 is crucial for the homeostasis of postnatal epidermis
To address how loss of Adam10 affects postnatal development and homeostasis of the skin, we analyzed the few surviving p18 Adam10epi–/– mice. These mice displayed a clear reduction in weight and body size (Fig. 4A,B), accompanied by a weak appearance. Another distinguishing feature of postnatal Adam10epi–/– (P18) mice is their complete lack of hair development (Fig. 4A) and the presence of short wavy whiskers (Fig. 4C, parts I, I′). The latter was already obvious in newborn mice (Fig. 1E). The P18 mice also display irregular tooth formation (Fig. 4C, parts II and II′, arrows) and patches of hyperkeratinized skin, especially on the upper dorsal side and the facial region (Fig. 4C, parts I and I′). Histology of the skin of P18 Adam10epi–/– mice showed a thickened epidermis, probably owing to hyperproliferation of basal cells (Fig. 4E, parts I and I′), and the presence of epidermal cysts in the dermis (Fig. 4D, parts I and II). In addition, these mice lacked obvious sebaceous glands (Fig. 4D, parts I, I′, II and II′), in line with the negative Nile Red and Scd1 staining in newborn Adam10epi–/– mice (Fig. 3E, parts I,I′).
Surprisingly, Adam10epi–/– (P18) mice developed splenomegaly, lymphadenopathy and thymic atrophy (Fig. 4C, parts III and IV). Further analysis revealed the presence of huge vacuoles in thymic epithelial cells while thymocytes appeared morphologically unaffected even though their number is reduced (Fig. 4D, parts III and III′). Although K14 promoter activity has been described in thymic epithelia (Fuchs, 1996), an exon-specific deletion PCR showed that in the P18 Adam10epi–/– mice Cre-mediated recombination occurred only to a very small extent in the thymus (Fig. 4F), whereas strong deletion was observed in the epidermis. This is in agreement with reports showing that K14 promoter activity is evident only in a subpopulation of thymic epithelial cells (Hafner et al., 2004; Kuraguchi et al., 2006; Rodewald, 2008), but not in spleen (Fig. 4F) or other organs (data not shown). Thus, epidermal loss of Adam10 has systemic consequences resulting in splenomegaly and lymphadenopathy.
Microarray data of epidermal splits from newborn mice (see Fig. S3A,B and Table S2A in the supplementary material) revealed that loss of Adam10 induced a 30-fold upregulation of the keratinocyte-derived chemokine thymic stromal lymphoprotein (TSLP) mRNA. ELISA analysis on serum of P9 and P18 mice confirmed the dramatic increase of TSLP at the protein level in Adam10epi–/– mice compared with control (Fig. 4G). In line with our observations, increased expression of TSLP has been shown to serve as an indicator of disturbed integrity of the murine epidermis and barrier dysfunction (Demehri et al., 2008; Moulson et al., 2003).
These postnatal phenotypes of P18 Adam10epi–/– mice could either be direct consequences of the loss of Adam10 in postnatal skin or indirect ones caused by the observed developmental skin phenotypes. To test directly whether Adam10 regulates skin homeostasis of IFE, hair follicles and sebaceous glands in adult mice, Adam10 was deleted in adult mice (Adam10epi-induc) by crossing Adam10 floxed mice with an inducible keratin 5 Cre driven tet-off system. This led to nearly complete loss of Adam10 expression, as judged by immunohistochemical staining of back skin samples (see Fig. S2B in the supplementary material). Deletion of Adam10 from P21 on resulted in a largely similar phenotype as the P18 Adam10epi–/– mice (Fig. 5A). Approximately 18 days after the induction of deletion first hair loss became apparent in Adam10epi-induc that gradually became worse with almost complete baldness after 40 days and no re-growth of hair (see Fig. S2C in the supplementary material). Additionally, we observed regions of severe hyperkeratosis (Fig. 5A, part II) in a similar pattern as observed in the Adam10epi–/– (P18) mice. As reported for the p18 Adam10epi–/–, postnatal deletion of Adam10 resulted in epidermal thickening characterized by an increased number of proliferating cells in the basal layer, and loss of K10 and increased K14 in the suprabasal layers (Fig. 5B-D), indicating hyperproliferation and disturbed differentiation. Hair follicles were destroyed accompanied by epidermal cyst formation, which were positively stained for loricirin (Fig. 5B, parts III and III′). Wholemount staining for Scd1 and Nile Red showed that inducible deletion of Adam10 did not lead to a significant loss of sebocytes at this time point, even though lipid production was already decreased in Adam10-deficient skin (Fig. 5E, parts I, I′, II and II′). Giemsa staining of back skin samples also revealed an increase of leukocyte infiltration (see Fig. S2D in the supplementary material), similar to what was observed in Adam10epi–/– (P18) skin (data not shown). Thymic atrophy (80% reduction in weight) was observed but, in contrast to P18 Adam10epi–/–, no signs of splenomegaly or lymphadenopathy were detected (see Fig. S2A in the supplementary material).
Fig. 5.
Time-controlled deletion of Adam10 using a doxycycline-inducible Tet-off system driven by the keratin 5 promoter. (A) Macroscopic appearance of inducible epidermal Adam10-deficient mice showed progressive hair loss starting ∼18 days after Dox withdrawal (I) together with hyperkeratinization and bloody lesions (shown by white arrowheads) (II). (B) Immunofluorescence analysis from paraffin sections of adult back skin revealed stratification of the epidermis, as detected with antibodies against the marker proteins keratin 14, keratin 10 and loricrin. The Adam10-deficient epidermis showed an increase in keratin 14-positive cells (I, I′) at the expense of keratin 10-positive compartments (II, II′) with loricrin-positive epidermal cyst formation (III′; arrowheads). Scale bars: 50 μm. (C) Ki67 staining showed hyperproliferation in the epidermis of Adam10-deficient mice (II) in contrast to control littermates (I). (D) Hematoxylin and Eosin staining of back skin sections demonstrated epidermal thickening, hyperkeratinization (indicated by black arrows), epidermal cyst formation (indicated by white asterisks) and reduction of lipid deposition in sebaceous glands (SB surrounded by broken lines in I) (I′) in contrast to control littermates (I). (E) Whole-mount preparations of inducible-deficient Adam10-deficient mice were stained with Nile Red (I,I′) or Scd1 antibodies (II,II′), and counterstained with DAPI. Polar lipids are stained red, non-polar lipids are stained green (I,I′). Representative pictures of sections from n=3 from each genotype are shown. Scale bars: 100 μm.
Adam10 regulates epidermal proliferation in a cell-autonomous manner
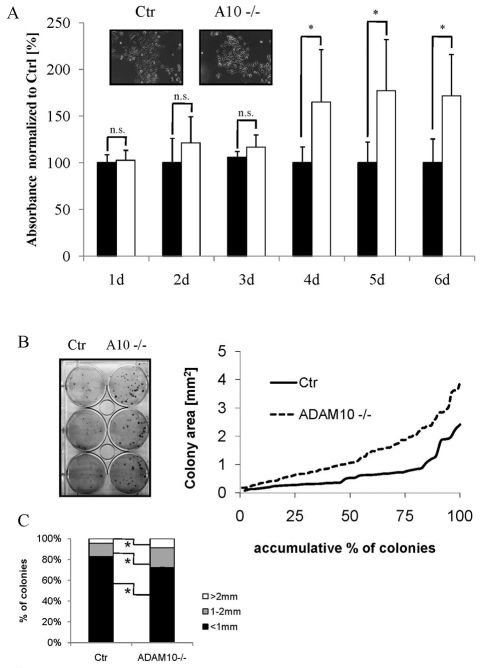
To examine whether the observed hyperproliferation in postnatal animals is a cell-autonomous consequence of loss of Adam10 or an indirect effect due to immune cell infiltration, primary keratinocytes were isolated from control and Adam10epi–/– mice. Loss of Adam10 did not obviously change the basic morphology of keratinocytes (Fig. 6A), in agreement with the ultrastructural analysis of the basal cells in newborn back skin sections (Fig. 3D). Adam10–/– cells consistently reached confluency earlier than control keratinocytes, suggesting increased growth rates. This was confirmed in proliferation assays (Fig. 6A). Colony forming capacity, a measure for the number of cells with high proliferative potential that probably represent epidermal progenitor cells (Jones and Watt, 1993; Kaur et al., 2004), was also increased in the absence of Adam10 as measured by both colony size and colony number (Fig. 6B,C). These observations indicate that the observed epidermal hyperproliferation in P18 Adam10epi–/– mice and Adam10Flox/Flox K5-tTA-Cre mice is probably due to cell-autonomous effects although an additional contribution of cytokines and growth factors secreted by infiltrated immune cells cannot be ruled out.
Fig. 6.
Adam10-deleted primary keratinocytes showed increased proliferative potential and delayed entry into differentiation. (A) Growth curve of primary keratinocytes isolated from control and mutant mice (Ctr, black bars; Adam10–/–, white bars). For each genotype, n=4 different keratinocyte cell lines were analyzed (*P<0.05). Results are mean±s.e.m. (B) Increased colony-size of Adam10-deficient keratinocytes. Quantification of the colony-forming assays is shown by plotting the increasing colony size as a percentage of the total number of colonies. The figure shows a representative example of n=4 mice for each genotype. (C) Percentage of keratinocyte colonies of control and Adam10-deficient mice with defined sizes (average from n=4 mice for each genotype; *P<0.05).
Impaired Notch signaling in conditional Adam10 skin is the trigger for defects in epidermal development and associated lipid metabolism in vivo
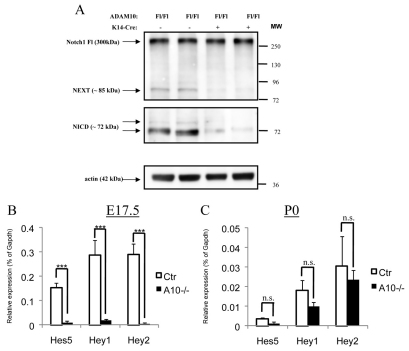
The data show that Adam10 is a crucial mediator of epidermal morphogenesis and of postnatal skin homeostasis, and raise the question of through which key pathways Adam10 exerts its physiological relevant skin function. Several key pathways, such as BMP, Notch and Wnt, have been implicated in skin morphogenesis and homeostasis. Only the phenotypes associated with epidermal deletion of different Notch pathway components were almost identical to those observed upon Adam10 deletion. To examine whether loss of Adam10 directly regulates Notch processing, we used a subset of different Notch1 antibodies to specifically detect the different cleavage fragments of the sequential Notch processing. As expected, no obvious difference was observed in Notch1 mRNA levels (data not shown) or in total full-length Notch1 between control and Adam10-deleted skin samples (Fig. 7A, upper panel). By contrast, in the absence of Adam10 only very small amounts of the Notch extracellular truncation (NEXT) and the Notch intracellular domain (NICD) fragments of Notch were found (Fig. 7A) when compared with control epidermal samples. The data thus show that Adam10 is an important protease for the generation of NEXT and that loss of Adam10 blocks NEXT generation and thus subsequent NICD production. Although we could see an accumulation of the furin S1 cleavage product in skin extracts from two mice (data not shown) the degree of this increase was highly variable among the different preparations, suggesting unspecific degradation of the remaining S1 cleavage fragment.
Fig. 7.
Adam10 is the major protease for Notch1 cleavage in murine epidermis. (A) Western Blot analysis of extracts from split epidermis from newborn mice show a strong decrease of C-terminal fragment generation demonstrated by the use of two different antibodies recognizing the NEXT and the NICD fragment of the shedded full-length receptor Notch1. (B) Regulation of Notch1 signaling in Adam10-deficient epidermis from E17.5 (n=5 for control mice, n=3 for A10–/– mice; ***P<0.0005) and newborn mice (n=3 for each genotype; n.s., not significant). (C) qRT-PCR of Notch target genes Hes5, Hey1 and Hey2. A strong reduction in the levels of Hes5, Hey1 and Hey2 was found in the Adam10 cKO epidermis in two different experiments. Results are mean±s.e.m.
A partial compensatory role for other Adam metalloproteases in the generation of NEXT cannot be completely excluded as very low levels of NEXT and NICD were still observed. Adam17 has also been proposed as a sheddase for Notch in murine cells (Mumm et al., 2000; Brou et al., 2000). However, no alterations in both mRNA (data not shown) and protein expression levels of the pro- and, most importantly, active form of Adam17 (see Fig. S2E in the supplementary material) were observed in Adam10-deficient epidermis. Microarray and quantitative real-time analysis identified Adam8 as another candidate that might partially compensate for the loss of Adam10. Real-time PCR analysis revealed upregulation of Adam8 at E17.5 and in newborn mice upon loss of Adam10 (see Fig. S2F in the supplementary material).
To examine whether the strong reduction in NICD generation has a functional impact on Notch signaling in Adam10–/– epidermis, we examined expression of Notch downstream genes such as Hes and Hey (Ehebauer et al., 2006) by qRT-PCR on epidermal splits from newborn mice. Surprisingly, control newborn epidermis showed relatively low mRNA levels of the Hes/Hey family when compared with control E17.5 epidermis (Fig. 7B,C), indicating that Notch signaling peaks at this embryonic time point during epidermal morphogenesis. At P0 these low Hes/Hey levels were even further reduced in the absence of Adam10, indicating that loss of Adam10 directly affects epidermal Notch signaling strength (Fig. 7C). Much more strikingly, the expression of Notch target mRNAs, with the exception of Hes1 (data not shown), was almost completely negative in E17.5 Adam10–/– compared with control (Fig. 7B), indicating that loss of Adam10 severely compromises Notch signaling at a time in development when signaling is at a peak in control animals.
In order to obtain a more general view on how loss of Adam10 affects Notch-dependent and -independent gene expression in newborn epidermis (P0) we employed a whole genome differential gene expression analysis (see Fig. S3A,B in the supplementary material). These analyses confirmed the Hes/Hey qRT-PCR results of P0 that showed that at this stage (P0) overall Notch target gene expression was only mildly affected (data not shown). An exception was a known negative-feedback regulator of Notch, NRARP (data not shown), which is activated when Notch signaling exceeds its normal magnitude (Lamar et al., 2001).
Direct comparison of these data with gene expression data published on other Notch-signaling skin mutant mice, showed strong overlaps in activated and inhibited targets between Adam10 and Notch pathway mutants. For example, 15 genes representing important members of epidermal lipid metabolism all exhibited at least twofold reduction in Adam10-deleted skin (see Table S1A in the supplementary material) as well as in Notch-1 knockouts (Demehri et al., 2009). This also fits with the observation that Adam10 mutant mice showed compromised sebaceous gland development accompanied by a loss of lipid production (Fig. 3E, Fig. 5E). A strong downregulation in expression of genes involved in retinol metabolism and retinoic acid signaling pathway was also observed in both Adam10 newborn mice and after Notch deletion (Vauclair et al., 2007) (see Table S1B in the supplementary material). In addition, a strong increase of genes involved in processes such as hyperkeratinization, hyperproliferation and wound healing was observed (see Table S2A in the supplementary material), which may be caused by the compromised barrier. Although we could not yet detect signs of hyperproliferation or hyperkeratinization in the IFE of newborn mice (Fig. 3A) (see Fig. S1E in the supplementary material), these alterations represent very early signs of impaired skin homeostasis in the epidermis of newborn mice and may partially explain the increased proliferation and hyperkeratinization seen later in Adam10epi–/– (P18) or in Adam10epi-induc mice. In conclusion, the in vivo mouse data together with the biochemical analysis and gene expression data strongly indicate that Adam10 is an important physiological regulator of Notch signaling in the epidermis through which it regulates skin morphogenesis and homeostasis.
DISCUSSION
Stratification of the epidermis is a tightly controlled process that requires the orchestrated action of different key signal transduction pathways, such as e.g. Wnt/β-catenin, Notch, TGF-β and NF-κB (Blanpain and Fuchs, 2009; Koster and Roop, 2007). Using epidermal specific Adam10 conditional knockout mice, we asked whether the protease Adam10 plays a decisive role in epidermal integrity and assessed the in vivo relevance of Adam10 as the crucial sheddase necessary to mediate epidermal-specific Notch signaling. Epidermal inactivation of Adam10 either during skin morphogenesis or in adult mice resulted in skin and visceral phenotypes that are largely identical to epidermal inactivation of key components of the Notch signaling pathway. Over 90% of the Adam10epi–/– mice die perinatally with compromised skin barrier function. Similar observations were made in different mice in which key components of the Notch transduction pathway were inactivated in the skin, for example in epidermal Notch1/Notch2, RBP-j, PS1/PS2 and Hes-1 conditional knockout mice (Demehri et al., 2009; Moriyama et al., 2008). Most strikingly, newborn Adam10epi–/– mice have an identical epidermal hypoplasia, owing to the same reduction of spinous epidermal layers as that found in mice with a disrupted Notch pathway (Blanpain et al., 2006; Segre, 2006). In addition, postnatal inactivation of either Notch signaling or Adam10 lead to epidermal thickening due to a hyperproliferative state of basal keratinocytes, as well as to degeneration of hair follicles associated with epidermal cyst formation (Lee et al., 2010; Nicolas et al., 2003; Vauclair et al., 2005). In line with these phenotypic similarities, epidermal loss of Adam10 did disturb Notch signaling, as judged by severely impaired Notch processing, a strong reduction in direct Notch target genes and overall overlap in alterations in global gene expression. Overall, the data identify Adam10 as a crucial regulator of skin morphogenesis and homeostasis.
The results indicate that Adam10 functions as the major sheddase for all three Notch receptors that are expressed in the epidermis, both during morphogenesis and in the adult epidermis. This is based on the observation that the phenotypes associated with epidermal deletion of Adam10 closely resemble phenotypes of mice with non-redundant mediators of Notch signaling, such as for example RBP-j, Hes1 or presenilin 1/presenilin 2 double knockout (Blanpain et al., 2006; Demehri et al., 2009; Moriyama et al., 2008), whereas single Notch receptor or ligand mice display milder phenotypes (Nicolas et al., 2003; Vauclair et al., 2005). Epidermal deletion of Notch1 during morphogenesis is similar to that of Adam10, indicating that Notch1 is the crucial Notch pathway during development. By contrast, the consequences of inducible deletion of Notch 1 alone in adult mice are much less pronounced than loss of Adam10 in adult mice (Demehri et al., 2009; Nicolas et al., 2003; Vauclair et al., 2005), indicating that other Notch receptors also play key roles during adult skin homeostasis.
Surprisingly, we could not detect a reduction in expression of Hes1, one of the major Notch target genes, in the skin at either E15.5, E17.5 or P0, even though we found a very strong reduction in NICD, the signaling fragment of Notch, and in the expression of several other Notch target genes upon loss of Adam10. Previous results indicated an important role for Hes1 in the development of the interfollicular epidermis (Ambler and Watt, 2007; Moriyama et al., 2008; Rangarajan et al., 2001). A potential explanation for this difference may be that Hes1 expression and function in the epidermis is largely independent of canonical Notch activation, as was shown in other systems (Berechid et al., 2002; Curry et al., 2006; Ingram et al., 2008; Zhang et al., 2010). In partial agreement with this are observations that showed an important role for Hes-independent Notch signaling in epidermal development (Blanpain et al., 2006; Moriyama et al., 2008). On the other hand, expression of Hes1 is tightly controlled by an autoregulatory feedback loop, e.g. by increased expression of Ascl2 (Moriyama et al., 2008).
Adam17 has also been claimed as a potential sheddase for the Notch signaling pathway (Brou et al., 2000; Mumm et al., 2000). However, the biochemical and expression data showing reduced Notch signaling in the absence of Adam10, together with the strong overlap in phenotypes and global gene expression between Adam10 and Notch mutants show that Adam17 or the upregulated Adam8 cannot functionally substitute for Adam10 in Notch processing. In agreement, epidermal deletion of Adam10 did not alter expression nor activity of Adam17 (see Fig. S2D in the supplementary material).
Interestingly, global gene expression analysis revealed a severe downregulation of genes involved in lipid and retinoid metabolism with a simultaneous upregulation of genes playing a role in calcium-mediated differentiation. Notch signaling has also been shown to regulate lipid (Demehri and Kopan, 2009; Nickoloff et al., 2002) and retinoid metabolism (Vauclair et al., 2007). These pathways are crucial for epidermal differentiation and barrier function, and alterations in these pathways may directly contribute to the observed barrier defects in Adam10epi–/– mice. In agreement with a reduction in these lipid regulatory pathways, altered sebaceous gland development associated with reduced lipid production is also observed in both Adam10 and Notch mutants (Demehri et al., 2008).
The Adam10epi-induc mice may serve as an interesting mouse model in which to study the development of skin diseases associated with hyperproliferation, alopecia and hyperkeratinization. These phenotypes are at least partially due to a cell-autonomous function of Adam10, as Adam10–/– keratinocytes showed increased proliferation in culture, in combination with an intrinsically compromised barrier. This in turn may alter the micro-environment, resulting in recruitment and activation of immune cells, and further aggravation of the phenotypes, as has been described for loss of Notch function in the skin (Demehri et al., 2008; Dumortier et al., 2010). Interestingly, overexpression or increased activation of Adam10 in humans has been associated with oncogenesis and chronic skin diseases such as psoriasis and eczema (Dittmer et al., 2009; Lee et al., 2010; Maretzky et al., 2008; Oh et al., 2008). As the inducible Notch1 mutants also showed increased susceptibility for spontaneous and chemically induced skin carcinogenesis (Demehri et al., 2009; Nicolas et al., 2003), it would be of great interest to examine a potential tumor suppressive effect of Adam10 in the skin.
Taken together, we show that the membrane-spanning metalloprotease Adam10 is indispensable for the morphogenesis and postnatal homeostasis of murine skin, and plays an essential role in the maintenance of skin appendages like hair follicle and sebaceous glands. The results identify Adam10 as the most important epidermal sheddase for Notch and show that the crucial role in the skin is mediated via the regulation of canonical Notch signaling pathways. As Adam10 is also the major protease for cleaving Notch receptors in postnatal skin it may serve as a potent target to control Notch activity in general.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft SFB877, 829 and 832, The Deutsche Krebshilfe, a Methusalem grant of the KU Leuven and IUAP P6/58 of the Belgian Federal Science Policy Office, DeZnit (EU-FP VI), the Center of Excellence `Inflammation at Interfaces', and NIH R01 grant GM64750 to C.P.B. We are grateful to Robert Hässler for support with the microarray experiments. We thank Tobias Lehmann, Dagmar Niemeier for technical assistance. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.055210/-/DC1
References
- Ambler C. A., Watt F. M. (2007). Expression of Notch pathway genes in mammalian epidermis and modulation by beta-catenin. Dev. Dyn. 236, 1595-1601 [DOI] [PubMed] [Google Scholar]
- Berechid B. E., Kitzmann M., Foltz D. R., Roach A. H., Seiffert D., Thompson L. A., Olson R. E., Bernstein A., Donoviel D. B., Nye J. S. (2002). Identification and characterization of presenilin-independent Notch signaling. J. Biol. Chem. 277, 8154-8165 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. (2009). Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10, 207-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Lowry W. E., Pasolli H. A., Fuchs E. (2006). Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022-3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkulak E. C., Weinmaster G. (2009). Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679-5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun K. M., Niemann C., Jensen U. B., Sundberg J. P., Silva-Vargas V., Watt F. M. (2003). Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development 130, 5241-5255 [DOI] [PubMed] [Google Scholar]
- Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israel A. (2000). A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207-216 [DOI] [PubMed] [Google Scholar]
- Curry C. L., Reed L. L., Nickoloff B. J., Miele L., Foreman K. E. (2006). Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab. Invest. 86, 842-852 [DOI] [PubMed] [Google Scholar]
- Dassule H. R., Lewis P., Bei M., Maas R., McMahon A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775-4785 [DOI] [PubMed] [Google Scholar]
- Demehri S., Kopan R. (2009). Notch signaling in bulge stem cells is not required for selection of hair follicle fate. Development 136, 891-896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S., Liu Z., Lee J., Lin M. H., Crosby S. D., Roberts C. J., Grigsby P. W., Miner J. H., Farr A. G., Kopan R. (2008). Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 6, e123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S., Turkoz A., Kopan R. (2009). Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell 16, 55-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond I., Owolabi T., Marco M., Lam C., Glick A. (2000). Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J. Invest. Dermatol. 115, 788-794 [DOI] [PubMed] [Google Scholar]
- Dittmer A., Hohlfeld K., Lutzkendorf J., Muller L. P., Dittmer J. (2009). Human mesenchymal stem cells induce E-cadherin degradation in breast carcinoma spheroids by activating ADAM10. Cell Mol. Life Sci. 66, 3053-3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A., Durham A. D., Di Piazza M., Vauclair S., Koch U., Ferrand G., Ferrero I., Demehri S., Song L. L., Farr A. G., et al. (2010). Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of notch signaling in the murine skin. PLoS ONE 5, e9258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehebauer M., Hayward P., Martinez-Arias A. (2006). Notch signaling pathway. Sci. STKE 2006, cm7 [DOI] [PubMed] [Google Scholar]
- Fuchs E. (1996). The cytoskeleton and disease: genetic disorders of intermediate filaments. Annu. Rev. Genet. 30, 197-231 [DOI] [PubMed] [Google Scholar]
- Gareus R., Huth M., Breiden B., Nenci A., Rosch N., Haase I., Bloch W., Sandhoff K., Pasparakis M. (2007). Normal epidermal differentiation but impaired skin-barrier formation upon keratinocyte-restricted IKK1 ablation. Nat. Cell Biol. 9, 461-469 [DOI] [PubMed] [Google Scholar]
- Hafner M., Wenk J., Nenci A., Pasparakis M., Scharffetter-Kochanek K., Smyth N., Peters T., Kess D., Holtkotter O., Shephard P., et al. (2004). Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis 38, 176-181 [DOI] [PubMed] [Google Scholar]
- Hardman M. J., Sisi P., Banbury D. N., Byrne C. (1998). Patterned acquisition of skin barrier function during development. Development 125, 1541-1552 [DOI] [PubMed] [Google Scholar]
- Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lubke T., Lena Illert A., von Figura K., et al. (2002). The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615-2624 [DOI] [PubMed] [Google Scholar]
- Hasler R., Begun A., Freitag-Wolf S., Kerick M., Mah N., Zvirbliene A., Spehlmann M. E., von Wurmb-Schwark N., Kupcinskas L., Rosenstiel P., et al. (2009). Genetic control of global gene expression levels in the intestinal mucosa: a human twin study. Physiol. Genomics 38, 73-79 [DOI] [PubMed] [Google Scholar]
- Hennings H., Michael D., Cheng C., Steinert P., Holbrook K., Yuspa S. H. (1980). Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell 19, 245-254 [DOI] [PubMed] [Google Scholar]
- Ingram W. J., McCue K. I., Tran T. H., Hallahan A. R., Wainwright B. J. (2008). Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene 27, 1489-1500 [DOI] [PubMed] [Google Scholar]
- Johnson E. N., Nanney L. B., Virmani J., Lawson J. A., Funk C. D. (1999). Basal transepidermal water loss is increased in platelet-type 12-lipoxygenase deficient mice. J. Invest. Dermatol. 112, 861-865 [DOI] [PubMed] [Google Scholar]
- Jones P. H., Watt F. M. (1993). Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73, 713-724 [DOI] [PubMed] [Google Scholar]
- Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L., Snellinx A., Craessaerts K., Thathiah A., Tesseur I., et al. (2010). The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 30, 4833-4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Li A., Redvers R., Bertoncello I. (2004). Keratinocyte stem cell assays: an evolving science. J. Investig. Dermatol. Symp. Proc. 9, 238-247 [DOI] [PubMed] [Google Scholar]
- Koster M. I., Roop D. R. (2004). Genetic pathways required for epidermal morphogenesis. Eur. J. Cell Biol. 83, 625-629 [DOI] [PubMed] [Google Scholar]
- Koster M. I., Roop D. R. (2007). Mechanisms regulating epithelial stratification. Annu. Rev. Cell Dev. Biol. 23, 93-113 [DOI] [PubMed] [Google Scholar]
- Kuraguchi M., Wang X. P., Bronson R. T., Rothenberg R., Ohene-Baah N. Y., Lund J. J., Kucherlapati M., Maas R. L., Kucherlapati R. (2006). Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2, e146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar E., Deblandre G., Wettstein D., Gawantka V., Pollet N., Niehrs C., Kintner C. (2001). Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 15, 1885-1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Schramme A., Doberstein K., Dummer R., Abdel-Bakky M. S., Keller S., Altevogt P., Oh S. T., Reichrath J., Oxmann D., et al. (2010). ADAM10 is upregulated in melanoma metastasis compared with primary melanoma. J. Invest. Dermatol. 130, 763-773 [DOI] [PubMed] [Google Scholar]
- Lin M. H., Kopan R. (2003). Long-range, nonautonomous effects of activated Notch1 on tissue homeostasis in the nail. Dev. Biol. 263, 343-359 [DOI] [PubMed] [Google Scholar]
- Lin M. H., Leimeister C., Gessler M., Kopan R. (2000). Activation of the Notch pathway in the hair cortex leads to aberrant differentiation of the adjacent hair-shaft layers. Development 127, 2421-2432 [DOI] [PubMed] [Google Scholar]
- Mao C., Obeid L. M. (2008). Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim. Biophys. Acta 1781, 424-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T., Scholz F., Koten B., Proksch E., Saftig P., Reiss K. (2008). ADAM10-mediated E-cadherin release is regulated by proinflammatory cytokines and modulates keratinocyte cohesion in eczematous dermatitis. J. Invest. Dermatol. 128, 1737-1746 [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Man W. C., Ntambi J. M. (2001). Targeted disruption of stearoyl-CoA desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J. Nutr. 131, 2260-2268 [DOI] [PubMed] [Google Scholar]
- Moriyama M., Durham A. D., Moriyama H., Hasegawa K., Nishikawa S., Radtke F., Osawa M. (2008). Multiple roles of Notch signaling in the regulation of epidermal development. Dev. Cell 14, 594-604 [DOI] [PubMed] [Google Scholar]
- Moulson C. L., Martin D. R., Lugus J. J., Schaffer J. E., Lind A. C., Miner J. H. (2003). Cloning of wrinkle-free, a previously uncharacterized mouse mutation, reveals crucial roles for fatty acid transport protein 4 in skin and hair development. Proc. Natl. Acad. Sci. USA 100, 5274-5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J. S., Kopan R. (2000). Notch signaling: from the outside in. Dev. Biol. 228, 151-165 [DOI] [PubMed] [Google Scholar]
- Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000). A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197-206 [DOI] [PubMed] [Google Scholar]
- Nickoloff B. J., Qin J. Z., Chaturvedi V., Denning M. F., Bonish B., Miele L. (2002). Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 9, 842-855 [DOI] [PubMed] [Google Scholar]
- Nicolas M., Wolfer A., Raj K., Kummer J. A., Mill P., van Noort M., Hui C. C., Clevers H., Dotto G. P., Radtke F. (2003). Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33, 416-421 [DOI] [PubMed] [Google Scholar]
- Oh S. T., Schramme A., Stark A., Tilgen W., Gutwein P., Reichrath J. (2008). Overexpression of ADAM 10 and ADAM 12 in lesional psoriatic skin. Br. J. Dermatol. 158, 1371-1373 [DOI] [PubMed] [Google Scholar]
- Okochi M., Fukumori A., Jiang J., Itoh N., Kimura R., Steiner H., Haass C., Tagami S., Takeda M. (2006). Secretion of the Notch-1 Abeta-like peptide during Notch signaling. J. Biol. Chem. 281, 7890-7898 [DOI] [PubMed] [Google Scholar]
- Okuyama R., Tagami H., Aiba S. (2008). Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases. J. Dermatol. Sci. 49, 187-194 [DOI] [PubMed] [Google Scholar]
- Oshima H., Rochat A., Kedzia C., Kobayashi K., Barrandon Y. (2001). Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 104, 233-245 [DOI] [PubMed] [Google Scholar]
- Pan Y., Lin M. H., Tian X., Cheng H. T., Gridley T., Shen J., Kopan R. (2004). gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell 7, 731-743 [DOI] [PubMed] [Google Scholar]
- Proweller A., Tu L., Lepore J. J., Cheng L., Lu M. M., Seykora J., Millar S. E., Pear W. S., Parmacek M. S. (2006). Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 66, 7438-7444 [DOI] [PubMed] [Google Scholar]
- Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J. C., Krishna S., Metzger D., Chambon P., et al. (2001). Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427-3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald H. R. (2008). Thymus organogenesis. Annu. Rev. Immunol. 26, 355-388 [DOI] [PubMed] [Google Scholar]
- Segre J. A. (2006). Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 116, 1150-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S., Hughes J. D., Campbell M. J., Cho R. J., Church G. M. (1999). Systematic determination of genetic network architecture. Nat. Genet. 22, 281-285 [DOI] [PubMed] [Google Scholar]
- Taylor G., Lehrer M. S., Jensen P. J., Sun T. T., Lavker R. M. (2000). Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102, 451-461 [DOI] [PubMed] [Google Scholar]
- Thelu J., Rossio P., Favier B. (2002). Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunggal J. A., Helfrich I., Schmitz A., Schwarz H., Gunzel D., Fromm M., Kemler R., Krieg T., Niessen C. M. (2005). E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 24, 1146-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttendaele H., Panteleyev A. A., de Berker D., Tobin D. T., Christiano A. M. (2004). Activation of Notch1 in the hair follicle leads to cell-fate switch and Mohawk alopecia. Differentiation 72, 396-409 [DOI] [PubMed] [Google Scholar]
- Vauclair S., Nicolas M., Barrandon Y., Radtke F. (2005). Notch1 is essential for postnatal hair follicle development and homeostasis. Dev. Biol. 284, 184-193 [DOI] [PubMed] [Google Scholar]
- Vauclair S., Majo F., Durham A. D., Ghyselinck N. B., Barrandon Y., Radtke F. (2007). Corneal epithelial cell fate is maintained during repair by Notch1 signaling via the regulation of vitamin A metabolism. Dev. Cell 13, 242-253 [DOI] [PubMed] [Google Scholar]
- Watt F. M., Estrach S., Ambler C. A. (2008). Epidermal Notch signalling: differentiation, cancer and adhesion. Curr. Opin. Cell Biol. 20, 171-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Tanigaki K., Han H., Hiai H., Honjo T. (2003). Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 13, 333-338 [DOI] [PubMed] [Google Scholar]
- Zhang P., Yang Y., Nolo R., Zweidler-McKay P. A., Hughes D. P. (2010). Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene 29, 2916-2926 [DOI] [PMC free article] [PubMed] [Google Scholar]