Abstract
The translational repressor Nanos is expressed in the germline and stem cell populations of jellyfish as well as humans. Surprisingly, we observed that unlike other mRNAs, synthetic nanos1 RNA translates very poorly if at all after injection into Xenopus oocytes. The current model of simple sequestration of nanos1 within germinal granules is insufficient to explain this observation and suggests that a second level of repression must be operating. We find that an RNA secondary structural element immediately downstream of the AUG start site is both necessary and sufficient to prevent ribosome scanning in the absence of a repressor. Accordingly, repression is relieved by small in-frame insertions before this secondary structure, or translational control element (TCE), that provide the 15 nucleotides required for ribosome entry. nanos1 is translated shortly after fertilization, pointing to the existence of a developmentally regulated activator. Oocyte extracts were rendered fully competent for nanos1 translation after the addition of a small amount of embryo extract, confirming the presence of an activator. Misexpression of Nanos1 in oocytes from unlocalized RNA results in abnormal development, highlighting the importance of TCE-mediated translational repression. Although found in prokaryotes, steric hindrance as a mechanism for negatively regulating translation is novel for a eukaryotic RNA. These observations unravel a new mode of nanos1 regulation at the post-transcriptional level that is essential for normal development.
Keywords: Xenopus, nanos family, Translational repression, RNA secondary structure, Germline
INTRODUCTION
The germ cell lineage and somatic fates of the early embryo are initially specified by maternal RNAs localized to the vegetal pole of Xenopus oocytes (King et al., 2005; Kloc et al., 2001; Zhang et al., 1998). These maternal determinants are active well before zygotic transcription begins, highlighting the importance of post-transcriptional mechanisms in regulating early development. Translation of the endoderm determinant VegT is initiated during maturation and appears to be regulated by cytoplasmic polyadenylation together with a factor that both stabilizes and promotes its translation (Souopgui et al., 2008; Stennard et al., 1999). Vg1, which is required for mesoderm formation and left-right asymmetry, is translated soon after its localization is complete during mid-oogenesis (reviewed by King et al., 2005). Prior to localization, Vg1 is bound by a trans-acting repressor at a 3′UTR site (Colegrove-Otero et al., 2005a). Thus, the translational regulation of essential somatic cell determinants is accomplished through trans-acting factors binding to the UTR via mechanisms that are common to eukaryotic RNAs (Johnstone and Lasko, 2001; Schier, 2007; Richter and Sonenberg, 2005). By contrast, little is known about how expression of the germ cell determinants is regulated, although such regulation is crucial for establishing the next generation.
One evolutionarily conserved component of germ cells that is required for maintenance and self-renewal is nanos (reviewed by Shen and Xie, 2010; Subramaniam and Seydoux, 1999; Wang and Lin, 2004). In vertebrate and invertebrate species, the Nanos family of proteins function as translational repressors that are essential to maintain the germline precursors – the primordial germ cells (PGCs) (Curtis et al., 1997; Kadyrova et al., 2007; Lai et al., 2011; Tsuda et al., 2003). Xenopus nanos RNA (nanos1; formerly called Xcat2) is transcribed during early oogenesis and becomes localized to the germ plasm, a subcellular compartment bearing the germ cell determinants. There, nanos1 is packaged into germinal granules, which are diagnostic structures of germ plasm that are considered to provide a mechanism for long-term storage of RNAs and proteins (Forristall et al., 1995; Kloc et al., 2002; Tsuda et al., 2003; Zhou and King, 1996). nanos1 is not translated during the 4 to 6 months required for oogenesis, but Nanos1 protein is detected during early development (Lai et al., 2011). Thus, nanos1 and other germline RNAs are activated sometime during development but the mechanisms remain unknown (reviewed by King et al., 2005; Kloc et al., 2001).
Although the repressive activity of Nanos is required for PGCs to maintain their identity in the presence of somatic determinants, the somatic cells must also require Nanos function to be restricted to the germline to allow somatic cell determination (Jadhav et al., 2008; Kobayashi et al., 1996; Köprunner et al., 2001; Tsuda et al., 2003). Therefore, one might expect a robust and overlapping translational repression of nanos transcripts, as best documented in Drosophila and C. elegans. Drosophila Nanos is restricted to the posterior pole of the oocyte and early embryo through both translational repression of unlocalized nanos mRNA and translational activation of localized nanos message at the posterior pole (Forrest and Gavis, 2003; Gavis and Lehmann, 1992; Gavis and Lehmann, 1994). The regulation of nanos germline expression in Drosophila, C. elegans and mouse requires the 3′UTR (Gavis et al., 1996; Jadhav et al., 2008; Suzuki et al., 2010). Interestingly, the 3′UTR translational control element (TCE) of Drosophila nanos forms a structure containing two hairpins, each of which binds a distinct repressor. These stem-loops act independently of each other to repress translation at different times in development: in embryonic somatic cells and in the oocyte, respectively (Forrest et al., 2004; Kalifa et al., 2006). Similarly, C. elegans nos-2 is translationally regulated to permit expression exclusively in the germline lineages by two independent stem-loops in the 3′UTR (Jadhav et al., 2008; Subramaniam and Seydoux, 1999; D'Agostino et al., 2006). Thus, in many respects, both the function and regulation of germline nanos have been conserved.
Surprisingly, we observed that unlike any other known mRNA, synthetic nanos1 RNA translates very poorly after injection into Xenopus oocytes. Clearly, the current model of simple sequestration of nanos1 within germinal granules is insufficient to explain this observation and suggests that a second level of repression must be operating. Here, we describe a novel mechanism for regulating nanos1 RNA expression. We present evidence that an RNA secondary structural element in the ORF is sufficient to repress the translational initiation of nanos1 RNA and the expression of a reporter. Consistent with this model, we find that insertions between the start codon and the TCE relieve repression. Misexpression of nanos1 in oocytes results in abnormal development, highlighting the importance of translational control mediated by the TCE during oogenesis. Such a structural mechanism that operates independently of a repressor for negative regulation of translation, although common in prokaryotes, is completely new for eukaryotic RNA (reviewed by Kozak, 2005). These observations unravel a novel mode of nanos1 regulation at the post-transcriptional level that is essential for normal development.
MATERIALS AND METHODS
Plasmids
Deletion, insertion and substitution mutants (see Fig. 2A, mutants S1-S4 and CS) of nanos1 (Mosquera et al., 1993) were generated using the QuickChange Mutagenesis Kit (Stratagene). Inserted nucleic acid sequences were from equivalent positions of the β-globin ORF. The β-globin transcripts were generated from pSPXβM (Krieg and Melton, 1984). Constructs of nanos1 and β-globin with exchanged UTRs were generated by two-step PCR. For Myc-nanos1, the nanos1 coding region was subcloned into the pCS2-Myc vector between EcoRI and XhoI sites. nanos1-Myc was generated by subcloning the 5′UTR and ORF of nanos1 into the pCS2-Myc vector between BamHI and ClaI sites. For primer sequences, see Table S1 in the supplementary material.
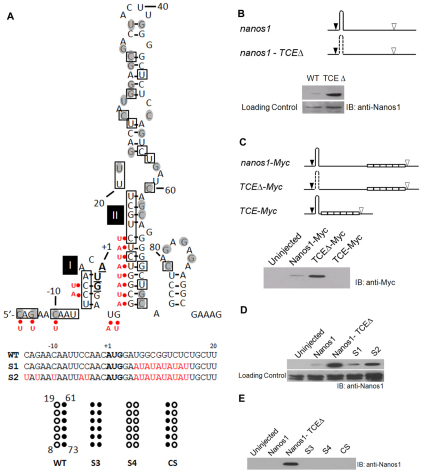
Fig. 2.
Identification of the nanos1 translational control element (TCE). (A) Summary of structural probing of the 5′ terminal region of nanos1 transcript with RNases A, T1 and V1. Secondary structure of the 5′ terminal region of nanos1 (–15 to +95) is shown as predicted by MFOLD (Zuker, 2003). The start codon nucleotides are in bold and underlined. The nucleotides sensitive to single-strand-specific RNase are within shaded circles and those sensitive to base-paired-specific RNase are in open boxes. AU substitutions are indicated by red dots. The S3, S4 and CS mutants are illustrated beneath, alongside the wild-type (WT) sequence. (B) Deletion of the TCE relieves repression. Translation of wild-type nanos1 and a TCE deletion mutant (TCEΔ) injected into Xenopus oocytes. The predicted nanos1 TCE (+8 to +73) is shown as a hairpin structure. Start (black arrowhead) and stop (white arrowhead) codons are indicated; dashed line signifies deletion. Immunoblot (IB) with anti-Nanos1 antibody. (C) The TCE is both necessary and sufficient for nanos1 repression. nanos1-Myc and deletion mutants, TCEΔ-Myc and TCE-Myc, were injected into oocytes and their translation analyzed by blotting with anti-Myc antibody. Myc tags are shown as open white. (D) Disruption of base-pairs in Stem-loops I and II promotes translation. Translation in oocytes of mutants S1 and S2, analyzed as detailed in B. (E) Substitution (S3, S4) or compensatory (CS) changes failed to relieve repression. See also Figs S2 and S3 in the supplementary material.
Oocytes, embryos and micro-injection
Oocytes and embryos were obtained as described (Sive et al., 2000). RNAs for injection were prepared from the plasmid templates described above using the mMESSAGE mMACHINE Kit (Ambion).
In vitro translation
In vitro translation with either reticulocyte lysates or wheat germ extracts was carried out according to the manufacturer's instructions (Promega). Translation-competent oocyte and embryo extracts were prepared essentially as described (Murray, 1991). For the translation assay, a mixture of amino acids (Promega), creatine kinase (1 mg/ml final concentration; Sigma) and capped mRNA was added to the extract.
Immunoprecipitation
Synthetic nanos1 transcripts were injected into Xenopus oocytes or 1-cell embryos. Samples were subjected to immunoprecipitation with goat anti-Nanos1 antibody and Protein G beads (Roche).
Western blotting
Western blots were performed as previously described with goat anti-Nanos1 (1:100) or rabbit anti-Myc-HRP (1:2000; Invitrogen) primary antibody and rabbit anti-goat-HRP secondary antibody (1:1000) (Venkatarama et al., 2010). All blots consistently showed a high molecular weight non-specific band, which was used as a loading control.
Sucrose gradient analysis
Wheat germ translation extract (50 μl; Promega) was mixed with 0.2 μg of radiolabeled transcripts and incubated at 20°C for 20 minutes. The reactions were stopped by addition of cold sucrose gradient buffer [(20 mM Hepes pH 7.5, 90 mM KCl, 1.5 mM MgCl2 and 500 μg/ml cycloheximide (Sigma) or 2 mM GMP-PNP (Sigma)] and layered onto a 10-30% linear sucrose gradient. The gradient was then centrifuged at 39,000 rpm (188,000 g) for 4 hours at 4°C in an SW41 rotor. The gradient was collected from the bottom in 0.5 ml fractions and the ribosomal RNA profile was determined by UV260. Radioactivity of each fraction was measured by scintillation.
RNase probing
RNase probing was performed as previously described (Darfeuille et al., 2007).
Immunofluorescence and confocal imaging
Immunofluorescence and confocal imaging were performed as described previously (Venkatarama et al., 2010).
Host transfer
Host transfer was performed as described by Mir and Heasman (Mir and Heasman, 2008).
RESULTS
nanos1 RNA is poorly translated after injection into oocytes
Xenopus oocytes have long been used to express injected mRNAs from a wide variety of sources, including viruses (Brown, 2004). Therefore, in experiments addressing the function of nanos1, we were surprised to find that although nanos1 RNA was translated in reticulocyte lysates, it was poorly translated, if at all, after injection into oocytes (Fig. 1A). One explanation is that nanos1 RNA and protein are degraded if present outside their normal cellular location within germ plasm. To explore this possibility, we injected radiolabeled nanos1 transcripts and isolated oocytes at different time points to assess the percentage that remained (see Fig. S1 in the supplementary material). We found that nanos1 RNA was comparable in stability to that of control β-globin mRNA (see Fig. S1A in the supplementary material). Next, we synthesized radiolabeled nanos1 protein by in vitro translation and injected it into oocytes. Nanos1 was still detected at least 6 hours after injection (see Fig. S1B in the supplementary material). These results showed that both nanos1 RNA and protein were stable, suggesting that injected nanos1 RNA is poorly translated in oocytes.
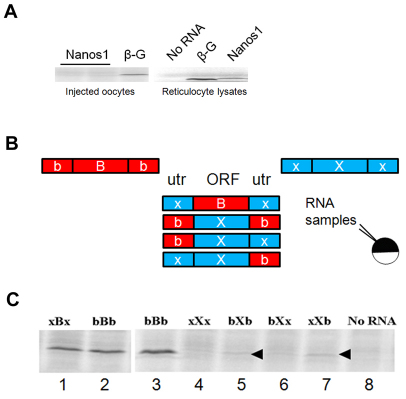
Fig. 1.
Repression of nanos1 translation in Xenopus oocytes is mediated by the ORF. (A) (Left) nanos1 transcripts were co-injected with [35S]methionine into oocytes and incubated overnight at 18°C. β-globin (β-G) served as control. (Right) nanos1 and β-globin were translated in reticulocyte lysates. (B) Schematic of the transcripts tested and the experimental design. ORFs (uppercase) and UTRs (lowercase) were from nanos1 [X] or β-globin [B]. (C) RNAs with UTR substitutions (labeled in accordance with B) were assayed in oocyte extracts. Arrowheads mark the position of nanos1. See also Fig. S1 in the supplementary material.
Translational repression of nanos1 is not dependant on UTRs
Repression of maternal transcripts is commonly regulated through TCEs in the UTRs that serve as binding sites for proteins or small RNAs. Previous work from our laboratory suggested that the TCE did not lie within the 3′UTR, as substitution with the 3′UTR of β-globin did not result in translation (MacArthur et al., 1999). To confirm these published results, and to identify which sequences constitute the nanos1 TCE, a set of constructs was generated representing the various combinations of UTRs and ORFs from Xenopus β-globin and nanos1 (Fig. 1B). These transcripts were tested for translation in stage VI oocyte extracts supplemented with radioactive methionine. β-globin is a particularly good control for these experiments as β-globin and Nanos1 have the same number of methionines, their UTRs are similar in length, and the proteins are of similar mass. Thus, we can judge translational efficiency directly from radiolabel-incorporation data. We first asked whether the nanos1 UTRs were sufficient to repress translation of the β-globin ORF in oocyte extracts. These hybrid constructs were equally well translated as the β-globin RNA itself (Fig. 1C, compare lane 1 with lanes 2 and 3). Similarly, substitution of the nanos1 UTRs with those of β-globin failed to relieve repression of the nanos1 ORF (Fig. 1C, compare lanes 5-7 with 4). Our results strongly suggested that the TCE lies within the nanos1 ORF itself.
The TCE lies within the first 75 nucleotides of the ORF
We reasoned that the TCE might lie in close proximity to the start codon and form a structure involved in repression. As a first step, we analyzed full-length nanos1 RNA using MFOLD RNA structure prediction software (Zuker, 2003) to compute the most probable structures based on the free energies of the folds. Of the 35 separate predictions returned, 16 (45.7%) revealed one large and two small stem-loop structures within the 5′UTR and first 93 nucleotides (nt) of the nanos1 ORF (Fig. 2A; see Fig. S2B in the supplementary material). Interestingly, the region 15 nt upstream of the AUG start site in X. laevis is identical to that in X. borealis and virtually identical to that in X. tropicalis, species that are separated from X. laevis by 50 and 120 million years, respectively (Graf, 1996), and that have much longer 5′UTRs (see Fig. S2A in the supplementary material). Structure predictions for nanos in X. borealis and X. tropicalis also revealed stem-loop structures around, and just downstream of, the start site (see Fig. S2B-D in the supplementary material).
We tested the functional significance of the large putative stem-loop by deleting these sequences (nt 8-73 in TCEΔ) and monitoring translation after injection into oocytes. Removal of these sequences completely relieved repression, as indicated by western blot analysis with anti-Nanos1 antibody (Fig. 2B). We then asked whether the nanos1 5′UTR plus the first 75 nt (TCE) placed upstream of Myc sequences would be sufficient to repress translation after injection into oocytes. Indeed, these sequences were sufficient to completely block translation (Fig. 2C). Taken together, these results reveal a TCE within the first 75 nt of the nanos1 ORF that is required to repress nanos1 translation and is sufficient to repress a reporter.
The TCE forms a secondary RNA structure
To determine whether the TCE we identified did indeed form a secondary structure as predicted by MFOLD, we performed an enzymatic structural analysis of nanos1 transcripts. RNases A and T1 cut at single-stranded residues 3′ of C and U and 3′ of G, respectively, whereas RNase V1 digests base-paired nucleotides. 32P-labeled nanos1 RNA was digested with one of the three RNases and the resulting products analyzed on sequencing gels (see Fig. S3 in the supplementary material). In general, the results of the enzymatic structural analyses were consistent with the MFOLD program prediction of unpaired RNA regions or loop structures (Fig. 2A, black circles). RNases A and T1 hit most of the C and G single-stranded nucleotides, including those in the loops, and a few of the base-paired nucleotides at the end of the stems of the predicted structure. It is likely that `breathing' at the end of stems exposed those nucleotides to attack by the single-strand-specific RNases. The predicted Stem I around the AUG site was validated in this assay as RNase V1 attacked four nucleotides within this region. Importantly, the large predicted stem in Stem-loop II was also recognized by RNase VI. Therefore, our analysis detected at least two stretches of nucleotides that appeared to form stable stems around the start codon.
To further probe the predicted secondary structures around the start codon, two mutants were made to disrupt the GC-rich stem regions by substituting AU sequences along one side of the predicted stems, as indicated in Fig. 2A (red dots). As shown by Fig. 2D, the AU mutants S1 and S2 were translated after injection into oocytes. The S1 mutant, with AU substitutions only downstream of the AUG site, was not as effective (translation was 27% of ΔTCE levels) in relieving repression as the S2 mutant with its additional substitutions in the 5′UTR (83.5% of ΔTCE levels). We conclude from these studies that the TCE forms a secondary RNA structure and that the conserved 5′UTR sequence plays a role in stabilizing it. We next made a compensatory change (Fig. 2A, CS) to this area, exchanging the left side (8-19 nt) of the predicted stem for the right (61-73 nt), thus preserving the predicted stem, but not its sequence. This mutant did not support translation (Fig. 2E). However, duplication of the sequence (i.e. two right or two left side sequences; Fig. 2A, S3, S4) in the GC-rich stem region did not support translation either (Fig. 2E). Taken together, we interpret these findings to suggest that the left half of the predicted Stem II is base-paired but not necessarily with the sequences predicted by MFOLD.
Translational repression is sensitive to the distance between the AUG and TCE
What role does the TCE play in nanos1 repression? nanos1 repression could act through a repressor that specifically binds the TCE. Alternatively, the structure alone could sterically prevent translational initiation events without the need for a repressor. In other studies defining the function of nanos1 in the germline, we noticed that epitope tags cloned in-frame 5′ to the nanos1 ORF were translated (Fig. 3A). We hypothesized that the 5′ tags might be disrupting the TCE structure and/or preventing a repressor from binding. Alternatively, the addition of extra in-frame sequences in the 5′ position effectively places the TCE at a distance from the start codon, eliminating any steric hindrance to initiation. As a first step, we asked whether the position of the Myc tag affected translation by placing it at the C-terminal end of the ORF. Although there are exceptions, in general a trans-acting repressor would be able to bind to the sequence and repress translation regardless of the location of the TCE. Either placement of the Myc tag was translated equally well in reticulocyte lysates (data not shown). However, only transcripts with the Myc tag 5′ of the nanos1 ORF were translated after injection into oocytes (Fig. 3A), a finding that is most consistent with a structure-based repression independent of a repressor.
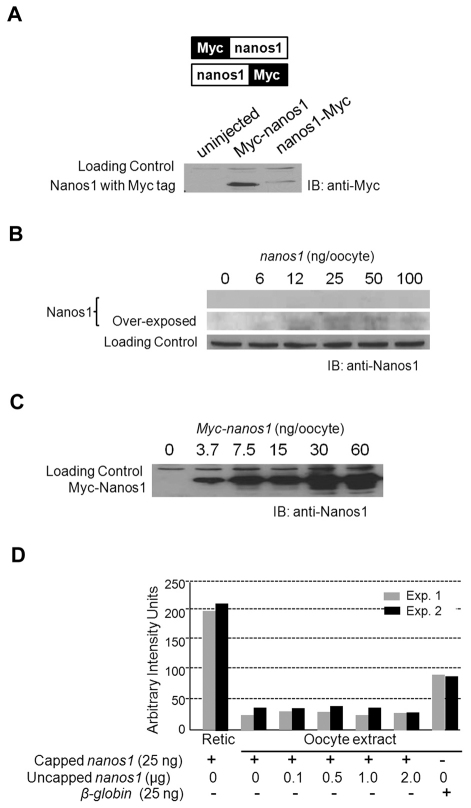
Fig. 3.
Repression of nanos1 in oocytes does not depend on trans-acting factors. (A) Xenopus oocytes were injected with either nanos1-Myc or Myc-nanos1 transcripts and incubated at 18°C overnight. Translation products were detected by blotting (IB) with anti-Myc antibody. (B,C) nanos1 repression is not relieved in vivo by excess levels of nanos1 mRNA. Oocytes were injected with increasing amounts of synthetic capped nanos1 (B) or capped Myc-nanos1 (C) as indicated, incubated at 18°C overnight, and products analyzed by blotting with anti-Nanos1 antibodies. Myc-nanos1 served as a positive control. Note that nanos1 repression was not relieved at any concentration of nanos1 mRNA. (D) Histogram showing the results from two independent in vitro competition experiments. Twenty-five nanograms of capped nanos1 transcripts were translated for 90 minutes at room temperature in oocyte extract containing [35S]methionine. Uncapped nanos1 transcripts were added to the translation mix in increasing amounts as indicated. As positive controls, 25 ng of capped nanos1 and β-globin RNAs were translated in reticulocyte lysates and oocyte extracts, respectively. Even in the presence of an 80-fold excess of uncapped nanos1 RNA, repression was not relieved.
Repression is not relieved by excess nanos1 RNA
To test the trans-acting repressor model for nanos1 repression, we asked whether an excess of nanos1 RNA could relieve repression either through competition or titration of the putative repressor. Two types of experiments were performed, one with capped and one with uncapped nanos1 transcripts in excess. Full-length capped nanos1 transcripts were injected into oocytes at doses ranging from 6 to 100 ng (20,000-fold excess over endogenous levels) and allowed to translate for 18 hours. To check that these excessive amounts were not causing a general inhibition of translation, we performed parallel injections with Myc-nanos1 (Fig. 3C). Although the Myc-Nanos1 protein was easily detected after 3.7 ng of Myc-nanos1 RNA was injected, Nanos1 protein was not detected even at 100 ng of nanos1 and was barely detected even after overexposure of the gel (Fig. 3B,C). These experiments rule out the presence of a soluble repressor as the cause of nanos1 translational repression in oocytes.
In a second approach, we used an in vitro oocyte translation system that mimics nanos1 repression in order to test higher concentrations of nanos1 RNA. Full-length uncapped nanos1 RNA was added in increasing amounts to oocyte extracts that contained a fixed amount of capped nanos1 RNA. Uncapped nanos1 RNA would compete for a putative oocyte repressor but not itself be translated (data not shown). It is highly unlikely that a putative repressor would require the cap for binding because excess capped message did not relieve repression (Fig. 3A,B). Increasing amounts of competitor transcripts, from 100 ng to 2 μg (20,000- to 400,000-fold excess), were added to the oocyte extracts. Even with the degradation of uncapped nanos1 transcripts during the course of the assay, a ∼80,000-fold excess over endogenous nanos1 remained after 2 hours (data not shown). Repression was not relieved at any concentration of competitor tested (Fig. 3D). Thus, translational repression of nanos1 was maintained at all concentrations of the competitor regardless of whether the competitor was capped or uncapped. We conclude from these data that repression is not mediated by a soluble repressor. Our results are consistent with the TCE preventing translation through steric hindrance and point to an activating factor(s) for nanos1 translation.
Insertion of 12 or more nucleotides relieves repression
The positioning of the 40S ribosomal subunit at the start codon requires ∼15 nt downstream of the first nucleotide at the P site (Pestova et al., 2001; Kozak, 1977). We propose that the structure-based TCE, lying just 4 nt downstream of the start codon, interferes with initiation events because there is insufficient space for the 40S ribosomal subunit to bind. Two predictions follow from this model. First, only insertions of 12 nt or longer before the TCE would be sufficient for ribosome loading and thus relief of repression. Subsequent elongation steps are predicted to overcome the negative ΔG of the TCE (Kozak, 2005; Takyar et al., 2005). Second, we would expect the nanos1 transcript to be severely impaired in binding to the 43S pre-initiation complex. To test the first prediction, 3, 6 and 15 nt were inserted in-frame between the start codon and the TCE of nanos1 (Fig. 4A). Transcripts containing the insertions were injected into oocytes and tested for their translational efficiency as before. As predicted by the model, insertions of 3 or 6 nt had no effect, whereas 15 nt relieved repression to levels comparable to that of the TCE deletion mutant (Fig. 4A).
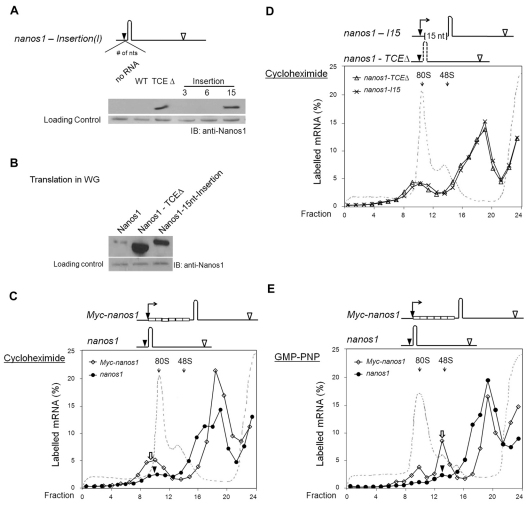
Fig. 4.
Translation of nanos1 RNA is blocked at initiation. (A) Translation of wild-type (WT) nanos1 and insertion mutants in Xenopus oocytes. WT nanos1 and TCEΔare described in Fig. 2B and the insertion mutants are as illustrated. Nucleotides from equivalent positions of β-globin were inserted in between the start codon and the TCE. Translation products from injected oocytes were analyzed by blotting with anti-Nanos1 antibody. (B) Translation of nanos1 and nanos1 mutants in wheat germ extracts. One microgram of synthetic mRNA was translated in wheat germ extracts and the resulting products analyzed as in A. (C-E) Ribosomal loading assay performed with radiolabeled Myc-nanos1, nanos1, nanos1-TCEΔ or nanos1-15nt-insertion (nanos1-I15) transcripts. Transcripts were incubated in wheat germ extract in the presence of cycloheximide (500 μg/ml) (C,D) or GMP-PNP (2 mM) (E) at 18°C for 15 minutes and the reaction mixture fractionated on 10-30% linear sucrose gradients. The labeled mRNA in the fractions is expressed as a percentage of total counts recovered and is plotted against the fraction number. The dashed line denotes the A254 absorption profile. Compare the regions indicated by the white arrows and black arrowheads (see text).
nanos1 translation is blocked at initiation
To test the second prediction of our model, we used a ribosomal loading assay to determine whether the 43S complex would associate with nanos1 mRNA. Initial experiments using oocyte extracts were not informative as the positive control did not accumulate in 80S complexes in the presence of cycloheximide, suggesting that translation was inefficient. However, wheat germ extract was found to mimic the results obtained in oocytes: nanos1 was only translated if the TCE was deleted or nucleotides were inserted before the TCE (Fig. 4B). Therefore, wild-type and mutant 35S-labeled nanos1 transcripts were incubated for 15 minutes in wheat germ extract with cycloheximide, followed by fractionation on a 10-30% linear sucrose gradient. As expected, in the presence of cycloheximide ∼10% (n=3) of the Myc-tagged nanos1 transcripts accumulated in the 80S complex (Fig. 4C, white arrow). By contrast, only 1% (n=3) of the nanos1 transcripts fractionated with the 80S peak (Fig. 4C, arrowhead), confirming that a step in initiation was blocked. Importantly, both the insertion and TCE deletion mutants of nanos1 showed increased association with the 80S complex (8%, n=2) that was comparable to the control Myc-nanos1 transcript (Fig. 4D).
To more precisely define the initiation step at which nanos1 translation was stalled, we tested the ability of the 43S complex to bind radiolabeled nanos1 transcripts in the presence of GMP-PNP, a non-hydrolysable GTP analog. The recruitment of the 60S subunit is dependent on GTP hydrolysis. Including the GMP-PNP in the translation mix prevents 60S subunit recruitment and increases the amount of the 43S complex that is associated with mRNA. Therefore, in the presence of GMP-PNP, mRNAs that are competent to initiate will accumulate within the 48S initiation complex. As expected, a significant proportion (14%, n=3) of the Myc-labeled nanos1 transcripts was associated with the 40S ribosomal subunit (Fig. 4E, white arrow). A small, but significant, amount (1%, n=3) of nanos1 RNA co-sedimented with the 43S peak, comparable to the amount observed in the 80S peak in the presence of cycloheximide (compare Fig. 4C with 4E, arrowhead). These results support the conclusion that nanos1 RNA expression is attenuated at a very early translational initiation step: loading of the 40S ribosomal subunit. Furthermore, as wheat germ extract is unlikely to contain a repressor that specifically recognizes the TCE, these findings also lend support to the conclusion that the TCE secondary structure alone is required for the observed repression.
Translation of injected nanos1 transcripts occurs prior to first cleavage
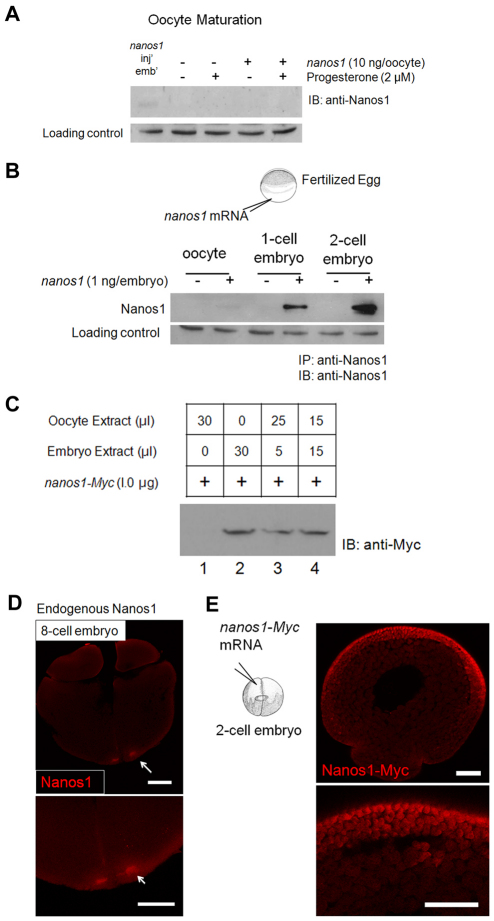
Oocyte maturation and fertilization are pivotal times for the activation of stored maternal mRNAs (Colegrove-Otero et al., 2005b; Richter, 2007; Standart and Minshall, 2008). To determine when nanos1 translation is initiated during development, nanos1 transcripts were injected into oocytes that were subsequently induced to mature with progesterone, or were injected into fertilized eggs. Embryos were collected 75 minutes post-fertilization and at each division cycle up to the 32-cell stage. Nanos1 protein levels were determined by immunoprecipitation and western blotting with anti-Nanos1 antibody. nanos1 RNA remained repressed during maturation events (Fig. 5A), but Nanos1 protein was detected before first cleavage and reached maximum levels by the 2-cell stage (Fig. 5B). These levels persisted through to the 32-cell stage, which was the last stage examined (data not shown).
Fig. 5.
nanos1 repression is relieved only after fertilization. (A) nanos1 remains repressed during oocyte maturation events. Xenopus oocytes were injected with 10 ng of nanos1 transcript per oocyte and incubated at 18°C. To trigger maturation events, oocytes were incubated with progesterone (2 μM) and collected until at least 50% displayed germinal vesicle breakdown. Samples were analyzed by blotting with anti-Nanos1 antibody. nanos1-injected embryos served as a positive control. (B) Injected nanos1 transcripts are efficiently translated in embryos before first cleavage. One nanogram of capped transcript was injected into 1-cell stage embryos soon after fertilization. Embryos were collected at the indicated stages and protein extracts analyzed by blotting after immunoprecipitation (IP) with anti-Nanos1 antibody. (C) nanos1-Myc was translationally active in the presence of embryo extract in a dose-independent fashion. One microgram of capped transcript was translated in vitro with oocyte and/or embryo extract. Samples were analyzed by blotting with anti-Myc antibody. (D) Endogenous Nanos1 was easily detected in the germ plasm by the 8-cell stage. Confocal immunofluorescence with anti-Nanos1 antibody. (E) Confocal immunofluorescence of embryos previously injected with nanos1-Myc RNA shows Nanos1 accumulation in somatic cells. Scale bars: 200 μm.
To gain further insight into the molecular mechanism that might govern nanos1 translational activity, we employed in vitro translation assays using oocyte and embryo extracts (Murray, 1991). Each extract translated β-globin RNA equally well (data not shown). We asked whether the addition of embryo to oocyte extract would render oocyte extract competent for nanos1 translation or whether oocyte extract would repress translation in embryo extracts. As shown in Fig. 5C (lanes 1 and 2), the efficiency of nanos1 translation was much greater in embryo extract than in oocyte extract, as expected from the in vivo assays (Fig. 5B). Strikingly, addition of 5 or 15 μl of embryo extract to oocyte extract supported nanos1 translation to levels that were indistinguishable from using embryo extract alone (Fig. 5C, compare lanes 3 and 4 with lane 2). These results are consistent with an activator being present in stage 2 embryos. Endogenous Nanos1 protein was found to accumulate to detectable levels as early as the 8-cell stage (Fig. 5D). These results are consistent with nanos1 translation being activated shortly after fertilization, but not at maturation, and suggest the presence of a developmentally regulated activator.
Can nanos1 RNA be translated in somatic cells or is its activation restricted to the germline? Immunofluorescence microscopy of nanos1-injected embryos revealed that nanos1 protein accumulated in somatic cells, especially at the animal pole (Fig. 5E), suggesting that either a common activator is present in somatic cells or that more than one activator is competent to activate nanos1 translation.
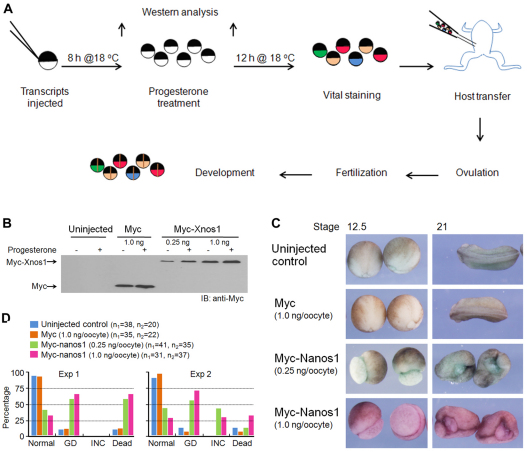
Misexpression of unlocalized nanos1 in oocytes results in abnormal development
Maternal nanos1 is transcribed and localized to the germ plasm during early oogenesis. The germ plasm containing nanos1 RNA is subsequently localized to the vegetal pole and asymmetrically distributed into the future germ cells (Mosquera et al., 1993). However, Nanos1 protein was not detected in germ plasm until early cleavage (the 8-cell stage), some 4 months after its RNA is localized (Fig. 5D) (MacArthur et al., 1999), consistent with nanos1 being under tight negative control. Our data suggest that unlocalized nanos1 RNA is not translated outside the context of germ plasm, pointing to a repressive mechanism in addition to physical sequestration in the germinal granules. Is this repressive mechanism essential for normal development or is premature Nanos1 expression from unlocalized mRNA tolerated in the oocyte? To address this, we forced Nanos1 expression in stage VI oocytes by placing Myc tags upstream of the nanos1 ORF (Myc-nanos1) and then assessed any developmental consequences by host transfer (Fig. 6A,B). Oocytes injected with Myc tags alone developed normally (Fig. 6C,D). The Myc-nanos1-injected embryos were indistinguishable from the uninjected controls until stage 11, at which time they displayed large blastopores that failed to close (data not shown). These embryos remained round, with their endodermal mass exposed. Those that survived gastrulation went on to display severely incomplete neural tube closure at tailbud stages (Fig. 6C,D). From these results, we conclude that Nanos1 expressed outside the germ plasm is tolerated by the oocyte (maturation is normal), but not by the somatic cells that inherit the ectopically expressed Nanos1 during development. These results underscore the importance of TCE structure-based repression to ensure that unlocalized maternal nanos1 RNA is not translated.
Fig. 6.
Misexpression of nanos1 in oocytes results in abnormal development. (A) Schematic of host transfer experiment. (B) Following the procedure in A, oocytes were injected with either Myc-nanos1 RNA or control Myc RNA and analyzed by blotting with anti-Myc antibody. RNAs were translated in a dose-dependent fashion. (C) Xenopus embryos from experiment 2 (see D) showing representative phenotypes that result from nanos1 expression in oocytes. Uninjected oocytes or Myc-injected oocytes served as controls and were normal. Embryos observed at stage 12.5 fail to close their blastopores. Surviving embryos display incomplete neural tube closure at stage 21. (D) The distribution of phenotypes from two independent experiments. Embryos with a gastrulation defect (GD) were counted at stage 12.5 and those with incomplete neural tube closure (INC) or that were dead were counted at stage 21. The key includes the total number of embryos in each category for each experiment.
DISCUSSION
We have identified a novel structure-based mechanism for the translational repression of germline nanos1 RNA that ensures its inactivity during oogenesis, a feature essential for normal development. We propose that ribosome scanning is sterically prevented by a structural element of the RNA without the involvement of a repressor. Several lines of evidence support this model. First, we have shown that a 73 nt TCE immediately downstream of the AUG site is both required to repress translational initiation of nanos1 RNA and sufficient to repress a reporter. Second, enzymatic probing and nucleotide substitutions revealed regions around the start site that were resistant to single-strand-specific RNase attack. Third, repression was relieved and initiation complexes were formed only after the insertion of sufficient nucleotides (15, but not 3 or 6 nt) between the start site and the TCE to permit ribosome scanning, a critical test of our model. Fourth, both in oocytes and in oocyte extracts, nanos1 repression persisted even in the presence of a vast excess of nanos1 RNA, but was relieved by the addition of a small amount of embryo extract, ruling out the involvement of a soluble repressor. After fertilization, endogenous nanos1 is translated, strongly suggesting the presence of a developmentally regulated activator.
In eukaryotes, the known mechanisms for mRNA-specific translational repression require trans-acting factors that interact with cis-regulatory regions (TCEs), which are most commonly found in the 3′UTR (reviewed by Livingstone et al., 2010). TCEs for which structure is a critical feature are not common, but have been described for nanos repression in Drosophila and C. elegans (D'Agostino et al., 2006; Gavis et al., 1996; Kalifa et al., 2006). Perhaps the best-known structural element controlling translation in eukaryotes is the iron-response element (IRE), a stem-loop in the 5′UTR of the ferritin gene and other genes that regulate iron homeostasis (Theil and Eisenstein, 2000). The IRE appears to be unique within eukaryotes as it does not inhibit initiation by interfering with the formation of the initiation complex, but rather is recognized by the iron-response protein, and together they sterically inhibit scanning of the 43S initiation complex.
Our model (Fig. 7) depends on the predicted structures in the TCE being sufficiently stable to prevent the initiation complex from forming. Stem-loops with a free energy of at least –50 kcal/mol inhibit ribosomal scanning in COS cells (Kozak, 1986). Stem-loops I and II in Xenopus laevis are predicted to have a free energy of –17 and –56 kcal/mol, respectively. Thus, based on thermodynamic considerations alone, the RNA secondary structures surrounding the AUG, including the 15 nt 5′UTR, are sufficient to prevent ribosome scanning and initiation events. However, disrupting the predicted Stem II by substituting a repeat of one side did not relieve repression, suggesting that this GC-rich region can form alternative structures of substantial stability (Fig. 2E). Consistent with this interpretation, disruption of the base-pairing by AU substitutions did `loosen' the TCE as revealed by increased translational efficiency. The predicted structure might simply not be accurate. Our data also suggest a role of the evolutionarily conserved nanos1 5′UTR in contributing to the repressive ability of the TCE, perhaps by further stabilizing the TCE. Low levels of nanos1 translation could be detected after RNA injection into oocytes. We cannot rule out the possibility that additional repressor-based mechanism(s) are operating, such as the physical sequestration of nanos within germinal granules. Although novel in eukaryotes, structural inhibition of translation has been demonstrated repeatedly in the regulation of prokaryotic mRNAs (Chowdhury et al., 2006; Serganov, 2009; Kozak, 2005).
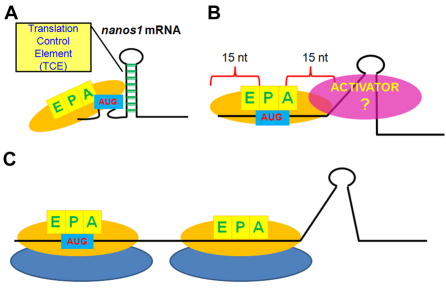
Fig. 7.
Model for nanos1 translational repression. (A) The secondary structure formed by the first 73 nt in the nanos1 ORF sterically prevents efficient loading of mRNA on the 43S pre-initiation complex, blocking translation in oocytes and in wheat germ extracts. (B,C) A developmentally regulated activator becomes available soon after fertilization, disrupts the secondary structure (B) and allows ribosome loading and efficient translation (C). Note that disruption of the secondary structure by deletion, insertion or substitution or by putative endogenous helicases allows loading of the P site and formation of the 48S initiation complex. 40S (yellow) and 60S (blue) ribosomal subunits. EPA, sites in 40S subunit required for initiation.
Toe-printing analysis has established that the binding cleft of the 40S subunit requires 15 nt downstream of the first nucleotide at the P site (Pestova et al., 2001). The proposed TCE within the nanos1 ORF is just 4 nt from the start codon. An insertion of 3 or 6 nt, which would result in only 7 or 10 nt between the start codon and the TCE, would not allow for 40S subunit positioning and efficient translation, as predicted from our model. However, an insertion of 15 nt would allow ribosome binding and efficient translation. An alternative, but less likely, explanation is that the addition of more nucleotides changed the structure so profoundly as to allow translation. We expect that such binding and subsequent translocation of the mRNA through the P site generates sufficient force to disrupt the TCE (Takyar et al., 2005).
The steric hindrance model predicts that an activator unwinds the TCE and permits an initiation complex to form (Fig. 7). Interestingly, nanos1 RNA injected into embryos was translated and its protein accumulated in somatic cells, indicating that the putative activator might not be restricted to the germline. We have ruled out three known germline helicases – DeadSouth, Centroid and Vasa – as well as eIF4F (Jaramillo et al., 1990) and the unwinding RNA-editing enzyme originally discovered in Xenopus embryos (Bass and Weintraub, 1988). Although we know of no examples, it is possible that the activator is an RNA that disrupts the TCE secondary structure. Fertilization initiates complex signaling events including calcium influx, activation of a kinase cascade and the translation of specific mRNAs (Richter, 2007). A possible scenario is that fertilization triggers the activity of a reserved cytoplasmic factor or stored mRNA within the germ plasm, which in turn activates nanos1 translation. The identity of such a putative activator is currently under intensive investigation.
Nanos1 synthesis occurs prior to the complete segregation of the germline during gastrulation, yet its presence in the somatic lineages is not tolerated and results in abnormal development. How then is Nanos1 activity confined to the germline? The germ plasm matrix could function as a scaffolding platform that anchors the germ cell determinants, including nanos1 RNA and Nanos1 protein. We have not observed nanos1 RNA or protein outside of PGCs or, for that matter, germ plasm (Lai et al., 2011) (Fig. 5D). Alternatively, any nanos1 RNA outside the germ plasm would have to be efficiently degraded, a mechanism described for nanos in Drosophila and zebrafish (Bashirullah et al., 1999; Mishima et al., 2006). Both Drosophila and zebrafish have inefficient modes of nanos RNA localization, necessitating a post-transcriptional control mechanism (Gavis et al., 2008; Giraldez et al., 2006). In zebrafish, maternal mRNAs are degraded by zygotic miR-430 during early embryogenesis. Xenopus miR-427 has been shown to resemble the orthologous zebrafish miR-430 in loss-of-function analyses and in its expression pattern and target specificity (Giraldez et al., 2006; Lund et al., 2009). However, we found ectopically expressed nanos1 RNA and protein to be stable within somatic cells (Fig. 5E; data not shown).
Interestingly, ectopic expression of Nanos1 protein in oocytes did not affect oocyte maturation or early cleavage stages. However, it did cause severe gastrulation defects and, at lower levels of lethality, incomplete neural tube closure. These are likely to be gain-of-function phenotypes. Nanos1 functions as a translational repressor of nanos-response element (NRE)-containing mRNAs such as VegT (Wharton et al., 1998; Nakahata et al., 2001) (our unpublished observations). We speculate that Nanos1 expressed in somatic cells represses regulatory pathways that are important for development, resulting in the observed embryonic abnormalities. Our results are consistent with a requirement for maternal nanos1 to be translationally repressed everywhere outside of the germ plasm. Identification of mRNAs targeted by Nanos1 for repression will be important in understanding the gain-of-function phenotype.
The model presented here is appealing because it offers a simple explanation for how somatic cells prevent translation of any unlocalized nanos1 mRNA. Nanos family members are potent repressors of somatic cell fates, functioning as repressors of translation and perhaps transcription as well (Curtis et al., 1997; Deshpande et al., 2005; Kadyrova et al., 2007; Lai et al., 2011). The need to restrict Nanos translation to the germline is crucial, with misexpression resulting in embryonic lethality. Structural inhibition of translation would provide a robust means of repression as it would be intrinsic to the transcript and independent of the proper localization of the message or the proper expression/stability of a repressor. Reliance on the stability of a repressor over long periods of time, as occurs during oogenesis, might present too high a biological risk. We predict that other mRNAs that encode regulatory proteins and are stored over extended periods of time would be excellent candidates for using a structure-based mechanism of translational repression. It will be of great interest to identify such mRNAs that contain complex RNA structures in close proximity to the start codon.
Supplementary Material
Acknowledgments
We thank Drs Lasko, Lipshitz, Gavis, Steitz, Rotundo and Malhotra for encouragement and advice and especially Drs Deutscher and Houston for their generosity in sharing reagents and methods. This work was supported by NIH grant GM33932 to M.L.K. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.056705/-/DC1
References
- Bashirullah A., Halsell S. R., Cooperstock R. L., Kloc M., Karaiskakis A., Fisher W. W., Fu W., Hamilton J. K., Etkin L. D., Lipshitz H. D. (1999). Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 18, 2610-2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. (1988). An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089-1098 [DOI] [PubMed] [Google Scholar]
- Brown D. D. (2004). A tribute to the Xenopus laevis oocyte and egg. J. Biol. Chem. 279, 45291-45299 [DOI] [PubMed] [Google Scholar]
- Chowdhury S., Maris C., Allain F. H., Narberhaus F. (2006). Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25, 2487-2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero L. J., Devaux A., Standart N. (2005a). The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol. Cell. Biol. 25, 9028-9039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero L. J., Minshall N., Standart N. (2005b). RNA-binding proteins in early development. Crit. Rev. Biochem. Mol. Biol. 40, 21-73 [DOI] [PubMed] [Google Scholar]
- Curtis D., Treiber D. K., Tao F., Zamore P. D., Williamson J. R., Lehmann R. (1997). A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 16, 834-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino I., Merritt C., Chen P. L., Seydoux G., Subramaniam K. (2006). Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 292, 244-252 [DOI] [PubMed] [Google Scholar]
- Darfeuille F., Unoson C., Vogel J., Wagner E. G. (2007). An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell 26, 381-392 [DOI] [PubMed] [Google Scholar]
- Deshpande G., Calhoun G., Jinks T. M., Polydorides A. D., Schedl P. (2005). Nanos downregulates transcription and modulates CTD phosphorylation in the soma of early Drosophila embryos. Mech. Dev. 122, 645-657 [DOI] [PubMed] [Google Scholar]
- Forrest K. M., Gavis E. R. (2003). Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13, 1159-1168 [DOI] [PubMed] [Google Scholar]
- Forrest K. M., Clark I. E., Jain R. A., Gavis E. R. (2004). Temporal complexity within a translational control element in the nanos mRNA. Development 131, 5849-5857 [DOI] [PubMed] [Google Scholar]
- Forristall C., Pondel M., Chen L., King M. L. (1995). Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development 121, 201-208 [DOI] [PubMed] [Google Scholar]
- Gavis E. R., Lehmann R. (1992). Localization of nanos RNA controls embryonic polarity. Cell 71, 301-313 [DOI] [PubMed] [Google Scholar]
- Gavis E. R., Lehmann R. (1994). Translational regulation of nanos by RNA localization. Nature 369, 315-318 [DOI] [PubMed] [Google Scholar]
- Gavis E. R., Lunsford L., Bergsten S. E., Lehmann R. (1996). A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122, 2791-2800 [DOI] [PubMed] [Google Scholar]
- Gavis E. R., Chatterjee S., Ford N. R., Wolff L. J. (2008). Dispensability of nanos mRNA localization for abdominal patterning but not for germ cell development. Mech. Dev. 125, 81-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., Enright A. J., Schier A. F. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75-79 [DOI] [PubMed] [Google Scholar]
- Graf J. D. (1996). Molecular approaches to the phylogeny of Xenopus. In The Biology of Xenopus (ed. Tinsley R. C., Kobel H. R.), pp. 379-389 Oxford: Clarendon Press; [Google Scholar]
- Jadhav S., Rana M., Subramaniam K. (2008). Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803-1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M., Browning K., Dever T., Blum S., Trachsel H., Merrick W., Ravel J., Sonenberg N. (1990). Translation initiation factors that function as RNA helicases from mammals, plants and yeast. Biochim. Biophys. Acta 1050, 134-139 [DOI] [PubMed] [Google Scholar]
- Johnstone O., Lasko P. (2001). Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35, 365-406 [DOI] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H., Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519-1527 [DOI] [PubMed] [Google Scholar]
- Kalifa Y., Huang T., Rosen L. N., Chatterjee S., Gavis E. R. (2006). Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev. Cell 10, 291-301 [DOI] [PubMed] [Google Scholar]
- King M. L., Messitt T. J., Mowry K. L. (2005). Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 97, 19-33 [DOI] [PubMed] [Google Scholar]
- Kloc M., Bilinski S., Chan A. P., Etkin L. D. (2001). Mitochondrial ribosomal RNA in the germinal granules in Xenopus embryos revisited. Differentiation 67, 80-83 [DOI] [PubMed] [Google Scholar]
- Kloc M., Dougherty M. T., Bilinski S., Chan A. P., Brey E., King M. L., Patrick C. W., Jr, Etkin L. D. (2002). Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Dev. Biol. 241, 79-93 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Yamada M., Asaoka M., Kitamura T. (1996). Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380, 708-711 [DOI] [PubMed] [Google Scholar]
- Köprunner M., Thisse C., Thisse B., Raz E. (2001). A zebrafish nanos-related gene is essential for development of primordial germ cells. Genes Dev. 15, 2877-2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1977). Nucleotide sequences of 5′-terminal ribosome-protected initiation regions from two reovirus messages. Nature 269, 391-394 [DOI] [PubMed] [Google Scholar]
- Kozak M. (1986). Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83, 2850-2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2005). Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13-37 [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. (1984). Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 12, 7057-7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F., Zhou Y., Luo X., Fox J., King M. L. (2011). Nanos1 functions as a translational repressor in the Xenopus germline. Mech. Dev. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone M., Atas E., Meller A., Sonenberg N. (2010). Mechanisms governing the control of mRNA translation. Phys. Biol. 7, 021001 [DOI] [PubMed] [Google Scholar]
- Lund E., Liu M., Hartley R. S., Sheets M. D., Dahlberg J. E. (2009). Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15, 2351-2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur H., Bubunenko M., Houston D. W., King M. L. (1999). Xcat2 RNA is a translationally sequestered germ plasm component in Xenopus. Mech. Dev. 84, 75-88 [DOI] [PubMed] [Google Scholar]
- Mir A., Heasman J. (2008). How the mother can help: studying maternal Wnt signaling by anti-sense-mediated depletion of maternal mRNAs and the host transfer technique. Methods Mol. Biol. 469, 417-429 [DOI] [PubMed] [Google Scholar]
- Mishima Y., Giraldez A. J., Takeda Y., Fujiwara T., Sakamoto H., Schier A. F., Inoue K. (2006). Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 16, 2135-2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera L., Forristall C., Zhou Y., King M. L. (1993). A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 117, 377-386 [DOI] [PubMed] [Google Scholar]
- Murray A. W. (1991). Cell cycle extracts. Methods Cell Biol. 36, 581-605 [PubMed] [Google Scholar]
- Nakahata S., Katsu Y., Mita K., Inoue K., Nagahama Y., Yamashita M. (2001). Biochemical identification of Xenopus pumilio as a sequence specific cyclin B1 mRNA-binding protein that physically interacts with a nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element binding protein. J. Biol. Chem. 276, 20945-20953 [DOI] [PubMed] [Google Scholar]
- Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., Hellen C. U. (2001). Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98, 7029-7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D. (2007). CPEB: a life in translation. Trends Biochem. Sci. 6, 279-285 [DOI] [PubMed] [Google Scholar]
- Richter J. D., Sonenberg N. (2005). Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477-480 [DOI] [PubMed] [Google Scholar]
- Schier A. F. (2007). The maternal-zygotic transition: death and birth of RNAs. Science 316,406-407 [DOI] [PubMed] [Google Scholar]
- Serganov A. (2009). The long and the short of riboswitches. Curr. Opin. Struct. Biol. 19, 251-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Xie T. (2010). NANOS: a germline stem cell's Guardian Angel. J. Mol. Cell. Biol. 2, 76-77 [DOI] [PubMed] [Google Scholar]
- Sive H., Grainger R. M., Harland R. M. (2000). Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Souopgui J., Rust B., Vanhomwegen J., Heasman J., Henningfeld K. A., Bellefroid E., Pieler T. (2008). The RNA-binding protein XSeb4R: a positive regulator of VegT mRNA stability and translation that is required for germ layer formation in Xenopus. Genes Dev. 22, 2347-2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standart N., Minshall N. (2008). Translational control in early development: CPEB, P-bodies and germinal granules. Biochem. Soc. Trans. 36, 671-676 [DOI] [PubMed] [Google Scholar]
- Stennard F., Zorn A. M., Ryan K., Garrett N., Gurdon J. B. (1999). Differential expression of VegT and Antipodean protein isoforms in Xenopus. Mech. Dev. 86, 87-98 [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G. (1999). nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861-4871 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Igarashi K., Aisaki K., Kanno J., Saga Y. (2010). NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc. Natl. Acad. Sci. USA 107, 3594-3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S., Hickerson R. P., Noller H. F. (2005). mRNA helicase activity of the ribosome. Cell 120, 49-58 [DOI] [PubMed] [Google Scholar]
- Theil E. C., Eisenstein R. S. (2000). Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs). J. Biol. Chem. 275, 40659-40662 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., Saga Y. (2003). Conserved role of nanos proteins in germ cell development. Science 301, 1239-1241 [DOI] [PubMed] [Google Scholar]
- Venkatarama T., Lai F., Luo X., Zhou Y., Newman K., King M. L. (2010). Repression of zygotic gene expression in the Xenopus germline. Development 137, 651-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lin H. (2004). Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303, 2016-2019 [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Sonoda J., Lee T., Patterson M., Murata Y. (1998). The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1, 863-872 [DOI] [PubMed] [Google Scholar]
- Zhang J., Houston D. W., King M. L., Payne C., Wylie C., Heasman J. (1998). The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell 94, 515-524 [DOI] [PubMed] [Google Scholar]
- Zhou Y., King M. L. (1996). Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development 122, 2947-2953 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406-3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.