Abstract
Here we show that presenilin-1 (PS1), a protein involved in Alzheimer's disease, binds directly to epithelial cadherin (E-cadherin). This binding is mediated by the large cytoplasmic loop of PS1 and requires the membrane-proximal cytoplasmic sequence 604–615 of mature E-cadherin. This sequence is also required for E-cadherin binding of protein p120, a known regulator of cadherin-mediated cell adhesion. Using wild-type and PS1 knockout cells, we found that increasing PS1 levels suppresses p120/E-cadherin binding, and increasing p120 levels suppresses PS1/E-cadherin binding. Thus PS1 and p120 bind to and mutually compete for cellular E-cadherin. Furthermore, PS1 stimulates E-cadherin binding to β- and γ-catenin, promotes cytoskeletal association of the cadherin/catenin complexes, and increases Ca2+-dependent cell–cell aggregation. Remarkably, PS1 familial Alzheimer disease mutant ΔE9 increased neither the levels of cadherin/catenin complexes nor cell aggregation, suggesting that this familial Alzheimer disease mutation interferes with cadherin-based cell–cell adhesion. These data identify PS1 as an E-cadherin-binding protein and a regulator of E-cadherin function in vivo.
Presenilin-1 (PS1) mutations are responsible for most cases of early-onset familial Alzheimer's disease (FAD). PS1 is a transmembrane protein expressed in many tissues, including brain, where it is enriched in neurons (1, 2). PS1 crosses the membrane eight times, with the N terminus, the C terminus, and the large hydrophilic loop all located in the cytoplasm. Most cellular PS1 is cleaved within the large cytoplasmic loop to yield N-terminal fragments (PS1/NTF) of approximately 30 kDa and C-terminal fragments (PS1/CTF) of approximately 20 kDa. After cleavage, the PS1 fragments form stable 1:1 heterodimers (3, 4). PS1 facilitates processing of Notch-1 receptor, and amyloid precursor protein (APP) (5, 6) stimulates production of Aβ peptide (6) and may play a role in neuroprotection (7, 8). Mice lacking PS1 (PS1−/−) die shortly after birth with skeletal malformations, impaired neurogenesis, and brain hemorrhage (7).
Recently, we reported that PS1 concentrates at synaptic and epithelial cell–cell contact sites, where it forms complexes with the cadherin/catenin adhesion system (9). Classic cadherins, including E-cadherin and neuronal cadherin (N-cadherin), are a family of type I transmembrane proteins that mediate Ca2+-dependent cell–cell adhesion and recognition, and control critical events in neurogenesis, tissue development, and tissue homeostasis (10, 11). These functions are mediated by homophilic interactions of the extracellular region of cadherins. The membrane-distal cytoplasmic sequence of cadherins binds either β-catenin or γ-catenin (plakoglobin), which in turn binds α-catenin. The latter protein binds polymerized actin, thus linking the cadherin/catenin adhesion complex to the cortical cytoskeleton. Cytoskeletal linkage of the cadherin/catenin complexes is crucial for the full expression of the adhesive functions of surface cadherins (12–14). β-Catenin and γ-catenin, two highly homologous members of the armadillo family of proteins (for a review see ref. 15), bind the same sequence of cytoplasmic cadherin in a mutually exclusive manner (16, 17). The juxtamembrane (membrane-proximal) region of cytoplasmic cadherins binds p120 (also called p120ctn), a cytosolic protein originally identified as a target for p60v-src kinase and later shown to regulate cadherin-mediated cell–cell adhesion and tumor metastasis (for a review see ref. 18).
Although PS1 binds to the cadherin/catenin adhesion complex (9), the mechanism and functional consequences of this association remain obscure. Here we show that PS1 binds directly to juxtamembrane cytoplasmic cadherin and inhibits cadherin binding of p120. Furthermore, PS1 stabilizes the cadherin/catenin complex, promotes its cytoskeletal association, and stimulates Ca2+-dependent cell–cell aggregation. In contrast, PS1 FAD mutant ΔE9 failed to stabilize the cadherin/catenin complex and did not stimulate cell–cell aggregation.
Materials and Methods
Antibodies.
Rabbit polyclonal antibody R222 specific for PS1/NTF amino acids 2–12 and mouse monoclonal antibody 33B10 specific for PS1/CTF sequence 331–350 were prepared as described (9). Anti-transferrin receptor antibody was from Zymed, and anti-epithelial cadherin (E-cadherin) antibody H108 (rabbit polyclonal) was from Santa Cruz Biotechnology. Other antibodies against E-cadherin, β-catenin, γ-catenin, α-catenin, and p120 were from Transduction Laboratories (Lexington, KY). Unless otherwise stated, “anti-E-cadherin antibody” refers to anti-cytoplasmic E-cadherin antibody.
Cell Cultures, Cell Aggregation, Immunoprecipitations (IPs), and Immunoblotting.
Unless otherwise stated, cell cultures were grown in DMEM plus 10% FBS, penicillin, and streptomycin in 5% CO2 at 37°C. Fibroblast cell lines were from wild-type (WT) (PS1 +/+) and PS1 knockout (PS1−/−) mice (9), and L cells were from the American Type Culture Collection. Stable transfections of E-cadherin cDNAs cloned into pcDNA3 vector (Invitrogen) were performed as described (2, 9). Transient transfections were performed with Lipofectamine Plus, and cells were processed for IP or cell aggregation 36 h later. Aggregation assays were performed as described (9, 12) in the presence or absence of 2 mM Ca2+. The degree of aggregation and size of aggregates was documented by photography. IPs were performed as described (9) and analyzed on Western blots (WBs). Mouse embryos at day 17 were rinsed in ice-cold PBS and frozen in a dry ice/methanol bath. Genotypes were determined by PCR amplification of tail DNA (7), and frozen PS1+/+ or PS1−/− embryos were homogenized in ice-cold TNE buffer [25 mM Tris⋅HCl (pH 7.6)/150 mM NaCl/1× complete protease inhibitor mixture/1 mM EDTA] plus 1% Triton X-100. Homogenates were centrifuged at 16,000 × g, and the supernatants were immunoprecipitated and immunoblotted. All experiments were repeated at least three times, except the crosslinking, which was repeated two times.
Crosslinking.
Confluent cells were rinsed twice with PBS minus Ca2+/Mg2+ and then incubated in the same buffer plus 200 μg/ml of dithiobis[succinimidylpropionate] (Pierce) for 20 min at room temperature. After that, cells were rinsed as above, incubated 5 min in quenching solution (50 mM glycine in PBS, pH 7.4), and solubilized in RIPA buffer [25 mM Hepes (pH 7.4)/1% Nonidet P-40/0.1% SDS/0.5% sodium deoxycholate/complete protease inhibitor mixture (Roche Molecular Biochemicals)].
Preparation of Triton-Insoluble, Urea-Soluble Fractions.
Confluent PS1 +/+ or PS1−/− fibroblast cell cultures in 60-mm plates were extracted with 300 μl of TNE buffer plus 1% Triton X-100 and centrifuged at 16,000 × g for 30 min, and the Triton X-100-insoluble fraction was solubilized in 100 μl SDS sample buffer plus urea [70 mM Tris⋅HCl (pH 6.8)/8 M urea/2.5% SDS/0.1 M DTT/10% glycerol]. Triton-soluble and insoluble fractions were analyzed on WBs.
Results
PS1/E-cadherin Binding Is Independent of Catenins.
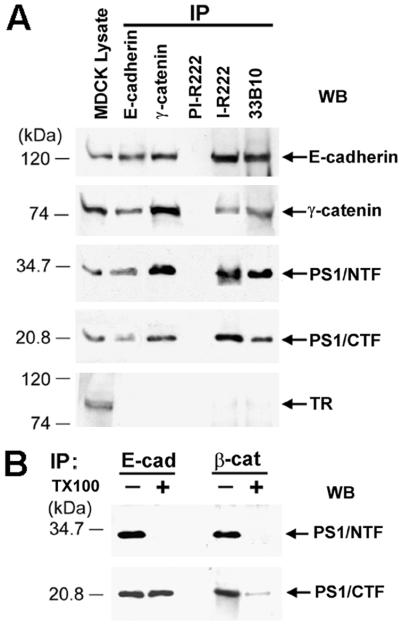
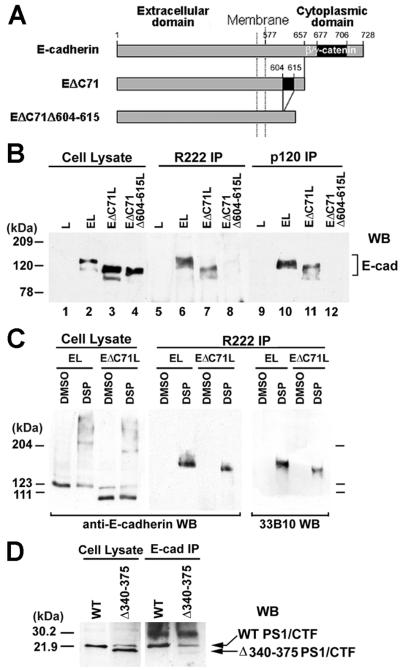
PS1 is a component of the E-cadherin/β-catenin adhesion complex (9). Examination of E-cadherin, γ-catenin, and PS1 IPs prepared from confluent Madin–Darby canine kidney (MDCK) cultures (9) revealed that PS1 is also a component of the E-cadherin/γ-catenin complex (Fig. 1A). Because PS1 binds β-catenin (19, 20), we attempted to determine whether PS1 binds E-cadherin independent of catenins. Triton X-100 extracted most of PS1/CTF from a β-catenin IP but failed to extract any PS1/CTF from an E-cadherin IP, indicating that the PS1/E-cadherin binding might be distinct from the PS1/β-catenin binding (Fig. 1B, Lower). To further test this hypothesis, we prepared PS1 IPs from L cells transfected with either full-length E-cadherin (EL cells) or with E-cadherin deletion construct EΔC71 that lacks the last (membrane-distal) 71 cytoplasmic amino acids that bind β- and γ-catenin (EΔC71L cells; see Fig. 2A). Untransfected L cells contain no detectable E-cadherin (21), and PS1 IPs from these cells did not react with anti-E-cadherin antibodies (Fig. 2B, lanes 1 and 5). In E-cadherin-transfected cells, however, PS1 formed complexes with both full-length and EΔC71 cadherins (Fig. 2B, lanes 2 and 3 and 6 and 7). Because construct EΔC71 lacks the catenin-binding sequence and binds neither β- nor γ-catenin (data not shown; see also refs. 14 and 22), our data show that PS1 associates with E-cadherin even when catenins are not bound to E-cadherin. Chemical crosslinking is used to analyze direct protein–protein contacts within protein complexes, including the cadherin/catenin complex (22, 23). PS1 IPs prepared from crosslinked EL or EΔC71L cells contained E-cadherin-reactive, SDS-resistant complexes of approximate apparent molecular mass 170 kDa and 160 kDa, respectively (Fig. 2C). The apparent molecular mass of the crosslinked products is in excellent agreement with the predicted molecular mass of a ternary complex containing one molecule each of PS1/CTF, PS1/NTF, and full length or truncated E-cadherin, respectively. These complexes were immunoprecipitated with anti-PS1/NTF antibody R222 and reacted with anti-PS1/CTF antibody 33B10, verifying that they contain both PS1 fragments (Fig. 2C). These data show that PS1 binds directly to E-cadherin at a sequence distinct from the catenin-binding site and suggest that the two proteins combine in a 1:1 ratio.
Figure 1.
(A) PS1 forms complexes with E-cadherin and γ-catenin. Extract of confluent MDCK cells in TNE buffer plus 1% digitonin (9) was treated with the antibodies indicated at the top of the figure, and the resulting IPs were probed on WBs with antibodies against the proteins indicated on the right. For reference, 20 μg of MDCK cell lysate was also probed. TR, transferrin receptor; PI-R222, rabbit 222 preimmune serum; I-R222, rabbit 222 antiserum; 33B10, anti-PS1/CTF monoclonal antibody. (B) Triton X-100 effects on PS1/β-catenin and PS1/E-cadherin complexes. Extracts of confluent MDCK cells in 1% digitonin were immunoprecipitated with antibodies against E-cadherin (E-cad) or β-catenin (β-cat), and the resulting IPs were washed in the presence (+) or absence (−) of 1% Triton X-100 (TX100). The remaining immunocomplexes were collected by centrifugation and probed on WBs with antibody R222 (PS1/NTF) or 33B10 (PS1/CTF).
Figure 2.
PS1 binds E-cadherin independently of β- or γ-catenin. Cytoplasmic residues 604–615 are required for PS1/E-cadherin binding. (A) Schematic representation of mouse E-cadherin and E-cadherin deletion constructs EΔC71 and EΔC71Δ604–615, which is a derivative of EΔC71 and lacks cytoplasmic residues 604–615 (see text). Constructs were prepared as previously described (25). (B) PS1 binding to E-cadherin constructs. Extracts (1% digitonin) from L cells (L, lanes 1, 5, and 9) or L cells transfected with mouse E-cadherin (EL, lanes 2, 6, and 10), construct EΔC71 (EΔC71L, lanes 3, 7, and 11), or construct EΔC71Δ604–615 (EΔC71Δ604–615L, lanes 4, 8, and 12) were treated with antibody R222 (lanes 5–8) or anti-p120 antibody (lanes 9–12), and the resulting IPs were probed on WBs with anti-E-cadherin antibody H108. For reference, 20 μg of cell lysates was probed (lanes 1–4). (C) Confluent EL or EΔC71L cells were incubated either with the crosslinking agent dithiobis (succinimidylpropionate) (DSP) in DMSO or with DMSO alone, and cell extracts were then prepared in RIPA buffer in the presence of SDS to inhibit noncovalent associations. Lysates were treated with antibody R222 (R222 IP), and the resulting IPs were probed on WBs with anti-E-cadherin (H108) or anti-PS1/CTF (33B10) antibodies. Twenty micrograms of cell lysate was probed with H108 antibody. (D) PS1 construct Δ340–375 does not bind E-cadherin. HEK293 cells expressing WT PS1 (WT) or a PS1 construct with amino acid deletion 340–375 (Δ340–375) were lysed in TNE plus 1% digitonin. Lysates were treated with anti-E-cadherin antibody (E-cad IP), and the resulting IPs were probed on WBs with antibody 33B10. Twenty micrograms of cell lysates was also probed.
PS1/CTF Mediates PS1 Binding to Juxtamembrane Cytoplasmic E-cadherin.
Triton X-100 treatment of a digitonin E-cadherin IP extracted the PS1/NTF but not the PS1/CTF, which remained associated with E-cadherin (Fig. 1B). This result suggests that PS1 binding to E-cadherin may be mediated by peptide PS1/CTF and is in agreement with recent data that the binding between PS1/NTF and PS1/CTF is sensitive to Triton X-100 extraction (20). Together with the chemical crosslinking data (see above), these results suggest that PS1/CTF, which contains PS1 sequence 299–467 (3), mediates the PS1/E-cadherin binding. In vitro blot overlay assays showed that PS1 sequence 256–410, which includes the entire large cytoplasmic loop, binds to the cytoplasmic sequence of E-cadherin (data not shown). Fig. 2D shows that, whereas full-length PS1/CTF bound E-cadherin in vivo, a PS1/CTF construct lacking sequence 340–375 did not. In contrast, PS1/CTF construct lacking sequence 299–330 bound E-cadherin (data not shown). Combined, these results show that PS1 sequence 340–375 of the large cytoplasmic loop is required for PS1 binding to cytoplasmic E-cadherin.
The observation that PS1 binds E-cadherin lacking the last 71 cytoplasmic residues, combined with in vitro experiments showing that the PS1 sequence binds cytoplasmic E-cadherin, suggested that PS1 binds within the remaining membrane proximal 80 residues of cytoplasmic E-cadherin (Fig. 2A). Because these residues also mediate E-cadherin binding of protein p120, a regulator of cadherin-mediated cell adhesion (21, 24), we asked whether there is an overlap between the E-cadherin sequences that mediate binding of both proteins. Indeed, deletion of E-cadherin cytoplasmic sequence 604–615, which is required for p120 binding to E-cadherin (Fig. 2B, lanes 4 and 12; and ref. 21), also abolished E-cadherin-PS1 binding (Fig. 2B, lanes 4 and 8), showing that this sequence is critical for E-cadherin binding of both PS1 and p120.
PS1 Competes with p120 for E-cadherin Binding in Vivo.
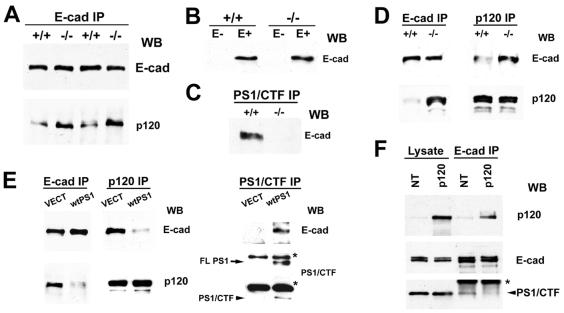
That the same E-cadherin sequence is required for binding of both PS1 and p120 suggests that these proteins might compete for E-cadherin binding. Indeed, E-cadherin IPs prepared from PS1−/− mouse embryos contained significantly more p120 than E-cadherin IPs from PS1+/+ embryos (Fig. 3A). To further explore this phenomenon, we used fibroblast cells derived from PS1−/− and PS1+/+ mice. Because these cells express undetectable levels of endogenous E-cadherin, we transfected both of them and selected lines expressing similar levels of exogenous human E-cadherin (Fig. 3B). As expected, transfected E-cadherin coimmunoprecipitated with peptide PS1/CTF in PS1+/+ cells but not in PS1−/− cells (Fig. 3C; see also Fig. 5A, Right, for the reverse experiment). E-cadherin IPs prepared from PS1−/− cells contained significantly higher levels of p120 than did E-cadherin IPs from PS1+/+ cells (Fig. 3D, Left), even though the two cell lines contain similar levels of cellular p120 (Fig. 3D, Lower Right). In the reverse experiments, p120 IPs from PS1−/− cells contained more E-cadherin than did p120 IPs from PS1+/+ cells (Fig. 3D, Upper Right). Reintroduction of PS1 into PS1−/− cells by transfection reduced the E-cadherin/p120 complex (Fig. 3E Left and Middle) and induced the appearance of the E-cadherin/PS1 complex (Fig. 3E Right). These data show that PS1 inhibits p120 binding to E-cadherin. Conversely, overexpression of p120 in EL cells decreased the E-cadherin/PS1 complex while it increased the E-cadherin/p120 complex (Fig. 3F, Lower and Upper, respectively). Thus, p120 is also capable of inhibiting PS1 binding to E-cadherin. In summary, our data show that PS1 binds E-cadherin and promotes the dissociation of the E-cadherin/p120 complex. The reverse is also true; p120 binds E-cadherin and promotes the dissociation of the E-cadherin/PS1 complex. Taken together, our results indicate that the two proteins compete for E-cadherin binding in vivo.
Figure 3.
PS1 inhibits p120 binding to E-cadherin. (A) E-cadherin IPs (E-cad IP) in TNE plus 1% Triton X-100 from either PS1−/− mouse embryos (−/−) or their PS1+/+ littermates (+/+) were probed with anti-E-cadherin (E-cad) or anti-p120 (p120) antibodies. (B) Extracts (1% SDS) of confluent nontransfected (E−) or E-cadherin-transfected (E+) PS1+/+ (+/+) and PS1−/− (−/−) mouse fibroblasts were probed with anti-E-cadherin antibody. (C) Extracts (1% Triton X-100) of confluent E-cadherin-expressing PS1+/+ (+/+) or PS1−/− (−/−) fibroblasts were immunoprecipitated with antibody 33B10 (PS1/CTF IP), and the resulting IPs were probed with anti-E-cadherin (E-cad) antibody. (D) Extracts (1% Triton X-100) of E-cadherin-transfected PS1 +/+ (+/+) or PS1−/− (−/−) fibroblasts were immunoprecipitated with anti-E-cadherin (E-cad IP) or anti-p120 (p120 IP) antibodies, and the resulting IPs were probed with anti-E-cadherin (E-cad) or anti-p120 (p120) antibodies. (E) E-cadherin-transfected PS1−/− fibroblasts were transiently transfected either with vector (VECT) or with WT PS1 (wtPS1), and Triton X-100 extracts were immunoprecipitated with anti-E-cadherin (E-cad IP), anti-p120 (p120 IP), or anti-PS1/CTF (PS1/CTF IP) antibodies. The resulting IPs were probed with anti-E-cadherin (E-cad), anti-p120 (p120), or anti-PS1/CTF antibodies. FLPS1, full-length PS1. The asterisks indicate IgGs. (F) Extracts (1% Triton X-100) of untransfected (NT) EL cells or EL cells transiently transfected with p120 (p120) were treated with anti-E-cadherin antibodies (E-cad IP), and the resulting IPs were probed with antibodies against the proteins indicated on the right. The asterisk indicates IgGs.
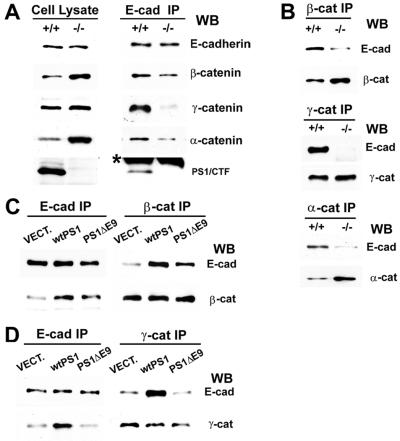
Figure 5.
PS1 stabilizes the cadherin/catenin complex. (A) Extracts of confluent E-cadherin-transfected PS1+/+ and PS1−/− fibroblasts in 1% Triton X-100 were treated with anti-E-cadherin antibody, and the resulting IPs (E-cad IP, Right), along with total cell lysate in 1% SDS (Left), were probed on WBs with antibodies against the proteins indicated at the right of the figure. The asterisk denotes low-molecular-weight IgGs. (B) Anti-β-catenin, anti-γ-catenin, or anti-α-catenin IPs (Top, Middle, and Bottom, respectively) prepared from E-cadherin-expressing PS1+/+ and PS1−/− fibroblasts were probed with anti-E-cadherin (E-cad), anti-β-catenin (β-cat), anti-γ-catenin (γ-cat), or anti-α-catenin (α-cat) antibodies. (C and D) Extracts (1% Triton X-100) of E-cadherin-expressing PS1−/− fibroblasts transiently transfected with vector (VECT), WT PS1 (wtPS1), or PS1 FAD mutant ΔE9 (PS1ΔE9) were treated with anti-E-cadherin (E-cad IP), anti-β-catenin (β-cat IP) (C), or anti-γ-catenin (γ-cat IP) (D) antibodies, and the resulting IPs were probed with antibodies against the proteins indicated at the right of the figure. E-cad, E-cadherin; β-cat, β-catenin; γ-cat, γ-catenin.
WT PS1, but Not PS1 ΔE9 Mutant, Stabilizes the Cadherin/Catenin Complex and Stimulates Ca2+-Dependent Cell–Cell Aggregation.
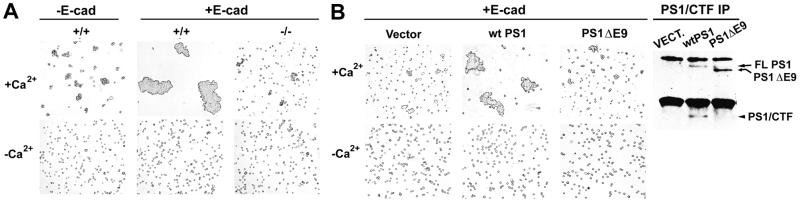
Recent reports suggest that PS1 regulates cell–cell adhesion, but the molecular basis of this regulation remains unknown (9). That PS1 competes with p120 for E-cadherin binding suggests that PS1 may function as an additional regulator of the cadherin-based adhesion system. Indeed, in the presence of Ca2+, E-cadherin-transfected PS1−/− cells aggregated poorly compared with E-cadherin-transfected PS1+/+ cells, suggesting that PS1 stimulates Ca2+-dependent cell–cell aggregation (Fig. 4A). Untransfected PS1+/+ cells expressing undetectable levels of E-cadherin showed little aggregation compared with E-cadherin-transfected PS1+/+ cells (Fig. 4A), indicating that aggregation is E-cadherin-mediated. Reintroduction of WT PS1 into PS1−/− cells substantially increased Ca2+-dependent cell–cell aggregation (Fig. 4B). In contrast to WT PS1 that was able to restore cadherin-dependent aggregation, PS1 mutant ΔE9 was inactive (Fig. 4B).
Figure 4.
PS1 stimulates Ca2+-dependent cell–cell aggregation. (A) Nontransfected (−E-cad) PS1 +/+ (+/+) or E-cadherin-transfected (+E-cad) PS1 +/+ (+/+) and PS1−/− (−/−) fibroblasts were allowed to aggregate for 4 h in the presence (+Ca2+) or absence (−Ca2+) of calcium (see Materials and Methods). (B) E-cadherin-expressing PS1−/− fibroblasts, transiently transfected with vector (Vector), WT PS1 (wtPS1), or PS1 FAD mutant ΔE9 (PS1ΔE9), were assayed for aggregation in the presence or absence of calcium. (Right) Extracts (1% Triton X-100) from the transiently transfected cultures shown in B were treated with antibody 33B10, and the resulting IPs (PS1/CTF IP) were analyzed for expression of the transfected PS1 proteins.
To further explore the possible mechanism by which PS1 stimulates cell–cell adhesion, we determined the PS1 effects on the stability of the cadherin/catenin complexes with the use of E-cadherin-transfected PS1−/− and PS1+/+ fibroblasts expressing similar levels of exogenous E-cadherin (see above). E-cadherin IPs prepared from confluent PS1+/+ cells contained significantly higher levels of γ-, β-, and α-catenin than E-cadherin IPs from PS1−/− cells (Fig. 5A). In the reverse experiments γ-, β-, and α-catenin IPs from PS1 +/+ cells contained higher levels of E-cadherin than the corresponding IPs from PS1−/− cells (Fig. 5B). PS1 had a more dramatic effect on the E-cadherin/γ-catenin association than on the E-cadherin/β-catenin association (Fig. 5 A and B). Reintroduction of WT PS1 in PS1−/− cells increased the E-cadherin complexes not only with PS1 (Fig. 3E, Right), but also with both β- and γ-catenin (Fig. 5 C and D, respectively). Interestingly, in contrast to WT PS1, FAD mutant ΔE9 failed to stimulate binding of γ-catenin to E-cadherin (Fig. 5D) and was less effective than the WT PS1 in stimulating the E-cadherin/β-catenin complex (Fig. 5C). Together with the data from the aggregation experiments, these results suggest that WT PS1, but not PS1 mutant ΔE9, stimulates Ca2+-dependent cell–cell adhesion by stabilizing the cadherin/catenin adhesion complex.
PS1 Promotes Cytoskeletal Association of the Cadherin/Catenin Complex.
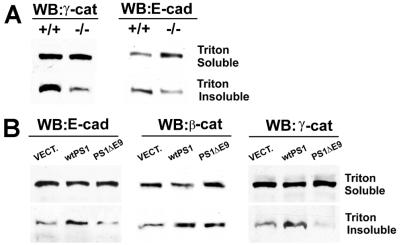
Stable cadherin-dependent cell–cell adhesion in intact cells requires linkage of the cadherin/β- and γ-catenin complexes to the actin cytoskeleton (12, 13, 23). This linkage is mediated by α-catenin (25) and is manifested by an increased insolubility of the cadherin/catenin complex components in Triton X-100 (23). Examination of E-cadherin and α-catenin IPs prepared from PS1+/+ and PS1−/− fibroblasts showed that the former cells contained higher levels of the E-cadherin/α-catenin complexes than the latter cells (Fig. 5 A and B), suggesting that PS1 may ultimately stimulate the cytoskeletal association of the cadherin/catenin adhesion complexes. Indeed, Fig. 6A shows that in PS1+/+ cells a significantly higher fraction of total γ-catenin was found in the Triton X-100-insoluble fraction compared with PS1−/− cells, where almost all of the γ-catenin was found in the Triton X-100-soluble fraction. E-cadherin also showed an increased distribution in the Triton X-100-insoluble fraction in PS1+/+ cells compared with PS1−/− cells, suggesting an increased cytoskeletal association (Fig. 6A). Similar results were obtained for β-catenin (data not shown). PS1 transfection of PS1−/− cells stimulated the cytoskeletal association of E-cadherin, β-catenin, and γ-catenin, whereas transfection with the FAD ΔE9 did not (Fig. 6B), in agreement with the inability of this FAD mutant to stabilize the cadherin/catenin complexes and to stimulate cell–cell adhesion.
Figure 6.
PS1 promotes cytoskeletal association of the cadherin/catenin complexes. (A) Confluent cultures of E-cadherin-transfected PS1+/+ or PS1−/− fibroblasts were extracted in 1% Triton X-100, and insoluble fractions were solubilized in urea buffer (see Materials and Methods). Sixty micrograms of protein of the Triton X-100-soluble fraction or 75 μg of the insoluble fraction was then probed with antibodies against γ-catenin (γ-cat) or E-cadherin (E-cad). (B) Extracts from E-cadherin-transfected PS1−/− fibroblasts, transiently transfected with vector (VECT), WT PS1 (wtPS1), or PS1 FAD mutant ΔE9 (PS1ΔE9), were prepared and probed as in A, plus anti-β-catenin (β-cat) antibodies.
Discussion
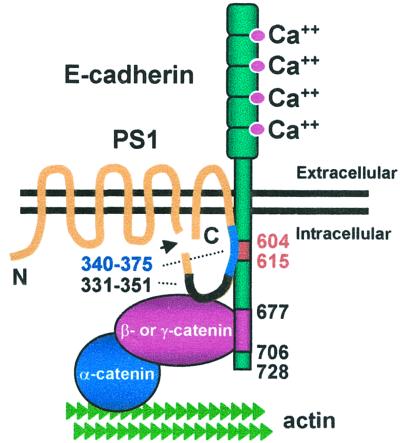
Cadherin-based cell–cell adhesion and communication are critical determinants of tissue development, function, and homeostasis (10, 11). The present study shows that PS1 binds directly to E-cadherin. Remarkably, this binding requires mature E-cadherin amino acids 604–615, which are also required for E-cadherin binding of p120, a regulator of cadherin-mediated cell–cell adhesion (21, 24). PS1 displaces p120 from E-cadherin, stabilizes the binding of β- and γ-catenins to E-cadherin, increases linkage of the cadherin/catenin complex to the cytoskeleton, and stimulates Ca2+- and cadherin-dependent cell–cell aggregation. These findings show that PS1 regulates the molecular composition and function of the cadherin-based adhesion system and suggest a possible mechanism for PS1-induced stabilization of the cadherin/catenin complex (Fig. 7). PS1/CTF sequence 340–375 binds E-cadherin sequence 604–615 and may tether the membrane-proximal cytoplasmic E-cadherin close to the plasma membrane. At a more distal site on E-cadherin are bound β- or γ-catenin (16, 17), which in turn may bind PS1 (19, 20). These interactions could result in the formation of a stable three-member complex, each member of which binds directly to the other two. β-Catenin or γ-catenin binds α-catenin, which in turn anchors the complexes to the cytoskeleton, consistent with the increased cytoskeletal association of E-cadherin, β-catenin, and γ-catenin induced by PS1.
Figure 7.
Schematic representation of hypothesized interactions between E-cadherin, β- οr γ-catenin, and PS1. β-Catenin or γ-catenin binds E-cadherin at amino acids 677–706. PS1 C-terminal fragment binds E-cadherin close to the membrane at amino acids 604–615 (required for PS1/E-cadherin complex formation). For more details see text.
Cadherins are expressed in specific patterns throughout the embryonic and adult life of vertebrates. At least 15 different classic cadherins have been detected in the central nervous system, where they function as guidance molecules in neurite target recognition and synapse formation and regulate synaptic plasticity (10, 11, 26, 27). The cadherin region critical for PS1 binding is conserved in classic cadherins including E-, N-, P- (placental), R- (retinal), and vascular endothelial cadherin (18). This region is crucial for cell adhesion during embryogenesis (28), for N-cadherin-mediated neurite outgrowth (29), cell motility (30), and regulation of N- and vascular endothelial-cadherin expression in endothelial cell junctions (31). Thus PS1 may regulate the function of multiple cadherin systems during development and adult life. That PS1 is important for central nervous system development is suggested by PS1 null mice that die shortly after birth with severe abnormalities of CNS development and impaired vascular integrity (7).
Members of the cadherin adhesion complex, including E-cadherin, β-catenin, and p120, have been identified as important oncogenes (15, 18, 32). That PS1 regulates their association suggests that PS1 itself might be involved in cancer development. E-cadherin is the main adhesion molecule of epithelia, and dysfunction of E-cadherin complexes has been implicated in tumorigenesis and metastasis of human epithelial cancer (32). That PS1 promotes cadherin adhesion raises the possibility that loss of PS1 function might decrease cell–cell adhesion and promote cancer development. This model suggests an explanation for the surprising observation that PS1 down-regulation leads to the formation of epidermal tumors in adult mice (33).
It is of great interest that in contrast to WT PS1, PS1 FAD mutant ΔE9 failed to increase cell–cell aggregation and stimulated neither formation of cadherin/catenin complexes nor the cytoskeletal association of cadherin and catenins. This mutant lacks the PS1 cleavage site and is not cleaved intracellularly (3, 4). Thus, the inability of this mutant to stimulate cell–cell adhesion may be due to a lack of functional PS1 fragments. We observed that uncleaved ΔE9 mutant forms complexes with E-cadherin (S.E. and N.K.R., unpublished observations), suggesting that incorporation of uncleaved PS1 into the cadherin/catenin complex cannot substitute functionally for the PS1 fragments. It remains an interesting question whether the lack of function of mutant ΔE9 contributes to the development of the FAD phenotype.
Recent evidence shows that PS1 forms complexes with APP and Notch-1 receptor and regulates production of Aβ (5, 6). Like E-cadherin, APP and Notch-1 are type I transmembrane proteins, but the mechanism(s) involved in the PS1 binding to APP and Notch-1 receptor is still not clear. Our findings on the sequence involved in E-cadherin/PS1 binding suggest a model for PS1 binding to APP and Notch-1 receptors. In addition, cadherin is involved in cell adhesion and signaling, and APP may have similar functions (34, 35). That PS1 forms complexes with the APP, Notch-1, and E-cadherin systems suggests that in addition to APP processing, PS1 FAD mutations may target other type I transmembrane protein systems, including Notch-1 and cadherins. That PS1 FAD mutants may affect an array of protein systems is in agreement with evidence that Alzheimer's disease is a heterogeneous disorder with multiple systems affected (36, 37).
Acknowledgments
We thank Drs. Tonegawa and Potter for PS1+/− Tg mice, Dr. P. Saftig for PS1−/− and PS1+/+ fibroblasts, and Dr. Gottardi for human E-cadherin cDNA. This work was supported by National Institute on Aging Grants AG-17926, AG-08200, and AG-05138, and the Alzheimer's Association.
Abbreviations
- PS1
presenilin-1
- PS1/NTF
PS1 N-terminal fragment
- PS1/CTF
PS1 C-terminal fragment
- APP
amyloid precursor protein
- E-cadherin
epithelial cadherin
- N-cadherin
neuronal cadherin
- FAD
familial Alzheimer's disease
- IPs
immunoprecipitations
- WBs
Western blots
- WT
wild type
References
- 1.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 2.Elder G A, Tezapsidis N, Carter J, Shioi J, Bouras C, Li H C, Johnston J M, Efthimiopoulos S, Friedrich V L, Jr, Robakis N K. J Neurosci Res. 1996;45:308–320. doi: 10.1002/(SICI)1097-4547(19960801)45:3<308::AID-JNR13>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Podlisny M B, Citron M, Amarante P, Sherrington R, Xia W, Zhang J, Diehl T, Levesque G, Fraser P, Haass C, et al. Neurobiol Dis. 1997;3:325–337. doi: 10.1006/nbdi.1997.0129. [DOI] [PubMed] [Google Scholar]
- 4.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, et al. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 5.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 6.De-Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 7.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 8.Giannakopoulos P, Bouras C, Kovari E, Shioi J, Tezapsidis N, Hof P R, Robakis N K. Am J Pathol. 1997;150:429–436. [PMC free article] [PubMed] [Google Scholar]
- 9.Georgakopoulos A, Marambaud P, Efthimiopoulos S, Shioi J, Cui W, Li H C, Schutte M, Gordon R, Holstein G R, Martinelli G, et al. Mol Cell. 1999;4:893–902. doi: 10.1016/s1097-2765(00)80219-1. [DOI] [PubMed] [Google Scholar]
- 10.Gumbiner B M. J Cell Biol. 2000;148:399–404. doi: 10.1083/jcb.148.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi T, Takeichi M. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 12.Nagafuchi A, Takeichi M. EMBO J. 1988;7:3679–3684. doi: 10.1002/j.1460-2075.1988.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagafuchi A, Takeichi M. Cell Regul. 1989;1:37–44. doi: 10.1091/mbc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozawa M, Baribault H, Kemler R. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Ze'ev A, Geiger B. Curr Opin Cell Biol. 1998;10:629–639. doi: 10.1016/s0955-0674(98)80039-2. [DOI] [PubMed] [Google Scholar]
- 16.Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- 17.Jou T S, Stewart D B, Stappert J, Nelson W J, Marrs J A. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anastasiadis P Z, Reynolds A B. J Cell Sci. 2000;113:1319–1334. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- 19.Murayama M, Tanaka S, Palacino J, Murayama O, Honda T, Sun X, Yasutake K, Nihonmatsu N, Wolozin B, Takashima A. FEBS Lett. 1998;433:73–77. doi: 10.1016/s0014-5793(98)00886-2. [DOI] [PubMed] [Google Scholar]
- 20.Levesque G, Yu G, Nishimura M, Zhang D M, Levesque L, Yu H, Xu D, Liang Y, Rogaeva E, Ikeda M, et al. J Neurochem. 1999;72:999–1008. doi: 10.1046/j.1471-4159.1999.0720999.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohkubo T, Ozawa M. J Biol Chem. 1999;274:21409–21415. doi: 10.1074/jbc.274.30.21409. [DOI] [PubMed] [Google Scholar]
- 22.Ozawa M, Kemler R. J Cell Biol. 1998;142:1605–1613. doi: 10.1083/jcb.142.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathke I S, Hinck L, Swedlow J R, Papkoff J, Nelson W J. J Cell Biol. 1994;125:1341–1352. doi: 10.1083/jcb.125.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoreson M A, Anastasiadis P Z, Daniel J M, Ireton R C, Wheelock M J, Johnson K R, Hummingbird D K, Reynolds A B. J Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rimm D L, Koslov E R, Kebriaei P, Cianci C D, Morrow J S. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka H, Shan W, Phillips G R, Arndt K, Bozdagi O, Shapiro L, Huntley G W, Benson D L, Colman D R. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 27.Tang L, Hung C P, Schuman E M. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- 28.Kintner C. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- 29.Riehl R, Johnson K, Bradley R, Grunwald G B, Cornel E, Lilienbaum A, Holt C E. Neuron. 1996;17:837–848. doi: 10.1016/s0896-6273(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Paradies N E, Fedor-Chaiken M, Brackenbury R. J Cell Sci. 1997;110:345–356. doi: 10.1242/jcs.110.3.345. [DOI] [PubMed] [Google Scholar]
- 31.Navarro P, Ruco L, Dejana E. J Cell Biol. 1998;140:1475–1484. doi: 10.1083/jcb.140.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perl A K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature (London) 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 33.Zheng X-F, Xia Y, Wu X. Neurobiol Aging. 2000;21:S133. [Google Scholar]
- 34.Williamson T G, Mok S S, Henry A, Cappai R, Lander A D, Nurcombe V, Beyreuther K, Masters C L, Small D H. J Biol Chem. 1996;271:31215–31221. doi: 10.1074/jbc.271.49.31215. [DOI] [PubMed] [Google Scholar]
- 35.Wu A, Pangalos M N, Efthimiopoulos S, Shioi J, Robakis N K. J Neurosci. 1997;17:4987–4993. doi: 10.1523/JNEUROSCI.17-13-04987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesulam M M. Neuron. 1999;24:521–529. doi: 10.1016/s0896-6273(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 37.Neve R L, Robakis N K. Trends Neurosci. 1998;21:15–19. doi: 10.1016/s0166-2236(97)01168-5. [DOI] [PubMed] [Google Scholar]