Abstract
The integration of genetic and physical maps of maize is progressing rapidly, but the cytogenetic maps lag behind, with the exception of the pachytene fluorescence in situ hybridization (FISH) maps of maize chromosome 9. We sought to produce integrated FISH maps of other maize chromosomes using Core Bin Marker loci. Because these 1 Kb restriction fragment length polymorphism (RFLP) probes are below the FISH detection limit, we used BACs from sorghum, a small-genome relative of maize, as surrogate clones for FISH mapping. We sequenced 151 maize RFLP probes and compared in silico BAC selection methods to that of library filter hybridization and found the latter to be the best. BAC library screening, clone verification, and single-clone selection criteria are presented along with an example of transgenomic BAC FISH mapping. This strategy has been used to facilitate the integration of RFLP and FISH maps in other large-genome species.
1. Introduction
Cytogenetics has proven invaluable in eukaryotic genome research, helping to elucidate genome structure in humans and model organisms such as fruit fly (Drosophila melanogaster) and maize (Zea mays) [1–6]. Historically, the field of cytogenetics, and in particular maize cytogenetics, was greatly advanced by Barbara McClintock's pioneering work in the 1920s and 1930s [7]. Her method for unequivocal identification of individual chromosomes permitted major discoveries regarding the structure and dynamic behavior of the maize genome while also establishing the connection between genetic and physical recombination [8–10]. Subsequent plant molecular cytology built on this foundation by using cloned DNA sequences as probes for in situ hybridization in order to visualize their location directly on the physical chromosomes [11–15]. In addition to single-locus detection, fluorescence in situ hybridization (FISH) with repetitive sequence probes has been used and combined with multicolor cocktails to characterize genome structure in polyploid or closely related plant species [16–19]. Modern plant cytogenetics has increasingly contributed to structural, functional, and comparative genomics especially when FISH probes from bacterial artificial chromosome (BAC) clones, which are 100- to 200-Kb genomic fragments, have been used. Individual clones can be arrayed and characterized by fingerprint contig (FPC) mapping and annotated with markers by filter hybridization [20]. Well-characterized libraries can then serve as a resource for whole genome sequencing as was recently done for maize [21].
Among plant crops, maize has great importance for both economics and research. In 2009, maize made up 27% of the United States crop harvest by acreage, more than cotton, sorghum, wheat, barley, and sunflower combined (National Corn Grower's Association, U.S.A. Figures, 2009). Maize is a powerful genetic system, because of its large families and ease of crossing, as evidenced by the historical advances in transmission genetics [22] and epigenetics [23–25], as well as more recent advances in genome structure and evolution [21, 26–34].
The maize genome has been charted in two fundamentally different ways: genetic/linkage maps and physical maps. Linkage mapping is based on recombination and cosegregation frequencies of markers, whereas maize physical maps comprise a larger array of maps most commonly based on molecular cytology or assemblies of contiguous DNA restriction fragments, overlapping clone-based contigs, or DNA sequence assemblies [2, 21, 32, 34–43]. In the last 20 years, the linkage maps and the BAC-based physical contig maps have demonstrated the greatest utility in maize. During the 1980s and 1990s the number of mapped loci on the linkage maps of maize increased markedly, as conventional mutant-based linkage maps were improved or replaced by high-density molecular marker maps [20, 31, 44]. These maps are based on DNA sequence polymorphisms detected with Southern blots of cloned restriction fragment length polymorphism (RFLP) probes or gel electrophoresis of PCR products of microsatellites and simple sequence repeats. These same molecular markers were mapped onto BACs in library arrays for annotation of the physical map with genetic markers [31, 37, 40].
The RFLP probes have featured prominently in the production of the genetic maps in maize (http://www.maizegdb.org/map.php). The polymorphic nature of the maize genome makes RFLPs ideal for use in mapping populations made from any two of the hundreds of divergent inbred lines of maize [45]. A select subset of evenly distributed RFLP markers, designated as Core Bin Markers (CBMs), provide a common and standardized set of linkage makers, which conveniently parcel out the maize genome into 100 linkage bins [20]. For example, the first bins on maize chromosome 1, starting from the telomere of the short arm (1S), are designated 1.00, 1.01, 1.02, and so on until the last bin, 1.11, at the telomere of the long arm (1L). By hybridization of the RFLP probes to the arrayed members of the maize BAC libraries used to build the contig maps, the genetic and physical maps have been integrated and used to guide the sequencing and assembly of the maize genome [21, 31, 32, 34, 37, 46].
Similarly, efforts to localize RFLP probes, like the CBM probes, on the maize cytogenetic maps have been undertaken but are hindered by the difficulty of detecting the small RFLP DNA segments by FISH. The cytogenetic maps are therefore among the least developed of all the maps of the maize genome, despite the early and prominent work in maize cytogenetics [22]. Although several studies have localized various types of repeat sequences on the chromosomes of maize, relatively few single-copy sequences have been cytologically mapped on any of the maize chromosomes other than chromosome 9 [15, 16, 36, 38, 42, 47–52]. Because of the extensive use of the CBM loci in maize genetics, along with other RFLPs, these markers have been selected for the development of a cytogenetic map of maize so that connections with the linkage and physical maps can be maximized [35, 53]. These efforts make use of meiotic pachytene-stage chromosomes because they are about 6–25 times longer, providing better axial resolution than their shorter somatic counterparts [2]. Although the location of the CBM probes on the meiotic chromosome karyotype of maize has been predicted from recombination nodule frequencies, the location of the genetic bins and their boundary CBM loci on the actual chromosomes remain largely unverified [35, 54–56].
The smallest probes and targets that can be localized and reliably detected on maize pachytene chromosomes have averaged 2.4–3.1 Kb in length [38, 42], but the maize CBM-RFLP probes were originally chosen to be small, less than 1 Kb. Their small size facilitated subsequent single-pass sequencing, but it put them well below the FISH detection limit [2, 20]. In addition, because of the complexity and low gene density of the maize genome, the simple use of maize BAC clones as FISH probes directly on maize chromosomes is not productive without additional steps for the amplification of single-copy sequences within each BAC [38, 52].
In the experiments reported here, we used an approach for indirect FISH mapping of the CBM loci that exploits the recent divergence of maize and sorghum by identifying sorghum BACs, homologous to maize-markers, for use as representative FISH probes [35, 53]. The procedures for the selection, verification, and use of sorghum BACs as transgenomic FISH probes for maize are described. The identity of homologous BAC clones for the CBM loci on several maize chromosomes is presented along with methods and criteria for selecting a single suitable sorghum BAC for each maize marker.
2. Materials and Methods
2.1. Plant Materials
Plants of a disomic maize chromosome addition line of oat, OMAd1.36, carrying a B73 maize chromosome 1 in an oat genome background [57, 58] were grown in the Mission Road Research Facility greenhouse (Florida State University, Tallahassee, FL, USA) or in Conviron growth chambers under short- and long-day conditions as described previously [57, 58].
2.2. Full-Length Insert Sequences for CBM and Other Maize RFLP Probes
Maize RFLP probes were obtained as freezer cultures from the UMC RFLP Lab (G. Davis, T. Muskett, University of Missouri-Columbia, http://www.maizegdb.org/cgi-bin/displaypersonrecord.cgi?id=97483). The full-length insert sequences (FLIS) were determined for 66 of the 90 maize CBM RFLP probes by primer walk sequencing across both strands (DNA Sequencing Facility, Department of Biological Science, Florida State University, Tallahassee, FL). The resulting sequence contigs were assembled and analyzed with Sequencher (Gene Codes Corporation, Ann Arbor, MI), inspected for accuracy, and verified by BLAST sequence-similarity searches. Vector sequences were removed, and detailed annotations along with the RFLP-FLIS sequences were submitted to GenBank. An additional 85 non-CBM maize RFLP probes were selected, arrayed as freezer cultures in 96-well plates, and fully sequenced on both strands by primer walking (SeqWrite, Houston, TX). Contig analysis, verification, and submission were carried out as described above. All of the maize RFLP-FLIS accessions can be retrieved from the supporting data available from the public NCBI Genbank database (http://www.ncbi.nlm.nih.gov/) with the query “Bass, maize, RFLP.”
2.3. Maize RFLP Probe-Based Selection of Sorghum Propinquum BACs
The Sorghum propinquum BAC library filter array [59] was screened by hybridization with RFLP probes as previously described [35, 53]. Briefly, insert sequences from plasmids carrying CBM-RFLP probes were obtained from PCR products amplified with insert-flanking vector primers. The PCR products were trimmed of vector sequences by restriction enzymes, gel purified, and used (50 ng) for random-primed labeling with 32P-dCTP (Rediprime II DNA Labeling System, GE Healthcare). The resulting probes were hybridized at a stringency of Tm-25°C to the YRL BAC filters (two 22 × 22-cm filters per hybridization). The resulting BAC hits were scored, and the individual BAC identities were examined within the S. propinquum contig map (http://www.stardaddy.uga.edu/fpc/WebAGCoL/propinquum/WebFPC/). Overlapping BACs were then grown, and their DNA was isolated with the Qiagen Spin Miniprep kit (no. 27104, Valencia, CA). The miniprepped BAC DNAs were digested with HindIII and subjected to Southern blot hybridization for homologous sequence verification with the same probe used in the BAC filter hybridizations.
2.4. Chromosome Spreading for FISH
Meiosis-stage florets from oat-maize addition lines were harvested, fixed, and stored, and anthers containing pachytene-staged meiocytes were identified as previously described [35]. Briefly, preemerged panicles were fixed in ethanol : acetic acid (3 : 1) solution for two days and stored in 70% ethanol until staging. For staging of the meiotic cells from fixed florets, single anthers were stained by the aceto-carmine squash method to identify pachytene-staged florets [60]. The remaining two pachytene-staged anthers were then stored in 70% ethanol at −20°C until their use for FISH. Meiotic chromosomes were prepared in the initial steps as previously described [61] with modifications used by Danilova and Birchler [38]. Breifly, individual anthers were washed three times with deionized H2O, washed twice for 5 minutes with citric buffer (10 mm sodium citrate, 10 mm EDTA, adjusted to pH 5 by addition of pH 5 citric acid), and then digested in a pectolytic enzyme solution (0.3% cytohelicase, 0.3% cellulase, 0.3% pectolyase in 30-mM citrate buffer, pH 4.5) for 2 hours. The tubes were then plunged into ice and immediately filled with 10 : 0.2 TE [(10 mM Tris-HCl (pH 7.6) : 0.2 mM EDTA-NaOH (pH 8.0)] to stop the reaction. The TE was aspirated, and the anthers were washed twice with 100% ethanol. The ethanol was then aspirated and replaced with 20 μl ice-cold acetic acid : methanol solution (9 : 1). The anthers were gently macerated in the tube with a straight probe and incubated them on ice for at least 15 minutes to clear the cytoplasm before chromosome spreading. Eight μl of the cell suspension was then dropped from a micropipettor onto two slides that had been placed in a humid chamber [16]. The chromosomes were gently spread as described by Koumbaris and Bass [53] by alternating the addition of ice-cold ethanol : acetic acid (3 : 1) solution with exposure to 75°C water vapor for a total of three rounds followed by the addition of 100% acetic acid and quick drying at 65°C for 3 minutes. The slides were then immediately FISH probed as described by Amarillo and Bass [35], but without the slide-aging step.
2.5. BAC FISH
Highly purified BAC DNA was isolated with the Qiagen Large Construct purification kit (Qiagen item No. 12462). The BAC DNA was digested with AluI, and 2 μg of the digested BAC DNA was direct-labeled with Alexa-Fluor-546-dCTP for FISH as previously described [35]. The probe cocktail also included centromere and whole chromosome painting probes, prepared and used as previously described [35, 53]. Image collection, chromosome straightening, and FISH chromosome locus determination were also carried out as previously described [53].
3. Results and Discussion
We previously developed a strategy using transgenomic BAC FISH to localize genetic marker sequences, such as RFLP probes, on maize chromosomes [53]. This strategy was initially developed to overcome the FISH detection limit so that small RFLP probes could be cytogenetically localized [38, 42]. It also avoided problems associated with direct use of maize BACs as FISH probes, namely, most maize BACs contain relatively few single-copy sequences for site-specific hybridization. The use of transgenomic FISH exploits the general colinearity and genic synteny between related species, supporting homologous cross-mapping in plants with large, complex genomes such as maize [53, 62]. In particular, the use of sorghum BACs as FISH probes on maize chromosomes works well because sorghum and maize diverged ∼12 mya, preserving genic sequences but not intergenic repetitive DNAs [30, 31, 35, 53, 63, 64].
3.1. RFLP Probes Are Ideal Markers for Integrating Physical and Linkage Maps of Maize
Having employed a filter hybridization method for selecting sorghum BACs that correspond to maize genetic markers, we attempted to accelerate the BAC selection process using an in silico approach. Many of the widely used maize RFLP probes are not sequenced or have only limited end sequence data. We therefore determined the FLIS of RFLP probes for markers that we selected for possible FISH mapping, including the CBM. Table 1 lists all 90 of the maize CBM-RFLPs along with their primary features and GenBank accession numbers for those that were completely sequenced. The table includes estimated insert sizes before our study as well as the vector-trimmed FLIS sizes that we obtained. A few of the CBM-RFLP probes were difficult to sequence. For example, CBM2.01 and CBM4.05 failed to yield good sequence traces with the vector primers, whereas the CBM4.01 insert is flanked by poly-A sequences on both sides. In addition, we determined the FLIS for an additional 85 non-CBM maize RFLPs, listed in Supplemental Table 1 in supplementary material available online at doi: 10.1155/2011/386862. For these markers, we chose numerous probes from the “CSU” set [65] because they were derived from B73 cDNA libraries and were therefore expected to be easy to place on the B73 reference genome and to provide excellent probes for high-stringency cross-hybridization to sorghum genomic DNA libraries.
Table 1.
Sequenced maize CBM-RFLPs.
| CBM | Marker/locus | Probe | Accession number | Type | Enzyme(s) | MaizeGDB insert size (bp) |
Sequenced RFLP insert size |
|---|---|---|---|---|---|---|---|
| 1.01 | tub1 | p-tub1 | AY987961 | Genomic | EcoRI/HindIII | 156 | 156 |
| 1.02 | umc157(chn) | p-umc157 | DQ123890 | Genomic | PstI | 1220 | 1250 |
| 1.03 | umc76(prob a) | p-umc76 | AY751079 | Genomic | PstI | 760 | 710 |
| 1.04 | asg45(ptk) | p-asg45 | AY771210 | Genomic | PstI | 350 | 332 |
| 1.05 | csu03 | p-csu03 | DQ123891 | cDNA | EcoRI/XhoI | 1200 | 1064 |
| 1.06 | umc67a | p-umc67 | AY771211 | Genomic | PstI | 650 | 644 |
| 1.07 | asg62 | p-asg62 | DQ001865 | Genomic | PstI | 500 | 465 |
| 1.08 | umc128 | p-umc128 | DQ123892 | Genomic | PstI | 740 | 755 |
| 1.09 | cdj2 | p-csu164 | DQ642431 | cDNA | EcoRI/XhoI | 700 | 466 |
| 1.10 | umc107a(croc) | p-umc107 | DQ642432 | Genomic | PstI | 1090 | 1105 |
| 1.11 | umc161a | p-umc161 | AY771212 | Genomic | PstI | 700 | 723 |
| 1.12 | bnl6.32a | p-bnl6.32 | Genomic | PstI | 2250 | ||
| 2.01 | bnl8.45ab | p-bnl8.45 | Genomic | PstI | 2100 | ||
| 2.02 | umc53a | p-umc53 | AY771214 | Genomic | PstI | 640 | 604 |
| 2.03 | umc06a | p-umc06 | AY771213 | Genomic | PstI | 590 | 608 |
| 2.04 | umc34 | p-umc34 | DQ001866 | Genomic | PstI | 970 | 934 |
| 2.05 | umc131 | p-umc131 | AY771215 | Genomic | PstI | 810 | 859 |
| 2.06 | umc255a | p-umc255 | DQ123893 | Genomic | PstI | 1050 | 1013 |
| 2.07 | umc005a | p-umc005 | AY771216 | Genomic | PstI | 850 | 830 |
| 2.08 | asg20 | p-asg20 | DQ123894 | Genomic | PstI | 550 | 337 |
| 2.09 | umc049a | p-umc049 | DQ123895 | Genomic | PstI | 630 | 627 |
| 2.10 | php20581b(tb) | p-php20581 | EU190456 | Genomic | PstI | 1400 | 1327 |
| 3.01 | umc032a | p-umc32 | DQ001867 | Genomic | PstI | 990 | 1019 |
| 3.02 | csu32 | p-csu32 | DQ123896 | Genomic | EcoRI/XhoI | 500 | 411 |
| 3.03 | asg24(gts) | p-asg24 | AY771217 | Genomic | PstI | 550 | 372 |
| 3.04 | asg48 | p-asg48 | DQ001868 | Genomic | PstI | 1600 | 1617 |
| 3.05 | umc102 | p-umc102 | DQ005498 | Genomic | PstI | 1010 | 1023 |
| 3.06 | bnl5.37ab | p-bnl5.37 | Genomic | PstI | 2300 | ||
| 3.07 | bnl6.16ab | p-bnl6.16 | Genomic | PstI | 2450 | ||
| 3.08 | umc17a | p-umc17 | AY771218 | Genomic | PstI | 850 | 840 |
| 3.09 | umc63a | p-umc63 | DQ123897 | Genomic | PstI | 620 | 881 |
| 3.10 | cyp1 | p-csu25 | DQ005499 | cDNA | EcoRI/XhoI | 1100 | 960 |
| 4.01 | agrr115c | p-agrr115 | Genomic | EcoRI/HindIII | 600 | ||
| 4.02 | php20725ab | p-php20725 | Genomic | PstI | 1650 | ||
| 4.03 | umc31a | p-umc31 | DQ123898 | Genomic | PstI | 550 | 582 |
| 4.04 | npi386(eks) | p-npi386 | DQ007988 | Genomic | PstI | 1200 | 1180 |
| 4.05 | agrr37bb | p-agrr37 | cDNA | EcoRI/HindIII | 949 | ||
| 4.06 | umc156a | p-umc156 | AY771219 | Genomic | PstI | 570 | 533 |
| 4.07 | umc66a(lcr) | p-umc66 | DQ007989 | Genomic | PstI | 1020 | 1036 |
| 4.08 | umc127c | p-umc127 | DQ642433 | Genomic | PstI | 1210 | 649 |
| 4.09 | umc52 | p-umc52 | DQ123899 | Genomic | PstI | 1500 | 824 |
| 4.10 | php20608a | p-php20608 | DQ007990 | Genomic | PstI | 780 | 1451 |
| 4.11 | umc169 | p-umc169 | DQ123900 | Genomic | PstI | 670 | 813 |
| 5.01 | npi409b | p-npi409 | Genomic | PstI | 710 | ||
| 5.02 | umc90 | p-umc90 | DQ642434 | Genomic | PstI | 1240 | 1226 |
| 5.03 | tub4 | p-tub4 | DQ007991 | cDNA | EcoRI/HindIII | 200 | 230 |
| 5.04 | bnl4.36 | p-bnl4.36 | DQ642435 | Genomic | PstI | 2300 | 2210 |
| 5.05 | csu93b | p-csu93 | DQ015673 | cDNA | EcoRI/XhoI | 800 | 677 |
| 5.06 | umc126a | p-umc126 | AY771220 | Genomic | PstI | 670 | 663 |
| 5.07 | umc108 | p-umc108 | DQ642436 | Genomic | PstI | 970 | 958 |
| 5.08 | bnl5.24aa | p-bnl5.24 | Genomic | PstI | 2500 | ||
| 5.09 | php10017 | p-php10017 | AY771221 | Genomic | PstI | 470 | 526 |
| 6.01 | umc85a | p-umc85 | AY772450 | Genomic | PstI | 600 | 561 |
| 6.02 | umc59aa | p-umc59 | Genomic | PstI | 930 | ||
| 6.03 | npi393 | p-G23A-06 | DQ015674 | Genomic | PstI | 1200 | 1249 |
| 6.04 | umc65a | p-umc65 | AY772451 | Genomic | PstI | 650 | 691 |
| 6.05 | umc21 | p-umc21 | DQ123901 | Genomic | PstI | 1050 | 1062 |
| 6.06 | umc38a | p-umc38 | DQ059316 | Genomic | PstI | 1010 | 1022 |
| 6.07 | umc132a(chk) | p-umc132 | DQ1238902 | Genomic | PstI | 500 | 472 |
| 6.08 | asg7a | p-asg7 | DQ642437 | Genomic | PstI | 550 | 353 |
| 7.01 | asg8(myb) | p-asg8 | DQ642438 | Genomic | PstI | 500 | 320 |
| 7.02 | asg34a(msd)a | p-asg34 | Genomic | PstI | 1350 | ||
| 7.03 | asg49 | p-asg49 | DQ059317 | Genomic | PstI | 400 | 400 |
| 7.04 | umc254 | p-umc254 | EU190457 | Genomic | PstI | 1050 | 1000 |
| 7.05 | umc245 | p-umc245 | AY772452 | Genomic | PstI | 700 | 665 |
| 7.06 | umc168 | p-umc168 | DQ059318 | Genomic | PstI | 1080 | 1072 |
| 8.01 | npi220a | p-G10F-01 | DQ059319 | Genomic | HindIII | 400 | 406 |
| 8.02 | bnl9.11a(lts)a | p-bnl9.11 | Genomic | PstI | 2400 | ||
| 8.03 | umc124a(chk) | p-umc124 | DQ059320 | Genomic | PstI | 1160 | 1162 |
| 8.04 | bnl7.08aa | p-bnl7.08 | Genomic | PstI | 2300 | ||
| 8.05 | bnl2.369a | p-bnl2.369 | cDNA | EcoRI | 700 | ||
| 8.06 | csu31aa | p-csu31 | cDNA | EcoRI/XhoI | 800 | ||
| 8.07 | npi268a | p-npi268 | DQ123903 | Genomic | PstI | 710 | 688 |
| 8.08 | npi414a | p-npi414 | AY772453 | Genomic | PstI | 870 | 893 |
| 8.09 | agrr21a | p-agrr21 | cDNA | EcoRI/HindIII | 899 | ||
| 9.01 | umc109 | p-umc109 | AY772454 | Genomic | PstI | 840 | 797 |
| 9.02 | bz1a | p-umc192 | cDNA | PstI | 1750 | ||
| 9.03 | wx1a | p-umc25 | cDNA | EcoRI | 2300 | ||
| 9.04 | csu147a | p-csu147 | cDNA | EcoRI/XhoI | 800 | ||
| 9.05 | umc95 | p-umc95 | AY772455 | Genomic | PstI | 680 | 660 |
| 9.06 | csu61a | p-csu61 | cDNA | EcoRI/XhoI | 500 | ||
| 9.07 | asg12 | p-asg12 | DQ123904 | Genomic | PstI | 700 | 647 |
| 9.08 | csu54ba | p-csu54 | cDNA | EcoRI/XhoI | 1400 | ||
| 10.01 | php20075a(gast) | p-php20075 | DQ059321 | Genomic | PstI | 1400 | 1311 |
| 10.02 | npi285a(cac)a | p-npi285 | Genomic | PstI | 1250 | ||
| 10.03 | umc130 | p-umc130 | DQ059322 | Genomic | PstI | 640 | 634 |
| 10.04 | umc64a | p-umc64 | Genomic | PstI | 710 | ||
| 10.05 | umc259a | p-umc259 | DQ123905 | Genomic | PstI | 550 | 579 |
| 10.06 | umc44a | p-umc44 | AY772456 | Genomic | PstI | 800 | 794 |
| 10.07 | bnl7.49a(hmd)a | p-bnl7.49 | Genomic | PstI | 2100 |
aFull length RFLP sequence not available.
bUnable to sequence with vector primers.
cPoly A tail at both ends of insert sequence.
Attempts to use the RFLP sequence information to identify homologous S. propinquum BACs in silico failed because the limited sequence data for characterized sorghum BAC libraries was insufficient for unambiguous selection of homologous sorghum BACs. The S. propinquum physical FPC map is very well annotated with markers, and the clones are available on arrayed filters, but relatively few genomic sequence data were available for BLAST searches [59]. The S. bicolor genome sequence could be searched by BLAST, but the sequence was based on whole-genome shotgun sequencing, not directly linked BACs that could be obtained and used for FISH [66]. Although the maize RFLP-FLIS data turned out to be useful for maize genome annotation, we found that the use of the sequence information alone was not as productive as the conventional filter-hybridization method for identification of corresponding sorghum BAC clones.
3.2. Purification of Maize Restriction Fragment Length Polymorphism (RFLP) Inserts
The overall procedure for identification, verification, and selection of sorghum BACs to be used for FISH is summarized in Table 2. It begins with the purification of maize RFLP probes for filter hybridization onto the arrayed sorghum BAC library and ends with the selection of a single BAC to be used as a FISH probe. This procedure can be completed for four different RFLP markers in approximately 4 weeks.
Table 2.
Major steps in the identification and maize marker-based selection of a sorghum BAC for use as a transgenomic FISH probe in maize. The figures and tables corresponding to the steps are indicated.
| Step | Procedure |
|---|---|
| 1 | Grow maize CBM-RFLP cultures and purify insert |
| (Figure 1; Table 1) | |
| 2 | Radiolabel CBM insert probes for use in filter library |
| (step 3) or Southern blot (step 5) hybridizations | |
| 3 | Hybridize probes (Tm-25°C) to Sorghum propinquum |
| BAC library (YRL, 36,000 BACs, ~6 × coverage; | |
| Figure 2) | |
| 4 | Score autoradiographs to identify homologous |
| sorghum BAC and examine location on the FPC map | |
| (Figure 2; Table 3) | |
| 5 | Grow identified overlapping BACs and verify them by |
| Southern blot (Figure 3) | |
| 6 | Select one BAC for use as a transgenomic FISH probe |
| on maize pachytene chromosome spreads from maize | |
| addition lines of oat (Figure 4) |
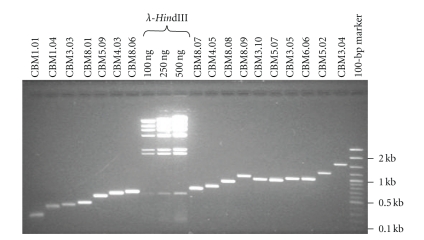
First, the bacterial cultures containing the maize CBM-RFLP probes were grown and their inserts amplified by PCR with vector primers. Then the flanking poly-linker sequences were trimmed with the appropriate restriction enzymes, and ethanol precipitated (Table 1, Table 2, step 2, and Supplemental Table 1). The resulting PCR products were redissolved in 10–20 μl of 10 : 0.2 TE and quantified by ethidium bromide (EtBr) staining in agarose gels as shown in Figure 1 for multiple different RFLP probes. The inserts from the maize RFLP probes were loaded from left to right in the order of expected fragment size, and the gel (Figure 1) included standards for mass (HindIII λ lanes) and size (100-bp marker lane). The preparations shown yielded inserts of the expected sized, except for CBM8.09, which was subsequently excluded because of additional difficulties in obtaining its FLIS.
Figure 1.
EtBr-stained agarose gel of CBM inserts. PCR amplification products for individual prepared CBM RFLP probes (indicated above the lanes), loaded from left to right in order of their predicted size. Mass standards (HindIII-digested λ DNA) are labeled, and the total amounts loaded appear above the lanes. Size standards (100-bp marker) are included, and selected band sizes are indicated at right.
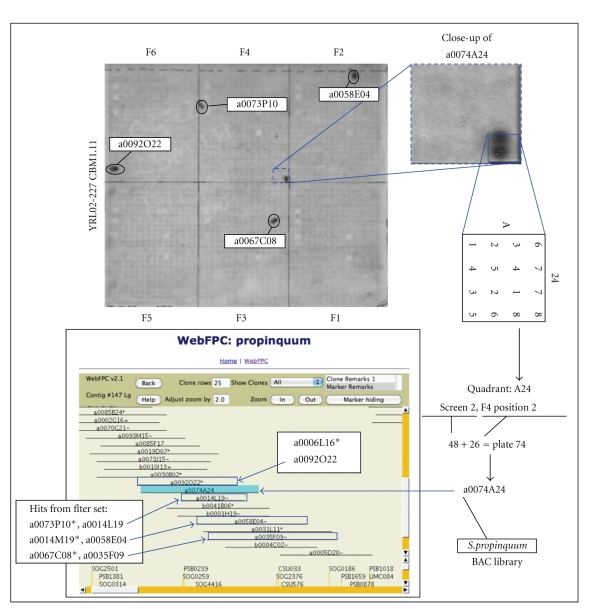
The CBM inserts were then radiolabeled with 32P-dCTP and hybridized to the pair of S. propinquum BAC filters at Tm-25°C, and the resulting hybridization signals were scored for identification of homologous Sorghum BACs. These were located, when possible, within the online FPC BAC map (WebFPC for S. propinquum, http://www.stardaddy.uga.edu/fpc/WebAGCoL/propinquum/WebFPC/) built from the same BAC library [64]. This procedure (Table 2, steps 3 and 4) is illustrated with CBM1.11 (umc161a) in Figure 2. A total of eight different sorghum BAC hits were detected, and those (5 BACs) identified on one of the two library filters are shown (Figure 2, circled double dots). All of these BACs were used to query the S. propinquum WebFPC, and five of the eight were overlapping in contig number 147 as shown in Figure 3(a) [64, 67]. This outcome exemplifies the successful detection of homologous BACs and is consistent with the sixfold genome coverage of this library [59].
Figure 2.
BAC filter hybridization, scoring of hits, and location in fingerprint contig (FPC) map for maize CBM1.11 (umc161). A representative filter-screen hybridization (Table 2, step 2) and the autoradiograph from one of the two filters are shown (top). A total of 5 positive signals are seen (circled or boxed), and the scoring scheme for one of them (a0074A24) is illustrated at the right. The autoradiograph is labeled to show the location of the 384-well plate fields 1–6 (F1–6), and the geometry duplicate spotting of the eight individual plates per field is indicated at the right for quadrant A24. A close-up of one hit (position 2, field 3) is shown with the resulting decoded BAC ID of plate 74 position A-24 (a0074A24). This BAC, along with four other contiguous (arrows) BACs, is indicated within contig number 113 of the Sorghum propinquum FPC map (screen capture bottom left). Four additional BACs (asterisks) were detected, but not found to belong to a contig.
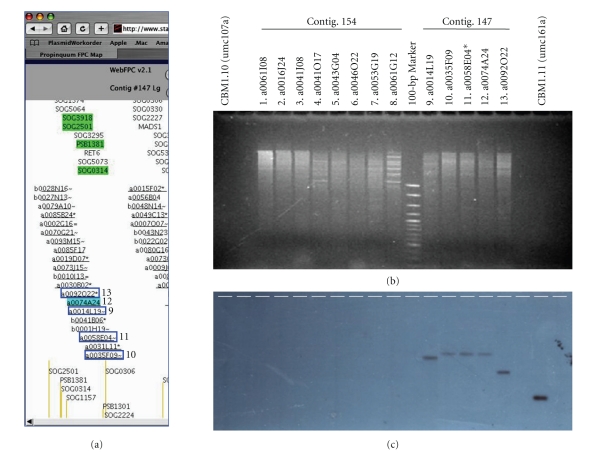
Figure 3.
Southern blot confirmation of sorghum BACs after filter hybridization. Step 5 (Table 2) of the BAC selection procedure is illustrated with CBM1.11 (umc161a). (a) Sorghum propinquum FPC contig map showing contig number 147 and the five contiguous BACs (boxed) detected by filter hybridization with the CBM1.11 probe. Lane numbers are indicated beside the BACs. (b) Ethidium bromide-stained agarose gel of HindIII-digested BAC minipreps. This gel includes the BACs under investigation (lanes 9–13) along with another set (lanes 1–8) serving as negative controls. For each confirmation blot, a positive control lane [CBM1.11 (umc161a)] is included. It contains a trace amount of the same insert preparation that was used in this experiment and in the preceding filter hybridization. (c) Autoradiograph after hybridization with the CBM1.11 probe. The asterisk indicates the BAC that was eventually selected as part of step 6 (Table 2).
3.3. Southern Blot Confirmation and Selection of Homologous Sorghum BACs for FISH
After filter hybridization, the overlapping BACs were grown from a freezer culture replica of the library. Next, BAC DNA was isolated by conventional plasmid minipreps, restriction enzyme digested, and subjected to Southern blot analysis for confirmation that each BAC contained RFLP-homologous sequences (Table 2, step 5). An example of this step is illustrated in Figure 3 with the BACs detected from the previous screen with CBM1.11 (umc161a). Confirmation blots also contain BACs from a separate selection experiment, providing multiple negative control BACs per blot (lanes 1–8, Figures 3(b) and 3(c)).
The confirmation blot for CBM1.11 resulted in the detection of a single cross-hybridizing band for each of the filter-hit BACs (Figure 3(c), lanes 9–13) but no bands for the negative control lanes. As expected, a common restriction fragment was observed for most of the five BACs in question, whereas two of them (Figure 3(c), lanes 9 and 13) showed bands of different sizes. This discrepancy probably resulted from the homologous sequences residing near one end or the other of the BAC, consistent with their size or location within the FPC BAC map. Consequently, these two BACs (a0092O22 and a0014L19) were demoted as candidate BACs to represent the maize CBM1.11 locus.
All together, 58 of the 59 maize CBM probes from maize chromosomes 1, 3–6, and 8 have been used to screen the sorghum BAC filter arrays. Of these, 47 (81%) resulted in the detection of overlapping BACs (Table 2, step 4), and 23 of them have passed the Southern blot confirmation test (Table 2, step 5) as summarized in Table 3. Previous work on maize chromosome 9 has shown that, with regard to this overall approach, once a marker has progressed through the point of Southern blot verification, the prospects for successful transgenomic BAC FISH is quite favorable [35].
Table 3.
Maize CBM-RFLP identified sorghum BACs.
| CBM | RFLP-probe | No. of BACS indentified | [contig] Member BACs scored | Southern blot comments |
|---|---|---|---|---|
| 1.01 | tub1 | 2 | [868] a0039M23 [154] a0053G19 |
Contiguous BACs not identified |
| 1.02 | umc157 | 3 | [154] a0045K08, a0064A06, a0055E08 | All BACs confirmed |
| 1.03 | umc076 | 4 | [193] a0046J07, a0086C09 [389] a0004B12 [?] a0071N02 |
BACs a0046J07, 86C09, and 71N02 confirmed |
| 1.04 | asg45 | 5 | [490] a0073P09 [536] a0080O07 [573] a0086J05, a0075I08 [967] a0036D16 |
Contiguous BACs not identified |
| 7 | [150] a0001K22 [187] a0061M20 [557] a0041B19 [?] a0029B11, a0036D16, a0043B12, a0052B23 |
|||
| 1.05 | csu003 | 4 | [179] a0001C04, a0026E17, a0052G17, a0094G22 | BACs a0001C04, 26E17, 94G22 confirmed |
| 1.06 | umc67a | 2 | [868] a0026H06, a0038A08 | |
| 1.07 | asg62 | 5 | [550] a0084J12 [537] a0084J14 [443] a0083B09 [542] a0095N15 [567] a0096N18 |
Contiguous BACs not identified |
| 1.08 | umc128 | 12 | [168] a0029P06, a0055L05, a0059H18, a0058G14, a0066G14, a0070C24, a0071B13, a0072H12, a0075H05, a0081P22, a0083A01, a0092C10 | Confirmation Hybe failed; only control seen |
| 1.09 | cjd2 | 8 | [159] a0011I10, a0023C11, a0039E21, a0056O22, a0081H24, a0091D22, a0093H06, a0093P21 | BACs 23C21-93P21 confirmed |
| 1.10 | umc107a | 8 | [154] a0006I08, a0016J24, a0041J08, a0041O17, a0043G04, a0046O22, a0053G19, a0061G12 | All BACs confirmed |
| 1.11 | umc161a | 5 | [147] a0014L19, a0035F09, a0058E04, a0074A24, a0092O22 | All BACs confirmed; the restriction fragments of BACs 14L19 & 35F09 were smaller than those of other BACs |
| 1.12 | bnl6.32 | 6 | [147] a0014L19, a0035F09, a0058E04 [?] a0014M19, a0067C08, a0006L16 |
|
| 3.01 | umc32a | 9 | [1] a0016H11, a0020E10, a0038G18, a0049H03, a0066I03, a0083B22, a0092J15, a0092J15, a0092K15 [293] a0048H03 [?] a0035E11, a0038G18, a0015L17, a0017E12 |
All BACs in contig 1 confirmed |
| 3.02 | csu32 | 6 | [1332] a0010D09, a0017F16, a0035I06, a0066B14, a0066B20 [?] a0015J10 |
|
| 3.03 | asg24a | Contiguous BACs not identified | ||
| 3.04 | asg48 | 0 | Contiguous BACs not identified | |
| 3.05 | umc102 | 9 | [898] a0016D10 [884] a0018B20 [394] a0055N13 [246] a0085B11 [894] a0083B12 [1257] a0090I03 [?] a007P18, a0012G07, a0027C12 |
Contiguous BACs not identified |
| 3.06 | bnl5.37a | 8 | [64] a0010K10, a0042I24, a0043H04, a0045D02, a0047F15, a0047J02, a0058O10, a0096N10 | |
| 3.07 | bnl6.16a | 7 (8) | [56] a0014M18, a0020L03, a0029M08 = a0029M07*, a0050D18, a0085M21 [143] a0051P09 [?] a0052D16 |
|
| 3.08 | umc17a | 6 | [49] a0002O18, a0041K14 [85] a0074H09 [?] a0074H10, a0096D08, a0035C01 |
|
| 3.09 | umc63a | 9 | [1292] a0021P11, a0025P06, a0046A02, a0043J05, a0044B15, a0055P13, a0029C04 [?] a0029C03 |
|
| 3.10 | cyp1 | 6 | [1118] a0029L19, a0067D01, a0078E10 [89] a0017J07, a0018N05 [?] a0096D16 |
All BACs in contig 1118 as well as BAC a0096D16 confirmed |
| 4.01 | agrr115a | PolyA at both ends | ||
| 4.02 | php20725a | 10 | [1] a0016H11, a0092J15, a0092K15, a0049H03 [1292] a0021P11, a0029C03, a0044B15, a0046A02, a0043J02, a0055P13 |
Confirmation Hybe failed |
| 4.03 | umc31a | 5 | [1320] a0001I16 [3] a0067O16 [?] a0057L09, a0084A23, a0007F05 |
Contiguous BACs not identified |
| 4.04 | npi386 | 7 | [232] a0002B15, a0019A12, a0035B24, a0058L10, a0071D21, a0078C04, a0082H03 | All BACs confirmed |
| 4.05 | agrr37b | 6 | [391] a0023C20, a0054H21, a0055K09, a0068N16, a0080D06, a0083G05 | All BACs except 55K09 confirmed |
| 4.06 | umc156a | 3 | [280] a0059B12, a0061G01, a0067K14, a0089D08 | |
| 4.07 | umc66a | Contiguous BACs not identified | ||
| 4.08 | umc127c | 3 | [290] a0082D10, a0059L11, a0033K19 | BACs 59L11 and 33K19 confirmed |
| 4.09 | umc32 | 6 | [352] a0030M10, a0045N24, a0050L19, a0056E02, a0069D03, a0072G04 | |
| 4.10 | php20608 | 6 | [313] a0008B07, a0015N19, a0019O20, a0026C03, a0063A16, a0064O16 | All BACs confirmed |
| 4.11 | umc169 | 5 | [316] a0012H04, a0017H01, a0038N14, a0040B11, a0094J01 | |
| 5.01 | npi409 | 2 | [483] a0006F01 [813] a0002O16 |
Contiguous BACs not identified |
| 5.02 | umc90 | 2 | [916] a0003P04 [1120] a0038C23 |
Contiguous BACs not identified |
| 4 | [154] a0045K08, a0055E08, a0064A06 [?] a0034N18 |
All BACs confirmed | ||
| 5.03 | tub4 | 0 | ||
| 5.04 | bnl4.36 | 8 | [118] a0025E21, a0024H23, a0046J09, a0045L08, a0041E24, a0069P23, a0058E11, a0051H03, a0052I15 [314] a0013B03, a0061I09, a0069K04 [385] a0045M19, a0078E12, a0078A08, a0066D12 |
All BACs confirmed except 13B03 |
| 5.05 | csu93b | 7 | [193] a0007P01, a0046J07, a0058O08, a0069L09, a0078B19, a0086C09 [382] a0009P04 |
|
| 5.06 | umc125a | 4 | [280] a0067K14, a0061G01, a0059B12, a0089D08 | All BACs confirmed |
| 5.07 | umc108 | 14 | [287] a0095P22, a0061G08, a0063D06, a0050A23, a0052I05, a0054L12, a0046N22 [1108] a0023P17, a0004F02, a006N04 [333] a0020D11, a0029N15 [1288] a0030E06, a0038E06 |
All BACs in contig 287 confirmed |
| 5.08 | bnl5.24a | 8 | [278] a0042O01, a0020K19, a0023D21, a0095O21, a0095O19, a0081C19, a0063M08, a0002O03 [1017] a0017F09, a0017E09, a0041G23, a0019G02, a0067B22 |
All BACs confirmed except 63M08 |
| 5.09 | php10017 | 6 | [275] a0028A23, a0033G16, a0043E09, a0057K02, a0060N20 [?] a0061I19 |
All BACs confirmed except 43E09 |
| 6.01 | umc085a | 10 | [426] a0006I03, a0009K09, a0072P24 [205] a0025E20, a0041O09, a0061M21, a0057H05, a0067N09, a0074E15, a0085D02 |
All BACs S. blot confirmed except 57H05 |
| 6.02 | umc059a | 9 | [366] a0004P17, a0006P16, a0019D19, a0054G04, a0063H16, a0069J20, a0080H11, a0082K02, a0089K05 | All BACs confirmed except 19D19 and 54G04 |
| 6.03 | npi393 | 6 | [759] a0004A06, a0017E10 [528] a0006D09, a0008G11, a0063A20 [?] a0039N21 |
All BACs confirmed |
| 6.04 | umc65a | Contiguous BACs not identified | ||
| 6.05 | umc21 | 5 | [382] a0006P21, a0030G09, a0036H03, a0039E04, a0061C05 | |
| 6.06 | umc38a | Contiguous BACs not identified | ||
| 6.07 | umc132a | 9 | [327] a0030L20, a0040J01, a0056F09, a0060H04, a0070I02, a0070K04, a0070K07, a0073H20 [?] a0020O17 |
All BACs except 20O17 confirmed with cross-Hybe to negative control |
| 6.08 | asg7a | 6 | [323] a0018I04, a0032J21, a0032K17, a0068H17, a0084B19 [486] a0015L19 |
Confirmation failed |
| 8.01 | npi220a | 6 | [22] a0027H19, a0063B11, a0080P02, a0084E01, a0096C21 [188] a0019O09 |
All BACs confirmed |
| 8.02 | bnl9.11 | 8 | [421] a0042P09, a0047M08, a0072L19, a0007N23, a0089I26 = 89I23* [908] a0074H18 [?] a0079F24, a0035C01 |
|
| 8.03 | umc124a | 7 | [6] a0014C07, a0017N03, a0040E17, a0078K17, a0002I13 = a0002I16(?), a0083P05 | |
| 8.04 | bnl7.08a | Contiguous BACs not identified | ||
| 8.05 | bnl2.369 | Contiguous BACs not identified | ||
| 8.06 | csu31a | 13 | [45] a0042J03, a0043C22, a0062K14, a0064A02 [233] a0089H10 = a0089G10* [359] a0078O19 = a0078N19* [915] a0085F10 = [866] 77F10* [?] a0035C01, a0044F14, a0053E10 = 51E10*, a0013N08, a0045C22, a0052D10, |
|
| 8.07 | npi268a | 10 | [61] a0033M15, a0073N21, a0039H13, a0045D11, a0059E23, a0065K05, a0068E24, a0072D01, a0078K07, a0089A17 | Confirmation failed; only positive control seen |
| 8.08 | npi414a | 9 | [58] a0007I19, a0007C07, a0033M05, a0073D02, [1019] a0002I19 = a0007I19* [1125] a0023G22 [223] a0046C21 [1278] a0090D06 [?] a0019G24 |
|
| 8.09 | agrr21 | Contiguous BACs not identified |
*Whenever BAC scoring was ambiguous, possible alternate identified BACs were also grown for S. blot verification.
The final steps involve the choice of a single BAC, primarily by a combination of four criteria: the BAC should (1) belong to a set of marker-detected, overlapping clones in a contig, (2) be centrally located within the group of detected BACs, (3) possess a cross-hybridizing restriction fragment that is common to other BACs in the group, and (4) grow well and yield at least 2 μg of high-quality DNA obtained using a BAC large-scale purification kit. In cases where the first three criteria are met by more than one BAC, we typically choose the first one that meets the fourth criterion, even though such BACs probably share considerable amounts of sequence and any of them may work for FISH.
3.4. Transgenomic BAC FISH Provides Locus-Specific Cytogenetic Mapping Data
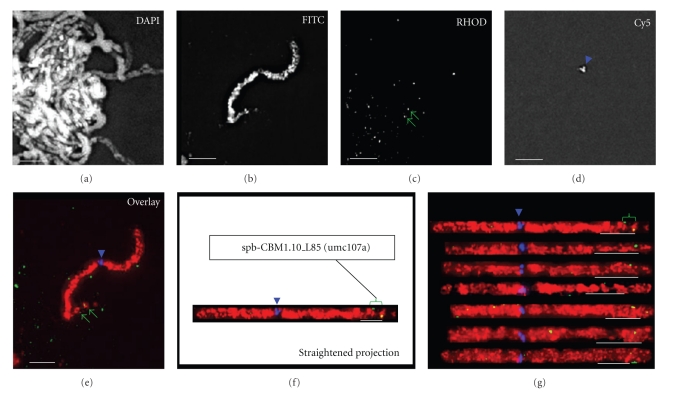
A maize-marker-selected sorghum BAC was direct-labeled for use as a FISH probe and hybridized, along with a multicolor probe mix, to pachytene spreads from maize chromosome addition lines of oat. These diploid (doubled haploid) lines contain the oat genome plus a single pair of maize chromosomes [57, 58]. Figure 4 illustrates S. propinquum BAC (a0053G04) FISH of the CBM1.10 locus on a maize B73 chromosome 1 addition line of oat. The multiprobe mix includes total DNA from the knobless Wilbur's Flint (KWF) line of maize, which paints the entire maize pachytene chromosome (Figure 4(b), FITC image). A centromere-specific probe, centC, first discovered by Ananiev et al. [68] is also included (Figure 4(d), Cy-5 image) along with the specific maize-marker-selected BAC probe (Figure 4(c), Rhodamine image). The nuclei were counterstained with DAPI (Figure 4(a), DAPI image), and 3D deconvolution images were collected as previously described [35, 53]. Images of the pachytene chromosomes were then traced and computationally straightened (Figure 4(f)). The cytogenetic position was measured in this way for multiple images (Figure 4(g)), and the average location (as fractional distance along the chromosome arm) was determined. In this example, the maize marker CBM1.10 was indirectly FISH mapped, with the sorghum BAC (a0053G04), to 85% of the distance along the long arm. This cytological locus is 15 centiMcClintocks from the telomere [55], and the cytogenetic position is denoted as spb-CBM1.10_L85 (umc107a) according to nomenclature first described by Koumbaris and Bass [53]. This location is in general agreement with that predicted from recombinant nodule frequency mapping in maize [54].
Figure 4.
FISH mapping of maize CBM1.10 with sorghum BAC a0053G04. (a) A DAPI-stained image of spread pachytene chromosomes from OMAd1.36. (b) FITC image showing maize chromosome 1 direct-labeled with Alexa-488-dUTP-KWF total maize DNA. (c) Rhodamine image from direct-labeled sorghum BAC FISH signals (green arrows). (d) Cy-5 image of centromere FISH signal (blue arrowhead) with direct-labeled CentC. (e–g) Three-color overlay of the FITC (red), rhodamine (green), and Cy-5 (blue) images. (f) Straightened projection of the maize chromosome from panel (e). The locations of the centromere (blue arrowhead) and CBM1.10 BAC FISH signals (green bracket) are indicated along with the resulting cytogenetic locus name (boxed). (g) Straightened projections of six additional chromosomes aligned at their centromeres for comparison. All scale bars represent 5 μm.
Sorghum BAC FISH mapping in maize is precise at the resolution of pachytene FISH even though the FISH signal is derived from sequences adjacent to the RFLP of interest. Furthermore, given the extensive segmental genome duplication in maize, we predict that these sorghum BACs will be useful as FISH probes for both the target CBM region and the corresponding duplicated segment. For example, sorghum BAC a0053G04 (Figure 4) was selected with a marker for chromosome arm 1L, but it is expected to hybridize to a region on chromosome arm 5S [21, 69].
4. Conclusions
Generation and integration of the cytological maps with the linkage maps of maize represents a valuable endeavor, contributing to positional cloning, allowing for accurate extrapolation of translocation breakpoints within linkage maps, and helping resolve discrepancies among various maps. In this study, we describe the method for using sorghum BACs to FISH map small widely used maize RFLP probe markers. We have shown that 44 of 59 target maize RFLP loci can be successfully used to screen for homologous and presumably syntenic-region sorghum BACs. So far, half of these have passed the Southern blot verification test, predictive of high success rate in subsequent transgenomic FISH mapping experiments. BACs from small-genome relatives of maize provide gene-rich reagents suitable for single-locus FISH mapping, a strategy that can be employed in other species. This approach hinges on the availability of arrayed BAC libraries and corresponding filters. Numerous such plant BAC libraries exist and are available as filter arrays (CUGI at http://www.genome.clemson.edu). This technique can be extended therefore to a large number of other plant species. Even in cases where the small-genome relative lacks an FPC physical map (i.e., unable to carryout the last part of step 4 of Table 2), one can still use shared restriction fragments (part of Step 5, Table 2) to identify overlapping BACs. Finally, in addition to solving the FISH detection limit, application of this scheme also generates evolutionarily meaningful links between the genomic and genetic maps of the two related species.
Supplementary Material
Supplemental Table 1 lists additional maize RFLP probes that were sequenced as part of this study. The GenBank Accession numbers, loci detected, vectors, and insert sizes are included.
Acknowledgments
The authors thank Amy M. Win, Katherine M. Miller, Tabatha M. McHill, and Roberta S. Hill for technical help on this project and Anne B. Thistle, Shaun P. Murphy, Amber N. Brown, and Elizabeth S. Howe for useful comments on the manuscript. This work was supported by the National Science Foundation (DBI-0321639).
References
- 1.Aulard S, Monti L, Chaminade N, Lemeunier F. Mitotic and polytene chromosomes: comparisons between Drosophila melanogaster and Drosophila simulans. Genetica. 2004;120(1–3):137–150. doi: 10.1023/b:gene.0000017637.10230.c4. [DOI] [PubMed] [Google Scholar]
- 2.Figueroa DM, Bass HW. A historical andmodern perspective on plant cytogenetics. Briefings in Functional Genomics and Proteomics. 2010;9(2):95–102. doi: 10.1093/bfgp/elp058. [DOI] [PubMed] [Google Scholar]
- 3.Gartler SM. The chromosome number in humans: a brief history. Nature Reviews Genetics. 2006;7(8):655–660. doi: 10.1038/nrg1917. [DOI] [PubMed] [Google Scholar]
- 4.Gatti M, Pimpinelli S. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. I. Organization of the fertility factors. Chromosoma. 1983;88(5):349–373. [Google Scholar]
- 5.Lozovskaya ER, Petrov DA, Hartl DL. A combined molecular and cytogenetic approach to genome evolution in Drosophila using large-fragment DNA cloning. Chromosoma. 1993;102(4):253–266. doi: 10.1007/BF00352399. [DOI] [PubMed] [Google Scholar]
- 6.Trask BJ. Human cytogenetics: 46 chromosomes, 46 years and counting. Nature Reviews Genetics. 2002;3(10):769–778. doi: 10.1038/nrg905. [DOI] [PubMed] [Google Scholar]
- 7.Fedoroff N, Botstein D. The Dynamic Genome: Barbara McClintock's Ideas in the Century of Genetics. Plainview, NY, USA: Cold Spring Harbor Press; 1992. [Google Scholar]
- 8.Creighton HB, McClintock B. A correlation of cytological and genetical crossing-over in Zea mays. Proceedings of the National Academy of Sciences of the United States of America. 1931;17(8):492–497. doi: 10.1073/pnas.17.8.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClintock B. Chromosome morphology in Zea mays. Science. 1929;69(1798):p. 629. doi: 10.1126/science.69.1798.629. [DOI] [PubMed] [Google Scholar]
- 10.McClintock B. The order of the genes C, Sh and Wx in Zea mays with reference to a cytologically known point in the chromosome. Proceedings of the National Academy of Sciences of the United States of America. 1931;17(8):485–491. doi: 10.1073/pnas.17.8.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros PF, Matzke MA, Matzke AJM. Detection of a 17 kb unique sequence (T-DNA) in plant chromosomes by in situ hybridization. Chromosoma. 1986;94(1):11–18. [Google Scholar]
- 12.Fransz PF, Stam M, Montijn B, et al. Detection of single-copy genes and chromosome rearrangements in Petunia hybrida by fluorescence in situ hybridization. Plant Journal. 1996;9(5):767–774. [Google Scholar]
- 13.Peacock WJ, et al. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(7):4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Earle ED. Direct and sensitive fluorescence in situ hybridization of 45S rDNA on tomato chromosomes. Genome. 1994;37(6):1062–1065. doi: 10.1139/g94-151. [DOI] [PubMed] [Google Scholar]
- 15.Shen DL, Wang ZF, Wu M. Gene mapping on maize pachytene chromosomes by in situ hybridization. Chromosoma. 1987;95(5):311–314. [Google Scholar]
- 16.Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(37):13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb JC, Birchler JA. Retroelement genome painting: cytological visualization of retroelement expansions in the genera zea and tripsacum. Genetics. 2006;173(2):1007–1021. doi: 10.1534/genetics.105.053165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lysak MA, Fransz PF, Ali HBM, Schubert I. Chromosome painting in Arabidopsis thaliana. Plant Journal. 2001;28(6):689–697. doi: 10.1046/j.1365-313x.2001.01194.x. [DOI] [PubMed] [Google Scholar]
- 19.Lysak MA, Pecinka A, Schubert I. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Research. 2003;11(3):195–204. doi: 10.1023/a:1022879608152. [DOI] [PubMed] [Google Scholar]
- 20.Davis GL, McMullen MD, Baysdorfer C, et al. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics. 1999;152(3):1137–1172. doi: 10.1093/genetics/152.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 22.Rhoades MM. The early years of maize genetics. Annual Review of Genetics. 1984;18:1–29. doi: 10.1146/annurev.ge.18.120184.000245. [DOI] [PubMed] [Google Scholar]
- 23.Arteaga-Vazquez MA, Chandler VL. Paramutation in maize: RNA mediated trans-generational gene silencing. Current Opinion in Genetics and Development. 2010;20(2):156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGinnis KM. RNAi for functional genomics in plants. Briefings in Functional Genomics and Proteomics. 2010;9(2):111–117. doi: 10.1093/bfgp/elp052. Article ID elp052. [DOI] [PubMed] [Google Scholar]
- 25.Springer NM. Small RNAs: how seeds remember to obey their mother. Current Biology. 2009;19(15):R649–R651. doi: 10.1016/j.cub.2009.06.049. [DOI] [PubMed] [Google Scholar]
- 26.Andorf CM, Lawrence CJ, Harper LC, Schaeffer ML, Campbell DA, Sen TZ. The Locus Lookup tool at MaizeGDB: identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Bioinformatics. 2010;26(3):434–436. doi: 10.1093/bioinformatics/btp556. [DOI] [PubMed] [Google Scholar]
- 27.Baucom RS, Estill JC, Chaparro C, et al. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genetics. 2009;5(11) doi: 10.1371/journal.pgen.1000732. Article ID e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckler ES, Gaut BS, McMullen MD. Molecular and functional diversity of maize. Current Opinion in Plant Biology. 2006;9(2):172–176. doi: 10.1016/j.pbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annual Review of Plant Biology. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- 30.Swigonova Z, Lai J, Ma J, et al. Close split of sorghum and maize genome progenitors. Genome Research. 2004;14(10):1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei F, Coe ED, Nelson W, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genetics. 2007;3(7):1254–1263. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei F, Zhang J, Zhou S, et al. The physical and genetic framework of the maize B73 genome. PLoS Genetics. 2009;5(11) doi: 10.1371/journal.pgen.1000715. Article ID e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Buckler ES. Genetic association mapping and genome organization of maize. Current Opinion in Biotechnology. 2006;17(2):155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Zhou S, Wei F, Nguyen J, et al. A single molecule scaffold for the maize genome. PLoS Genetics. 2009;5(11) doi: 10.1371/journal.pgen.1000711. Article ID e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amarillo FIE, Bass HW. A transgenomic cytogenetic sorghum (Sorghum propinquum) bacterial artificial chromosome fluorescence in situ hybridization map of maize (Zea mays L.) pachytene chromosome 9, evidence for regions of genome hyperexpansion. Genetics. 2007;177(3):1509–1526. doi: 10.1534/genetics.107.080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CC, Chen CM, Hsu FC, Wang CJ, Yang JT, Kao YY. The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theoretical and Applied Genetics. 2000;101(1-2):30–36. [Google Scholar]
- 37.Cone KC, McMullen MD, Bi IV, et al. Genetic, physical, and informatics resources for maize. On the road to an integrated map. Plant Physiology. 2002;130(4):1598–1605. doi: 10.1104/pp.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danilova TV, Birchler JA. Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: resolution, sensitivity, and banding paint development. Chromosoma. 2008;117(4):345–356. doi: 10.1007/s00412-008-0151-y. [DOI] [PubMed] [Google Scholar]
- 39.Hans De Jong J, Fransz P, Zabel P. High resolution FISH in plants—techniques and applications. Trends in Plant Science. 1999;4(7):258–263. doi: 10.1016/s1360-1385(99)01436-3. [DOI] [PubMed] [Google Scholar]
- 40.Gardiner J, Schroeder S, Polacco ML, et al. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiology. 2004;134(4):1317–1326. doi: 10.1104/pp.103.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang J, Gill BS. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome. 1994;37(5):717–725. doi: 10.1139/g94-102. [DOI] [PubMed] [Google Scholar]
- 42.Wang CJR, Harper L, Cande WZ. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell. 2006;18(3):529–544. doi: 10.1105/tpc.105.037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J, Gill BS. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome. 2006;49(9):1057–1068. doi: 10.1139/g06-076. [DOI] [PubMed] [Google Scholar]
- 44.Helentjaris T, Slocum M, Wright S, Schaefer A, Nienhuis J. Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theoretical and Applied Genetics. 1986;72(6):761–769. doi: 10.1007/BF00266542. [DOI] [PubMed] [Google Scholar]
- 45.Flint-Garcia SA, Thuillet AC, Yu J, et al. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant Journal. 2005;44(6):1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 46.Coe EH, Schaeffer ML. Genetic, physical, maps, and database resources for maize. Maydica. 2005;50(3-4):285–303. [Google Scholar]
- 47.Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(10):4487–4491. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang J, Hulbert SH, Gill BS, Ward DC. Interphase fluorescence in situ hybridization mapping: a physical mapping strategy for plant species with large complex genomes. Molecular and General Genetics. 1996;252(5):497–502. doi: 10.1007/BF02172395. [DOI] [PubMed] [Google Scholar]
- 49.Koo DH, Jiang J. Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant Journal. 2009;59(3):509–516. doi: 10.1111/j.1365-313X.2009.03881.x. [DOI] [PubMed] [Google Scholar]
- 50.Lamb JC, Meyer JM, Corcoran B, Kato A, Han F, Birchler JA. Distinct chromosomal distributions of highly repetitive sequences in maize. Chromosome Research. 2007;15(1):33–49. doi: 10.1007/s10577-006-1102-1. [DOI] [PubMed] [Google Scholar]
- 51.Adawy SSM, Stupar RM, Jiang J. Fluorescence in situ hybridization analysis reveals multiple loci of knob-associated DNA elements in one-knob and knobless maize lines. Journal of Histochemistry and Cytochemistry. 2004;52(8):1113–1116. doi: 10.1369/jhc.4B6335.2004. [DOI] [PubMed] [Google Scholar]
- 52.Lamb JC, Danilova T, Bauer MJ, et al. Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics. 2007;175(3):1047–1058. doi: 10.1534/genetics.106.065573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koumbaris GL, Bass HW. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant Journal. 2003;35(5):647–659. doi: 10.1046/j.1365-313x.2003.01829.x. [DOI] [PubMed] [Google Scholar]
- 54.Anderson LK, Doyle GG, Brigham B, et al. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics. 2003;165(2):849–865. doi: 10.1093/genetics/165.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson LK, Salameh N, Bass HW, et al. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics. 2004;166(4):1923–1933. doi: 10.1534/genetics.166.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence CJ, Seigfried TE, Bass HW, Anderson LK. Predicting chromosomal locations of genetically mapped loci in maize using the Morgan2McClintock Translator. Genetics. 2006;172(3):2007–2009. doi: 10.1534/genetics.105.054155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kynast RG, Riera-Lizarazu O, Vales MI, et al. A complete set of maize individual chromosome additions to the oat genome. Plant Physiology. 2001;125(3):1216–1227. doi: 10.1104/pp.125.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rines HW, Phillips RL, Kynast RG, et al. Addition of individual chromosomes of maize inbreds B73 and Mo17 to oat cultivars Starter and Sun II: maize chromosome retention, transmission, and plant phenotype. Theoretical and Applied Genetics. 2009;119(7):1255–1264. doi: 10.1007/s00122-009-1130-2. [DOI] [PubMed] [Google Scholar]
- 59.Lin YR, Zhu L, Ren S, Yang J, Schertz KF, Paterson AH. A Sorghum propinquum BAC library, suitable for cloning genes associated with loss-of-function mutations during crop domestication. Molecular Breeding. 1999;5(6):511–520. [Google Scholar]
- 60.Singh RJ. Plant Cytogenetics. 2nd edition. Boca Raton, Fla, USA: CRC Press LLC; 2003. [Google Scholar]
- 61.Zhong XB, De JH, Zabel P. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH) Chromosome Research. 1996;4(1):24–28. doi: 10.1007/BF02254940. [DOI] [PubMed] [Google Scholar]
- 62.Zwick MS, Islam-Faridi MN, Czeschin DG, et al. Physical mapping of the liguleless linkage group in sorghum bicolor using rice RFLP-selected sorghum BACs. Genetics. 1998;148(4):1983–1992. doi: 10.1093/genetics/148.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaut BS, D’Ennequin MLT, Peek AS, Sawkins MC. Maize as a model for the evolution of plant nuclear genomes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(13):7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bowers JE, Abbey C, Anderson S, et al. A high-density genetic recombination map of sequence-tagged sites for Sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics. 2003;165(1):367–386. doi: 10.1093/genetics/165.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keith CS, Hoang DO, Barrett BM, et al. Partial sequence analysis of 130 randomly selected maize cDNA clones. Plant Physiology. 1993;101(1):329–332. doi: 10.1104/pp.101.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paterson AH, Bowers JE, Bruggmann R, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 67.Draye X, Lin YR, Qian XY, et al. Toward integration of comparative genetic, physical, diversity, and cytomolecular maps for grasses and grains, using the sorghum genome as a foundation. Plant Physiology. 2001;125(3):1325–1341. doi: 10.1104/pp.125.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ananiev EV, Phillips RL, Rines HW. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(22):13073–13078. doi: 10.1073/pnas.95.22.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaut BS. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Research. 2001;11(1):55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 lists additional maize RFLP probes that were sequenced as part of this study. The GenBank Accession numbers, loci detected, vectors, and insert sizes are included.