Abstract
We introduced disulfide bonds to lock the integrin αLβ2 I domain in predicted open, ligand binding or closed, nonbinding conformations. Transfectants expressing αLβ2 heterodimers containing locked-open but not locked-closed or wild-type I domains constitutively adhered to intercellular adhesion molecule-1 (ICAM-1) substrates. Locking the I domain closed abolished constitutive and activatable adhesion. The isolated locked-open I domain bound as well as the activated αLβ2 heterodimer, and binding was abolished by reduction of the disulfide. Lovastatin, which binds under the conformationally mobile C-terminal α-helix of the I domain, inhibited binding to ICAM-1 by αLβ2 with wild-type, but not locked-open I domains. These data establish the importance of conformational change in the αL I domain for adhesive function and show that this domain is sufficient for full adhesive activity.
Integrins are large heterodimeric adhesive glycoproteins that bind extracellular ligands and communicate to the cytoskeleton and signaling pathways. The integrin αLβ2 [lymphocyte function-associated antigen (LFA)-1, CD11a/CD18] is important in almost all functions of lymphocytes, monocytes, granulocytes, and NK cells, including recognition of specific antigen, emigration from the vasculature, and cell migration (1–3). αLβ2 binds to intercellular adhesion molecules (ICAMs) that are cell surface glycoproteins containing Ig superfamily domains. Like many integrins, the adhesiveness of LFA-1 can be dynamically regulated on a time scale of <1 s by signals in the cytoplasm (inside–out signaling) (4).
The integrin α- and β-subunits are each type I transmembrane glycoproteins with a large extracellular domain, a single transmembrane region, and a short cytoplasmic tail (5). The N-terminal region of the integrin α-subunits contains seven repeats of about 60 amino acids each, and has been predicted to fold into a seven-bladed β-propeller domain (6). Half of the 18 known vertebrate integrin α-subunits, including αL, contain an inserted domain or I domain of about 200 amino acids (3). The I domain is predicted to be inserted between β-sheets 2 and 3 of the β-propeller domain (6). The I domain is critical for ligand binding by I domain-containing integrins (5). Integrin β-subunits contain an N-terminal cysteine-rich plexin semaphorin integrin (PSI) domain (7), a highly evolutionarily conserved I-like domain (8), and a C-terminal cysteine-rich region. Electron microscopy of integrins reveals a globular head that is connected to the membrane by two rod-like stalks corresponding to the C-terminal regions of the α- and β-subunits (9). The globular head region binds ligand, and contains the α-subunit β-propeller and I domains and the β-subunit I-like domain.
Both conformational change in extracellular domains and clustering in the membrane have been proposed as mechanisms to regulate adhesiveness of integrins; however, there is little evidence that conformational change can regulate cell adhesion (10, 11). Three-dimensional structures of the I domain show that it adopts the dinucleotide-binding fold like small G proteins, and has a unique divalent cation coordination site designated the metal ion-dependent adhesion site (MIDAS) in which a Mg2+ forms five coordinations to I domain residues (12). Two different crystal forms of the αM I domain, “open” and “closed,” have been obtained, and it has been hypothesized that the two structures represent the “high-affinity” and “low-affinity” conformations, respectively (12, 13). The two conformers differ in the sidechains that coordinate the metal ion in the MIDAS. Also, in the open conformation, there is a 10-Å movement of the C-terminal helix down the body of the I domain and a large rearrangement and downward movement of the loop connecting this helix to the preceding β-strand (13). In the open αM I domain, a glutamate from a neighboring I domain provides the sixth metal coordination site in a ligand-mimetic lattice contact, whereas, in the closed αM I domain, a water molecule completes the metal coordination sphere, and the coordination of the metal to the I domain is altered. A recent structure of the α2 I domain bound through the metal of the MIDAS to a triple helical collagen peptide provides a second example of the open I domain conformation and demonstrates an almost identical structural rearrangement compared with the closed α2 I domain (14). Structures have been determined for the αL I domain only in the closed conformation (15–17).
To examine the relationship between the conformation and function of the αL I domain, we have introduced cysteines to lock it in the closed or the predicted open conformation with disulfide bonds. The effect of these mutations on binding of soluble I domains to ICAM-1 is examined elsewhere.** Here, we examine the consequences for cell adhesion. We find that: (i) Locking the I domain in the open conformation activates cell adhesion to ICAM-1 and locking it closed prevents activation of cell adhesion by Mn2+ or an activating mAb that binds to the β2-subunit, proving that the β6–α6 loop where the disulfide is introduced is critical for transduction of signals that regulate ligand binding by the I domain. (ii) The I domain by itself is sufficient to give fully active cell adhesion to ICAM-1. (iii) Lovastatin inhibits αLβ2 by altering I domain conformation, because it does not inhibit when the I domain is locked open.
Materials and Methods
Modeling of the Open αL I Domain and Prediction of Disulfide Bonds.
Modeling of the open conformation of the αL I domain on the open αM I domain (12) and identification of mutations to cysteines to introduce conformation-specific disulfide bonds (19) are described elsewhere.**
Monoclonal Antibodies.
The mouse anti-human αL (CD11a) mAbs TS1/22, TS2/4, the activating anti-β2 (CD18) mAb CBR LFA-1/2, and the nonbinding myeloma IgG X63 have been described previously (20, 21).
cDNA Construction and Expression.
Overlap extension PCR (22, 23) was used to generate cysteine substitution mutations in the αL I domain. Wild-type human αL cDNA in vector AprM8 (24) was used as template. The outer left primer for PCR extension was complementary to the vector sequence 5′ to the EcoRI site, and the outer right primer was 3′ to the EcoRI site in the αL cDNA. The inner primers were designed for each individual mutation and contained overlapping sequences. The final PCR product with outer primers was digested with EcoRI and ligated into the same site in the wild-type αL cDNA in AprM8. All mutations were confirmed by DNA sequencing.
To construct the isolated, cell surface-expressed I domains, DNA that encodes the signal peptide of αL was ligated to the sequence encoding residues E124–A338 that contains the αL I domain. HindIII and SalI sites were introduced immediately adjacent to the 5′ and 3′ ends of this fragment, respectively. The HindIII–SalI fragment was subcloned in frame 5′ to the c-myc tag and the platelet-derived growth factor receptor (PDGFR) transmembrane domain and the first five residues of the PDGFR cytoplasmic domain in the vector pDisplay (Invitrogen). The DNA encoding this fusion was excised with HindIII and NotI and further subcloned into the HindIII/NotI site of pcDNA3.1/Hygro. The final constructs were verified by DNA sequencing.
Transient transfection of 293T and stable transfection of K562 cells were described previously (25, 26). Stable K562 cell lines that express αLβ2 were maintained in RPMI 1640/10% FBS supplemented with 4 μg/ml puromycin, and stable cells that express the isolated I-domain were maintained in RPMI 1640/10% FBS containing 100 μg/ml hygromycin B.
Flow Cytometry.
Flow cytometric analysis was performed as previously described (26). The nonbinding myeloma IgG X63 was used at a final concentration of 1:20 hybridoma supernatant, and mAbs TS1/22, TS2/4, and CBR LFA-1/2 were used at 10 μg/ml purified IgG.
Cell Adhesion to Immobilized ICAM-1.
Cell adhesion to immobilized ICAM-1 in L15 medium supplemented with 2.5% FBS (L15/FBS) was assayed by using fluorescently labeled cells (26). The activating mAb CBR LFA-1/2 was used at 10 μg/ml, and the control nonbinding IgG X63 at 1:20 hybridoma supernatant. For testing the effect of divalent cations, cells were washed two times with 20 mM Tris, pH 7.5, 150 mM NaCl (TS buffer) containing 5 mM EDTA, followed by two washes with TS buffer. Cells were then resuspended in TS buffer supplemented with 1 mM MnCl2 or 5 mM EDTA and added to ICAM-1-coated plates. After incubation at 37°C for 30 min, unbound cells were washed off and the bound cells were quantitated (26).
Lovastatin was kindly provided by Merck. Lovastatin dissolved in DMSO at 50 mM was diluted in assay buffer (L15/FBS). Cells were preincubated with lovastatin (0–50 μM) at 37°C for 15 min, and then transferred to the 96-well plate coated with ICAM-1 and further incubated at 37°C for 30 min in the presence of mAb CBR LFA1/2 (10 μg/ml) or 1 mM MnCl2.
Binding of Soluble, Dimeric ICAM-1 to K562 Transfectants.
The soluble IC1–5D/IgA chimera containing the five Ig domains of human ICAM-1 fused to the Fc portion of IgA was purified from the culture supernatant of stable CHO transfectants by mAb R6.5 affinity chromatography as previously described (27). K562 transfectants were washed once with L15/FBS, and resuspended in the same buffer at 107/ml. Cells (25 μl) were mixed with 25 μl L15/FBS containing ICAM-1-IgA fusion protein or control human IgA at a final concentration of 100 μg/ml, and incubated at 37°C for 30 min. After incubation, cells were washed once in L15/FBS and incubated with FITC-conjugated anti-human IgA (Sigma) at room temperature for 20 min. After two washes, cells were subjected to flow cytometry.
Results
In αLβ2 Heterodimers, the Open I Domain Mutant Is Constitutively Active, and the Closed Mutant Lacks Ligand Binding Activity and Resists Activation.
Pairs of residues were identified that when mutated to cysteine could form disulfide bonds only in the closed structure of the αL I domain or in a model of the open, putative high affinity conformation of the αL I domain (Fig. 1). The double cysteine substitution mutants K287C/K294C and L289C/K294C were constructed to lock the αLβ2 I domain in putative open and closed conformations, respectively. As a control for the introduction of cysteines, the single cysteine substitution mutants K287C, L289C, and K294C were constructed. αL-subunits containing mutated I domains were transiently coexpressed with the integrin β2-subunit in 293T cells, and cell surface expression was determined by flow cytometry with mAb TS2/4 that only recognizes the αL-subunit when present in an αLβ2 complex (Fig. 2A). The double cysteine open and closed I domain mutants, and the single cysteine mutants were expressed as αLβ2 complexes at similar levels as wild-type αLβ2. Furthermore, the double and single cysteine mutants reacted with a panel of mAbs to the αL-subunit β-propeller domain and the β2-subunit I-like domain and cysteine-rich region as well as wild-type αLβ2 (28), indicating that the structural integrity of the αLβ2 molecule was not disturbed by the mutations.
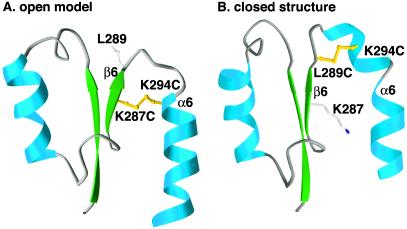
Figure 1.
Predicted disulfide bonds that are selective for open or closed conformers of the αL I domain. The K287C/K294C mutation is modeled in the open conformer (A), and the L289C/K294C mutation is modeled in the closed conformer (B). For clarity, ribbon diagrams show only residues 254 to 305 of the I domain. The models were superimposed by using residues not involved in conformational shifts and are shown in exactly the same orientation. The downward shift in the α6 helix in A compared with B is readily apparent. The remodeling of the loop connecting β6 and α6 is accompanied by a reversal in the orientation of the sidechain of residue 289. Sidechains for residues 287, 289, and 294 are shown as ball and stick. Prepared with ribbons.**
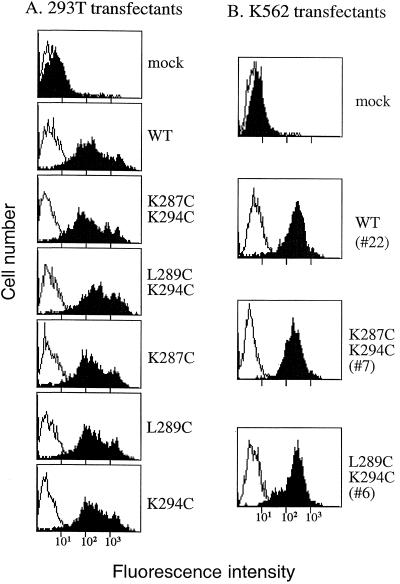
Figure 2.
Cell surface expression of αLβ2 cysteine substitution mutants. (A) 293T transient transfectants. (B) K562 stable transfectants. Wild-type or mutant αL cDNA was cotransfected with β2 cDNA in 293T cells or K562 cells. Cell surface expression of the αLβ2 complex was determined by immunofluorescent flow cytometry of the transfectants with mAb TS2/4 (shaded histogram) specific for αL in the αLβ2 complex, or the nonbinding IgG X63 (open histogram). Numbers in parentheses are clone numbers of the K562 stable transfectants.
Ligand binding activity of the mutants was first examined with αLβ2 heterodimers transiently expressed in 293T cells, in which wild-type β2 integrins, including αMβ2 and αLβ2, are basally active (29). 293T transient transfectants that express wild-type αLβ2, αLβ2 with the open K287C/K294C mutant I domain, and αLβ2 with single cysteine mutations showed constitutive binding to immobilized ICAM-1 (Fig. 3A). By contrast, αLβ2 with the L289C/K294C mutant closed I domain did not bind to ICAM-1. The αLβ2 with wild-type, open, and single cysteine substitution I domains showed equivalently high levels of binding to ICAM-1 when stimulated with the activating mAb CBR LFA-1/2 to the β2 cysteine-rich region. By contrast, CBR LFA-1/2 was not able to activate binding to ICAM-1 of αLβ2 with the mutant closed L289C/K294C I domain (Fig. 3A).
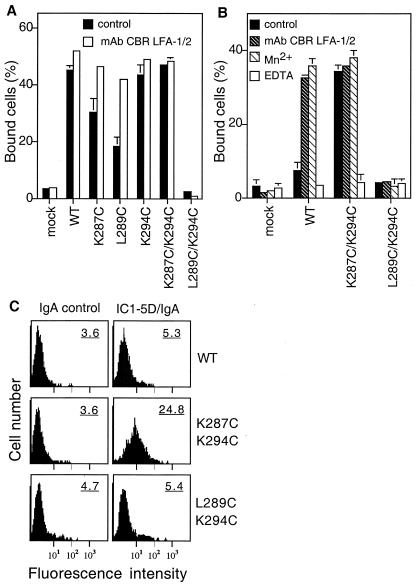
Figure 3.
Ligand binding activity of αLβ2 cysteine substitution mutants. (A) Binding of 293T transient transfectants to immobilized ICAM-1. (B) Binding of K562 stable transfectants to ICAM-1. Binding of the transfectants to immobilized ICAM-1 was determined in L15/FBS that contains Ca2+ and Mg2+ in the presence of the activating mAb CBR LFA-1/2 (10 μg/ml) or the control nonbinding IgG X63 (control), or in the absence of Ca2+ and Mg2+ and presence of 1 mM Mn2+ or 5 mM EDTA. Results are mean ± SD of triplicate samples and are representative of at least three experiments; some error bars are too small to be visible. (C) Binding of soluble ICAM-1-IgA Fc fusion protein (IC1–5D/IgA) to K562 stable transfectants. K562 transfectants were incubated with 100 μg/ml IC1–5D/IgA (Right) or control human IgA (Left), followed by incubation with FITC-labeled anti-human IgA and flow cytometry. Numbers on the upper right corner of each histogram plot are mean fluorescent intensity. Results are representative of three experiments.
The function of αLβ2 with mutant open or closed I domains was further studied in stable K562 transfectants in which wild-type αLβ2 shows little basal activity. Representative transfectant clones were selected that expressed similar levels of αLβ2 complex, as determined by flow cytometry with mAb TS2/4 (Fig. 2B). K562 transfectants expressing wild-type αLβ2 showed low basal binding to ICAM-1, and binding was greatly increased by the activating mAb CBR LFA-1/2 or Mn2+ (Fig. 3B). By contrast, cells expressing αLβ2 with the open K287C/K294C mutant I domain strongly bound to ICAM-1, even in the absence of activation. This binding appeared to be maximal, because it was not further increased with mAb CBR LFA-1/2 or Mn2+. In further contrast, αLβ2 with the closed L289C/K294C mutant I domain did not bind to ICAM-1, and binding could not be activated with mAb CBR LFA-1/2 or Mn2+. Ligand binding by wild type and the open K287C/K294C mutant αLβ2 was abolished by EDTA, demonstrating dependence on divalent cations (Fig. 3B).
We further examined binding of soluble, dimeric ICAM-1 to K562 transfectants. Soluble ICAM-1-IgA fusion protein bound to K562 transfectants that express αLβ2 with the open K287C/K294C I domain, but not wild-type αLβ2 or the closed L289C/K294C mutant (Fig. 3C). Thus, αLβ2 with the open mutant K287C/K294C I domain appears to be in a high affinity state.
Function of Mutant Open or Closed I Domains in Isolation from Other Domains.
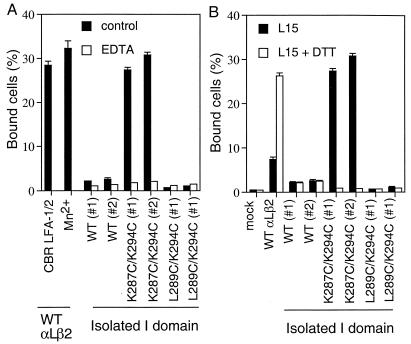
Because shape-shifting in I domains is proposed to be related to conformational movements elsewhere in integrins, it was important to examine ligand binding by the mutant I domains in isolation from other integrin domains. To express isolated I domains, the αL signal sequence and PDGFR transmembrane domain were fused to an αL I domain construct encoding residues 124 to 338. Two clones from each K562 cell transfection that expressed different levels of the I domain, as shown by flow cytometry with mAb TS1/22 to the I domain (Fig. 4), were selected for functional comparisons. Transfectants that expressed the isolated wild-type I domain and the L289C/K294C mutant closed I domain did not bind to ICAM-1. By contrast, cells that expressed the K287C/K294C mutant open I domain showed strong binding to ICAM-1 (Fig. 5A). Identical results were obtained with the two clones representative of each transfectant. Notably, the binding of the isolated open αL I domain was comparable to that obtained with intact αLβ2 that was activated with mAb CBR LFA-1/2 or Mn2+ (Fig. 5A). Binding of the K287C/K294C mutant open I domain to ICAM-1 was divalent cation dependent, as EDTA abolished the binding (Fig. 5A).
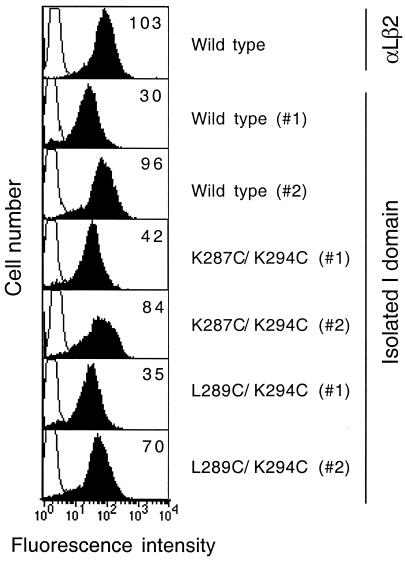
Figure 4.
Cell surface expression of the isolated αL I domains. The wild-type αL I domain, the open mutant K287C/K294C I domain, and the closed mutant L289C/K294C I domain were expressed on the surface of K562 transfectants with the PDGFR transmembrane domain. The level of cell surface expression was determined by immunofluorescent flow cytometry using mAb TS1/22 to the I domain (filled histograms). Binding of the control X63 IgG is shown as open histograms. The mean fluorescent intensity of TS1/22 binding is shown in the upper right corner of each plot. Two individual clones (#1 and #2) from each I domain transfectant are shown.
Figure 5.
Ligand binding activity of the isolated αL I domains. (A) Comparison of binding to immobilized ICAM-1 of activated wild-type αLβ2 and isolated, mutant αL I domains expressed in K562 transfectants. Binding of K562 transfectants expressing isolated I domains was in the presence of Ca2+ and Mg2+ (control), or in absence of Ca2+ and Mg2+ and presence of 1 mM EDTA. Binding of K562 transfectants expressing wild-type αLβ2 was in the presence of Ca2+ and Mg2+ and mAb CBR LFA-1/2 (10 μg/ml), or in absence of Ca2+ and Mg2+ and presence of 1 mM Mn2+. (B) Effect of disulfide reduction by DTT on binding of K562 transfectants to immobilized ICAM-1. Binding was performed in L15/FBS in the presence or absence of 10 mM DTT.
To confirm that a disulfide bond was present and required for the ligand binding activity of the open K287C/K294C mutant, the disulfide was disrupted with a reducing agent. Transfectants expressing wild-type αLβ2, or isolated wild-type or mutant I domains, were treated with 10 mM DTT and then tested for binding to ICAM-1 (Fig. 5B). Binding of the isolated, open I domain K287C/K294C mutant to ICAM-1 was completely abolished after DTT treatment. The wild-type I domain, which contains no disulfide, and the L289C/K294C closed mutant failed to bind to ICAM-1 either with or without DTT treatment. In contrast to the results with isolated I domains, DTT increased binding of intact wild-type αLβ2 (Fig. 5B). This served as a positive control to show that abolition of ligand binding by the mutant open I domain was not because of nonspecific effects of DTT. DTT treatment presumably disrupts disulfide bonds in intact αLβ2 that constrain the integrin in an inactive conformation, as demonstrated previously for other integrins (30, 31). These data strongly suggest that Cys287 and Cys294 introduced in the open I domain K287C/K294C mutant form a disulfide bridge that constrains the I domain in the open, high affinity conformation, and that after reduction the conformation shifts to the closed conformation. Elsewhere, we demonstrate formation of the disulfide bond in soluble, locked open and closed I domains.**
The Mutant, Open I Domain Is Resistant to Inhibition by the Small Molecule Antagonist Lovastatin.
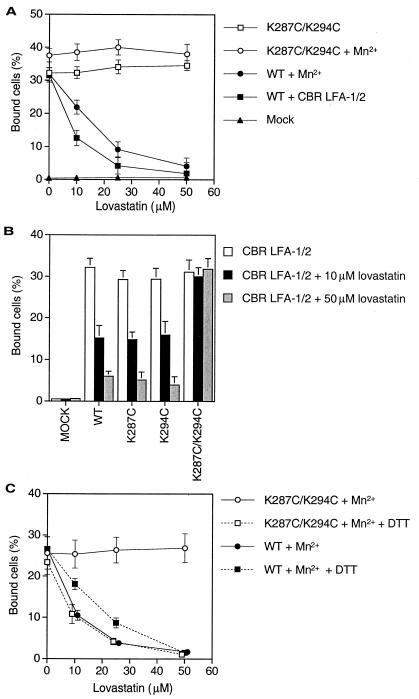
Lovastatin is a small drug molecule that inhibits β-hydroxy-β-methylglutaryl CoA reductase and is used clinically to lower cholesterol; however, it also inhibits ligand binding by αLβ2 (17). Structural studies show that lovastatin binds to a hydrophobic cavity in the αL I domain between the central β-sheet and the C-terminal α-helix. Because this site is distal from the MIDAS, it was hypothesized that binding of lovastatin might stabilize the αL I domain in an inactive conformation (17). If so, then the I domain locked in the open conformation with a disulfide bond should be resistant to inhibition by lovastatin. We confirmed that binding of wild-type αLβ2 activated by either mAb CBR LFA-1/2 or Mn2+ was inhibited by lovastatin, with an IC50 near the previously reported value of 25 μM (Fig. 6A) (17). By contrast, the αLβ2 heterodimer containing the open mutant K287C/K294C I domain was not inhibited by lovastatin in the absence or presence of Mn2+ (Fig. 6A). Residue Lys287 is part of the lovastatin binding pocket; however, the single cysteine substitution mutants K287C and K294C were fully susceptible to inhibition by lovastatin (Fig. 6B). Reduction with DTT rendered the K287C/K294C mutant fully susceptible to inhibition by lovastatin (Fig. 6C), demonstrating that locking the I domain open with a disulfide, rather than double substitution to cysteine per se, was responsible for lovastatin resistance.
Figure 6.
Lovastatin, a small molecule inhibitor of LFA-1, inhibits the function of wild-type αLβ2 but not of αLβ2 with a mutant, open I domain. (A and C) K562 transfectants. (B) 293T transfectants. Cells expressing wild-type αLβ2 or αLβ2 with mutant I domains, or mock-transfected cells, were preincubated with different concentrations of lovastatin at 37°C for 15 min before addition to ICAM-1-coated 96-well plastic plates. In C, cells were incubated in the presence or absence of 2 mM DTT at 37°C for 15 min before lovastatin treatment. Results are expressed as mean ± SD of three independent experiments in duplicate.
Discussion
Locking the Open and Closed Conformations of the I Domain with Disulfide Bonds.
We have demonstrated that locking the integrin αL I domain in two alternative conformations with a disulfide bond has profound consequences for its adhesive function, whether it is expressed in an intact αLβ2 heterodimer or as an isolated I domain. We have further demonstrated that the isolated αL I domain is sufficient for full adhesive function when locked in the open conformation, because it binds to ligand equivalently to the fully activated αLβ2 heterodimer. Moreover, we have shown that locking the I domain in the open conformation renders it resistant to inhibition by the small molecule antagonist lovastatin, demonstrating that lovastatin inhibits by an indirect mechanism.
Thus far, structures for the αL I domain have been determined only in the closed conformation (15–17); however, on binding to ICAM-1, NMR signals are altered for β-strand 6, the C-terminal α-helix 6, and the loop connecting them, as well as residues in the MIDAS (32). We hypothesized that the αL I domain should have an open conformation similar to that observed for the αM I domain, and that the αL I domain could be locked into the open or closed conformation by introducing a disulfide bond. To trap the I domain in the open or closed conformations, respectively, we introduced cysteines in place of K287 and K294 that could form a disulfide bond only in the open conformer, and cysteines in place of L289 and K294 that could form a disulfide only in the closed conformer. Our results demonstrate that the locked open I domain has adhesive activity equivalent to that of maximally activated wild-type αLβ2 when expressed not only within the intact αLβ2 receptor but also in isolated form. By contrast, neither the wild-type or locked closed I domain was adhesive to ICAM-1 when stably expressed in K562 cells as intact αLβ2 receptors or in isolated form. Whereas ligand binding by wild-type αLβ2 could be activated by an activating mAb or Mn2+, αLβ2 with the locked closed I domain was resistant to activation. The increased adhesiveness of the mutant open I domain was because of the formation of the disulfide bond, because disruption of the disulfide bond by reduction with DTT completely abolished ligand binding activity by the isolated I domain. Thus, after reduction of the disulfide bond, the mutant open I domain appears to shift to the closed conformation, which is strongly favored by its lower energy (29). In agreement with this, we have expressed soluble versions of the mutant I domains in Escherichia coli, and demonstrated disulfide bond formation after refolding of both the locked open and closed I domains.** Reduction with DTT markedly decreased the mobility in SDS/PAGE of the open and closed I domains but not that of the wild-type I domain. Furthermore, the affinity for ICAM-1 of the locked open I domain was increased 10,000-fold relative to the wild-type, locked closed, and DTT-treated locked open I domains.**
The I Domain by Itself Is Sufficient for the Full Adhesiveness of αLβ2 for ICAM-1.
Although the I domain is implicated as an important site for ligand binding in those integrins that contain it, we found that the isolated wild-type αL I domain did not support K562 transfectant binding to immobilized ICAM-1. Previous work using a glycosylphosphatidylinositol-anchored, isolated wild-type αL I domain has shown that ligand binding by the isolated I domain is much weaker than that by the intact integrin, and is only detected when the I domain is expressed at very high levels (33). It was suggested that the I domain alone might not be sufficient to mediate strong and stable interaction with ligand. However, we find that when the I domain is locked in the open conformation, it gives as robust binding to ICAM-1 as fully activated intact αLβ2. Either alterations in integrin conformation (affinity), clustering on the cell surface (avidity), or both of these mechanisms may be important in regulating cell adhesion through integrins (4, 10). Our work with αLβ2 and isolated I domains locked in different conformational states demonstrates the profound importance of integrin conformation for regulating cell adhesion. It is particularly remarkable that fully activated αLβ2 and the locked open, isolated I domain were equally adhesive. It is unlikely that receptor clustering and/or postligand binding events proposed for enhancing cell adhesion by intact integrin receptors could operate for an isolated I domain expressed using the transmembrane domain of the PDGFR.
Our work demonstrates that the I domain is sufficient for full adhesion of αLβ2 to ICAM-1. This suggests that the I domain contains the complete binding site for the ligand ICAM-1. In agreement with the current findings, deletion of the I domain abolishes binding to ICAM-1 and ICAM-3 (34, 35). By contrast, previous work had suggested that direct contact sites for ligand were present in repeats 5–7 of the predicted α-subunit β-propeller domain (36), and that the β2-subunit I-like domain contributed either directly or indirectly to ligand binding by αLβ2 (8, 37). Elsewhere, we report that mAbs to the β2 I-like domain cannot inhibit ligand binding by αLβ2 when the αL I domain is locked open, suggesting that the β2 I-like domain has an indirect role in ligand binding (28).
The αM I domain has been stabilized in the open and closed conformations by mutationally repacking the hydrophobic core (29). The stabilized open conformer expressed on the cell surface binds better than wild-type to the ligand iC3b, and shows enhanced expression of an activation epitope that maps near the MIDAS (29, 38). In contrast to results here with the αL I domain and ICAM-1, the isolated, stabilized, open αM I domain adheres to iC3b less well than the stabilized, open αMβ2 heterodimer (29). In agreement, αMβ2 with the I domain deleted shows diminished but significant binding activity to iC3b, and binding is inhibited by a mAb to the β-propeller domain of the αM subunit (35). Thus, the I domain is not sufficient for full binding of all I domain-containing integrins to all ligands.
Discrimination Between Direct and Indirect Mechanisms of Inhibition by Small Drug Molecules.
Lovastatin inhibits αLβ2 by binding to a pocket under the C-terminal α-helix of the I domain. Lovastatin binds distal to the MIDAS and thus has been thought to inhibit by an indirect mechanism (17). We found that αLβ2 with a locked open I domain was completely resistant to inhibition by lovastatin. Resistance was reversed by disulfide reduction with DTT. Thus, the mode of action by lovastatin is to stabilize the I domain in the closed conformation and block the downward shift of the C-terminal α-helix along the side of the I domain that occurs in the transition to the open conformation. Recently, other structural classes of small molecules have been described that antagonize the function of αLβ2 (39, 40). It will be interesting to determine their mechanism of action. It is notable that antagonists with direct/competitive and indirect/noncompetitive modes of inhibition can be discriminated by using the locked open I domain. Thus, locked open I domains not only provide important insights into allosteric regulation in integrins, but also can be used to advance pharmaceutical development of integrin antagonists.
Acknowledgments
We thank M. J. Humphries, Y. Takada, and J. S. Bennett for reviewing the manuscript. This work is supported by National Institutes of Health Grant CA31798. C.L. was supported by a Cancer Research Institute Fellowship.
Abbreviations
- ICAM
intercellular adhesion molecule
- LFA
lymphocyte function-associated antigen
- MIDAS
metal ion-dependent adhesion site
- PDGFR
platelet-derived growth factor receptor
Footnotes
M.S., C.L., R. Paframan, U. H. von Andrian, J.K., and T.A.S., unpublished work.
References
- 1.Larson R S, Springer T A. Immunol Rev. 1990;114:181–217. doi: 10.1111/j.1600-065x.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 2.Springer T A. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 3.Gahmberg C G, Tolvanen M, Kotovuori P. Eur J Biochem. 1997;245:215–232. doi: 10.1111/j.1432-1033.1997.00215.x. [DOI] [PubMed] [Google Scholar]
- 4.Diamond M S, Springer T A. Curr Biol. 1994;4:506–517. doi: 10.1016/s0960-9822(00)00111-1. [DOI] [PubMed] [Google Scholar]
- 5.Humphries M J. Biochem Soc Trans. 2000;28:311–339. [PubMed] [Google Scholar]
- 6.Springer T A. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bork P, Doerks T, Springer T A, Snel B. Trends Biochem Sci. 1999;24:261–263. doi: 10.1016/s0968-0004(99)01416-4. [DOI] [PubMed] [Google Scholar]
- 8.Huang C, Zang Q, Takagi J, Springer T A. J Biol Chem. 2000;275:21514–21524. doi: 10.1074/jbc.M002286200. [DOI] [PubMed] [Google Scholar]
- 9.Weisel J W, Nagaswami C, Vilaire G, Bennett J S. J Biol Chem. 1992;267:16637–16643. [PubMed] [Google Scholar]
- 10.Stewart M, Hogg N. J Cell Biochem. 1996;61:554–561. doi: 10.1002/(sici)1097-4644(19960616)61:4<554::aid-jcb8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 11.Bazzoni G, Hemler M E. Trends Biochem Sci. 1998;23:30–34. doi: 10.1016/s0968-0004(97)01141-9. [DOI] [PubMed] [Google Scholar]
- 12.Lee J-O, Rieu P, Arnaout M A, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 13.Lee J-O, Bankston L A, Arnaout M A, Liddington R C. Structure (London) 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 14.Emsley J, Knight C G, Farndale R W, Barnes M J, Liddington R C. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 15.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu A, Leahy D J. Structure (London) 1996;4:931–942. doi: 10.1016/s0969-2126(96)00100-1. [DOI] [PubMed] [Google Scholar]
- 17.Kallen J, Welzenbach K, Ramage P, Geyl D, Kriwacki R, Legge G, Cottens S, Weitz-Schmidt G, Hommel U. J Mol Biol. 1999;292:1–9. doi: 10.1006/jmbi.1999.3047. [DOI] [PubMed] [Google Scholar]
- 18.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 19.Hazes B, Dijkstra B W. Protein Eng. 1988;2:119–125. doi: 10.1093/protein/2.2.119. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Madrid F, Krensky A M, Ware C F, Robbins E, Strominger J L, Burakoff S J, Springer T A. Proc Natl Acad Sci USA. 1982;79:7489–7493. doi: 10.1073/pnas.79.23.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petruzzelli L, Maduzia L, Springer T A. J Immunol. 1995;155:854–866. [PubMed] [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Horton R M, Cai Z, Ho S N, Pease L R. BioTechniques. 1990;8:528. [PubMed] [Google Scholar]
- 24.Huang C, Springer T A. J Biol Chem. 1995;270:19008–19016. doi: 10.1074/jbc.270.32.19008. [DOI] [PubMed] [Google Scholar]
- 25.Lu C, Oxvig C, Springer T A. J Biol Chem. 1998;273:15138–15147. doi: 10.1074/jbc.273.24.15138. [DOI] [PubMed] [Google Scholar]
- 26.Lu C, Springer T A. J Immunol. 1997;159:268–278. [PubMed] [Google Scholar]
- 27.Martin S, Casasnovas J M, Staunton D E, Springer T A. J Virol. 1993;67:3561–3568. doi: 10.1128/jvi.67.6.3561-3568.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, Shimaoka M, Zang Q, Takagi J, Springer T A. Proc Natl Acad Sci USA. 2001;98:2393–2398. doi: 10.1073/pnas.041618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimaoka M, Shifman J M, Jing H, Takagi J, Mayo S L, Springer T A. Nat Struct Biol. 2000;7:674–678. doi: 10.1038/77978. [DOI] [PubMed] [Google Scholar]
- 30.Davis G E, Camarillo C W. J Immunol. 1993;151:7138–7150. [PubMed] [Google Scholar]
- 31.Lynam E B, Rogelj S, Edwards B S, Sklar L A. J Leukocyte Biol. 1996;60:356–364. doi: 10.1002/jlb.60.3.356. [DOI] [PubMed] [Google Scholar]
- 32.Huth J R, Olejniczak E T, Mendoza R, Liang H, Harris E A, Lupher M L, Jr, Wilson A E, Fesik S W, Staunton D E. Proc Natl Acad Sci USA. 2000;97:5231–5236. doi: 10.1073/pnas.97.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knorr R, Dustin M L. J Exp Med. 1997;186:719–730. doi: 10.1084/jem.186.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitinger B, Hogg N. Mol Biol Cell. 2000;11:677–690. doi: 10.1091/mbc.11.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yalamanchili P, Lu C, Oxvig C, Springer T A. J Biol Chem. 2000;275:21877–21882. doi: 10.1074/jbc.M908868199. [DOI] [PubMed] [Google Scholar]
- 36.Stanley P, Bates P A, Harvey J, Bennett R I, Hogg N. EMBO J. 1994;13:1790–1798. doi: 10.1002/j.1460-2075.1994.tb06447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman T G, Bajt M L. J Biol Chem. 1996;271:23729–23736. doi: 10.1074/jbc.271.39.23729. [DOI] [PubMed] [Google Scholar]
- 38.Oxvig C, Lu C, Springer T A. Proc Natl Acad Sci USA. 1999;96:2215–2220. doi: 10.1073/pnas.96.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly T A, Jeanfavre D D, McNeil D W, Woska J R, Jr, Reilly P L, Mainolfi E A, Kishimoto K M, Nabozny G H, Zinter R, Bormann B J, Rothlein R. J Immunol. 1999;163:5173–5177. [PubMed] [Google Scholar]
- 40.Liu G, Link J T, Pei Z, Reilly E B, Leitza S, Nguyen B, Marsh K C, Okasinski G F, von Geldern T W, Ormes M, et al. J Med Chem. 2000;43:4025–4040. doi: 10.1021/jm0002782. [DOI] [PubMed] [Google Scholar]