Abstract
A major challenge for non-viral gene delivery is gaining a mechanistic understanding of the rate-limiting steps. A critical barrier in polyplex-mediated gene delivery is the timely unpacking of polyplexes within the target cell to liberate DNA for efficient gene transfer. In this study, the component plasmid DNA and polymeric gene carrier were individually labeled with quantum dots (QDs) and Cy5 dyes, respectively, as a donor and acceptor pair for fluorescence resonance energy transfer (FRET). The high signal-to-noise ratio in QD-mediated FRET enabled sensitive detection of discrete changes in polyplex stability. The intracellular uptake and dissociation of polyplexes through QD-FRET was captured over time by confocal microscopy. From quantitative image–based analysis, distributions of released plasmid within the endo/lysosomal, cytosolic, and nuclear compartments formed the basis for constructing a three-compartment first-order kinetics model. Polyplex unpacking kinetics for chitosan, polyethylenimine, and polyphosphoramidate were compared and found to correlate well with transfection efficiencies. Thus, QD-FRET-enabled detection of polyplex stability combined with image-based quantification is a valuable method for studying mechanisms involved in polyplex unpacking and trafficking within live cells. We anticipate that this method will also aid the design of more efficient gene carriers.
INTRODUCTION
A major challenge for non-viral gene delivery is gaining a mechanistic understanding of the rate-limiting steps in the intracellular trafficking process in order to facilitate the rational design of novel delivery systems that address them.1,2 Cationic polymers, which bind and condense DNA to form polyplexes or nanocomplexes offer an attractive alternative and have tremendous potential for optimization. One of the critical barriers in polyplex-mediated gene delivery is the timely unpacking of polyplexes within the target cell to liberate the DNA for efficient gene transfer.3 The binding stability between the polymer and DNA must be optimized since either pre-mature dissociation or overly stable binding would be detrimental to the overall transfection efficiency. Modifications that alter these electrostatic interactions have been shown to affect transfection efficiencies for chitosan,4–7 polyethylenimine (PEI),8–10 and polyphosphoramidate (PPA).11,12 The methods used in these studies to characterize polyplex stability are typically performed in fixed cells and non-physiological conditions.
Fluorescently labeled DNA and its polymer or lipid carrier have been used to determine their intracellular stability and trafficking behavior.13–15 Colocalization of the fluorescent markers may indicate that the plasmid (pDNA) and its carriers are associated, but such detection methods do not provide sufficient sensitivity to detect the onset of dissociation, as the components must diffuse far enough away. Fluorescence fluctuation spectroscopy improved the sensitivity for detecting dissociation,16 but it relies on diffusion of complexes through a stationary excitation volume within the cell after microinjection. Thus, this approach is not amenable to monitoring the dynamic behavior of polyplexes as they are trafficked through different cellular compartments.
The stability of lipoplexes and polyplexes have been studied with a pair of organic fluorophores for fluorescence resonance energy transfer (FRET).9,17,18 However, these labeling strategies require additional ratiometric analysis, and organic fluorophores are often susceptible to photobleaching,19 hampering their use in time-lapse studies of intracellular trafficking. Quantum dots (QDs) have emerged as more efficient FRET donors with proximal organic acceptors.20,21 QDs are monodisperse semiconductor nanocrystals that have unique photophysical properties such as broad absorption and narrow symmetric emission spectra, which permit efficient energy transfer while significantly reducing direct acceptor excitation or spectral cross-talk.22,23 Their high photo-stability further promotes the application of QDs as biological probes and expands our capabilities in studying single molecule and cell behavior over an extended period of time with minimal cytotoxicity.22–24
Previously, we developed a QD-FRET-based method to study the intracellular stability of polyplexes.25 We demonstrated that QD-FRET provided a digital (on/off) indication of polyplex stability. In this study, we apply QD-FRET to compare three model polymers which are promising gene carriers: (i) chitosan, a biodegradable polysaccharide that is modestly effective in vitro and in vivo,4,26,27 (ii) PEI, a widely studied carrier that is efficient but relatively cytotoxic,28,29 and (iii) PPA, a new carrier based on a polyphosphate backbone that shows promising transfection efficiency but with a significantly different chemical structure from chitosan and PEI.12 The QD-FRET detection of intracellular dissociation formed the basis for quantitatively determining compartmental distributions of released DNA and to construct a mathematical model of polyplex unpacking kinetics. By correlating these kinetics to transfection efficiencies, the quantitative analysis herein provides new insights into the contributing roles of polyplex stability and intracellular trafficking during gene transfer.
RESULTS
Physical characterization of QD-FRET polyplexes
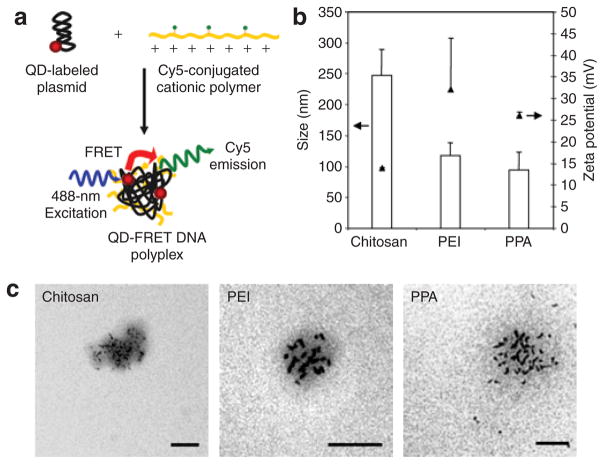
Component pDNA and polymers were individually labeled with QD605 and Cy5, respectively, as a donor and acceptor pair for FRET.21 Nanocomplexes were subsequently formed by vigorously mixing these labeled components (Figure 1a). Amine to phosphate charge ratios (N/P ratios) previously shown to exhibit high transfection efficiencies for chitosan,27 PEI,28 and PPA12 were used to formulate nanocomplexes. The size and zeta potential of QD-FRET polyplexes (Figure 1b) were found to be comparable to reported values for unlabeled polyplexes12,27,30 and confirmed by transmission electron microscopy (Figure 1c), indicating that labeling did not significantly affect their physical properties. The transmission electron microscopy images also showed that multiple QD-labeled pDNA, appearing as dark elliptically shaped objects due to the electron-dense QDs, were encapsulated within a single nanocomplex. By gel electrophoresis, similar DNA retention characteristics of unlabeled and QD-FRET polyplexes verified that DNA condensation or unpacking was not significantly affected (Supplementary Figure S1). Having conserved the physical and DNA retention properties after labeling, these QD-FRET polyplexes are expected to have similar cellular trafficking and unpacking behavior as unlabeled polyplexes.
Figure 1. Synthesis and physical characterization of quantum dot-fluorescence resonance energy transfer (QD-FRET) polyplexes.
(a) Schematic of polyplex synthesis by condensing QD-labeled anionic pDNA with Cy5-conjugated cationic polymeric gene carriers. Excitation of the QD at 488 nm results in energy transfer to nearby Cy5 dyes on the polymer, indicating an intact polyplex. (b) Sizes (open bars, mean + SD) and zeta potentials (filled triangles, mean + SD) of QD-FRET chitosan, polyethylenimine (PEI) and polyphosphoramidate (PPA) nanocomplexes were determined by dynamic light scattering. (c) Representative transmission electron microscopy (TEM) images of QD-FRET polyplexes show the encapsulation of multiple pDNA labeled with electron-dense QDs within each polyplex. Sizes by TEM were consistent with the hydrodynamic size measured in b. Bar = 100 nm.
Intracellular trafficking and unpacking
QD-FRET polyplexes were transfected into HEK293 cells and imaged by confocal microscopy to evaluate their intracellular distribution at time points between 1 and 48 hours. With a calculated Forster distance of 6.9 nm,19,21 the QD605 and Cy5 pair is ideally suited to probe the initial onset of polyplex dissociation which can include swelling or loosening of the carriers and subsequent unpacking (Supplementary Figure S2). At 4 hours post-transfection, intact polyplexes (indicated by FRET-mediated Cy5 emission) and free QD-labeled pDNA (QD emission only) were observed in the cytoplasm and in the perinuclear space (Figure 2c–k). Most of the polyplexes were not colocalized within acidic vesicles, suggesting that these polyplexes have escaped from endo/lysosomes. This behavior has been previously reported13,14 and confirmed that their trafficking behavior was not affected by labeling. The photostability of QDs allowed extended intracellular and intranuclear tracking of the free pDNA up to 72 hours.24,25,31 Interestingly, polyplexes were observed to be associated with the nuclear membrane and within the nucleus (Figure 2d), providing evidence that intact polyplexes may cross the nuclear membrane.
Figure 2. Live-cell imaging of quantum dot-fluorescence resonance energy transfer (QD-FRET) polyplexes.
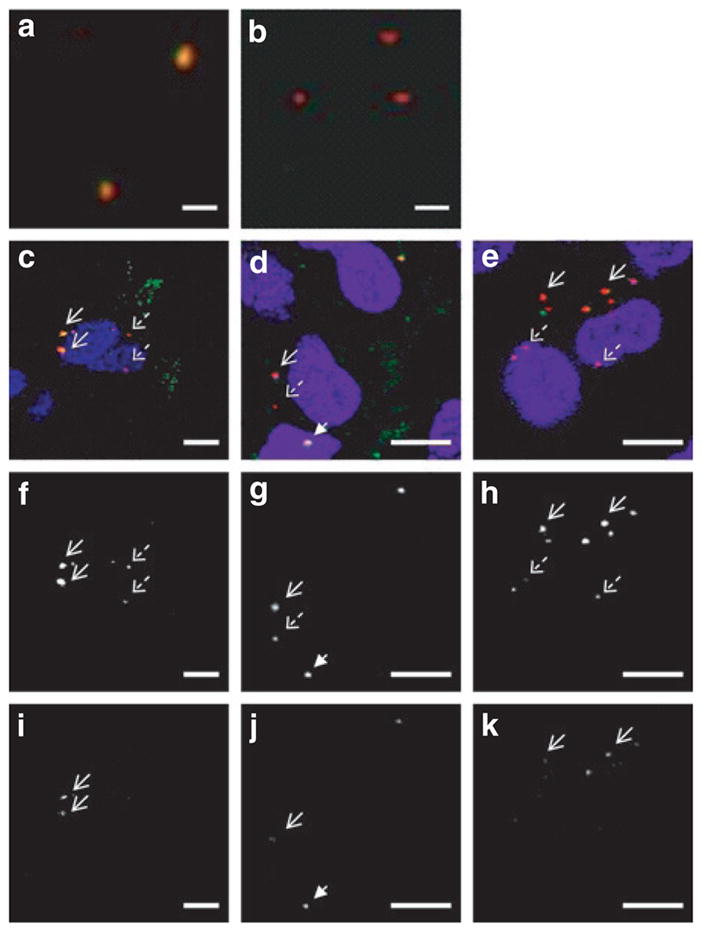
(a) Fluorescent image of intact QD-FRET chitosan polyplexes mounted on a coverslip. Upon excitation of the QDs, individual polyplexes exhibited energy transfer as indicated by the colocalization (orange/yellow) of QD (red) and Cy5 (green) signals. (b) After complete disruption by treating with heparin and chitosanase, energy transfer is abrogated and the QD signal is recovered. Bar in a and b = 1 μm. At 4 hours post-transfection with QD-FRET polyplexes, (c–e) composite images from confocal imaging of live HEK293 cells, and the corresponding gray scale images of the individual (f–h) QD and (i–k) Cy5 channels are shown separately. Immediately prior to imaging, cells were labeled with nuclear (blue) and endo/lysosome specific dyes (green). Colocalization of QD and FRET-mediated Cy5 signals are shown in yellow. (c,f,i) Chitosan polyplexes. Internalized stable chitosan polyplexes (solid arrows) exhibited FRET as indicated by the corresponding signals in f QD and i Cy5 channels. Free QD-labeled pDNA (dashed arrows) were observed in the perinuclear space and exhibited only QD and no Cy5 emission. (d,g,j) Polyethylenimine (PEI) polyplexes. Similarly, intact PEI polyplexes (solid arrow) and free pDNA (dashed arrow) were observed in the cytosol. An intact polyplex was found in the nucleus (arrowhead), providing evidence that polyplexes may cross the nuclear membrane. (e,h,k) Polyphosphoramidate (PPA) polyplexes. Stable PPA polyplexes (solid arrows) remained in the cytosol while released pDNA (dashed arrows) were closely associated with the nucleus. Bar for c–k = 10 μm.
Quantitative analysis of overall plasmid release
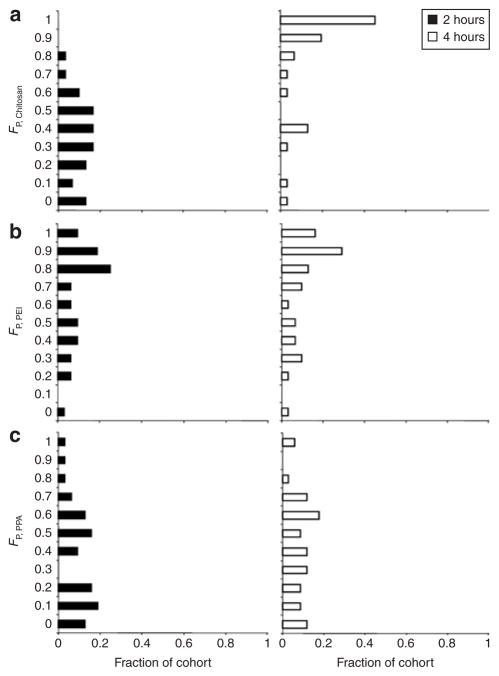
From confocal image stacks at each time point, the distributions of intact and unpacked polyplexes in the cytosol, endo/lysosomes, and nucleus were quantified with a particle-specific voxel-based method adapted from a previous report using a pixel-based algorithm.32 Using this image-based method to quantify free QD-labeled pDNA and QD-FRET polyplexes mounted onto coverslips showed that volume (in voxels) and DNA concentration were strongly correlated (R2 = 0.90, data not shown), and that these volumetric results may be used as an alternative metric of DNA content. Images from transfection experiments with QD-FRET chitosan, PEI, and PPA polyplexes were similarly analyzed to generate an overall (i.e., sum of all compartments) quantitative distribution of intracellular dissociation over time. In Figure 3, histograms were generated to compare the fractions of cohorts of cells having specific volume fractions of free pDNA (FP) at 2 and 4 hours post-transfection with polyplexes of each polymer. Each cohort consisted of at least 30 randomly selected cells per time point. There is a striking difference in the overall unpacking or free pDNA levels between PEI and chitosan polyplexes from 2 to 4 hours. At 2 hours, the majority of cells transfected with chitosan had a FP < 0.6, or in other words, <60% of the internalized polyplexes had unpacked and released pDNA (Figure 3a). A shift in the histogram toward higher FP values (FP > 0.8) at 4 hours clearly indicated the continuing dissociation of chitosan polyplexes (Figure 3a). Having quantitatively analyzed ≥30 cells, the representative unpacking behavior for chitosan polyplexes is likely to be observed here than what was previously reported.25 In contrast, more than half of the cells transfected with PEI polyplexes already had a FP > 0.8 at 2 hours (Figure 3b), which suggests that PEI dissociated more readily relative to chitosan and PPA. Among the polymers tested, PPA formed the most stable complexes since only moderate FP values were observed and there was only a modest shift in the histogram to higher FP values from 2 to 4 hours (Figure 3c). Previous studies reported that most PEI polyplexes remained stable up to 4 hours in a qualitative manner by colocalization14 or by conventional FRET where photobleaching was cited to affect their analysis.9 In this study, the potential effects of fluorophore photobleaching over time were found to be minimal since the photostabilities of QDs and Cy5 dyes in cells were confirmed by observation of QDs and by inspection of Cy5 under direct excitation up to 72 hours (Supplementary Figure S3). This difference underscores the benefit of quantitative analysis and the higher sensitivity and photostability of QD-FRET to detect dissociation over time.
Figure 3. Overall distribution of free plasmids.
The overall (i.e., sum of all compartments) intracellular distributions of released pDNA from (a) chitosan, (b) polyethylenimine (PEI), and (c) polyphosphoramidate (PPA) polyplexes at 2 hours (filled bar) and 4 hours (open bar) post-transfection. At each time point, histograms show the fraction of an analyzed cohort of cells (30–34 cells) having a particular binned volume fraction of free pDNA (FP) which was determined by quantitative image-based analysis and Eq. 3.
Quantitative analysis of compartmental plasmid release
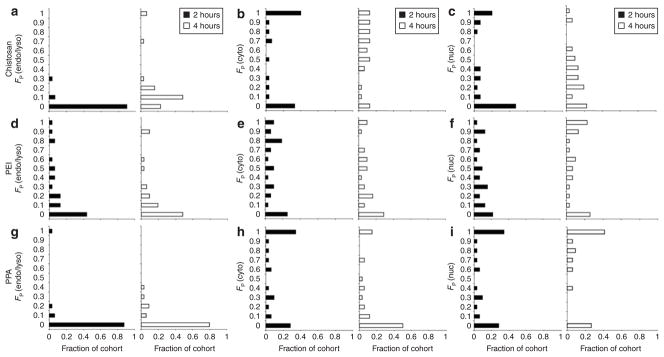
The intracellular compartment where polyplexes dissociate also impacts gene transfer. To evaluate the compartmental dissociation of polyplexes, the overall free pDNA distribution was parsed into three subcellular compartments: endo/lysosomes, cytosol, and nucleus. In Figure 4, histograms showed the fractions of cohorts of cells having a particular binned volume fraction of free pDNA in each compartment (k) normalized to the total free pDNA in the cell [FP(k)] and in Figure 5, to the total free and complexed pDNA within the compartment [F′P(k)]. The low volume fraction of free pDNA in endo/lysosomes represented by FP(endo/lyso) for all the polymers showed that free pDNA were generally not localized in this compartment (Figure 4a,d,g). For chitosan, a shift toward higher FP(endo/lyso) values at 4 hours suggested that these polyplexes began to unpack within acidic vesicles (Figures 4a and 5a). Endosomal escape with PEI polyplexes, likely due to the proton sponge effect,33,34 was shown by the shift in the histogram from high to low FP(endo/lyso) values, meaning less free pDNA was localized within the endo/lysosomes (Figure 4d). As a side effect of this buffering capacity, the accumulation of endo/lysosomal dyes within PEI-containing vesicles may lessen and lead to a slight overestimation of the DNA content in the cytosolic compartment.
Figure 4. Distribution of free plasmids across compartments.
The distributions of released pDNA across compartments from (a–c) chitosan, (d–f) polyethylenimine (PEI), and (g–i) polyphosphoramidate (PPA) polyplexes at 2 hours (filled bar) and 4 hours (open bar) post-transfection. At each time point, histograms show the fraction of an analyzed cohort of cells (30–34 cells) having a particular binned volume fraction of free pDNA [FP(k)] in the a,d,g endo/lysosomal [FP(endo/lyso)], b,e,h cytosolic [FP(cyto)], and c,f,i nuclear compartments [FP(nuc)] as determined by quantitative image-based analysis and normalized to the total free pDNA in the cell (Eq. 4).
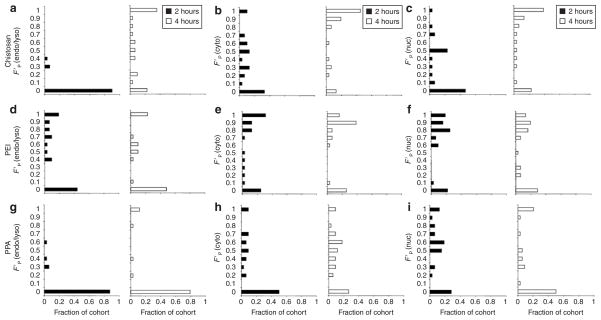
Figure 5. Distribution of free plasmids within compartments.
The distributions of pDNA released within each compartment from (a–c) chitosan, (d–f) polyethylenimine (PEI), and (g–i) polyphosphoramidate (PPA) polyplexes at 2 hours (filled bar) and 4 hours (open bar) post-transfection. At each time point, histograms show the fraction of an analyzed cohort of cells (30–34 cells) having a particular binned volume fraction of free pDNA [F′P(k)] in the a,d,g endo/lysosomal [F′P(endo/lyso)], b,e,h cytosolic [F′P(cyto)], and c,f,i nuclear compartments [F′P(nuc)] as determined by quantitative image-based analysis and normalized to the total free and complexed pDNA within the compartment (Eq. 5).
The majority of free pDNA for all the polymers was localized in the cytosolic compartment as most cells have generally high FP(cyto) values (Figure 4b,e,h). Chitosan polyplexes continued to release DNA which generally remained in the cytosol since the histogram shifted to higher F′P(cyto) and FP(cyto) values (Figures 5b and 4b, respectively). By contrast, the histograms for PEI and PPA showed a shift to lower FP(cyto) values (Figure 4e and h), which was accompanied by an opposite shift to higher FP(nuc) values (Figure 4f and i). Taken together, these results suggested that PEI- and PPA-mediated transfection allowed for more efficient transport toward the nucleus.14 More importantly, high F′P(nuc) values for PEI after 2 hours resulted from rapid release and high levels of pDNA in the nucleus (Figure 5f). Although these factors may have contributed to the higher transfection efficiency for PEI as determined by luciferase activity (Figure 6b), transfection by PPA was limited by its high stability in the nucleus as indicated by its lower F′P(nuc) values (Figure 5i).
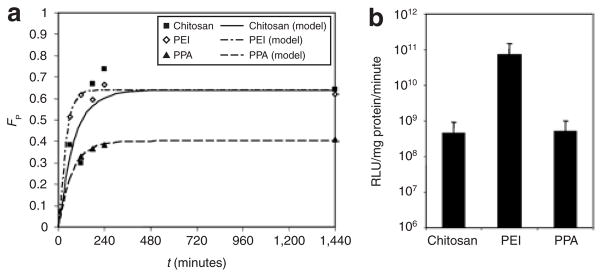
Figure 6. Fitted model of overall unpacking kinetics and transfection efficiency.
(a) The kinetics of unpacking based on the overall volume fractions of free pDNA (FP). Experimental data for chitosan (filled squares), polyethylenimine (PEI) (open diamonds), and polyphosphoramidate (PPA) (filled triangles), and the corresponding fitted mathematical model (Eq. 7) for chitosan (solid line), PEI (dash-dot line), and PPA (dashed line) are plotted over the first 24 hours. Models were fitted to at least four time points (1–48 hours) selected based on gene expression rates for each polymer. (b) Transfection efficiencies of chitosan, PEI, and PPA polyplexes containing luciferase-encoding pDNA in HEK293 cells. Luciferase activity was assayed 48 hours post-transfection and expressed as relative light units (RLU) normalized to the total protein content (mean + SD, n ≥ 3).
Model of intracellular unpacking kinetics
The unpacking process of polyplexes within cells was modeled as a first-order reaction. The kinetic parameters, steady-state volume fraction of free pDNA (FP,ss) and the unpacking time constant (τ), were determined by fitting the analytical solution (Eq. 7) to the average overall or compartmental volume fractions, which were determined from a cohort of at least 30 randomly selected cells per time point ranging from 1 to 48 hours (Table 1, RFigure 6a). The goodness-of-fit (2) for the overall unpacking kinetics ranged from 0.50 to 0.96. Both chitosan and PEI polyplexes had similar steady-state stabilities or FP,ss, indicating that although most have unpacked, some intact polyplexes may persist for ≥24 hours, but more importantly, PEI dissociated faster than chitosan as indicated by its shorter unpacking time constant. With the lowest FP,ss value, PPA released the least amount of pDNA overall and in the nucleus among the polymers tested. The high stability of PPA and slow unpacking rate of chitosan appeared to adversely affect their transfection efficiencies. By contrast, PEI exhibited rapid dissociation kinetics and delivered more free pDNA to the nucleus, resulting in the highest transgene expression. Comparing the overall unpacking time constants to the compartmental time constants and free pDNA distributions revealed a compartmental dependence of unpacking. PEI polyplexes exhibited similar time constants and release distributions for endo/lysosomes and the cytosol (Figure 5d and e), suggesting a similar stability in both compartments perhaps due to the high buffering capacity of PEI which facilitates endosomal escape.34 PPA polyplexes were highly stable in endo/lysosomes (Figure 5g), and they predominantly dissociated in the cytosol (Figure 5h) where the unpacking time constant was nearly identical to the overall time constant. Having buffering capacity or maintaining stability within endo/lysosomes, PEI and PPA may provide more protection from DNA degradation than chitosan, which begins to slowly dissociate within this compartment (Table 1, Figure 5a).
Table 1.
Unpacking rate constants from the fitted model
| Polymer | Overall |
Endosome/lysosome |
Cytosol |
||||
|---|---|---|---|---|---|---|---|
| FP,ss | τ (minutes) | k1c (minutes−1) | τ (minutes) | k1c (minutes−1) | τ (minutes) | k1c (minutes−1) | |
| Chitosana | 0.64 | 83.4 | 0.012 | 232.2 | 0.004 | 63.6 | 0.016 |
| PEIb | 0.64 | 36.6 | 0.027 | 54.6 | 0.018 | 52.2 | 0.019 |
| PPAb | 0.40 | 72.6 | 0.014 | 1,662 | 6 × 10−4 | 76.2 | 0.013 |
Abbreviations: PPA, polyphosphoramidate; PEI, polyethylenimine.
Parameters were derived by fitting average volume fractions at least four time points ranging between a1–48 hours and b1–24 hours. At least 30 randomly selected cells were analyzed per time point.
The unpacking rate constant (k1) was calculated assuming that recondensation (k2) was negligible; thus, k1≈1/τ.
DISCUSSION
We have developed a new method that combines the advantages of sensitive detection of polyplex stability by QD-FRET and image-based quantitative analysis of the intracellular distribution of polyplexes or released pDNA to gain a better mechanistic understanding of the unpacking process during polyplex-mediated gene transfer. Previously, image-based data have been used to compare the intracellular distributions of pDNA delivered by liposomes, polypeptide vectors,32 and adenoviruses.35 As these studies were focused on compartmental distributions, no models to determine transport kinetics were presented. Previous compartmental models have been used to simulate transport of DNA based on biological half-lives36 or to quantitatively assess rate constants for cellular uptake and nuclear translocation of lipoplex-mediated transfection,37 although pDNA release kinetics was not included in the latter model. A more complex first-order mass action model that included vector unpacking as a kinetic parameter was developed.38 Based on data for polylysine taken from the literature, this model predicted an unpacking rate constant threshold of 10−3 minutes−1, above which gene expression would be significantly increased.38
In this study, the unpacking process was modeled as a reversible first-order reaction. Dissociation, which is proportionate to the unpacking rate constant (k1), is assumed to be the dominant reaction. Recondensation after unpacking is very slow and unlikely in the intracellular milieu where an excess of competing polyanions and anionic proteins can shield the dissociated cationic gene carrier, reducing the probability of efficiently rebinding to pDNA. A recent report revealed that 20 of the 28 proteins bound to PEI polyplexes were negatively charged at cytosolic pH,39 including actin and tubulin whose concentrations are typically in the micromolar range.40 Thus, we consider k1 to be much larger than k2, and as a result, the unpacking time constant and unpacking rate constant are inversely related: τ ≈ 1/k1. The overall k1 determined by the model for chitosan, PEI, and PPA range from 0.012 to 0.027 minutes−1 (Table 1), and all exceed the reported 10−3 minutes−1 threshold, verifying the use of these polymers as effective gene carriers. In a recent study, the k1 for PEI was determined to be 0.05 minutes−1 by fitting a mathematical model to intracellular pDNA copy numbers.41 This rate constant is comparable to the k1 value of 0.027 minutes−1 for the same PEI polymer, as determined by fitting our first-order model to the volumetric distributions of free pDNA. This again serves as corroboration that image-based volumetric quantification may be an alternative metric for mass detection of DNA and that labeling did not significantly affect unpacking rates. The lower unpacking rate derived from our model may be attributed to the differences in polyplex formulation (N/P ratio of 10 in this study versus 20), size (118 versus 217 nm), and cell line (HEK293 versus C3A hepatoblastoma).41 To our knowledge, the results presented here are the first to report on the modeling of intracellular unpacking kinetics for chitosan and PPA polyplexes. Thus, the implications based on PEI are that intracellular unpacking kinetics, being dependent on polyplex formulation and cell line differences, must be considered when selecting the appropriate gene carrier.
Correlating unpacking kinetics and transfection efficiencies for the different gene carriers can give further insights into the mechanisms of rate-limiting steps involved in polyplex-mediated gene delivery. At steady state, chitosan and PEI have similar stabilities since they have the same steady-state volume fraction of free pDNA (FP,ss) (Table 1), but the transfection efficiency of PEI is much greater (Figure 6b). This difference can be attributed to the uptake of DNA which is 98 ± 1% of the initial dose for PEI compared to 68 ± 16% for chitosan due to the smaller size and high amine content of PEI polyplexes42 and the higher unpacking levels within the nucleus. Another significant factor is the onset of DNA degradation within endo/lysosomes where chitosan begins to dissociate. When intact, chitosan polyplexes have been shown previously to protect DNA from degradation after exposure to nucleases.27 The endosomal buffering and escape properties of PEI allow complexed pDNA to avoid degradation. Although the high stability of PPA polyplexes offers more protection, it also limits efficient DNA release, especially in the nucleus where it can critically affect downstream expression levels. Consequently, the transfection efficiency for PPA is less than PEI and only slightly higher than chitosan, despite having 92 ± 1% uptake of the initial DNA dose. From this comparison, an efficient gene carrier must be designed to either escape from or be stable within endo/lysosomes to avoid DNA degradation, while at the same time, facilitate the rapid release of DNA after reaching the nucleus. In terms of gene carrier design, factors such as molecular weight, charge density, biodegradability, and buffering capacity influence unpacking rates within different compartments, which then affect expression.
To better reflect the experimental data obtained by QD-FRET detection of dissociation, the kinetic parameters determined by the model in this study combined multiple biological processes into a single mechanistic step.37 Specifically, enzymatic degradation of the polymer and/or competitive binding with cytosolic proteins are potential mechanisms of dissociation.43 Despite this lumped-parameter approach, a comparison of the overall and compartmental distributions of free pDNA and unpacking rate constants provides valuable insights into the intracellular mechanisms that induce pDNA release from polyplexes. Unpacking for chitosan slowly begins in endo/lysosomes and continues in the cytosol, whereas PPA dissociates mainly in the cytosol, given the similar cytosolic and overall k1 values and the very low k1 for endo/lysosomes (Table 1). By contrast, PEI has a high buffering capacity and it dissociated at about the same rate in both endo/lysosomes and the cytosol. A recent report showed the release of DNA from PEI polyplexes when mixed with cytosolic fractions, albeit at a much slower rate.39 This discrepancy in DNA release rate underscores the importance of studying intracellular dissociation within the native microenvironments of live cells. With this model, the lumped-parameter approach may better describe dissociation by a single mechanism such as competitive binding which is expected to be the primary mode for PEI and PPA. It appears to be less apt in describing the potentially combined enzymatic and competitive binding mechanisms of dissociation. For example, the onset of dissociation for chitosan polyplexes was detected within endo/lysosomes, where lysozymes may initiate the degradation of chitosan,44 and then followed by displacement of DNA by competing polyanions in the cytosol. Consequently, the average R2 value for chitosan was 0.37 compared to PEI and PPA, which were 0.61 and 0.76, respectively. In general, the modest R2 values may be attributed to the intrinsic heterogeneities of the primary data, which consist of individual cells and polyplexes. In addition to differences in cell cycle and metabolism, there are heterogeneities in polyplex size and composition. Characterizing the variations in polyplex stability will be useful for understanding the effect of such heterogeneity on intracellular trafficking and transfection efficiency.
In summary, we have shown that QD-FRET-enabled detection of polyplex stability combined with image-based volumetric quantification is a useful method to study polyplex unpacking and trafficking within live cells. This quantification forms the basis for constructing a first-order model, from which unpacking kinetics can be determined and compared for three current gene carriers. This study contributes to the very limited data for polyplex unpacking kinetics in the current literature. Additional work is necessary to incorporate parameters in the model that will reflect potential enzymatic degradative pathways leading to polyplex dissociation and DNA degradation. We anticipate that this methodology will facilitate the development of more efficient gene carriers in the future and advance our understanding of the intracellular mechanisms involved in polyplex dissociation and trafficking.
MATERIALS AND METHODS
Labeling of plasmid and polymeric gene carriers
Plasmid DNA (pEGFP-C1, 4.9 kilobase, Clontech, Mountain View, CA) was labeled with streptavidin-functionalized QD605 (Qdot 605 ITK; Invitrogen, Carlsbad, CA) via a biotin–streptavidin linkage as described previously.25 Briefly, pDNA were biotinylated using polyethylene oxide-psoralen-biotin (Pierce, Rockford, IL) and exposure to UV light (320–500 nm) for 30 minutes or by covalent modification with guanine-specific biotin labels as described by the manufacturer (Mirus Bio, Madison, WI) but scaled to have ~1–2 biotin labels per pDNA. Biotinylated DNA was purified from unreacted biotin cross-bridges by ethanol or isopropanol precipitation and centrifugation following standard protocols.45 The molar ratio of pDNA to QD was kept in excess to ensure complete conjugation of QDs to pDNA. The number of QDs labeled onto each pDNA is estimated to be ~1–3.25
The primary amines of the cationic polymeric gene carriers were labeled with N-hydroxy-succinimide–functionalized Cy5 (Amersham Biosciences, Piscataway, NJ) through carbodiimide chemistry. A solution of Cy5 dye was gradually added to aqueous solutions of chitosan (390 kd, 83.5% deacetylated; Vanson, Redmond, WA), PEI (25 kd, branched; Sigma Aldrich, St. Louis, MO), or PPA (PPA-DPA, 40 kd, synthesized as described in ref. 12) and stirred continuously for at least 12 hours. For chitosan, the reaction mixture was maintained at pH ~6.5 to keep chitosan soluble. To facilitate complete conjugation of Cy5 dye, the molar ratio of Cy5 to free amines was kept at 1:200. Any remaining free dye was removed by dialysis to obtain only Cy5-labeled polymer. The level of Cy5 labeling on the polymer was determined by comparing fluorescence intensities of labeled polymer to a standard curve of Cy5 dye, resulting in ~2–6 Cy5 dyes per chitosan molecule,25 ~2 dyes per PEI molecule, and ~4 dyes per PPA molecule.
Synthesis and physical characterization of QD-FRET polyplexes
Nanocomplexes were prepared from chitosan as described previously.25,27 Briefly, Cy5-chitosan (pH 5.5–5.7; 0.1% in 25 mmol/l acetic acid solution) and QD-labeled pDNA in 50 mmol/l of sodium sulfate solution were both heated to 55 °C separately. Equal volumes of both solutions were quickly mixed under high vortexing. Chitosan nanocomplexes were formed at an N/P ratio (primary amine to phosphate ratio) of 4:1 (refs. 26,27). The synthesis of nanocomplexes formed with PEI29 or PPA11 are described elsewhere. Briefly, pDNA was added to 5% glucose solutions of PEI or aqueous solution of PPA. Complexes were formed by adding equal volumes of polymer solution to the DNA solution and then vigorously mixing by pipetting or brief vortexing. The N/P ratios used for PEI and PPA nanocomplexes were 10:1 and 16:1, respectively. All polyplexes were then used without further purification.
Polyplex size and zeta potential were measured by dynamic light scattering (Zetasizer 3000, Malvern Instruments, Southborough, MA). Size measurements were performed at 25 °C at 90° scattering angle in automatic mode, and zeta potential measurements were performed using the aqueous flow cell in the automatic mode at 25 °C. The ultrastructure of polyplexes was examined by transmission electron microscopy (Philips EM 420 TEM; Philips Electron Optics, Eindhoven, The Netherlands).
Transfection and imaging of live cells
HEK293 cells (American Type Culture Collection, Manassas, VA) were seeded (1 × 105 cells/well) and grown on chambered coverglasses (4-well Labtek II; Nalge Nunc, Rochester, NY) in complete media (minimal essential medium supplemented with 10% fetal bovine serum, 2 mmol/l L-glutamine, 50 U/ml penicillin, and 50 U/ml streptomycin; Gibco Invitrogen, Carlsbad, CA). Duplicate wells of 60–70% confluent cells were transfected with QD-FRET polyplexes containing 0.5 μg DNA in 0.5 ml of reduced-serum media (Opti-MEM; Gibco Invitrogen, Carlsbad, CA) for 1–4 hours at 37 °C. At specific time points after transfection (1–48 hours), the transfection media was replaced and live cells were imaged with a confocal microscope (LSM 510 Meta; Carl Zeiss, Thornwood, NY) equipped with a ×63 objective (numerical aperture 1.40) and heated stage. Immediately prior to imaging, cells were stained with nuclear dye (Hoechst 33342, Molecular Probes, Carlsbad, CA) and a dye specific for acidic vesicles (LysoTracker Green DND-26; Molecular Probes, Carlsbad, CA). Imaging was performed in the multi-track mode using 405-nm excitation for Hoechst (emission bandpass 420–480 nm) and 488-nm excitation for LysoTracker (bandpass 505–530 nm), QD605 (Meta 593–615 nm), and Cy5 (Meta 636–754 nm). The main dichroic was HauptFarbTeiler 405/488/543 and the secondary dichroic was Neben Farb Teiler 545. For all cells, image stacks were composed of at least 20 z-sections, which was optimal for image-based analysis.32 Methods to measure transfection efficiencies and cellular uptake are described in Supplementary Materials and Methods.
Quantification of distributions of complexed or free plasmid
An image-based quantification method was adapted from a previous report using a pixel-based algorithm.32 The approach used in this study takes full advantage of the 3D image data stacks obtained by confocal microscopy from which particles are isolated as individual 3D objects. For each time point, at least 30 cells containing at least one particle were randomly selected. This number of cells was determined to be statistically sufficient and representative for image-based analysis.32 Each individual cell was outlined with a corresponding phase image and the volumes of all internalized objects were determined in voxels where v, the volume per ith particle, was summed over of N total particles to obtain V, the total volume per cellular compartment (Eq. 1). The type of each object [t: complexed (C) or free pDNA (P)] was identified digitally by QD-FRET,25 without needing to calculate or normalize FRET efficiencies. Specifically, objects exhibiting FRET-mediated Cy5 signals were considered as condensed pDNA within polyplexes, and objects exhibiting only QD signals were considered as released or free pDNA. Threshold levels were based on background intensities of the cell. The subcellular location [k: cytosol (cyto), endo/lysosome (endo/lyso), or nucleus (nuc)] of each object was determined by colocalization with nuclear and endo/lysosomal-specific dyes and mass balance for the entire cell (Eq. 2). Image processing and volume determinations were performed with a custom script using ImageJ (v1.37; http://rsb.info.nih.gov/ij). Volume fractions (Ft) of complexed or free pDNA for the entire cell were calculated overall using Eq. 3, which was normalized to all internalized particles. Volume fractions across or in specific compartments [Ft(k) or F′t(k)] were calculated using Eq. 4, which was normalized to all internalized particles of the same type in the cell, or using Eq. 5, which was normalized to all particles in the same compartment, respectively. At each time point, the fraction of cells within an analyzed cohort having a particular binned volume fraction of free pDNA (FP) was calculated to generate histograms.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
First-order model of polyplex unpacking
A mathematical model was constructed based on the volumetric distributions resulting from image-based quantification as an alternative measure of the amount of DNA. The dissociation of polyplexes (C) to release free pDNA (P) was modeled as a first-order reversible reaction38:
Dissociation of polyplexes was described by the unpacking rate constant (k1) and assumed to be the dominant reaction. After unpacking, an excess of competing polyanions or anionic proteins can bind to the cationic gene carrier39 and shield it from efficiently rebinding to pDNA. Thus, the recondensation rate is assumed to be much slower, and the probability of recondensation occurring is much smaller in the intracellular milieu. With this model, the release of free pDNA is described by the differential equation:
| (6) |
and the corresponding analytical solution:
| (7) |
The parameters of the model, steady-state volume fraction of free pDNA (FP,ss) and the unpacking time constant (τ), were determined by least-squares fitting of the average overall or compartmental volume fractions for at least four time points selected based on prior knowledge of gene expression rates for each model polymer (Origin v6.0; OriginLab, Northampton, MA). At least 30 randomly selected cells were analyzed to find the average volume fraction at each time point. This number of cells was determined to be statistically sufficient and representative for image-based analysis.32
Supplementary Material
DNA retention with unlabeled and QD-FRET PEI polyplexes.
Emission spectra of QD-FRET polyplexes.
Long-term visualization of Cy5-labeled polymer under direct excitation.
Acknowledgments
This work was supported by NIH EB002849, NIH DK068399, and NSF DBI-0552063.
References
- 1.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 2.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 3.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 4.Kiang T, Wen J, Lim HW, Leong KW. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials. 2004;25:5293–5301. doi: 10.1016/j.biomaterials.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 5.Koping-Hoggard M, Varum KM, Issa M, Danielsen S, Christensen BE, Stokke BT, et al. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11:1441–1452. doi: 10.1038/sj.gt.3302312. [DOI] [PubMed] [Google Scholar]
- 6.Danielsen S, Strand S, de Lange Davies C, Stokke BT. Glycosaminoglycan destabilization of DNA-chitosan polyplexes for gene delivery depends on chitosan chain length and GAG properties. Biochim Biophys Acta. 2005;1721:44–54. doi: 10.1016/j.bbagen.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Lavertu M, Methot S, Tran-Khanh N, Buschmann MD. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials. 2006;27:4815–4824. doi: 10.1016/j.biomaterials.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Pun SH, Bellocq NC, Liu A, Jensen G, Machemer T, Quijano E, et al. Cyclodextrin-modified polyethylenimine polymers for gene delivery. Bioconjug Chem. 2004;15:831–840. doi: 10.1021/bc049891g. [DOI] [PubMed] [Google Scholar]
- 9.Gabrielson NP, Pack DW. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules. 2006;7:2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- 10.Doody AM, Korley JN, Dang KP, Zawaneh PN, Putnam D. Characterizing the structure/function parameter space of hydrocarbon-conjugated branched polyethylenimine for DNA delivery in vitro. J Control Release. 2006;116:227–237. doi: 10.1016/j.jconrel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Gao SJ, Zhang PC, Wang S, Mao HQ, Leong KW. Polyphosphoramidate gene carriers: effect of charge group on gene transfer efficiency. Gene Ther. 2004;11:1001–1010. doi: 10.1038/sj.gt.3302248. [DOI] [PubMed] [Google Scholar]
- 12.Mao HQ, Leong KW. Design of polyphosphoester-DNA nanoparticles for non-viral gene delivery. Adv Genet. 2005;53PA:275–306. doi: 10.1016/S0065-2660(05)53011-6. [DOI] [PubMed] [Google Scholar]
- 13.Kiang T, Bright C, Cheung CY, Stayton PS, Hoffman AS, Leong KW. Formulation of chitosan-DNA nanoparticles with poly(propyl acrylic acid) enhances gene expression. J Biomater Sci Polym Ed. 2004;15:1405–1421. doi: 10.1163/1568562042368112. [DOI] [PubMed] [Google Scholar]
- 14.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci USA. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci USA. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas B, Remaut K, Sanders NN, Braeckmans K, De Smedt SC, Demeester J. Studying the intracellular dissociation of polymer-oligonucleotide complexes by dual color fluorescence fluctuation spectroscopy and confocal imaging. Biochemistry. 2005;44:9905–9912. doi: 10.1021/bi0476883. [DOI] [PubMed] [Google Scholar]
- 17.Itaka K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Evaluation by fluorescence resonance energy transfer of the stability of nonviral gene delivery vectors under physiological conditions. Biomacromolecules. 2002;3:841–845. doi: 10.1021/bm025527d. [DOI] [PubMed] [Google Scholar]
- 18.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat Mater. 2005;4:460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 19.Lakowicz JR. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum; New York: 1999. [Google Scholar]
- 20.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. J Am Chem Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-quantum-dot-based DNA nanosensor. Nat Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 22.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 23.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 24.Srinivasan C, Lee J, Papadimitrakopoulos F, Silbart LK, Zhao M, Burgess DJ. Labeling and intracellular tracking of functionally active plasmid DNA with semiconductor quantum dots. Mol Ther. 2006;14:192–201. doi: 10.1016/j.ymthe.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Ho YP, Chen HH, Leong KW, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J Control Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy K, Mao HQ, Huang SK, Leong KW. Oral gene delivery with chitosan—DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nat Med. 1999;5:387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- 27.Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, et al. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 28.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Ma N, Gao SJ, Yu H, Leong KW. Transgene expression in the brain stem effected by intramuscular injection of polyethylenimine/DNA complexes. Mol Ther. 2001;3:658–664. doi: 10.1006/mthe.2001.0324. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Wang J, Lee CG, Wang CY, Gao SJ, Tang GP, et al. CNS gene transfer mediated by a novel controlled release system based on DNA complexes of degradable polycation PPE-EA: a comparison with polyethylenimine/DNA complexes. Gene Ther. 2004;11:109–114. doi: 10.1038/sj.gt.3302135. [DOI] [PubMed] [Google Scholar]
- 31.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 32.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Haensler J, Szoka FC., Jr Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug Chem. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 34.Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 35.Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T, Harashima H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol Ther. 2006;13:786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Ledley TS, Ledley FD. Multicompartment, numerical model of cellular events in the pharmacokinetics of gene therapies. Human Gene Ther. 1994;5:679–691. doi: 10.1089/hum.1994.5.6-679. [DOI] [PubMed] [Google Scholar]
- 37.Banks GA, Roselli RJ, Chen R, Giorgio TD. A model for the analysis of nonviral gene therapy. Gene Ther. 2003;10:1766–1775. doi: 10.1038/sj.gt.3302076. [DOI] [PubMed] [Google Scholar]
- 38.Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4:438–446. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- 39.Iida T, Mori T, Katayama Y, Niidome T. Overall interaction of cytosolic proteins with the PEI/DNA complex. J Control Release. 2007;118:364–369. doi: 10.1016/j.jconrel.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 40.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. Garland Publishing, Inc; New York: 1994. [Google Scholar]
- 41.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery. Structural effects on cellular internalization, buffering capacity, and gene expression. Bioconjug Chem. 2007;18:19–30. doi: 10.1021/bc060029d. [DOI] [PubMed] [Google Scholar]
- 43.Okuda T, Niidome T, Aoyagi H. Cytosolic soluble proteins induce DNA release from DNA—gene carrier complexes. J Control Release. 2004;98:325–332. doi: 10.1016/j.jconrel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Aiba S. Studies on chitosan: 4. Lysozymic hydrolysis of partially N-acetylated chitosans. Int J Biol Macromol. 1992;14:225–228. doi: 10.1016/s0141-8130(05)80032-7. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA retention with unlabeled and QD-FRET PEI polyplexes.
Emission spectra of QD-FRET polyplexes.
Long-term visualization of Cy5-labeled polymer under direct excitation.