Abstract
A TaqMan-based real-time polymerase chain reaction (PCR) assay was devised for the detection of porcine parvovirus (PPV). Two primers and a TaqMan probe for the non-structural protein NS1 gene were designed. The detection limit was 1 × 102 DNA copies/μL, and the assay was linear in the range of 1 × 102 to 1 × 109 copies/μL. There was no cross-reaction with porcine circovirus 2 (PCV2), porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (PRV), classical swine fever virus (CSFV), or Japanese encephalitis virus (JEV). The assay was specific and reproducible. In 41 clinical samples, PPV was detected in 32 samples with the real-time PCR assay and in only 11 samples with a conventional PCR assay. The real-time assay using the TaqMan-system can therefore be practically used for studying the epidemiology and management of PPV.
Introduction
Porcine parvovirus (PPV) is an autonomous parvovirus belonging to the genus parvovirus, subfamily Parvovirinae, family Parvoviridae. It is one of the major etiological agents of reproductive failure in pigs. Reproductive failure caused by PPV is characterized by embryonic and foetal death, mummification, stillbirth, and delayed return to oestrus [1]. In addition, PPV has been implicated as the causative agent of diarrhea, skin disease, and arthritis in swine [2]. PPV has been reported from many different countries [3-5].
PPV is composed of a linear single-stranded segment of DNA approximately 5 kb long (Molitor, T.W., 1983), and its genome has more than two open reading frames (ORF) [6]. The 3' end of ORF1 encodes nonstructural proteins (NS proteins), and the 5' end of ORF2 encodes structural proteins (VP proteins).
For diagnostic purposes, PPV can be rapidly and sensitively detected with polymerase chain reaction (PCR) assays [7,8]. However, current PCR assays for PPV often require multiple steps and do not provide quantitative data. In contrast, real-time PCR using SYBR Green and TaqMan is rapid, specific, and efficient for the large-scale screening, strain identification, and quantification of PPV [9].
NS1, which is encoded by the NS1 gene, is a main nonstructural protein of PPV and is associated with the early and late transcription of the virus. Given that inactivated virus used in current vaccines have only little NS1 protein which could not produce antibody, the presence or absence of antibody against NS1 protein could be used in an NS1-based diagnostic kit for determining in clinical settings whether pigs have been vaccinated with the inactivated-PPV or infected with wild-type PPV, and the test would give a negative result for for vaccinated/noninfected pigs. NS proteins are also important in virus research because they play an important regulatory role in viral replication even though they do not directly participate in the assembly of virus particles.
In this study, a TaqMan-based real-time PCR assay was developed for the rapid and quantitative detection of PPV with a probe specific for the PPV NS1 gene. The results of the real-time PCR assays were compared with those of previously established, conventional PCR assays.
Materials and methods
Primers and probes
PCR primers and a TaqMan probe, which were designed with the program DNAStar and synthesized by Saituo Matrix Biotechnology (Haerbin) Co., Ltd, were used to amplify a 123-bp fragment of the NS1 gene. The sequences of the primers and probe were:
NS1-FP (forward primer): 5'-GAAGACTGGATGATGACAGATCCA-3',
NS1-RP (reverse primer): 5'-TGCTGTTTTTGTTCTTGCTAGAGTAA-3'.
NS1-P (probe): FAM-AATGATGGCTCAAACCGGAGGAGA-BHQ1. The probe was labeled with 6-carboxyfluorescein (FAM) at the 5'-end and with BHQ1 at the 3'-end.
Preparation of standard plasmid DNA
PCR amplification of the NS1 gene was carried out in a reaction mix of 25 μL: 16.0 μL sterilized water, 2.5 μL of 10× buffer, 3.0 μL of dNTP, 1 μL of each primer (NS1-FP and NS1-RP), 1 μL of BQ strain DNA, and 0.5 μL of Ex Taq™ DNA Polymerase (Ex taq). The thermal conditions were as follows: one cycle at 94 C for 5 min; followed by 30 cycles at 94 C for 30 s, 58 C for 45 s, and 72 C for 30 s; with a final extension at 72 C for 7 min.
The PCR product was inserted into a vector, pMD18-T (TaKaRa Biotechnology (Dalian) Co., Ltd.). After the culture was increased in DH5a host bacteria (TaKaRa Biotechnology Co., Ltd), the recombinant plasmid was purified using a commercial test kit (Watson Biotechnologies, Inc.). The products were kept at -20°C for later use.
Establishment of real-time PCR
The real-time PCR amplifications of the NS1 gene used 25-μL reaction mixtures containing 2.5 μL of 10× buffer, 3.5 μL of dNTP (TaKaRa Biotechnology Co., Ltd), 3 μL of MgCl2, 1 μL of each primer (10 pM/μL of NS1-FP and NS1-RP), 1 μL of the recombinant plasmid, 0.5 μL of the probe (PPV-P), 0.5 μL of Hotstar taq (TaKaRa Biotechnology Co., Ltd), and 12.0 μL sterile water. The reactions were carried out in an Rotor-Gene Thermocycler (Corbett Research. Co. Ltd.). The conditions were as follows: one cycle at 95°C for 30 s followed by 40 cycles at 95°c for 10 s, 58°c for 20 s, and 72°C for 20 s. The data were analyzed with the Rotor-Gene software.
Sensitivity of the real-time PCR
To determine the detection limit and efficiency of the assay, recombinant plasmid of standard DNA was used as a template and was 10-fold serially diluted with sterile water, giving 6.00 × 109 to 6.00 × 101 copies/μL. The sensitivity of the real-time PCR was compared with conventional PCR (Yue et al., 2009).
Specificity of the real-time PCR
To determine the specificity of the real-time PCR, the standard plasmid-positive template and different strains of PPV, five different viruses (PRRSV, CSFV, PRV, JEV, and PCV-2), and control (sterile water) were processed with the real-time PCR.
Reproducibility of real-time PCR
To determine the reproducibility of the real-time PCR, the standard plasmid was diluted to 6.00 × 108, 6.00 × 106, 6.00 × 104, and 6.00 × 102 copies/μL. To obtain variation within an assay (within a block), each dilution was processed four different times, i.e., in four blocks, with the real-time PCR assay. Each block contained four repeated determinations for each dilution, giving 16 total determinations for each dilution. Coefficients of variation (CVs) for Ct values within each block and among blocks (using the mean values from each block) were determined.
Detection of the clinical samples
Forty-one clinical samples (20% organ suspensions stored at -70°C) that were suspected of being infected with PPV were subjected to the real-time PCR and conventional PCR.
Results
Establishment of the standard plasmid-positive template
The amplified NS1 gene fragment was about 123 bp long. The plasmid DNA concentration was 0.189 μg/μL before dilution. So, the DNA concentration was equivalent to 6.00 × 1010 copies/μL before dilution.
Establishment of a standard curve for the real-time PCR
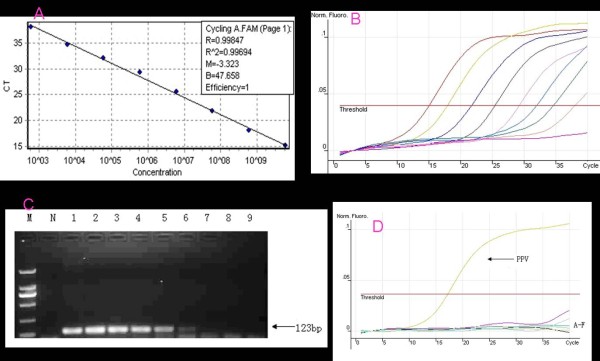
The standard curve was generated with a range of 6.00 × 109 to 6.00 × 102 copies/μL (Figure 1A). The assays were linear in a dilution range of template DNA from 6.00 × 102 to 6.00 × 109 copies/μL, with R2 values of 0.996 and reaction efficiencies of 100% for NS1. Quantitative data for Cycling A. FAM are shown in Figure 1B. The amplification product was about 123 bp long, and no false amplification was observed.
Figure 1.
Establishment of real-time PCR. A. Standard curves (based on plasmid DNA) indicating the linearity and efficiency for detecting NS1 by real-time PCR. The x-axis represents copies of plasmid DNA in 10-fold dilutions, and the y-axis represents the fluorescence data used for cycle threshold (Ct) determinations. The assays were linear in the range of 109 to 102 template copies/μL, with an R2 of 0.996 and a reaction efficiency of 100% for NS1. B. The results of the Quantitation data for Cycling A. FAM. The amplification product was about 123 bp long, and no false amplification was observed. The detection limit of the real-time PCR for the NS1 gene of PPV was 1.00 × 102 copies/μL. C. Sensitivity of normal PCR. N: Negative Control. M: DNA marker DL2000. Lane 1-9: The standard plasmid was 10-fold serially diluted as template. D. Specificity of the real-time PCR assay. PPV:Positive sample; A-F:Negative control, PCV2, PRV, PRRSV, CFSV, JEV, and H2O control.
Sensitivity of the real-time PCR
The detection limit of the real-time PCR for the NS1 gene of PPV was 1.00 × 102 copies/μL(Figure 1B). The conventional PCR assay showed a negative result when the solution was diluted to 1.00 × 104 copies/μL. These results indicate that, based on direct observation, the sensitivity of the real-time PCR is 100 times greater than that of the conventional PCR (Figure 1C).
Specificity of the real-time PCR
The real-time PCR gave positive results for the standard plasmid of PPV strains (6.00 × 108 copies/μL) and negative results for the other porcine viruses involved in reproductive disorders (PCV2, PRV, PRRSV, CFSV, and JEV) and for the sterile water control (Figure 1D).
Reproducibility of the real-time PCR
When the serially diluted standard plasmid (6.00 × 108, 6.00 × 106, 6.00 × 104, and 6.00 × 102 copies/μL) was subjected to the real-time PCR, the within-block CV value (four replicate assays for each dilution performed at one time) and the among-block CV value (the means of four replicates from each of four times) were relatively small (Table 1).
Table 1.
Coefficients of variation for the real-time PCR for PPV-NS1 performed for four concentrations of the standard plasmid at one time (one block with four replications per concentration) or at four different times (four blocks).
| Within one block or among four blocks | Concentration of standard plasmid (copies/μL) | n | Ct (mean) |
S.D. | CV (%) |
|---|---|---|---|---|---|
| Within | 6.0 × 108 | 4 | 17.67 | 0.38 | 2.15 |
| Within | 6.0 × 106 | 4 | 24.38 | 0.53 | 2.17 |
| Within | 6.0 × 104 | 4 | 30.23 | 0.44 | 1.46 |
| Within | 6.0 × 102 | 4 | 37.18 | 0.36 | 0.97 |
| Among | 6.0 × 108 | 4 | 17.35 | 0.04 | 0. 21 |
| Among | 6.0 × 106 | 4 | 24.30 | 0.14 | 0. 57 |
| Among | 6.0 × 104 | 4 | 30.83 | 0.14 | 0. 46 |
| Among | 6.0 × 102 | 4 | 37.80 | 0.16 | 0. 42 |
Detection of PPV in clinical samples by real-time PCR and conventional PCR
Real-time PCR and conventional PCR were used simultaneously with 41 samples that had been collected from several swine herds for diagnostic purposes. Real-time PCR detected PPV in 32 samples and conventional PCR detected PPV in 11 samples; therefore, they were 100% sensitivity and 100% specificity (Table 2).
Table 2.
Detection of PRRSV in 41 clinical samples by real-time PCR vs. conventional PCR.
| Conventional PCR | ||
| Positive | Negative | |
| Real-time PCR | ||
| Positive | 11a | 21c |
| Negative | 0b | 9d |
a, d indicate the number of samples that generated similar data for both assays.
b, dindicate the number of samples that generated different data for the assays.
Discussion
This study describes a real-time PCR assay for PPV based on detection of the NS1 gene. The real-time PCR assay was 100 times more sensitive than conventional PCR. The assay was specific in that it provided positive results with different strains of PPV but negative results with other viruses (PCV2, PRV, PRRSV, CFSV, JEV) associated with reproductive disorders of swine. The real-time PCR assay was also highly reproducible.
The real-time PCR method has several advantages for detection of PPV. First, the real-time PCR is more sensitive than conventional PCR, and high sensitivity is required for early diagnosis of PPV in the clinic. Second, the real-time PCR is faster than conventional PCR because it does not require gel electrophoresis. Third, the TaqMan real-time PCR described here is less likely to produce a false positive than a conventional PCR assay or the SYBR Green I-based real-time PCR. TaqMan-based real-time PCR may be more specific than SYBR Green I-based real-time PCR because the former requires specific probes that bind with PPV DNA template, while the latter uses probes that can bind to any double-stranded DNA, even non-specific amplicons.
Design of the primers and probe was based on the NS1 region of the PPV genome because this region is highly conserved. NS1 protein is believed to be a bifunctional protein with ATPase and helicase activities. In all parvoviral DNAs, both the 5'- and 3'-terminal palindromic sequences act as primers during replication [10].
Conclusion
In conclusion, the TaqMan real-time PCR assay has been shown to be rapid, sensitive, specific, and reproducible for the detection and quantification of PPV. It should be an excellent tool for laboratory detection of PPV in tissue-culture samples as well as in field samples. The TaqMan real-time PCR assay will be useful for studying the molecular epidemiology of PPV infections in swine populations. The assay will also be very useful for early diagnosis of PPV and therefore for management of PPV.
Contributor Information
Cuiping Song, Email: song_cuiping@189.cn.
Chao Zhu, Email: zhuchao2046@126.com.
Chaofan Zhang, Email: chaofanzhang@163.com.
Shangjin Cui, Email: cuishangjin@yahoo.cn.
Acknowledgements
The study was supported in part by funding from the National High-tech R&D Program (863 Program-2007AA100606) and the Chinese National Key Laboratory of Veterinary Biotechnology Fund (NKLVBP201002).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CPS and CZhu carried out the molecular probe studies, and drafted the manuscript. CZha participated in the design of the study and performed the statistical analysis. SC conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
References
- Mengeling WL. In: Diseases of Swine. Ninth. Straw BE, Zimmerman JJ, D'Allaire S, Taylor DJ, editor. Black-well Publishing, Ames, IA; 2006. Porcine parvovirus; pp. 93–1400. [Google Scholar]
- Hong-Ying Chen, Xiao-Kang Li, Bao-An Cui, Zhan-Yong Wei, Xin-Sheng Li, Yan-Bin Wanga, Li Zhaoa, Zhen-Ya Wanga. A TaqMan-based real-time polymerase chain reaction for the detection of porcine parvovirus. Journal of Virological Methods. 2009. pp. 84–88. [DOI] [PubMed]
- Mengeling WL, Lager KM, Zimmerman JK, Samarikermani N, Beran GW. A current assessment of the role of porcine parvovirus as a cause of fetal porcine death. J Vet Diagn Invest. 1991;3:33–35. doi: 10.1177/104063879100300107. [DOI] [PubMed] [Google Scholar]
- Kim J, Chae C. A comparison of virus isolation, polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine circovirus 2 and porcine parvovirus in experimentally and naturally coinfected pigs. J Vet Diagn Invest. 2004;16(1):45–50. doi: 10.1177/104063870401600108. [DOI] [PubMed] [Google Scholar]
- Zeeuw EJ, Leinecker N, Herwig V, Selbitz HJ, Truyen U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J Gen Virol. 2007;88:420–427. doi: 10.1099/vir.0.82302-0. [DOI] [PubMed] [Google Scholar]
- Shackelton LA, Hoelzer K, Parrish CR, Holmes EC. Comparative analysis reveals frequent recombination in the parvoviruses. J Gen Virol. 2007;88:3294–3301. doi: 10.1099/vir.0.83255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor TW, Joo HS, Collet MS. Porcine parvovirus Virus purification and structural and antigenic properties of viron polypeptides. Virology. 1983;45:842–854. doi: 10.1128/jvi.45.2.842-854.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares RM, Durignon EL, Bersano JG, Richtzenhain LJ. Detection of porcine parvovirus DNA by the polymerase chain reaction assay using primers to the highly conserved nonstructural protein gene, NS-1. J Virol Methods. 1999;78:191–198. doi: 10.1016/S0166-0934(98)00177-3. [DOI] [PubMed] [Google Scholar]
- Wilhelm S, Zeeuw EJ, Selbitz HJ, Truyen U. Real-time PCR protocol for the detection of porcine parvovirus in field samples. J Virol Methods. 2006;134:257–260. doi: 10.1016/j.jviromet.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Astell CR, Thomson M, Chow MB, Ward DC. Structure and replication of minute virus of mouse DNA. Cold Spring Harb Sym Quant Biol. 1983;47:751–762. doi: 10.1101/sqb.1983.047.01.086. [DOI] [PubMed] [Google Scholar]