Abstract
Background
Vibrio Pathogenicity Island-2 (VPI-2) is a 57 kb region present in choleragenic V. cholerae isolates that is required for growth on sialic acid as a sole carbon source. V. cholerae non-O1/O139 pathogenic strains also contain VPI-2, which in addition to sialic acid catabolism genes also encodes a type 3 secretion system in these strains. VPI-2 integrates into chromosome 1 at a tRNA-serine site and encodes an integrase intV2 (VC1758) that belongs to the tyrosine recombinase family. IntV2 is required for VPI-2 excision from chromosome 1, which occurs at very low levels, and formation of a non-replicative circular intermediate.
Results
We determined the conditions and the factors that affect excision of VPI-2 in V. cholerae N16961. We demonstrate that excision from chromosome 1 is induced at low temperature and after sublethal UV-light irradiation treatment. In addition, after UV-light irradiation compared to untreated cells, cells showed increased expression of three genes, intV2 (VC1758), and two putative recombination directionality factors (RDFs), vefA (VC1785) and vefB (VC1809) encoded within VPI-2. We demonstrate that along with IntV2, the RDF VefA is essential for excision. We constructed a knockout mutant of vefA in V. cholerae N16961, and found that no excision of VPI-2 occurred, indicating that a functional vefA gene is required for excision. Deletion of the second RDF encoded by vefB did not result in a loss of excision. Among Vibrio species in the genome database, we identified 27 putative RDFs within regions that also encoded IntV2 homologues. Within each species the RDFs and their cognate IntV2 proteins were associated with different island regions suggesting that this pairing is widespread.
Conclusions
We demonstrate that excision of VPI-2 is induced under some environmental stress conditions and we show for the first time that an RDF encoded within a pathogenicity island in V. cholerae is required for excision of the region.
Background
Vibrio cholerae is the etiological agent of the severe diarrheal disease cholera. Similar to many Gram-negative enteric pathogens, horizontal gene transfer and recombination plays a significant role in the evolution and emergence of new pathogenic strain of this species [1-12]. The main cause of the explosive rice water diarrhea characteristic of cholera is the cholera toxin (CT), an AB type enterotoxin, which is encoded within the ssDNA filamentous phage CTXɸ [13,14]. The B subunit of CT binds to the GM1 gangliosides, which are exposed when higher order gangliosides found in the intestinal mucus are cleaved by sialidase/neuraminidase (NanH). This protein is encoded within a 57 kb region named Vibrio Pathogenicity Island-2 (VPI-2) [15,16]. In addition to encoding sialidase, VPI-2 also encodes the sialic acid catabolism (SAC) gene cluster (Figure 1A) [16-19]. The SAC cluster was shown to be present only in pathogenic isolates of V. cholerae and enables the bacterium to grow on sialic acid as a sole carbon source [18,20]. Recently, we demonstrated that the ability to catabolize sialic acid gives V. cholerae a competitive advantage in vivo [19]. In non-O1/O139 pathogenic isolates, in addition to the SAC cluster are the genes required for a type 3 secretion system which is important for virulence [21-25]. The toxin co-regulated pilus (TCP), an essential intestinal colonization factor for V. cholerae, is encoded within the 40 kb Vibrio Pathogenicity Island-1 (VPI-1 or TCP Island) region [26,27].
Figure 1.
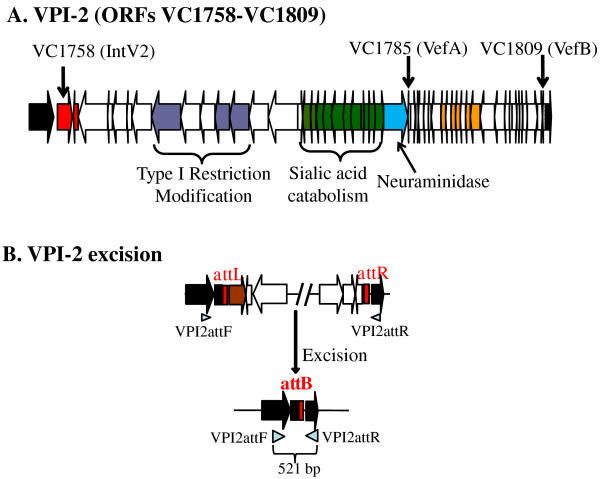
Vibrio Pathogenicity Island-2 (VPI-2) ORFs and primers used in this study. A. Schematic representation of VPI-2. Small black vertical arrows mark ORFs VC1758 (IntV2), VC1785 (VefA), or VC1809 (VefB). Block arrows represent ORFs and direction of transcription. Black arrows represent core genome ORFs (VC1757 and VC1810) present in all V. cholerae isolates; red arrow indicates an integrase, purple arrows indicate type 1 restriction modification ORFs, green arrows represent sialic acid catabolism, transport and regulator ORFs, blue arrow indicates sialidase/neuraminidase and yellow arrows represent transposase and Mu-phage like proteins. B. Schematic of VPI-2 excision mechanism and primer pair VPI2attF and VPI2attR used to detect the VPI-2 attB locus after excision of the entire region.
VPI-1 and VPI-2 do not share any genes in common but do share some functional characteristics such as the ability to integrate into the chromosome, specifically at a tRNA site using an integrase belonging to the tyrosine recombinase family [16,18,23,26,28]. VPI-2 integrates into chromosome 1 at a tRNA-serine locus, whereas VPI-1 is located at the tmRNA locus. Both regions are flanked by direct repeats (DRs) named attL and attR [16,18,23,26,28]. These integrases, IntV1 (VC0847) and IntV2 (VC1758), are believed to mediate insertion into the host chromosome through site specific recombination between an attachment site attP, present in the pathogenicity island, and attB, present in the bacterial chromosome.
Pathogenicity islands have been shown to excise from their host genome in pathogenic Escherichia coli and Yersinia species [29-36]. In E. coli strain 536, a uropathogenic isolate, Hacker and colleagues have identified six PAIs, all of which encode a tyrosine recombinase integrase and are flanked by DRs [31,33,36-39]. They demonstrated that PAI-I, II, III and V can excise from the chromosome by site-specific recombination involving their respective DRs (attL and attR) [31,33]. The PAIs were shown to excise at different frequencies depending on the growth conditions [31,33]. Likewise, both VPI-1 and VPI-2 have been shown to excise from their host chromosome [23,28]. Rajanna and colleagues demonstrated that VPI-1 can excise from V. cholerae N16961 at very low rates [28]. They determined that the integrase IntV1 (VC0847) was not essential for excision since a transposase within the region appeared to compensate for an IntV1 knockout [28]. Recently, Murphy and Boyd demonstrated that VPI-2 from V. cholerae N16961 can excise from chromosome 1, which also occurred at very low frequency under optimal growth conditions [23]. Their study showed that IntV2 (VC1758) was essential for excision and the formation of a circular intermediate (CI) [23]. Pathogenicity islands from both E. coli and V. cholerae are non-self mobilizable, they do not encode any proteins such as those for phage structural proteins or conjugation systems needed for cell to cell mobility [23,28,31,33,36-39]. The mechanism of transfer for most pathogenicity islands remains to be elucidated but likely involves hitchhiking with plasmids, conjugative transposons, Integrative and Conjugative Elements (ICEs), or generalized transducing phages or uptake by transformation.
It is known that for some mobile and integrative genetic elements (MIGEs) the presence of a recombination directionality factor (RDF)/excisionase is required for excision [40,41]. For instance, Xis is required for the excision of the ICE SXT from V. cholerae [41], Hef from the High Pathogenicity Island of Yersinia pestis [32], and Rox from the Shigella Resistance Locus (SRL) of Shigella flexneri [42]. RDFs are small basic proteins that bind and bend DNA on the recombination sites attL and attR triggering excision by coordinating the assembly of the excisive intasome [43-45]. In addition, some RDFs have been found to inhibit reintegration of the CI by converting attP into a catalytically inactive structure and are thought to stabilize the appropriate positioning of the integrase within the excisive intasome [46-48]. To date, no RDFs have been identified in E. coli or V. cholerae pathogenicity islands.
Here, we report the environmental conditions that induce excision of VPI-2. We examined the VPI-2-encoded factors that are required for VPI-2 excision, determining that V. cholerae cells subjected to stress conditions showed an increase in the excision levels of VPI-2 compared to cell grown at optimal conditions. Bioinformatic analysis of the VPI-2 region identified two open reading frames (ORFs) VC1785 and VC1809 that show homology to previously described RDFs, which we named VefA and VefB. We examined the role of these genes in VPI-2 excision.
Methods
Bacterial strains and growth conditions
The strains and plasmids used in this study are listed in table 1. Bacteria were grown in lysogeny broth more commonly known as Luria-Bertani broth (LB), LB agar, or LB agar 10% sucrose without NaCl (LB-Suc) [49]. Strains harboring the pBAD33 expression vector were grown on LB supplemented with 0.02% W/V of L-Arabinose (LB-Ara). Bacteria were incubated overnight at 37°C with aeration unless otherwise indicated. When required, ampicillin (Amp, 100 μg/ml), streptomycin (Sm, 200 μg/ml), or chloramphenicol (Cm, 25 μg/ml) were added to the media.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains/plasmids | Genotype and/or phenotype | Reference |
|---|---|---|
| V. cholerae | ||
| N16961 | O1 El Tor, VPI-2 +, SmR | [57] |
| RAM-1 | N16961, ΔVC1758, SmR | [23] |
| SAM-1 | RAM-1, pIntV2, SmR CmR | This study |
| SAM-3 | N16961, ΔVC1785, SmR | This study |
| SAM-4 | N16961, ΔVC1809, SmR | This study |
| SAM-5 | SAM3, pVefA, SmR CmR | This study |
| SAM-11 | N16961, pBAD33, SmR CmR | This study |
| SAM-12 | RAM-1, pBAD33, SmR CmR | This study |
| SAM-13 | SAM-3, pBAD33, SmR CmR | This study |
| Plasmids | ||
| pDS132 | Suicide plasmid, CmR, SacB | [59] |
| pBAD33 | Expression plasmid, Ara, CmR | [60] |
| pIntV2 | vc1758 cloned into pBAD33 | This study |
| pD1785 | ΔVC1785 cloned into pDS132 | This study |
| pD1809 | ΔVC1809 cloned into pDS132 | This study |
| pVefA | vc1785 cloned into pBAD33 | This study |
Determination of VPI-2 excision rate
Excised circular VPI-2 DNA containing attP is expected to be a very rare event given the predicted low excision rate under normal conditions and the inability of VPI-2 to replicate after excision [23]. Therefore, we quantified the excision rates of VPI-2 by measuring the presence of attB, the locus present on the V. cholerae chromosome after VPI-2 excises (Figure 1B), in different strains under different conditions, and comparing it with the presence of attB in cultures of V. cholerae N16961 grown under standard optimal conditions: 12 hours in LB at 37°C with aeration. Using the O.D. values of 1 mL of a culture of V. cholerae N16961 grown for 12 hours in LB at 37°C with aeration as a reference, 750 μL to 4 mL were pelleted by centrifugation and genomic DNA was extracted using ABI PrepMan Ultra reagent from the test cultures. We took 50 μL from each DNA extraction and diluted each with 200 μL of sterile ddH2O. A 5 μL aliquot of DNA after dilution was used as template for Real-Time quantitative PCR (QPCR) reactions. The QPCR assay calculated the percentage of cells in a culture that contained an unoccupied VPI-2 attB site. We quantified attB sites present in cell grown under different growth conditions and normalized to the amount of attB present in N16961 grown for 12 hours at 37°C. The gene-specific primers were designed using Primer3 software according to the real-time PCR guidelines, and are listed in Table 2. The Applied Biosystems 7000 system was used for RT fluorescence detection of PCR products that resulted from binding of the dye SYBR Green to double stranded DNA and the results were examined with Applied Biosystems SDS software V 1.3. The reference gene mdh was assayed both separately and in the same reaction. To confirm that primer pairs only amplified target genes to assure accurate quantification of the results, non-template controls were included in each replicate. The attB and mdh PCR products were visually checked on agarose gels. The melting curves of PCR products were used to ensure the absence of primer dimers, contamination with genomic DNA and non-specific homologous sequences. PCR reactions were performed in 10 uL volumes containing 5 uL of 2X SYBR Green PCR Master Mix (Applied Biosystems), 900 nm of each primer, and 1 uL of DNA template. PCR cycling conditions were 30 sec at 95°C followed by 40 cycles of 15 sec at 95°C and 30 sec at 60°C. Serial doubling dilutions were used as templates for QPCR to generate standard curves for each PCR reaction by plotting relative DNA concentrations versus log (Ct) value (Ct is the PCR cycle at which fluorescence rises beyond background). The Ct value for mdh was 15 cycles and for attB 30 cycles. Every sample was assayed in triplicate and each experiment was performed using a minimum of three different samples. Differences in the attB ratio were extrapolated using the delta-delta Ct method as developed by Pfaffl [50].
Table 2.
Oligonucleotide primers used in this study.
| Oligo name | Sequence (5'-3'). Restriction site underscored | Target gene |
|---|---|---|
| SOE PCR | ||
| VC1785A | GAGCTCAATGGTGCATCGGCATATTCT | vefA (A-D) |
| VC1785B | CAGCGATGATGGCGTGATTA | |
| VC1785C | TAATCACGCCATCATCGCTGGGATGT TCTCCTATGTCTTGT | |
| VC1785D | TCTAGACGCGCACATAACGCTGTTC | |
| VC1809A | CTGCAGTGAGAGCAAGGGAAGTGATCGT | vefB(A-D) |
| VC1809B | CGAGAGCTGGTTTACTTGTGTG | |
| VC1809C | CACACAAGTAAACCAGCTCTCGGTGCATGATTGTCAAGTCATGCA | |
| VC1809D | GAGCTCGATGGCTATGAATTAGCTCAGGA | |
| Flanking | ||
| VC1785FF | GATGCTTCTATTACTCGGTT | vefA (F-R) |
| VC1785FR | TCACCGCTGCTGCGTTAA | |
| VC1809FF | GATTGATAGTAACAACACGCG | vefB (F-R) |
| VC1809FR | GTAATGCGCTATTGCTAAGTG | |
| VPI2attF | AGAGTGAAAGTCGCCAAAGC | attB (F-R) |
| VPI2attR | GGGTGCAATTTCGCATGTTGC | |
| Complements | ||
| VC1758CF | GAGCTCGAGTCCTCATGCTCTAGCCAG | intV2 (F-R) |
| VC1758CR | TCTAGAGGCATGCTGGTGTGTTACTAC | |
| VC1785CF | GAGCTCGCTTTGAATATAGGTAAGGGACTG | vefA (F-R) |
| VC1785CR | TCTAGACTATAGTACATGACGCATGTATAATC | |
| Real-Time PCR | ||
| VPI2attBQF | GATTCGGTGAGTTGTCCGAGT | attB (VPI-2) (F-R) |
| VPI2attBQR | GTGTTGGTGCAATGCTCAGTC | |
| VC1758QF | TGCATGATCTTATACTCACCG | intV2 (F-R) |
| VC1758QR | ACGTGCTCGCGGTTCATCTTC | |
| VC1785QF | CCGCACTAAGCCGTTCAGCAA | vefA (F-R) |
| VC1785QR | CCATCCACTCATCCACTTCGC | |
| VC1809QF | CTGAGAGGTGTGAATATGCCAG | vefB (F-R) |
| VC1809QR | GTGATCCGTTGAGCAATCCAC | |
| VC0432QF | TGTATGATATTGCGCCTGTCACAC | mdh(F-R) |
| VC0432QR | CCAGAACCACATCCGCACCTTC |
Underlined sequence represents restriction enzyme sites SacI, XbaI and Pst1.
Bioinformatic analysis
BLAST search was performed using Xis (ABA87014), an RDF from V.cholerae SXT ICE element required for excision, AlpA, a well known RDF from E. coli (AAA18418) and the Hef protein (NP_405464) from Y. pestis pathogenicity island, as seeds on the genome sequence of V. cholerae N16961 [51]. DNA sequences from putative RDFs were downloaded from GenBank and the sequences were aligned using ClustalW [52]. Next, the protein sequences of characterized RDFs were used as seeds to perform BLASTN and BLASTP searches against Vibrio genomes sequences in the database [51]. The retrieved sequence must give an e-value below 10-3, relative to known RDFs.
RNA extraction and Real-Time quantitative PCR (QPCR)
Total RNA from V. cholerae N16961 was extracted 12 hours post-inoculation in LB broth from one group treated with sub-lethal UV-irradiation and one group untreated as follows. The cells from 5 mLs 11 hours growth cultures were pelleted and resuspended in 5 mLs of PBS. A 100 uL aliquot was taken from each sample prior to treatment to calculate colony forming units (CFUs). Each 5 mL sample was placed in a plastic Petri dishes without a cover and one set of samples was irradiated with a sublethal dose of 25 J/m2 of UV irradiation in a Fisher Scientific UV cross linker (FB-UVXL-1000) and the other 5 mL set of samples was left untreated as previously described by others [53]. The cells from both UV treated and untreated samples were recovered, pelleted, resuspended in 5 mLs of LB broth and grown for 1 hour at 37°C. A 100 uL aliquot was taken from each sample to calculate CFUs post treatment from both sets of samples. The CFU counts pre and post treatment were identical at ~9.75 × 109/ml as expected. Every experiment was performed in triplicate. Total RNA was extracted from each culture using RNAprotect Bacteria reagent (Qiagen, Valencia, CA) and an RNeasy mini kit (Qiagen) according to the manufacturer's protocols. RNA purity and the presence of genomic DNA were assessed using an ND-1000 NanoDrop UV-Vis spectrophotometer (NanoDrop Technologies) giving values of A260/A280 > 2.0 and A260/A230 > 2.0 indicating of no protein and solvent contamination, respectively. In addition, 1 μg of each sample of RNA was run on a 1% agarose gel in 1× TBE buffer to examine quality of the samples. RNA was measured to calculate the volume of sample to be added to perform a reverse transcriptase (RT) reaction using SuperScript II Reverse Transcriptase and random hexamers following manufacturer's instructions (Invitrogen). The purity and quantity of cDNA was examined using an ND-1000 NanoDrop UV-Vis spectrophotometer as above. QPCR was performed using standard protocol using primer pairs for vc1758, vc1785, vc1809 and vc0432 (intV2, vefA, vefB and mdh, respectively) listed in Table 2 using SYBR green PCR Master Mix (Invitrogen) on an Applied Biosystems 7000 Real Time PCR System (Foster City, CA). To confirm that primer pairs only amplified target genes to assure accurate quantification of the results, non-template controls were included in each replicate. The intV2, vefA, vefB and mdh PCR products were visually checked on agarose gels. The melting curves of PCR products were used to ensure the absence of primer dimers, contamination with genomic DNA and non-specific homologous sequences. The data was analyzed using ABI PRISM 7000 SDS software (Applied Biosystems). Differences in the gene ratios were extrapolated using the delta-delta Ct method [50]. Every sample was assayed in triplicate and each experiment was performed using a minimum of three different samples.
Construction of mutant strains
To construct the mutant strains, primers were designed to conduct Splice Overlap Extension (SOE) PCR followed by allelic exchange [54]. SOE PCR primers were designed to produce non-functioning constructs of the 204-bp vefA and the 228-bp vefB genes. The size of the regions removed from vefA and vefB is 169-bp and 191-bp, respectively and were constructed in V. cholerae strain N16961 to create mutant strains V. cholerae SAM-3 and SAM-4, respectively (Table 1). Primer pairs SOEVC1785A/SOEVC1785B and SOEVC1785C/SOEVC1785 D were used to amplify PCR products from VC1785 from V. cholerae strain N16961 (Table 2). The ligated product was amplified with primer pair SOEVC1785A and SOEVC1785 D, which was restricted with enzymes, XbaI and SacI and ligated with pDS132 (New England Biolabs) resulting in pΔ1785. pΔ1785 was transformed into E. coli strain DH5αλpir, plasmid purified and then transformed into E. coli β2155 cells. E. coli β2155 transformants were conjugated with N16961. V. cholerae cells were passaged in LB-suc to cure them of the integrated pΔ1785. PCR was used to screen for V. cholerae strains in which the wild type gene was replaced by the mutant gene, which was confirmed by sequencing. The Δ1785 strain was designated V. cholerae strain SAM-3. A knockout mutant of VC1809 was constructed in N16961 as described above using primer pairs listed in Table 2. Complementation of RAM-1 and SAM-3 mutant strains were generated by creating pIntV2 and pVefA, by cloning intV2 (VC1758) and vefA (VC1785), respectively into the SacI/XbaI sites of the expression plasmid pBAD33 (New England Biolabs) using standard cloning protocol (Table 1 and 2).
Results and Discussion
VPI-2 excision rates under different growth conditions
It was previously shown that the four pathogenicity islands identified in V. cholerae N16961 can excise from chromosome 1 and form circular intermediates (CI) [23,28]. The excision of VPI-1 and VPI-2 occurs at very low levels suggesting that excision is tightly controlled, although it may also suggest that the excision event is inefficient, possibly due to poor expression of the regulatory genes, an altered regulatory circuit, or mutations that might occur in these sequences as the region become evolutionarily integrated into the host chromosome [23,28]. First, we quantified the excision levels of VPI-2 in cultures of V. cholerae N16961 grown for 12 hours in LB at 37°C (standard conditions) by measuring the presence of attB, the locus present on the chromosome after VPI-2 excises (Figure 1), and comparing it with the housekeeping gene mdh using QPCR. We used attB as a surrogate for VPI-2 excision measurements since the copy number of attP in the CI is minuscule compared to attB, which replicates along with the rest of the chromosome unlike excised VPI-2. We compared the presence of attB with mdh since all cells encode one functional copy of the latter. PCR products of attB and mdh were visually checked on an agarose gel and their melting temperature analyzed to ensure we had the correct PCR products. The reference gene was assayed both separately and in the same reaction. Both primer pairs used were tested by comparing the results obtained using previously quantified cloned copies of mdh and attB and gave comparable results. We found that attB was present in 1 in every 1.6 (±0.2) × 106 V. cholerae cells under optimal growth conditions.
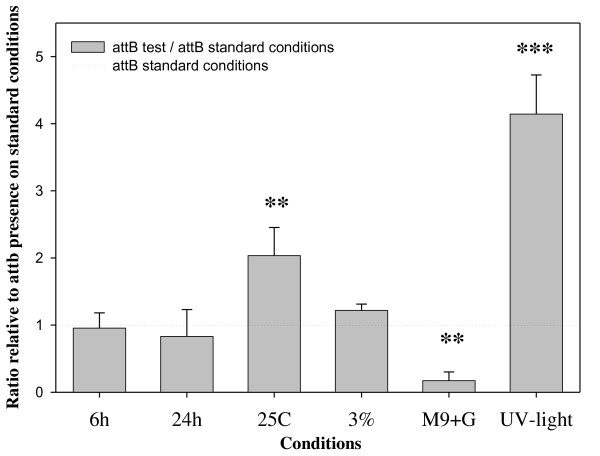
Next, we measured the presence of attB from V. cholerae cells cultured under different conditions compared with the presence of attB under our standard condition, growth for 12 hours at 37°C. We determined that incubation time does not affect the excision levels of VPI-2 indicating that excision does not occur in a growth phase dependent manner (Figure 2). However, V. cholerae cultures grown at 25°C showed a 2-fold increase in the presence of the attB site compared to cells grown at the optimum temperature 37°C (Figure 2). In addition, we found that nutrient limitation affected the excision level showing over a 5-fold decrease in the presence of attB when compared to the growth on LB at the same temperature (Figure 2). Furthermore, we found that sub-lethal UV-light irradiation of cell cultures compared to untreated cells, resulted in a significant increase in the level of excision of VPI-2, over 4-fold compared to untreated cells grown under the same conditions (Figure 2). Taken together, these data indicate that environmental factors can affect the induction of excision and circularization of VPI-2, which is probably the first step required for the horizontal transfer of the region. These results are consistent with what was previously shown for other mobile and integrative genetic elements as well as PAIs from E. coli, where excision occurs upon exposure to stress conditions such as sub-lethal UV-light irradiation [53,55,56].
Figure 2.
Detection of VPI-2 excision by using real time quantitative PCR (QPCR) of attB levels in cell cultures grown under different conditions. The X-axis specifies culture conditions: 6 h, incubation time of 6 h; 24 h, incubation time of 24 h; 25C, incubation temperature of 25°C; 3%, the LB broth contained 3% NaCl; M9+G, cell grown on minimal media supplemented with glucose; UV-light, bacterial cultures were UV-light irradiated. The Y-axis represent the ratio of the attB presence in the cultures tested compared with cultures grown on standard conditions 12 h at 37°C in LB. Unpaired t-test was used in order to infer statistical significance for the differences in VPI-2 excision. ***, p < 0.005. **, p < 0.05. Error bars indicate standard deviation. Each experiment was performed in triplicate a minimum of three times.
VPI-2 encodes two novel recombination directionality factors
Both the high pathogenicity island HPI from Y. pestis and ICE SXT from V. cholerae encode small accessory proteins called recombination directionality factors (RDFs) or excisionases (Xis) that are required for efficient excision of these elements [29,41]. In order to identify candidate RDFs within VPI-2 from V. cholerae N16961, we performed BLAST and PSI-BLAST searches on the V. cholerae N16961 genome using RDFs, the V. cholerae Xis protein (ABA87014) from SXT, the Y. pestis Hef protein (NP_405464) from HPI and E. coli K12 AlpA protein (AAA18418) from λ phage as queries [57]. The most significant BLAST result in these searches was ORF VC0497, which is annotated as a transcriptional regulator, and is encoded within Vibrio Seventh Pandemic island-II (VSP-II). VSP-II also encodes a tyrosine recombinase integrase at ORF VC0516 (IntV3) [58]. ORFs VC1785 and VC1809 encoded within VPI-2 were the second and third most significant hits retrieved from these BLAST searches, which we termed VefA (for Vibrio excision factor A) and VefB, respectively (Figure 3). The VefA and VefB proteins share 46% amino acid identity/72% similarity. VefA shares 37% amino acid identities with AlpA, 46% identity with Hef and 29% with Xis from the V. cholerae SXT element as was previously shown [53] (Figure 3). The vefB gene is located at the 3' end of VPI-2 at ORF VC1809 marking the end of the island, and vefA (VC1785) is adjacent to neuraminidase gene, nanH (VC1784) in the middle of the island (Figure 1A).
Figure 3.
Alignments of VPI-2 RDFs VefA and VefB with other known RDFs: AlpA (AAA18418), Hef (NP_405464), Xis (ABA87014). For both alignments black shading highlights residues shared by all the sequences and grey shading highlights residues shared by all but one sequences. The numbers on the right indicate the number of amino acids of the predicted protein.
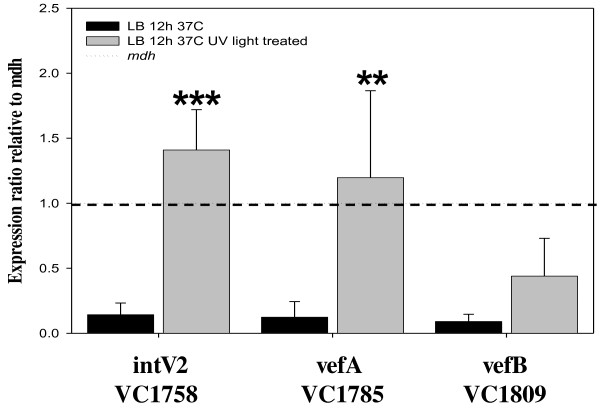
As shown in Figure 2, UV-light irradiation increased excision of VPI-2 over 4-fold. In order to investigate this further, we determined the effect of UV-light irradiation on the expression of intV2, vefA and vefB in V. cholerae N16961 (Figure 4). We examined transcript levels of intV2, vefA and vefB in cells grown for 12 h in LB and in cells grown for 12 h in LB followed UV-light irradiation treatment. We found that all three genes showed negligible levels of transcription under standard optimum growth conditions but after UV-light treatment both intV2 and vefA show a 10-fold and vefB a 5-fold increase in expression levels (Figure 4). These results indicate that UV-light induces expression of factors potentially involved in VPI-2 excision.
Figure 4.
Expression of intV2 (VC1758), vefA, and vefB from cultures grown in standard (black bars) or UV-light irradiated cultures (grey bars). The Y-axis represents the expression ratio of the genes relative to the expression of mdh. Unpaired t-test was used in order to infer statistical significance for the differences in gene expression between cultures of V. cholerae N16961 with or without UV-light treatment. **, p < 0.05; ***, p < 0.005. Error bars indicate standard deviation. Each experiment was performed in triplicate a minimum of three times.
IntV2 and VefA are essential for the excision of VPI-2
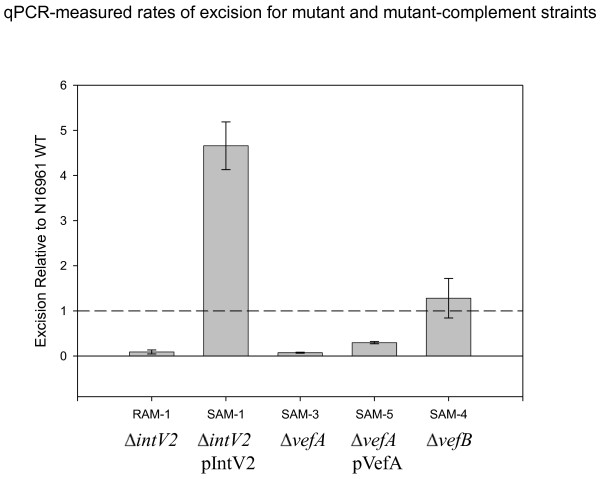
To determine in more detail the role of intV2, vefA and vefB in VPI-2 excision, we created deletion mutations in each gene and measured excision levels of VPI-2 by determining attB levels in cells. In V. cholerae RAM-1, an intV2 mutant, we did not detect any VPI-2 attB products, demonstrating that intV2 is essential for excision as was previously shown (Figure 5) [23]. We complemented RAM-1 with a functional copy of intV2 by transforming V. cholerae RAM-1 with pIntV2 creating strain SAM-1. In our SAM-1 strain, we found that excision of VPI-2 was restored in addition, attB levels were approximately four-fold higher than wild-type levels which is represented by the dotted broken horizontal line in Figure 5. These data demonstrate that over expressing intV2 ectopically induces excision of VPI-2. In our control experiments, transformation of either wild-type N16961 or RAM-1 with pBAD33 alone (strains SAM-11 and SAM-12 respectively) did not affect attB levels (data not shown).
Figure 5.
Excision levels of VPI-2 in mutant strains and strains complemented with intV2 (VC1758), and vefA (VC1785). Excision levels of ΔintV2 mutant (RAM-1), ΔintV2 mutant complemented (SAM-1), ΔvefA mutant (SAM-3), ΔvefA mutant complemented (SAM-5), and ΔvefB mutant (SAM-4). Unpaired t-test was used in order to infer statistical significance for the differences in VPI-2 excision between V. cholerae N16961 and test strains. **, p < 0.05; ***, p < 0.005. Error bars indicate standard deviation. Each experiment was performed in triplicate a minimum of three times.
Next, we determined whether one, both, or neither of the putative RDFs uncovered by our bioinformatic analysis are required for VPI-2 excision. To do this, we constructed in-frame deletion mutations in each gene to create mutant strain SAM-3 (ΔvefA) and SAM-4 (ΔvefB). The two mutant strains and the wild-type N16961 were each inoculated into LB and all three strains grew similarly indicating that the mutant constructs did not have any general growth defect (data not shown). We determined the attB levels using QPCR in strain SAM-3 compared to the wild-type strain grown under the same conditions. We found that no VPI-2 excision occurs in SAM-3 cells when compared with the wild type, indicating that a functional copy of vefA is essential for efficient excision of VPI-2 (Figure 5). We complemented SAM-3 with a functional copy of vefA (SAM-5) and measured attB levels in these cells with the wild type levels both under standard conditions, to find that some excision occurred, but it was less than in wild-type cells (Figure 5). In our vefB mutant strain (SAM-4), we found no difference in VPI-2 excision levels compared to wild-type grown under the same conditions, which demonstrates that vefB is not essential for excision (Figure 5). From these data it appears that vefA is the cognate RDF for VPI-2 excision. In our control experiments, transformation of SAM-3 with pBAD33 alone (resulting in strain SAM-13) did not affect attB levels (data not shown).
Vibrio species island-encoded integrases with corresponding RDFs
Given that our initial search for RDFs within one V. cholerae genome (strain N16961) yielded three putative RDFs (VC0497, VC1785, and VC1809), we decided to investigate further the occurrence of RDFs among Vibrio species whose genome sequence is available in the database. We performed BLAST searches against the 20 Vibrio species in the genome database, and we uncovered a total of 27 putative RDFs (Table 3). Next, we identified putative integrases within the genomes of the RDF homologues using BLAST search analysis by using IntV2 as a seed. For each of the RDFs identified among the 27 genomes encompassing 10 different Vibrio species (V. cholerae, V. coralliilyticus, V. furnissii, V. harveyi, V. parahaemolyticus, V. splendidus, V. vulnificus, Vibrio sp. Ex25, RC341, and MED222), we identified a corresponding integrase with greater than 40% amino acid identities to IntV2 (VC1758) (Table 3). We examined the gene context of each RDF and integrase within each of the 20 strains to determine whether the RDF and integrase were located on the same region within a strain. From these analyses, we found that each of the 27 RDFs has a corresponding integrase within approximately 100 kb of each other (Table 3). It should be noted that from table 3, only three of the strains have been annotated completely and for many of the strains examined their ORF annotation numbering is not consecutive. Within V. cholerae, integrases and RDFs located in the same region of the genome in different strains had the same gene content indicating the same island is present in different strains. Among the different species, however, integrases and RDFs associated with the same insertion site did not have the same gene content indicating a novel island region in the different species (data not shown).
Table 3.
Locus tags for integrases and corresponding RDFs identified in this study.
| Integrases | RDFs | |
|---|---|---|
| Species Strain | Locus tag | Locus tag |
| Vibrio cholerae N16961* | VC1758 | VC1785/VC1809 |
| Vibrio cholerae TM 11079-80 | VIF_001175 | VIF_000799 |
| Vibrio cholerae TMA21 | VCB_002798 | VCB_002857 |
| Vibrio cholerae 12129(1) | VCG_002315 | VCG_002259 |
| Vibrio cholerae V51 | VCV51_1204 | VCV51_0550 |
| Vibrio cholerae 1587 | A55_1986 | A55_2025 |
| Vibrio cholerae CT 5369-93 | VIH_002346 | VIH_002364 |
| Vibrio cholerae RC385 | VCRC385_0574 | VCRC385_3603 |
| Vibrio cholerae TMA 21 | VCB_000212 | VCB_000197 |
| Vibrio cholerae MZO-3 | A51_B0496 | A51_B0476 |
| Vibrio cholerae 12129(1) | VCG_003155 | VCG_003160 |
| Vibrio cholerae N16961* | VC0516 | VC0497 |
| Vibrio cholerae MZO-3 | A51_B0965 | A51_B0948 |
| Vibrio vulnificus YJO16* | VV2262 | VV2261 |
| Vibrio vulnificus YJ016* | VV0817 | VV0810 |
| Vibrio vulnificus YJO16* | VV0560 | VV0515 |
| Vibrio furnissii CIP 102972 | VFA_001916 | VFA_001914 |
| Vibrio furnissii CIP 102972 | VFA_000464 | VFA_000468 |
| Vibrio coralliilyticus ATCC BAA-450 | VIC_001980 | VIC_001987 |
| Vibrio sp. Ex25 | VEA_004301 | VEA_004310 |
| Vibrio sp. RC341 | VCJ_000330 | VCJ_000314 |
| Vibrio sp. MED222 | MED222_15534 | MED222_15529 |
| Vibrio splendidus 12B01 | V12B01_04993 | V12B01_05053 |
| Vibrio parahaemolyticus AQ3810 | A79_5467 | A79_5463 |
| Vibrio parahaemolyticus K5030 | VparK_010100010115 | VparK_010100010135 |
| Vibrio parahaemolyticus AQ3810 | A79_2546 | A79_2541 |
| Vibrio harveyi HY01 | A1Q_2023 | A1Q_2003 |
* indicates a genome that is completely annotated
From our analysis, no RDF was identified within the VPI-1 or the VSP-I regions in N16961 or within homologous regions in the other 27 sequenced strains of V. cholerae in the database. Both the VPI-1 and VSP-I regions have been shown to excise from their chromosome location, and VPI-1 encodes a tyrosine recombinase with homology to IntV2, thus they may therefore use an alternative mechanism of excision or perhaps co-opt an RDF from another region on the genome. Overall our data indicates that the presence of both an integrase and a cognate RDF pairing is a relatively conserved feature but not an essential one.
Conclusions
In this study, we analyzed the excision dynamics of VPI-2 encoded within V. cholerae N16961. Our results indicate that excision is controlled by at least two conserved factors within the island, an integrase encoded by intV2 and an RDF encoded by vefA, whose expression is induced by environmental stimuli similar to other MIGEs such as prophages, ICEs and integrons. We identified two putative RDFs and found that of the two we identified, only one VefA is essential for the efficient excision of VPI-2. We determined the occurrence of RDFs among the genomes of sequenced Vibrio species and found 27 putative RDFs that also had a homologue of IntV2 associated with it, which suggests that requirement for both an RDF and a corresponding integrase is a relatively common feature.
Authors' contributions
EFB designed the research; SA-M and MGN performed the research; SA-M, MGN and EFB analyzed data; SA-M, MGN and EFB wrote the paper.
Contributor Information
Salvador Almagro-Moreno, Email: Salvador.Almagro-Moreno@Dartmouth.EDU.
Michael G Napolitano, Email: lakhim@gmail.com.
E Fidelma Boyd, Email: fboyd@udel.edu.
Acknowledgements
This research was supported by National Science Foundation CAREER award DEB-0844409 to E.F.B.
The authors declare no conflicts of interest.
References
- Faruque SM, Sack DA, Sack RB, Colwell RR, Takeda Y, Nair GB. Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci USA. 2003;100(3):1304–1309. doi: 10.1073/pnas.0337468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Chowdhury N, Kamruzzaman M, Dziejman M, Rahman MH, Sack DA, Nair GB, Mekalanos JJ. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc Natl Acad Sci USA. 2004;101(7):2123–2128. doi: 10.1073/pnas.0308485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Quezada-Calvillo R, Marrero J, Waldor M. SXT-related integrating conjugative element in New World Vibrio cholerae. Appl Environ Microbiol. 2006;72:3054–3057. doi: 10.1128/AEM.72.4.3054-3057.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrin S, Gil AI, Bhuiyan NA, Safa A, Asakura M, Lanata CF, Hall E, Miranda H, Huapaya B, Vargas GC. et al. Peruvian Vibrio cholerae O1 El Tor strains possess a distinct region in the Vibrio seventh pandemic island-II that differentiates them from the prototype seventh pandemic El Tor strains. J Med Microbiol. 2009;58:342–354. doi: 10.1099/jmm.0.005397-0. [DOI] [PubMed] [Google Scholar]
- Tay C, Reeves P, Lan R. Importation of the major pilin TcpA gene and frequent recombination drive the divergence of the Vibrio pathogenicity island in Vibrio cholerae. FEMS Microbiol Lett. 2008;289:210–218. doi: 10.1111/j.1574-6968.2008.01385.x. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Nair GB, Tang L, Morris JG, Sharma NC, Ballal M, Garg P, Ramamurthy T, Stine OC. Epidemiological study of Vibrio cholerae using variable number of tandem repeats. FEMS Microbiol Lett. 2008;288(2):196–201. doi: 10.1111/j.1574-6968.2008.01352.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Fraga S, Pichel M, Binsztein N, Johnson JA, Morris JG Jr, Stine OC. Lateral gene transfer of O1 serogroup encoding genes of Vibrio cholerae. FEMS Microbiol Lett. 2008;286(1):32–38. doi: 10.1111/j.1574-6968.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Lee JH. et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106(36):15442–1547. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim CJ, Hasan NA, Taviani E, Haley B, Chun J, Brettin TS, Bruce DC, Detter JC, Han CS, Chertkov O. et al. Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J Bacteriol. 2010;192(13):3524–3533. doi: 10.1128/JB.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C, Octavia S, Reeves P, Wang L, Lan R. Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America. Emerg Infect Dis. 2010;16:1130–1132. doi: 10.3201/eid1607.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Ohnishi M, Arakawa E, Yamamoto S, Nair GB, Matsushita S, Yokoyama K, Kai A, Seto K, Watanabe H. et al. Emergence and genetic diversity of El Tor Vibrio cholerae O1 that possess classical biotype ctxB among travel-associated cases of cholera in Japan. J Med Microbiol. 2010;59(Pt 6):708–712. doi: 10.1099/jmm.0.017624-0. [DOI] [PubMed] [Google Scholar]
- Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18(1):46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- De SN. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature. 1959;183(4674):1533–1534. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- Galen JE, Ketley JM, Fasano A, Richardson SH, Wasserman SS, Kaper JB. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60(2):406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn WS, Boyd EF. Molecular evolution of Vibrio pathogenicity island-2 (VPI-2): mosaic structure among Vibrio cholerae and Vibrio mimicus natural isolates. Microbiology. 2005;151(Pt 1):311–322. doi: 10.1099/mic.0.27621-0. [DOI] [PubMed] [Google Scholar]
- Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99(3):1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn WS, Boyd EF. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology. 2002;148(Pt 11):3681–3693. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- Almagro-Moreno S, Boyd EF. Sialic Acid Catabolism Confers a Competitive Advantage to Pathogenic Vibrio cholerae in the Mouse Intestine. Infect Immun. 2009;77(9):3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9(1):118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L. et al. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci USA. 2005;102(9):3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Johnson JA, Pusch GD, Morris JG Jr, Stine OC. The genome of non-O1 Vibrio cholerae NRT36 S demonstrates the presence of pathogenic mechanisms that are distinct from those of O1 Vibrio cholerae. Infect Immun. 2007;75(5):2645–2647. doi: 10.1128/IAI.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol. 2008;190(2):636–647. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A, Tam V, Hamilton E, Dziejman M. vttRA and vttRB Encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect Immun. 2010;78(6):2554–2570. doi: 10.1128/IAI.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe. 2007;1(2):95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, Reeves PR. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95(6):3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84(9):2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanna C, Wang J, Zhang D, Xu Z, Ali A, Hou YM, Karaolis DK. The vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J Bacteriol. 2003;185(23):6893–6901. doi: 10.1128/JB.185.23.6893-6901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30(5):965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180(9):2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol. 2006;61(3):584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- Lesic B, Bach S, Ghigo JM, Dobrindt U, Hacker J, Carniel E. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol Microbiol. 2004;52(5):1337–1348. doi: 10.1111/j.1365-2958.2004.04073.x. [DOI] [PubMed] [Google Scholar]
- Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol. 2004;186(10):3086–3096. doi: 10.1128/JB.186.10.3086-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellaris H, Luck SN, Al-Hasani K, Rajakumar K, Turner SA, Adler B. Regulated site-specific recombination of the she pathogenicity island of Shigella flexneri. Mol Microbiol. 2004;52(5):1329–1336. doi: 10.1111/j.1365-2958.2004.04048.x. [DOI] [PubMed] [Google Scholar]
- Schubert S, Dufke S, Sorsa J, Heesemann J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol Microbiol. 2004;51(3):837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- Wilde C, Mazel D, Hochhut B, Middendorf B, Le Roux F, Carniel E, Dobrindt U, Hacker J. Delineation of the recombination sites necessary for integration of pathogenicity islands II and III into the Escherichia coli 536 chromosome. Mol Microbiol. 2008;68(1):139–151. doi: 10.1111/j.1365-2958.2008.06145.x. [DOI] [PubMed] [Google Scholar]
- Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62(2):606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23(6):1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70(11):6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29(11):2205–2216. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Control of SXT integration and excision. J Bacteriol. 2003;185(17):5045–5054. doi: 10.1128/JB.185.17.5045-5054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck SN, Turner SA, Rajakumar K, Adler B, Sakellaris H. Excision of the Shigella resistance locus pathogenicity island in Shigella flexneri is stimulated by a member of a new subgroup of recombination directionality factors. J Bacteriol. 2004;186(16):5551–5554. doi: 10.1128/JB.186.16.5551-5554.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman W, Thompson JF, Vargas L, Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992;256(5054):198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Moitoso de Vargas L, Nunes-Duby SE, Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990;63(4):773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- Franz B, Landy A. The Holliday junction intermediates of lambda integrative and excisive recombination respond differently to the bending proteins integration host factor and excisionase. Embo J. 1995;14(2):397–406. doi: 10.1002/j.1460-2075.1995.tb07014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L, Landy A. A switch in the formation of alternative DNA loops modulates lambda site-specific recombination. Proc Natl Acad Sci USA. 1991;88(2):588–592. doi: 10.1073/pnas.88.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam MD, Cascio D, Johnson RC, Clubb RT. Crystal structure of the excisionase-DNA complex from bacteriophage lambda. J Mol Biol. 2004;338(2):229–240. doi: 10.1016/j.jmb.2004.02.053. [DOI] [PubMed] [Google Scholar]
- Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Quinones M, Kimsey HH, Waldor MK. LexA cleavage is required for CTX prophage induction. Mol Cell. 2005;17(2):291–300. doi: 10.1016/j.molcel.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77(1):61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427(6969):72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbe J, Ploy MC, Mazel D. The SOS response controls integron recombination. Science. 2009;324(5930):1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L. et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea YA, Finnan S, Reen FJ, Morrissey JP, O'Gara F, Boyd EF. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology. 2004;150(Pt 12):4053–4063. doi: 10.1099/mic.0.27172-0. [DOI] [PubMed] [Google Scholar]
- Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51(3):246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]