Abstract
Background
Gastrointestinal stromal tumors (GIST) represent the most common mesenchymal tumors of the gastrointestinal tract. About 85% carry an activating mutation in the KIT or PDGFRA gene. Approximately 10% of GIST are so-called wild type GIST (wt-GIST) without mutations in the hot spots. In the present study we evaluated appropriate reference genes for the expression analysis of formalin-fixed, paraffin-embedded and fresh frozen samples from gastrointestinal stromal tumors. We evaluated the gene expression of KIT as well as of the alternative receptor tyrosine kinase genes FLT3, CSF1-R, PDGFRB, AXL and MET by qPCR. wt-GIST were compared to samples with mutations in KIT exon 9 and 11 and PDGFRA exon 18 in order to evaluate whether overexpression of these alternative RTK might contribute to the pathogenesis of wt-GIST.
Results
Gene expression variability of the pooled cDNA samples is much lower than the single reverse transcription cDNA synthesis. By combining the lowest variability values of fixed and fresh tissue, the genes POLR2A, PPIA, RPLPO and TFRC were chosen for further analysis of the GIST samples. Overexpression of KIT compared to the corresponding normal tissue was detected in each GIST subgroup except in GIST with PDGFRA exon 18 mutation. Comparing our sample groups, no significant differences in the gene expression levels of FLT3, CSF1R and AXL were determined. An exception was the sample group with KIT exon 9 mutation. A significantly reduced expression of CSF1R, FLT3 and PDGFRB compared to the normal tissue was detected. GIST with mutations in KIT exon 9 and 11 and in PDGFRA exon 18 showed a significant PDGFRB downregulation.
Conclusions
As the variability of expression levels for the reference genes is very high comparing fresh frozen and formalin-fixed tissue there is a strong need for validation in each tissue type. None of the alternative receptor tyrosine kinases analyzed is associated with the pathogenesis of wild-type or mutated GIST. It remains to be clarified whether an autocrine or paracrine mechanism by overexpression of receptor tyrosine kinase ligands is responsible for the tumorigenesis of wt-GIST.
Background
Gastrointestinal stromal tumors (GIST) are the most common mesenchymal tumors of the gastrointestinal tract and are characterized by the expression of the KIT receptor (stem cell factor receptor, CD117) and to a lesser extent of PDGFRA (platelet derived growth factor receptor alpha), representing two closely related receptor tyrosine kinases (RTK) [1,2]. The majority of GIST shows oncogenic mutations either in KIT or PDGFRA [3,4]. Mainly, mutations in exon 9 or 11 of the KIT gene or in exon 18 of PDGFRA lead to ligand independent, constitutive activation of the kinase function [5]. About 60% of all GIST carry an exon 11 mutation of KIT which encodes the juxtamembrane domain of the receptor possessing an autoinhibitory function [6,7]. Less common mutations in PDGFRA (~ 10%) are detected in GIST that often display gastric location and epithelioid morphology [2].
In a minority of cases (10-15%) no mutations in the known KIT or PDGFRA hot spots are detected although these tumors express the KIT protein. This subgroup is called wild type GIST (wt-GIST) and comprises tumors in pediatric patients, in patients affected by the Carney triad, neurofibromatosis type 1 (NF1) associated GIST and a subset of sporadic adult GIST [8-11]. The pathogenetic mechanisms underlying wt-GIST are poorly understood and there is limited benefit of imatinib therapy in these patients [12]. Therefore the identification of additional genetic factors contributing to the pathogenesis of GIST may help to find new concepts of individualized therapy.
Recently, the BRAF mutation p.V600E was found in 4-13% of wt-GIST [13-15]. For another subgroup of wt-GIST including pediatric tumors, a strong IGF1R expression combined partly with gene amplification was described [16-18]. Two other alternative RTK probably involved in the pathogenesis of GIST are AXL and MET. Both kinases have been shown to be upregulated in GIST resistant to treatment [19]. AXL is a member of the Ufo/AXL subfamily and activates the same signaling pathway as KIT. The tyrosine kinase domain of MET is mutated in sporadic papillary renal carcinomas. Some mutations in the MET gene are located in codons homologous to those in KIT and it is suggested that these missense mutations lead to constitutive activation of the MET protein [20]. To develop additional therapy approaches it would be of interest to know whether this RTK also plays a role in wt-GIST.
Besides KIT and PDGFRA, CSF1R (colony stimulating factor 1 receptor), FLT3 (fms like tyrosine kinase 3) and PDGFRB (platelet derived growth factor receptor β) belong to the same family of type III RTK. These five tyrosine kinases show a homologous structure and a comparable function in activation, proliferation and suppressing apoptosis [21-23].
Aberrant expression and mutations in either CSF1R, FLT3 and PDGFRB or their ligands have been described in several malignant diseases. Overexpression of CSF1R is found in epithelial tumors such as breast and ovarian cancer [24]. The translocation t(1;2) of its ligand CSF1 leads to the development of the tenosynovial giant cell tumor [25].
Aberrantly expressed FLT3 is observed at high levels in a spectrum of hematologic malignancies [26]. Additionally, in AML an internal tandem duplication in the transmembrane domain of FLT3 was identified which leads to constitutive activation of its kinase domain. It seems that this mutation is not present or very rare in GIST [27].
PDGFRB is overexpressed in malignant peripheral nerve sheath tumors (MPNST) and chordomas [28,29].
To analyse expression profiles of certain tumors for research and diagnostic purposes, qPCR (quantitative PCR) is frequently applied because of its reproducibility and high sensitivity [30,31]. This method is based on the normalization of the target gene expression on stably expressed internal reference genes. A major challenge is the application of suitable reference genes which have to be tested and verified under defined experimental conditions [32,33]. Ideal reference genes have to be non-regulated, stable and not affected by biological or experimental conditions. The target gene is amplified together with the reference gene in order to minimize experimental variability concerning reverse transcription enzymatic efficiencies, PCR efficiency, amount of starting material and differences between human tissues. The reference gene and the target gene should have very robust and stable expression profiles to ensure accurate normalization and interpretation of results. The most stable expressed gene from a set of genes can be identified by geNorm [34], a software program which additionally provides the number of genes required to calculate a robust normalization factor based on the geometric mean of these genes.
Typical reference genes regulate basic and ubiquitous cellular functions and are responsible for the cellular maintenance, e.g. GAPDH or ß-actin. However, these commonly used reference genes vary considerably in different tissue types or under different experimental conditions [35,36]. There is no standard reference gene for all kinds of tissue types. To our best knowledge only few studies investigated the alteration of stability of reference genes in different mesenchymal tumour entities.
In the present study we attempted to identify suitable reference genes in gastrointestinal stromal tumors by using a set of sixteen reference genes which are currently applied in qPCR procedures. Furthermore we evaluated the gene expression of KIT as well as the alternative RTK FLT3, CSF1-R, PDGFRB, AXL and MET in mutated and non-mutated gastrointestinal stromal tumors by qPCR using the identified reference genes. The study was focussed on wt-GIST compared to samples with mutations in KIT exon 9 and 11 and PDGFRA exon 18 in order to evaluate whether overexpression of these alternative RTK might contribute to the pathogenesis of wt-GIST.
Methods
Samples
A total of 107 samples were included into this study. All specimens were obtained in the years 2005 and 2006 under approved ethical protocols and with informed consent from each patient. All samples were fixed in neutral-buffered formalin prior to paraffin embedding. 20 samples from normal tissue (i.e., muscularis propria of stomach and gut) as control group and 87 GIST representing different mutational subgroups were evalutated. 20 samples of wt-GIST, 7 samples of wt-GIST associated with neurofibromatosis type 1, 20 samples with exon 9 mutation in KIT, 20 samples with exon 11 mutation in KIT and 20 samples with exon 18 mutation in PDGFRA). Sequence analysis of KIT (exons 8, 9, 11, 13, 14, 15 and 17) and PDGFRA (exons 12, 14 and 18) was carried out as described earlier [1,37,38]. Additionally, in all samples the wild type status of the BRAF gene was ascertained. All GIST samples were stained immunohistochemically for CD117. Four normal tissues from the gastrointestinal region were available as fresh frozen and formalin-fixed, paraffin-embedded samples.
RNA extraction and cDNA synthesis
Prior to RNA extraction, paraffin-embedded tissues were cut into 10 μm sections and mounted on glass-slides. Six slides of each specimen were used for RNA extraction. The sections were deparaffinized by extracting twice in xylene for 10 min at room temperature. Rehydration was done in 100% ethanol, 90% ethanol, 80% ethanol and 70% ethanol made with DEPC-water for 10 min each. Tumor areas previously marked on a H&E slide were scraped from the sections with a sterile scalpel. Sections were transferred into a sterile 1.5 ml tube. Extraction and purification of RNA was done using the RNeasy FFPE KIT (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. After tumor localization by H&E staining fresh frozen material was rasped into 10 μm thick pieces and RNA purification was carried out using the RNeasy Kit (Qiagen, Hilden, Germany). Both RNeasy Kits contain a step of DNase treatment. Finally RNA from fresh and from fixed tissue was eluted in water. The quantification was done spectrophotometrically (NanoDrop, PeqLab Technology, Erlangen, Germany). 500 ng of RNA from each sample was reverse-transcribed using a random-hexamer primer and Avian Myoblastosis Virus reverse transcriptase (AMV-RT) according to the manufacturer's protocol (Qiagen). cDNA from those four fresh frozen and corresponding formalin-fixed, paraffin-embedded (FFPE) control samples was generated in only one reverse transcription and loaded on the reference low density arrays (see below). Further cDNA samples were generated from the four FFPE control tissues, pooled within one patient and loaded onto a second independent reference low density array.
Identification of reference genes
The reference genes used here were preselected because of their constitutive, non-regulated stable expression over a wide spectrum of tissues. But nevertheless the preselected reference genes are not suitable for every kind of tissue and therefore need to be analyzed prior to use in a certain study. The detection of suitable reference genes was carried out using TaqMan Low Density Arrays (TLDAs, Microfluidic Cards, Applied Biosystems, Darmstadt, Germany). These arrays are prefabricated 384-well cards where gene-specific primer and probe sets are spotted in small reaction chambers during manufacturing. The cards have 8 separate loading ports leading into 48 wells each. In this study, arrays with triplicates of 16 putative reference genes were used, so 8 different samples could be analyzed. The list of assays is given in table 1. For each sample, 500 ng cDNA was mixed with 2× TaqMan Universal PCR Master Mix (Applied Biosystems). 100 μl of this mixture was loaded into each port and distributed into the reaction chambers by centrifugation. The card was sealed and the quantitative PCR (qPCR) was performed on an ABI PRISM HT 7900 (Applied Biosystems) sequence detection system. After pre-incubation for 2 min at 50°C and 10 min at 95°C, the PCR reaction was performed (15 s at 94°C followed by 60 s at 60°C, 40 cycles). The fluorescent signal was measured in each cycle.
Table 1.
Selected candidate reference genes for gene expression analysis.
| Abbreviation | Gene name | Cellular function | |
|---|---|---|---|
| 18S rRNA | 18S ribosomal RNA | ribosome subunit | X03205.1 |
| ACTB | ß-actin | cytoskeleton | NM_001101.3 |
| B2M | ß-2-microglobulin | major histocompatibility complex | NM_004048.2 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | glycolysis enzyme | NM_002046.3 |
| GUSB | ß-glucuronidase | glycosaminoglycan degradation | NM_000181.2 |
| HMBS | hydroxymethylbilane synthase | heme production | NM_000190.3 |
| HPRT1 | hypoxanthine ribosyltransferase | metabolic salvage of purines | NM_000194.2 |
| IPO8 | importin 8 | intracellular protein transport | NM_006390.2 |
| PGK1 | phosphoglycerate kinase 1 | glycolysis enzyme | NM_000291 |
| POLR2A | DNA-dependent RNA polymerase II | transcription | NM_000937.3 |
| PPIA | peptidyl-prolyl isomerase A | protein folding | NM_021130 |
| RPLPO | ribosomal phosphoprotein, large, P0 | ribosome | NM_001002 |
| TBP | TATA-box-binding protein | transcription factor | NM_003194 |
| TFRC | transferrin receptor | cellular iron uptake | NM_021009.4 |
| UBC | ubiquitin C | posttranslational modification | NM_003406.3 |
| YWHAZ | tyrosine 3-/tryptophan 5-Monooxygenase-activation protein, zeta isoform | signal transduction | NM_003404 |
Analyses of gene expression by qPCR
qPCR analysis was performed using the assays-on-demand products (Applied Biosystems) listed in table 2. These gene-specific qPCR assays consist of a pair of unlabeled PCR primers and a FAM labeled specific probe. According to the manufacturer of these assays, probe and primer sets that would amplify pseudogenes are excluded in the process of development. Reactions were carried out in a reaction volume containing 5 μl PCR Master Mix (Applied Biosystems), 0.5 μl forward and reverse primer mix, 500 ng cDNA ad 10 μl A.dest. Triplicate reactions were carried out for each transcript. Control reactions were performed using a minus RT preparation and a sample with A.dest instead of RNA. PCR conditions were the same as for the TaqMan Low Density Arrays.
Table 2.
Gene-specific qPCR assays.
| Gene symbol | Name | Chr | Function | Assay-on-demand |
|---|---|---|---|---|
| CSF1R | colony stimulating factor 1 receptor | 5 | tyrosine protein kinase receptor | Hs00234617_m1 |
| FLT3 | fms-related tyrosine kinase 3 | 13 | tyrosine protein kinase receptor | Hs00174690_m1 |
| KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 4 | tyrosine protein kinase receptor | Hs00174029_m1 |
| PDGFRß | platelet-derived growth factor receptor, beta | 5 | tyrosine protein kinase receptor | Hs00387362_m1 |
| MET | met proto-oncogene | 7 | hepatocyte growth factor receptor | Hs00179845_m1 |
| AXL | AXL receptor tyrosine kinase | 19 | tyrosine protein kinase receptor | Hs00242357_m1 |
Statistical analyses
The geNorm applet for Microsoft Excel was used to determine the most stable genes among the sixteen candidate reference genes. Raw Cq values were converted into relative quantities for analysis with geNorm, where the highest relative quantity for each gene is set to 1. The program selects from a panel of candidate reference genes the two most stable genes or a combination of multiple stable genes for normalization. The gene expression stability (M) value is based on the combined estimate of intra- and intergroup expression variations of the genes studied and takes the PCR efficiency into account. The limited M-value is 1.5. The most stable genes are stepwise selected from the investigated gene panel to estimate how many reference genes should be used. The normalization factors define the optimal number of reference genes required for a precised normalization design. The analysis of the expression data of tumor samples compared to control samples was performed with the REST software (Relative Expression Software Tool) [39]. The software normalizes the measured Cq-values of the target genes with those of the reference genes and compares the expression data of tumor and control samples by considering the PCR efficiency and the mean crossing point deviation.
Results
RNA quality
RNA quality of the samples was inspected on a 1% agarose gel. The concentration and purity of the RNA was characterized by the mean A260/280 ratio and was on average 1.99 for fresh frozen as well as for formalin fixed, paraffin-embedded samples and reflected pure and protein-free RNA.
Expression variability of cDNA synthesis
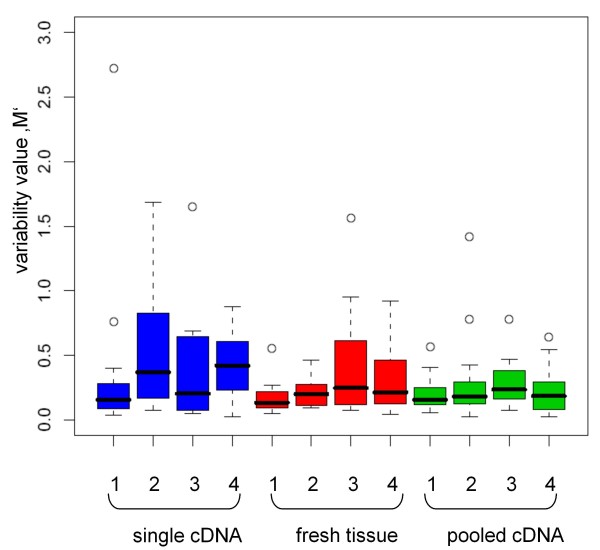
Four normal control FFPE tissue samples from the gastric and bowel wall were used to determine suitable reference genes. One TLDA was done with cDNA from only one reverse transcription for each sample. As it is assumed that cDNA synthesis from FFPE tissue varies strongly in efficiency, pooled cDNA synthesis samples from the same FFPE tissue were used for a second TLDA. The pooled cDNA preparation showed an essentially lower variance in contrast to the cDNA samples from only one reverse transcription (Figure 1). The median of the M-values ranges from 0.1 to 0.2, thus the variability of the technical replicates is low. Within a single specimen the range is also very small. The median of the cDNA samples from a single reverse transcription ranges from 0.1 to 0.4. However, each cDNA sample from one reverse non-pooled transcription showed a high variance of the values (median of M-values 0.1 to 0.8, Figure 1). Therefore the variance of the samples can be lowered by cDNA pooling. The median of the cDNA samples from one reverse transcription from fresh frozen tissue ranged from 0.1 to 0.2 and can be compared to the median of the pooled cDNA from the FFPE tissue. Within a single specimen the range is higher than in the pooled cDNA samples, but smaller than in the cDNA from only one reverse transcription from the FFPE samples.
Figure 1.
Variability of technical replicates. Expression variability of cDNA from a single reverse transcription synthesis (blue), cDNA from pooled reverse transcription synthesis (green) and fresh tissue cDNA (red) from a single reverse transcription synthesis. The whiskers are representing the range of the data, the median is shown as a black line within a bar. Circles are representing the outliers.
Identification of suitable reference genes for normalization
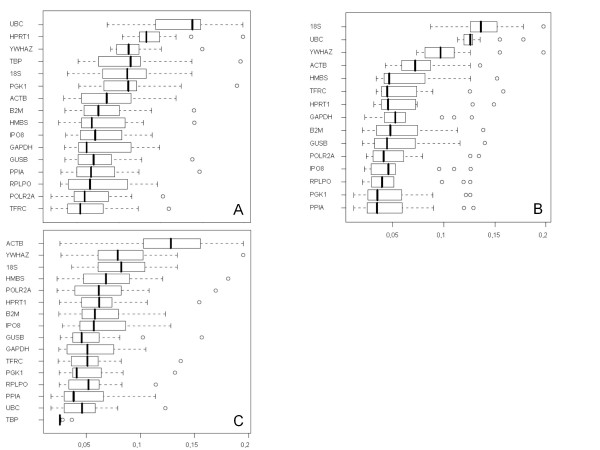
All 16 putative reference genes had a high expression stability and the 'M'-value (0.02-0.06) was clearly below the 'M'-cutoff-value of 1.5. By stepwise exclusion of genes, the expression stability value 'M' is calculated (data not shown) and the expression stability rises with the exclusion of further genes. Samples generated from only one reverse transcription, showed POLR2A, TFRC, RPLPO and GAPDH as the most stable genes (Figure 2a). The average stability value was between 0.05 and 0.07. The range of the values was 0.01-0.15. The most stable genes for pooled cDNA samples were PGK1, PPIA, RPLPO and IPO8 with a median of 0.04-0.05 (Figure 2b). The range of the values was lower and varied between 0.01 and 0.09. All in all the gene expression variability of the pooled cDNA samples is much lower compared to the single reverse transcription cDNA synthesis. The gene UBC is the most stable one in fresh frozen tissue with the smallest value range (0.01-0.8), but RPLPO and PPIA belong also to the most stable genes in fresh frozen tissue with a median of 0.03-0.045 and the range of the values was 0.1-0.12 (Figure 2c). TBP was not expressed in the tissue. By combining the lowest variability values of fixed and fresh tissue, the genes POLR2A, PPIA, RPLPO and TFRC were detected (Figure 3). Hence these four genes were used for further analysis with the GIST samples.
Figure 2.
Expression variability of reference genes. Analysis of reference gene expression on TaqMan Low Density Arrays. a) cDNA from a single RT-transcription. b) pooled cDNA samples from normal tissue. The genes POLR2A, TFRC, PPIA and RPLPO had the lowest expression variability. c) cDNA from fresh frozen tissue. RPLPO, PPIA and UBC had the lowest expression variability, TBP was not expressed in the tissue. On the x-axis, the stability values are plotted. Whiskers represent range of data of 4 samples.
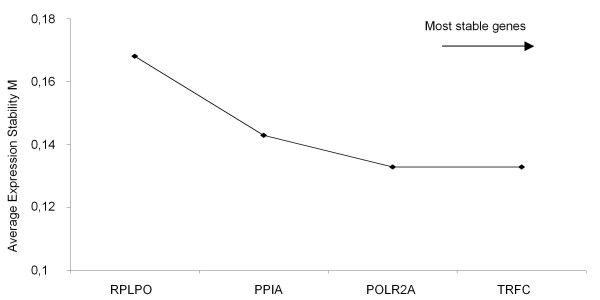
Figure 3.
GeNorm analysis of selected reference genes. Calculation of the average expression stability of the remaining 4 candidate reference genes for normalization in GIST by geNorm analysis. The least stable gene with the highest 'M'-value is indicated from left to right on the x-axis. After combining the data of the single RT-transcription cDNA and the pooled cDNA the stepwise exclusion of the least stable reference genes resulted in POLR2A and TRFC.
Expression profiles of KIT, FLT3, CSF1-R, PDGFRB, AXL and MET
Six patient groups (NT = normal tissue -control, WT = wild type, Ex9 = KIT Exon 9 mutated, Ex11 = KIT Exon 11 mutated, NF1 = wild type and neurofibromatosis type 1, Ex18 = PDGFRA Exon 18 mutated) were used to analyze the gene expression of KIT, FLT3, CSF1-R, PDGFRB, AXL and MET. Here, the REST analysis with POLR2A is shown exemplarily. The REST analysis of the target genes KIT, CSF1R, FLT3, PDGFRB, AXL and MET with the reference genes RPLPO, PPIA and TRFC showed the same significant results for the differential expression or at least the same trend in mutated and non-mutated GIST (data not shown). As suspected, we found a significant overexpression of KIT in exon 9 and exon 11 mutated GIST in comparison with normal tissue. Also a significantly lowered expression of PDGFRB in both groups compared to normal tissue was shown. The same effect was observed in PDGFRA exon 18 mutated GIST compared to normal tissue (table 3).
Table 3.
Expression analysis of target genes.
| Gene | Factor | p-value | Gene | Factor | p-value | ||
|---|---|---|---|---|---|---|---|
| NT↔WT | KIT | 5.9+ | 0.095 | NT↔NF1 | KIT | 17+ | 0.003 |
| CSF1R | 3.99- | 0.217 | CSF1R | 2.99- | 0.294 | ||
| FLT3 | 2.43- | 0.498 | FLT3 | 3.985- | 0.179 | ||
| PDGFRB | 5.79- | 0.087 | PDGFRB | 2.827- | 0.196 | ||
| AXL | 2.1- | 0.66 | AXL | 2.5+ | 0.53 | ||
| MET | 8.7- | 0.034 | MET | 6.5- | 0.13 | ||
| NT↔Ex11 | NT↔Ex18 | ||||||
| KIT | 11.7+ | 0.011 | KIT | 1.66+ | 0.373 | ||
| CSF1R | 4.65- | 0.155 | CSF1R | 4.7- | 0.051 | ||
| FLT3 | 2.383- | 0.41 | FLT3 | 2.113- | 0.263 | ||
| PDGFRB | 6.3- | 0.046 | PDGFRB | 4.925- | 0.007 | ||
| AXL | 3- | 0.27 | AXL | 1.35- | 0.58 | ||
| MET | 17.81- | 0.001 | MET | 7.7- | 0.001 | ||
| NT↔Ex9 | Ex11↔WT | ||||||
| KIT | 5.261+ | 0.05 | KIT | 1.981- | 0.232 | ||
| CSF1R | 13.98- | 0.003 | CSF1R | 1.164+ | 0.861 | ||
| FLT3 | 5.38- | 0.022 | FLT3 | 1.023- | 0.971 | ||
| PDGFRB | 12.78- | 0.001 | PDGFRB | 1.087+ | 0.920 | ||
| AXL | 3.4- | 0.08 | AXL | 1.4+ | 0.6 | ||
| MET | 9.766- | 0.001 | MET | 2+ | 0.33 |
Expression analysis of KIT, CSF1R, FLT3, PDGFRB, AXL and MET was carried out in six GIST cohorts (NT = normal tissue, WT = wild type, Exon 9, Exon 11, NF1 = neurofibromatosis type 1, Exon 18). Significant variances are in bold. The expression data were calculated using the REST software with POLR2A as reference gene. NT: normal tissue; p < 0.05; +/- indicates up- or downregulation
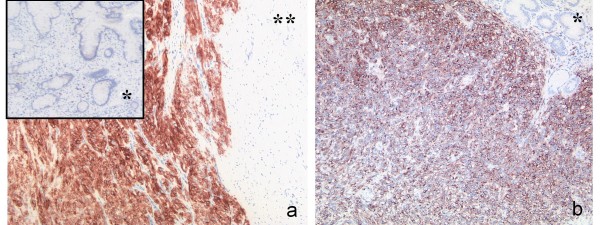
On closer inspection of normal tissue and wt-GIST a clear overexpression of KIT in the tissue of wild type tumors was shown. The same effect, but even stronger, could be detected by comparing NF1-associated wt-GIST with normal tissue. Concerning KIT expression, the tumors without mutation behave like KIT mutated GIST (table 3). This was in accordance with immunohistochemical staining (Figure 4).
Figure 4.
Immunohistochemical staining for CD117 in wild type and mutant GIST. a) NF1 associated wild type GIST located in the small intestine with corresponding normal mucosa (*) and muscularis propria (**). b) GIST located in the stomach with a mutation in exon 11 of the KIT gene with corresponding normal mucosa (*).
The other target genes, FLT3, CSF1R, PDGFRB, AXL and MET showed lower expression levels in the tumor tissue compared to normal tissue except AXL in NF1 associated GIST. MET was even significantly reduced in wt-GIST, KIT exon 9, 11 and PDGFRA exon 18 mutated GIST. No changes in expression levels were detected comparing normal tissue and wt-GIST with KIT exon 11 mutated GIST, indicating that both groups possess similar expression levels for the analyzed receptors. None of the groups showed significant expression alterations for CSF1R and FLT3. An exception is the group of KIT exon 9 mutated GIST. A lower expression of CSF1R, FLT3, PDGFRB and MET compared to normal tissue was identified in contrast to KIT, which showed a significant overexpression. In summary, overexpression of KIT was detected in each GIST subgroup compared to the corresponding normal tissue by using the preassigned reference gene POLR2A (table 3). GIST with a mutation in exon 18 of PDGFRA did not show a significant upregulation of KIT compared to normal tissue. GIST with a mutation in KIT exon 9, 11 and in the PDGFRA exon 18 showed significant PDGFRB downregulation.
Discussion
Most gastrointestinal stromal tumors exhibit mutations in exon 11 of the KIT gene. This exon encodes for the juxtamembrane domain of the receptor which possesses an autoinhibitory function. In wt-GIST, activating mutations are found neither in the KIT nor in the PDGFRA gene. Clinical treatment with the tyrosine kinase inhibitor imatinib targets the ATP binding site in the kinase domain of the KIT receptor. In wt-GIST, response to treatment is often poor [12] although most of them overexpress the KIT protein. A screen for activating mutations in the juxtamembrane domain of the alternative receptor tyrosine kinases CSF1R, FLT3, PDGFRB in 30 wt-GIST samples yielded only wild type sequences (data not shown) in agreement with previously published results [40]. Therefore we investigated in this study whether the expression of alternative receptor tyrosine kinases may contribute to the pathogenesis of wt-GIST and thus may help to identify wt-GIST subgroups with different response to imatinib and elucidate novel therapeutic targets.
The pathogenesis of several malignant tumors is associated with overexpression of CSF1R and PDGFRB [24,28]. Imatinib inhibits cell invasion in malignant peripheral nerve sheath tumors by blocking PDGFRB [41] and it has been found to have antitumor activity in patients with chordoma [42]. In our study, the gene-expression level of CSF1R, FLT3 and PDGFRB was determined in a cohort of 87 GIST samples. Furthermore, we assessed the expression of AXL and MET, two receptor tyrosine kinases which were found to be alternatively activated in therapy resistant GIST [19].
Gene expression analysis by qPCR requires suitable reference genes. The expression of reference genes like GAPDH or BETA-ACTIN is regulated differentially depending on the tissue type. Therefore they are not suited as univocal reference genes [35,43]. The determination of reference genes with stable expression in the experimental system used is essential to ensure accurate normalization and interpretation of results.
Whereas fresh tissue is frequently not available for genetic analysis, FFPE material is the standard. After formalin-fixation and paraffin-embedding of tissue, the isolated RNA is often heavily fragmented. In our study we used fresh as well as fixed material from the gastrointestinal tract to determine suitable reference genes and to analyze whether their expression levels are comparable. We then validated the reference genes by qPCR in our GIST cohort.
As reported also by others [44,45] the variability of expression levels for the reference genes was very diverse comparing fresh frozen and formalin-fixed tissue. Therefore, we decided to validate separate reference genes for each tissue type.
The genes TRFC, POLR2A, PPIA and RPLPO were validated as appropriate reference genes for FFPE tissue. For fresh frozen tissue, PPIA and RPLPO were also found to be suitable. Additionally, UBC is suited as a reference gene in fresh frozen tissue. The variability of the reference genes for fresh frozen tissue was lower than for fixed tissue. To overcome the problem of high variability in FFPE tissue, we pooled two independent cDNA syntheses from one sample as recommended in the MIQE guidelines [30,46]. The MIQE guidelines give considerations for a consistent application of the qPCR technology including experimental details, data analysis and reporting principles. Unequal efficiency of cDNA synthesis might be a reason for deviation. Additionally, we tried to select consistent patient material for our cohort by choosing paraffin blocks having the same age. It was shown by Bibikova [45], that Cq-values in qPCR experiments depend on the age and condition of the tissue blocks. Because the expression value depends also on amplicon length [30,44], only primer sets generating amplicons of about 100 bp were chosen for qPCR. This corresponds to the fragment length of degraded RNA between 100 and 200 bp. The application of the MIQE guidelines results in a minimum variability for reference genes. In summary, considering the MIQE guidelines FFPE material can be used reliably for expression analysis in GIST, but the use of separate reference genes for FFPE tissue is indispensable.
Our qPCR analysis included GIST with wild type sequences in the hot-spot regions of KIT and PDGFRA (wt-GIST), KIT exon 9 and exon 11 mutated samples, PDGFRA exon 18 mutated samples, NF-1-associated wt-GIST and normal tissue controls. The wt-GIST and the samples with the two different KIT mutations showed a significantly increased expression of KIT in contrast to the normal tissue. The results correspond to the immunohistochemical stainings of our samples and were in agreement with published data [47]. It was shown immunohistochemically that PDGFRA mutated GIST have only a slightly increased protein expression of KIT [13]. Our data revealed the same trend on RNA level compared to the normal tissue. Additionally, we could show in all groups of mutated GIST a significantly reduced expression of PDGFRB compared to the normal tissue. This could be due to the concomitant increase of KIT expression. When comparing NF-1-associated GIST without KIT mutation with wt-GIST without NF-1 association the latter showed a lower expression of KIT. Thus, the results of the two wt-GIST groups give a heterogenous profile, which suggests that different genomic events may be responsible for the development of these tumors.
Comparing our sample groups with each other, no significant difference in the gene expression levels of FLT3, CSF1R and AXL were determined. An exception was seen in the sample group with KIT exon 9 mutation. Here, a significantly reduced expression of CSF1R, FLT3 and PDGFRB compared to the normal tissue was detected. The results lead us to the assumption that KIT exon 9 mutated GIST play a special role compared to GIST carrying other mutations. Interestingly, KIT exon 9 mutated GIST need a double daily dose of the tyrosine kinase inhibitor imatinib to be effectively treated [48]. Furthermore, they develop preferentially in the small intestine but only rarely in the stomach where the majority of GIST are detected [49].
All qPCR data were calculated four times with TRFC, POLR2A, PPIA and RPLPO as reference genes using the REST software. Concordant results with the four reference genes are based on the extensive and complex preselection of our cohort and the preparation of cDNA synthesis according to the MIQE guidelines.
Conclusions
In summary, we conclude that none of the alternative receptor tyrosine kinases analyzed here are associated with the pathogenesis of wild type or mutated GIST. It remains to be clarified whether an increased expression of receptor tyrosine kinase ligands is responsible for tumorigenesis of wt-GIST as it is described for dermatofibrosarcoma protuberans (DFSP) and tenosynovial giant cell tumor (TGCT) [25,50]. Further studies are needed to elucidate the role of ligand-driven pathogenesis in wt-GIST.
Authors' contributions
MCB, JF and HK carried out the molecular genetic studies and JF wrote and drafted the manuscript. SMB and HUS developed the design of the study. SMB coordinated the study together with EW and HUS. BB and MZ performed the statistical analysis of the expression data. EW and HUS participated in the diagnosis and selection of tumor material. RB revised the manuscript for important intellectual content and approved the final manuscript. All authors read and approved the manuscript.
Contributor Information
Jana Fassunke, Email: jana.fassunke@ukb.uni-bonn.de.
Marie-Christine Blum, Email: marie-christine.blum@med.ovgu.de.
Hans-Ulrich Schildhaus, Email: hans-ulrich.schildhaus@ukb.uni-bonn.de.
Marc Zapatka, Email: m.zapatka@dkfz.de.
Benedikt Brors, Email: b.brors@dkfz.de.
Helen Künstlinger, Email: hkuenstl@uni-bonn.de.
Reinhard Büttner, Email: Reinhard.Buettner@ukb.uni-bonn.de.
Eva Wardelmann, Email: eva.wardelmann@ukb.uni-bonn.de.
Sabine Merkelbach-Bruse, Email: sabine.merkelbach-bruse@ukb.uni-bonn.de.
Acknowledgements and Funding
This study was supported by a grant of the BONFOR programme of the Bonn Medical centre to HUS.
References
- Pauls K, Merkelbach-Bruse S, Thal D, Buttner R, Wardelmann E. PDGFRalpha- and c-kit-mutated gastrointestinal stromal tumours (GISTs) are characterized by distinctive histological and immunohistochemical features. Histopathology. 2005;46:166–175. doi: 10.1111/j.1365-2559.2005.02061.x. [DOI] [PubMed] [Google Scholar]
- Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M. CD117: a sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol. 1998;11:728–734. [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A. et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M. et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53:245–266. doi: 10.1111/j.1365-2559.2008.02977.x. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA. et al. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Wardelmann E, Losen I, Hans V, Neidt I, Speidel N, Bierhoff E, Heinicke T, Pietsch T, Buttner R, Merkelbach-Bruse S. Deletion of Trp-557 and Lys-558 in the juxtamembrane domain of the c-kit protooncogene is associated with metastatic behavior of gastrointestinal stromal tumors. Int J Cancer. 2003;106:887–895. doi: 10.1002/ijc.11323. [DOI] [PubMed] [Google Scholar]
- Lasota J, Miettinen M. KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs) Semin Diagn Pathol. 2006;23:91–102. doi: 10.1053/j.semdp.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Medeiros F, Corless CL, Duensing A, Hornick JL, Oliveira AM, Heinrich MC, Fletcher JA, Fletcher CD. KIT-negative gastrointestinal stromal tumors: proof of concept and therapeutic implications. Am J Surg Pathol. 2004;28:889–894. doi: 10.1097/00000478-200407000-00007. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Fetsch JF, Sobin LH, Lasota J. Gastrointestinal stromal tumors in patients with neurofibromatosis 1: a clinicopathologic and molecular genetic study of 45 cases. Am J Surg Pathol. 2006;30:90–96. doi: 10.1097/01.pas.0000176433.81079.bd. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J, Sobin LH. Gastrointestinal stromal tumors of the stomach in children and young adults: a clinicopathologic, immunohistochemical, and molecular genetic study of 44 cases with long-term follow-up and review of the literature. Am J Surg Pathol. 2005;29:1373–1381. doi: 10.1097/01.pas.0000172190.79552.8b. [DOI] [PubMed] [Google Scholar]
- Debiec-Rychter M, Dumez H, Judson I, Wasag B, Verweij J, Brown M, Dimitrijevic S, Sciot R, Stul M, Vranck H. et al. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer. 2004;40:689–695. doi: 10.1016/j.ejca.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Hostein I, Faur N, Primois C, Boury F, Denard J, Emile JF, Bringuier PP, Scoazec JY, Coindre JM. BRAF mutation status in gastrointestinal stromal tumors. Am J Clin Pathol. 2010;133:141–148. doi: 10.1309/AJCPPCKGA2QGBJ1R. [DOI] [PubMed] [Google Scholar]
- Martinho O, Gouveia A, Viana-Pereira M, Silva P, Pimenta A, Reis RM, Lopes JM. Low frequency of MAP kinase pathway alterations in KIT and PDGFRA wild-type GISTs. Histopathology. 2009;55:53–62. doi: 10.1111/j.1365-2559.2009.03323.x. [DOI] [PubMed] [Google Scholar]
- Agaimy A, Terracciano LM, Dirnhofer S, Tornillo L, Foerster A, Hartmann A, Bihl MP. V600E BRAF mutations are alternative early molecular events in a subset of KIT/PDGFRA wild-type gastrointestinal stromal tumours. J Clin Pathol. 2009;62:613–616. doi: 10.1136/jcp.2009.064550. [DOI] [PubMed] [Google Scholar]
- Janeway KA, Zhu MJ, Barretina J, Perez-Atayde A, Demetri GD, Fletcher JA. Strong expression of IGF1R in pediatric gastrointestinal stromal tumors without IGF1R genomic amplification. Int J Cancer. 2010;127:2718–2722. doi: 10.1002/ijc.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo MA, Astolfi A, Di Battista M, Heinrich MC, Paterini P, Scotlandi K, Santini D, Catena F, Manara MC, Nannini M. et al. Insulin-like growth factor 1 receptor expression in wild-type GISTs: a potential novel therapeutic target. Int J Cancer. 2009;125:2991–2994. doi: 10.1002/ijc.24595. [DOI] [PubMed] [Google Scholar]
- Tarn C, Rink L, Merkel E, Flieder D, Pathak H, Koumbi D, Testa JR, Eisenberg B, von Mehren M, Godwin AK. Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc Natl Acad Sci USA. 2008;105:8387–8392. doi: 10.1073/pnas.0803383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D, Cooke L, Riley C, Swart R, Simons B, Della Croce K, Wisner L, Iorio M, Shakalya K, Garewal H. et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene. 2007;26:3909–3919. doi: 10.1038/sj.onc.1210173. [DOI] [PubMed] [Google Scholar]
- Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, Scherer SW, Zhuang Z, Lubensky I, Dean M. et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- Coussens L, Yang-Feng TL, Liao YC, Chen E, Gray A, McGrath J, Seeburg PH, Libermann TA, Schlessinger J, Francke U. et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science. 1985;230:1132–1139. doi: 10.1126/science.2999974. [DOI] [PubMed] [Google Scholar]
- Lavagna-Sevenier C, Marchetto S, Birnbaum D, Rosnet O. The CBL-related protein CBLB participates in FLT3 and interleukin-7 receptor signal transduction in pro-B cells. J Biol Chem. 1998;273:14962–14967. doi: 10.1074/jbc.273.24.14962. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Escobedo JA, Kuang WJ, Yang-Feng TL, Daniel TO, Tremble PM, Chen EY, Ando ME, Harkins RN, Francke U. et al. Structure of the receptor for platelet-derived growth factor helps define a family of closely related growth factor receptors. Nature. 1986;323:226–232. doi: 10.1038/323226a0. [DOI] [PubMed] [Google Scholar]
- Kacinski BM. CSF-1 and its receptor in breast carcinomas and neoplasms of the female reproductive tract. Mol Reprod Dev. 1997;46:71–74. doi: 10.1002/(SICI)1098-2795(199701)46:1<71::AID-MRD11>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- West RB, Rubin BP, Miller MA, Subramanian S, Kaygusuz G, Montgomery K, Zhu S, Marinelli RJ, De Luca A, Downs-Kelly E. et al. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc Natl Acad Sci USA. 2006;103:690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG. Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells. Leukemia. 1996;10:588–599. [PubMed] [Google Scholar]
- Shiozawa E, Takimoto M, Makino R, Hirayama Y, Ota H. Flt3/Itd mutation is not associated with gastrointestinal stromal tumors (gists) J Gastroenterol Hepatol. 2005;20:1132–1134. doi: 10.1111/j.1440-1746.2005.03834.x. [DOI] [PubMed] [Google Scholar]
- Perrone F, Da Riva L, Orsenigo M, Losa M, Jocolle G, Millefanti C, Pastore E, Gronchi A, Pierotti MA, Pilotti S. PDGFRA, PDGFRB, EGFR, and downstream signaling activation in malignant peripheral nerve sheath tumor. Neuro Oncol. 2009;11:725–736. doi: 10.1215/15228517-2009-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborini E, Miselli F, Negri T, Lagonigro MS, Staurengo S, Dagrada GP, Stacchiotti S, Pastore E, Gronchi A, Perrone F. et al. Molecular and biochemical analyses of platelet-derived growth factor receptor (PDGFR) B, PDGFRA, and KIT receptors in chordomas. Clin Cancer Res. 2006;12:6920–6928. doi: 10.1158/1078-0432.CCR-06-1584. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Ohl F, Jung M, Radonic A, Sachs M, Loening SA, Jung K. Identification and validation of suitable endogenous reference genes for gene expression studies of human bladder cancer. J Urol. 2006;175:1915–1920. doi: 10.1016/S0022-5347(05)00919-5. [DOI] [PubMed] [Google Scholar]
- Saviozzi S, Cordero F, Lo Iacono M, Novello S, Scagliotti GV, Calogero RA. Selection of suitable reference genes for accurate normalization of gene expression profile studies in non-small cell lung cancer. BMC Cancer. 2006;6:200. doi: 10.1186/1471-2407-6-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR--a perspective. J Mol Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/S0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Wardelmann E, Neidt I, Bierhoff E, Speidel N, Manegold C, Fischer HP, Pfeifer U, Pietsch T. c-kit mutations in gastrointestinal stromal tumors occur preferentially in the spindle rather than in the epithelioid cell variant. Mod Pathol. 2002;15:125–136. doi: 10.1038/modpathol.3880504. [DOI] [PubMed] [Google Scholar]
- Wardelmann E, Thomas N, Merkelbach-Bruse S, Pauls K, Speidel N, Buttner R, Bihl H, Leutner CC, Heinicke T, Hohenberger P. Acquired resistance to imatinib in gastrointestinal stromal tumours caused by multiple KIT mutations. Lancet Oncol. 2005;6:249–251. doi: 10.1016/S1470-2045(05)70097-8. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihto H, Franssila K, Tanner M, Vasama-Nolvi C, Sarlomo-Rikala M, Nupponen NN, Joensuu H, Isola J. Platelet-derived growth factor receptor family mutations in gastrointestinal stromal tumours. Scand J Gastroenterol. 2006;41:805–811. doi: 10.1080/00365520500483207. [DOI] [PubMed] [Google Scholar]
- Aoki M, Nabeshima K, Koga K, Hamasaki M, Suzumiya J, Tamura K, Iwasaki H. Imatinib mesylate inhibits cell invasion of malignant peripheral nerve sheath tumor induced by platelet-derived growth factor-BB. Lab Invest. 2007;87:767–779. doi: 10.1038/labinvest.3700591. [DOI] [PubMed] [Google Scholar]
- Casali PG, Messina A, Stacchiotti S, Tamborini E, Crippa F, Gronchi A, Orlandi R, Ripamonti C, Spreafico C, Bertieri R. et al. Imatinib mesylate in chordoma. Cancer. 2004;101:2086–2097. doi: 10.1002/cncr.20618. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Antonov J, Goldstein DR, Oberli A, Baltzer A, Pirotta M, Fleischmann A, Altermatt HJ, Jaggi R. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab Invest. 2005;85:1040–1050. doi: 10.1038/labinvest.3700303. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, Wickham E, Atkins D, Barker D, Chee M. et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol. 2004;165:1799–1807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Why the need for qPCR publication guidelines?-The case for MIQE. Methods. 2010;50:217–226. doi: 10.1016/j.ymeth.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sobin LH, Sarlomo-Rikala M. Immunohistochemical spectrum of GISTs at different sites and their differential diagnosis with a reference to CD117 (KIT) Mod Pathol. 2000;13:1134–1142. doi: 10.1038/modpathol.3880210. [DOI] [PubMed] [Google Scholar]
- Reichardt P. Optimal use of targeted agents for advanced gastrointestinal stromal tumours. Oncology. 2010;78:130–140. doi: 10.1159/000312655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory TS, Sobin LH, Lukes L, Lee DH, O'Leary TJ. Prognosis of gastrointestinal smooth-muscle (stromal) tumors: dependence on anatomic site. Am J Surg Pathol. 1999;23:82–87. doi: 10.1097/00000478-199901000-00009. [DOI] [PubMed] [Google Scholar]
- Simon MP, Pedeutour F, Sirvent N, Grosgeorge J, Minoletti F, Coindre JM, Terrier-Lacombe MJ, Mandahl N, Craver RD, Blin N. et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]