Abstract
Background
Epigenetic silencing can extend to whole chromosomal regions in cancer. There have been few genome-wide studies exploring its involvement in tumorigenesis.
Methods
We searched for chromosomal regions affected by epigenetic silencing in cancer by using Affymetrix microarrays and real-time quantitative polymerase chain reaction to analyze RNA from 57 bladder tumors compared with normal urothelium. Epigenetic silencing was verified by gene re-expression following treatment of bladder cell lines with 5-aza-deoxycytidine, a DNA demethylating agent, and trichostatin A, a histone deacetylase inhibitor. DNA methylation was studied by bisulfite sequencing and histone methylation and acetylation by chromatin immunoprecipitation. Clustering was used to distinguish tumors with multiple regional epigenetic silencing (MRES) from those without and to analyze the association of this phenotype with histopathologic and molecular types of bladder cancer. The results were confirmed with a second panel of 40 tumor samples and extended in vitro with seven bladder cancer cell lines. All statistical tests were two-sided.
Results
We identified seven chromosomal regions of contiguous genes that were silenced by an epigenetic mechanism. Epigenetic silencing was not associated with DNA methylation but was associated with histone H3K9 and H3K27 methylation and histone H3K9 hypoacetylation. All seven regions were concordantly silenced in a subgroup of 26 tumors, defining an MRES phenotype. MRES tumors exhibited a carcinoma in situ–associated gene expression signature (25 of 26 MRES tumors vs 0 of 31 non-MRES tumors, P < 10−14), rarely carried FGFR3 mutations (one of 26 vs 22 of 31 non-MRES tumors, P < 10−6), and contained 25 of 33 (76%) of the muscle-invasive tumors. Cell lines derived from aggressive bladder tumors presented epigenetic silencing of the same regions.
Conclusions
We have identified an MRES phenotype characterized by the concomitant epigenetic silencing of several chromosomal regions, which, in bladder cancer, is specifically associated with the carcinoma in situ gene expression signature.
CONTEXT AND CAVEATS
Prior knowledge
Although the epigenetic silencing of isolated genes has often been reported and studied in detail in cancer, there have been few studies on the silencing of entire chromosomal regions and its association with tumor progression.
Study design
Microarrays, reverse transcription–polymerase chain reaction, inhibitors of DNA modification, and chromatin immunoprecipitation were used to evaluate epigenetic silencing in 28 chromosomal regions among two sets of bladder tumors (N = 57 + 40) and seven bladder cancer cell lines. Clustering software was used to identify tumors with “multiple regional epigenetic silencing” (MRES) and the association of this phenotype with molecular gene signatures and histological types of bladder cancer.
Contribution
Seven stretches of contiguous genes were found to be concurrently silenced in 26 of 57 tumors by a mechanism involving histone methylation and hypoacetylation. Among bladder tumors and cell lines, the MRES phenotype was tightly associated with what was reported to be a carcinoma in situ (CIS) gene expression signature, the absence of FGFR3 mutations, and a more aggressive phenotype.
Implications
A new epigenetic phenomenon the MRES phenotype, is described in cancer. It appears to often be associated with carcinoma in situ among bladder cancers.
Limitations
More studies will be necessary to verify the reported CIS gene signature and to evaluate the relationship of the MRES phenotype with carcinoma in situ progression and with patient prognosis.
From the Editors
Cancer development not only depends on genetic alterations but also on epigenetic changes (1–3). These changes modify gene expression through DNA methylation, histone modifications, chromatin remodeling, and/or the expression of noncoding RNA (4–6). The reversibility of epigenetic gene silencing provides new opportunities for treatment based on the use of DNA methyltransferase inhibitors, like zebularine (7), or histone deacetylase inhibitors, such as suberoylanilide hydroxamic acid (8,9).
Until very recently, epigenetic gene silencing in cancer was thought to be restricted to focal events that silenced isolated genes (10). However, recent findings have indicated that epigenetic silencing can extend to a whole chromosomal region and has been reported to involve DNA methylation and/or histone modification in various cancers (bladder, breast, colorectal, and prostate cancer) (11–14).
The goal of this study was to take a global view, at the genome scale, of regional epigenetic silencing in malignant vs normal urothelium and to assess its clinical relevance to tumor progression. We previously developed a bioinformatics method that combines transcriptome and comparative genome hybridization array data from the same set of tumors to obtain an overview of the regional transcriptional deregulation that occurs independently of DNA copy number changes (11). The application of this method to a series of 57 bladder tumors led to the identification of 28 regions that harbored groups of neighboring genes with correlated expression independent of copy number changes. Epigenetic silencing affecting multiple neighboring genes was one possible mechanism that could account for this regional correlated expression (11).
In this current report, we have determined which of the 28 regions are epigenetically silenced by 1) identifying regions that harbor stretches of adjacent genes with decreased expression in tumors with respect to normal urothelium, and 2) searching for an epigenetic mechanism, such as DNA methylation and/or histone modification, that might be responsible for this decreased expression. We then investigated whether the epigenetically silenced regions were randomly distributed among tumors or whether they occurred together in a particular subset of tumors, defining a multiple regional epigenetic silencing (MRES) phenotype. Finally, we analyzed whether this silencing mechanism was related to one of the two pathways of bladder tumor progression: the carcinoma in situ (CIS) pathway or the low-grade Ta tumor pathway.
Methods
Patients and Tissue Samples
The first set of 57 bladder carcinomas were obtained from 53 patients who were included between 1988 and 2001 in the prospective database established in 1988 at the Department of Urology of Henri Mondor Hospital (Créteil, France). These cancers were selected randomly from a consecutive set, to cover the different stages of bladder cancer as follows: 16 Ta, eight T1, seven T2, 13 T3, and 13 T4 tumors. An additional criterion for inclusion was the availability of both tumor RNA and DNA. Three patients had multiple bladder tumors, two with one additional tumor and one with two additional tumors. Five normal urothelial samples were also used for transcriptome analysis. They were obtained from fresh urothelial cells scraped from the normal bladder wall and dissected from the lamina propria during organ procurement from cadaveric donors for transplantation, as previously described (15). A second independent set of 40 bladder carcinomas was obtained from tumor tissue banks at Henri Mondor Hospital (n = 18), Institut Gustave Roussy (n = 9) (Villejuif, France), and Foch Hospital (n = 13) (Suresnes, France). These tumors, sampled from 40 patients treated between 1993 and 2006, were selected so as to obtain a distribution of tumor stages similar to that for the first set of 57 cancers: 10 Ta, six T1, six T2, seven T3, and 11 T4. The characteristics of the patients and the tumors in the two sets are summarized in Table 1. All patients provided written informed consent and the study was approved by the ethics committees of the different hospitals.
Table 1.
Patient and tumor characteristics
| First set of tumors* | Second set of tumors | |
| Patients | N = 53 | N = 40 |
| Sex | ||
| Males, No. (%) | 43 (81) | 32 (80) |
| Females, No. (%) | 10 (19) | 8 (20) |
| Mean age at surgery, yr ± SD | 62.7 ± 11.0 | 66.9 ± 12.5 |
| Mean follow-up, mean ± SD | 73 ± 44.6 | 44 ± 38.5 |
| Bladder tumors | ||
| Clinical presentation | (N = 57) | (N = 40) |
| Incident tumors, No (%) | 44 (77) | 33 (82.5) |
| Recurrent tumors, No (%) | 13 (23) | 7 (17.5) |
| Stage† | ||
| Ta | 16 | 10 |
| T1 | 8 | 6 |
| T2 | 7 | 6 |
| T3 | 13 | 7 |
| T4 | 13 | 11 |
| Grade† | ||
| G1 | 6 | 2 |
| G2 | 16 | 9 |
| G3 | 35 | 29 |
| FGFR3 mutated tumors, No (%) | 23 (40.5) | 13 (32.5) |
For the three patients from whom we studied several tumors, only the first surgical intervention was taken into account for the age at surgery and follow-up calculations.
RNA and DNA Extraction from Tissues
Immediately after surgery, the samples were frozen in liquid nitrogen and stored at −80°C until nucleic acid extraction. RNA and DNA were extracted from the surgical samples by cesium chloride density centrifugation (18). Briefly, the frozen samples were homogenized in 4 M guanidium thiocyanate with an Ultraturrax T25 homogenizer (Janke & Kunkel, IKA-Labortechnik, Staufen, Germany), overlaid on a 5.7 M cesium chloride cushion and centrifuged. RNA was found in the pellet, whereas DNA was found on the top of the cesium chloride cushion. RNA and DNA were further purified by phenol–chloroform extraction and ethanol precipitation. The concentration, integrity and purity of each RNA sample were determined with the RNA 6000 LabChip Kit (Agilent Technologies, Massy, France) and an Agilent 2100 bioanalyzer. DNA purity was also assessed from the ratio of absorbances at 260 and 280 nm. DNA concentration was determined with a Hoechst dye-based fluorescence assay (19).
Affymetrix Array Analyses
We have previously reported the Affymetrix gene expression profile of the 57 urothelial bladder carcinomas (53 tumors from 53 patients plus four additional tumors from these patients) (11). For all Affymetrix array expression analyses, Affymetrix MAS5 signal values were log2-transformed and normalized by removing chip-specific and probe set–specific effects (the mean signal for all probe sets across one chip and the mean signal for one probe set across all chips, respectively).
Clustering Analyses
The Cluster 3.0 program (20) was used for hierarchical clustering. Results were displayed with the TreeView program (20). Cluster analyses were used to identify, from Affymetrix expression data, regions of correlated increased or decreased expression independent of copy number changes, in subsets of tumor samples. For the identification of regions that had decreased (or increased) expression independent of copy number losses (or gains) in a subset of tumors, the clustering approach was applied to those tumors without copy number losses (or gains). Cluster analyses using Affymetrix or TaqMan low-density array data were also used to identify tumors with the multiple region epigenetic silencing (MRES) phenotype and to identify tumors with the CIS signature using Affymetrix or TaqMan low-density array data.
Cell Culture
The human bladder cancer cell line CL1207 was derived from tumor T1207 of the first tumor set, as previously described (21). The human bladder cancer cell lines TCCSUP, HT1376, RT112, T24, MGHU3, and CL1207 were cultured in Dulbecco's modified Eagle medium F-12 Glutamax medium (Invitrogen, Cergy Pontoise, France) supplemented with 10% fetal bovine serum (Lonza Verviers, Verviers, Belgium); the human bladder cancer cell line JMSU1 was cultured in Roswell Park Memorial Institute (RPMI) Glutamax medium (Invitrogen) supplemented with 10% fetal bovine serum. HT1376, RT112, T24, and JMSU1 were obtained from DKFZ (Heidelberg, Germany). TCCSUP was obtained from the laboratory of D. Chopin (Hôpital Henri Mondor, Créteil, France) and MGHU3 from the laboratory of Y. Fradet (University of Laval, Québec, Canada). To verify the identity of the various cell lines used, we analyzed the genomic alterations with comparative genomic hybridization (CGH) arrays (22) and FGFR3, TP53, HRAS, and KRAS mutations with the SNaPshot technique (for FGFR3, see below) or classical sequencing for the other genes. CGH array profiles were found to be similar to published CGH profiles and/or consistent with previously reported genomic alterations, and mutations were found to be identical to the reported mutations (data not shown).
Normal human urothelial (NHU) cells were established as finite cell lines from surgical specimens of human urothelium obtained with the approval of the local research ethics committee and full written consent from patients with no history of bladder cancer undergoing open urological procedures. A representative piece of each specimen was examined to confirm that it was histologically normal and the remaining tissue was used to establish cultures, as detailed elsewhere (23,24). Briefly, the urothelium was detached from the stroma by incubation in Ca+2- and Mg+2-free Hanks balanced salt solution containing 10 mM HEPES pH 7.6, 2 × 104 IU/mL aprotinin and 0.1% (w/v) EDTA. The urothelium was collected, disaggregated in 200 U/mL collagenase type IV (Sigma-Aldrich, St. Quentin Fallavier, France) and used to seed Primaria tissue culture flasks (Becton Dickinson, Le Pont de Claix, France). NHU cell cultures were routinely maintained in complete keratinocyte serum-free medium (KSFMc), consisting of KSFM supplemented with bovine pituitary extract and endothelial growth factor, at the concentrations recommended by the manufacturer (Invitrogen), together with 30 ng/mL cholera toxin (Sigma-Aldrich). NHU cell cultures were subcultured each time that they started to become confluent by incubation in phosphate-buffered saline supplemented with 0.1% (w/v) EDTA for 5 minutes at 37°C, followed by treatment with trypsin–versene (containing 0.25% [w/v] trypsin and 0.02% [w/v] EDTA in phosphate-buffered saline) to detach the cells, which were harvested in KSFMc supplemented with 50 mg/mL soybean trypsin inhibitor (Sigma-Aldrich) (23). In these experiments, two independent NHU cell lines were used at passage 4.
RNA and DNA Extraction from Cell Lines
RNA and DNA were extracted from cell lines with Qiagen extraction kits (Qiagen, Courtaboeuf, France): the RNeasy Mini Kit for RNA extraction and the QIAamp DNA Mini Kit for DNA extraction. For RNA to be considered suitable for DNA array analysis, it had to fulfill two of the following three criteria: 1) RIN (RNA integrity number) above 7; 2) 28S to 18S ratio greater than 1.5; and 3) intact profile with a flat baseline. RNA samples with 260/280 nm absorbance ratios greater than 1.8 were considered suitable for RT-qPCR analysis. DNA was considered suitable for methylation or CGH analyses if it gave a visible band on an agarose gel with a smear above 2 kb when 300 ng was loaded, and if the 260/280 nm absorbance ratio was greater than 1.8.
Analysis of the Effect of Epigenetic Drugs on Gene Expression
For analyses of the effect of the histone deacetylase inhibitor trichostatin A (TSA) (Calbiochem, Fontenay-sous-Bois, France) and/or the DNA demethylating agent 5-aza-deoxycytidine (Calbiochem) on transcript levels, normal and tumor cells were seeded in 25 cm2 dishes at a density of 8 × 105 cells per dish. Cultures were treated the next day with 300 nM TSA for 16 hours, 5 μM 5-aza-deoxycytidine for 72 hours, or 5 μM 5-aza-deoxycytidine for 48 hours followed by 300 nM TSA for 10 hours. These experiments were repeated twice and each time, each condition was tested in duplicate. The efficiency of the TSA treatment was controlled by measuring the expression of the SMPD3 gene. We have found this gene to be repressed in normal bladder and cancer cell lines by histone methylation and hypoacetylation but not by DNA methylation (data not shown). The efficiency of the 5-aza-deoxycytidine treatment was controlled by monitoring the re-expression of several methylated genes: CDH1 in T24, TCCSUP, and HT1376 cells; TMEFF2 in MGHU3, RT112, and JMSU1 cells; and ANKRD30A (25) in NHU cells.
Quantitative Reverse Transcription–Polymerase Chain Reaction (RT-qPCR)
One microgram of total RNA was used for reverse transcription with 20 pmol of random hexamers and 200 units of Moloney Murine Leukemia Virus (MMLV) reverse transcriptase. To assess mRNA levels by real-time quantitative PCR (RT-qPCR), we used either individual assays or the TaqMan low-density array on an ABI PRISM 7900 real-time thermal cycler (Applied Biosystems, Courtaboeuf, France). With both methods, all samples were run in duplicate. For all experiments that involved RNA from T1207 and CL1207 cells, both methods were used. For individual assays, we used the SYBR Green kit (Applied Biosystems) to measure the expression of the RNAs of interest (for primer sequences, see Supplementary Table 1, available online) and the TaqMan kit (assays-on-demand; Applied Biosystems) for the reference RNAs (18S rRNA). For TaqMan low-density array, the same reference 18S rRNA was used; predesigned TaqMan probe and primer sets for the different genes were chosen from the Applied Biosystems catalog. The amounts of mRNAs of the genes of interest were normalized to that of the reference gene according to the 2−ΔCt method (26).
DNA Methylation Analysis
We investigated the methylation status of the promoters by bisulfite sequencing or combined bisulfite restriction analysis (COBRA) assay (27). We used the Epitect kit (Qiagen) for bisulfite treatment of 2 μg of genomic DNA and its subsequent purification. This modified DNA was amplified as follows: initial incubation at 94°C for 4 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at Tm for 30 seconds and extension at 72°C for 30 seconds, with Biolabs Taq Polymerase (Ozyme, Saint-Quentin-en-Yvelines, France). For bisulfite sequencing, we cloned the purified PCR product using the TA cloning kit (Invitrogen, Cergy Pontoise, France) and sequenced 10 clones for each sample and gene. For the COBRA assay, the PCR products were digested for 16 hours with a restriction enzyme recognizing a restriction site containing a CpG dinucleotide. When the corresponding CpG site is methylated, the PCR product is digested.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out in duplicate in three 150 cm2 dishes for untreated CL1207, TCCSUP, RT112, and NHU cells and for CL1207 cells treated with 300 nM TSA for 16 hours. Using an Active Motif kit (Active Motif, Rixensart, Belgium), chromatin was extracted from cell nuclei and sheared enzymatically. An extract of the original chromatin was kept as an internal standard (input DNA). The complexes were immunoprecipitated with 4 μg of rabbit polyclonal antibodies against trimethyl histone H3 lysine 27 (Upstate Biotechnology, Santa Cruz, CA), trimethyl histone H3 lysine 9 (Abcam, Cambridge, UK) or acetyl histone H3 lysine 9 (Abcam) and Protein A/G PLUS-Agarose beads (Santa Cruz, Heidelberg, Germany) in dilution buffer containing 1% Triton X-100, 150 mM NaCl, 2 mM EDTA, 20 mM Tris–HCl at pH 8.0, and protease inhibitors. The amount of immunoprecipitated target was determined by real-time PCR, in duplicate, using the ABI PRISM 7900HT Sequence Detection System and appropriate primer sequences (Supplementary Table 1, available online). For each sample and each promoter, an average Ct value was obtained for immunoprecipitated material and for the input chromatin. The amount of immunoprecipitated material was defined as 2Ct(input DNA) - Ct(immunoprecipitated DNA).
Defining the MRES Phenotype
Two methods were used to define the MRES phenotype. The first method used individual clustering (Figure 1): expression of a region was considered to be decreased in a given sample if, in the individual clustering for this region, the sample belonged to the cluster arm of tumors with decreased expression. The second method used the regional expression score: for each sample and each region, expression levels of all the genes in the region were log2-transformed and normalized; the regional expression score of a sample was calculated as the average of the differences between the expression in this sample and the average expression in normal samples of the genes in the region. Tumors were clustered according to these regional expression scores.
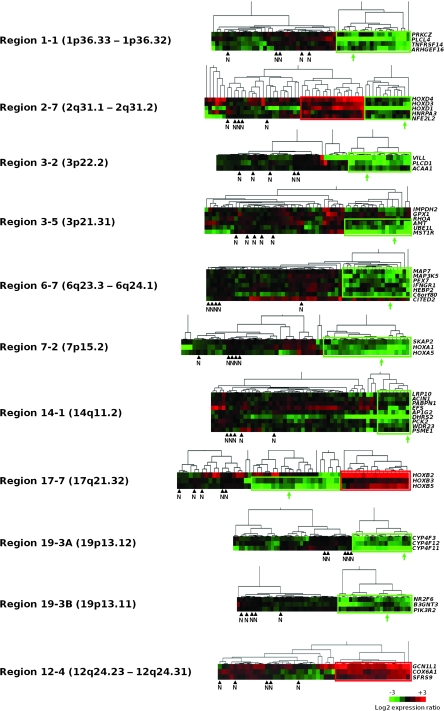
Figure 1.
Clustering analyses to identify coordinated gene expression patterns independent of copy number changes in bladder tumors. For each of the 28 chromosomal regions that contained genes with correlated expression independent of copy number changes (11), tumors were subjected to hierarchical clustering based on gene expression levels. Each row represents such a gene, ordered according to its chromosomal position in the region, and each column represents a tumor or normal tissue sample. Only genes for which normal samples had a “present” signal according to the Affymetrix MAS5 algorithm were included. As shown in the color bar at the bottom of the figure, green indicates decreased expression, red indicates increased expression, and black indicates no change of gene expression, with respect to average expression in normal urothelium (n = 5). For each region, the subset of tumors with coordinately decreased gene expression in the region is boxed in green and the subset of tumors with coordinately increased gene expression is boxed in red. N indicates the position of the normal samples and the green arrows indicate the position of the T1207 bladder tumor from which the CL1207 cell line was derived. Different tumor expression patterns were identified among the 28 chromosomal regions containing genes with coordinated expression. Ten regions (shown here) (1-1, 2-7, 3-2, 3-5, 6-7, 7-2, 14-1, 17-7, 19-3A, and 19-3B) contained a subset of tumor samples with coordinated decreased expression. These regions were potentially affected by a regional epigenetic silencing mechanism. Two of the 10 regions (2-7 and 17-7) also contained a subset of tumors with coordinated increased expression. Although region 19-3 was initially identified as a single region (11), it was separated here into two regions (regions 19-3A and 19-3B) because we found that it comprised two well-spaced clusters of genes with decreased expression that were separated by several genes whose expression was not decreased in tumors compared to normal urothelium (data not shown). Among the 19 remaining regions, six presented a subset of tumors with coordinated increased expression (one of these six region, region 12-4 is shown) and 13 presented no clear expression pattern.
FGFR3 Mutation Analysis
FGFR3 mutations were studied with the SNaPshot technique (28). Briefly, three regions that encompassed all of the FGFR3 mutations reported in bladder cancer were amplified simultaneously. After the removal of excess primers and dNTPs, SnaPshot primers to detect the following FGFR3 mutations (R248C, S249C, G372C, Y375C, A393E, K652E, K652Q, K652M, K652T) were annealed to the PCR products and extended with a labeled dideoxynucleotide (see [28] for the primer sequences). The extended primers were analyzed on an automatic sequencer (ABI Prism 3100 genetic analyzer; Applied Biosystems).
Statistical Analysis
Statistical analysis and numerical calculations were carried out with R 2.6 (R Foundation for Statistical Computing, http://www.r-project.org/) and Amadea (Isoft, Gif-sur-Yvette, France). Fisher exact test was used to test the relationship between the MRES phenotype and various histopathologic and molecular traits of bladder cancer. We calculated 95% confidence intervals for qRT-PCR expression data for normal samples. All statistical tests were two-sided and P values less than .05 were considered to be statistically significant.
Results
Identification of Chromosomal Regions With Decreased Expression in Bladder Tumors
We previously developed a large scale bioinformatics approach that combined paired transcriptome and CGH array data for the identification of regions of neighboring genes with correlated expression that was not dependent upon changes in copy number. The application of this method to a series of 57 bladder tumors led to the identification of 28 such regions on 12 different chromosomes with one to four regions per chromosome. These regions were candidates for control by regional epigenetic mechanisms (11). Because we were principally interested in epigenetically silenced regions, we first searched for those regions that harbored genes with decreased expression with respect to normal urothelial samples. For each gene in the 28 regions, we calculated the ratio between its level of expression in each tumor sample and its mean expression in five samples of mRNA from normal urothelium, as measured on Affymetrix arrays. We then used these expression ratios to cluster, for each region, all normal and tumor samples. This cluster analysis identified several types of regions. For eight regions (regions 1-1, 3-2, 3-5, 6-7, 7-2, 14-1, 19-3A, and 19-3B), we observed coordinately decreased gene expression in a subset of tumors (Figure 1). For six other regions (regions 2-3, 5-3, 5-4, 6-3, 6-5, and 12-4), we observed coordinately increased gene expression in a subset of tumors (Figure 1 and data not shown). For two regions (2-7 and 17-7), one subset of tumors presented decreased gene expression and a different subset showed increased gene expression (Figure 1). The remaining regions displayed no clear expression pattern. Because we were interested in regions that were candidates for epigenetic silencing, we focused our subsequent analysis on the 10 regions that had decreased gene expression (1-1, 2-7, 3-2, 3-5, 6-7, 7-2, 14-1, 17-7, 19-3A, and 19-3B) in a subset of tumors (Figure 1).
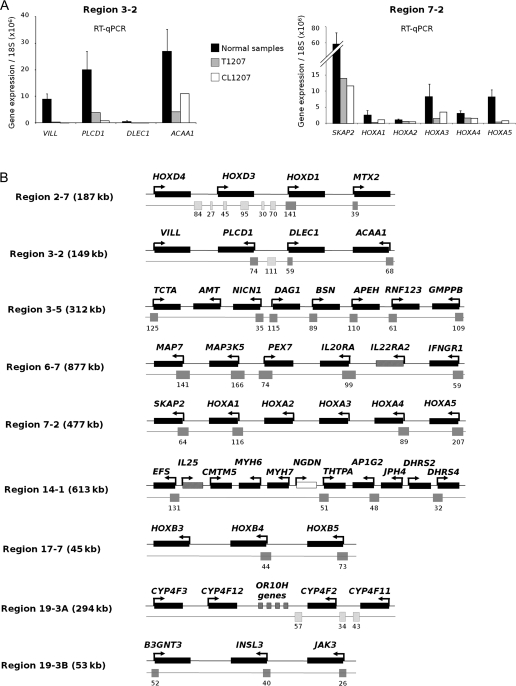
To determine whether coordinated decreased expression affected contiguous genes in these 10 regions, we extensively studied the expression of all genes within these regions by RT-qPCR, including both the genes that were present and absent on the Affymetrix array (Figure 2, A and Supplementary Figure 1, available online). For this analysis, we used mRNA from a T4G2 bladder tumor, T1207, and from a cell line derived from this tumor, CL1207. We chose to study tumor T1207 because it showed decreased gene expression in all 10 regions on Affymetrix arrays (Figure 1), it did not show any genetic loss in these regions, and a cell line derived from this tumor, CL1207 was available, allowing subsequent functional analyses. We found stretches of contiguous genes with coordinately decreased or nonexpression within all of the regions except region 1-1. Both tumor T1207 and the cell line that was derived from it displayed identical stretches of genes with reduced expression (Figure 2, A and Supplementary Figure 1, available online). A schematic representation of the stretches found in nine of the 10 regions is given in Figure 2, B.
Figure 2.
Further analysis of gene expression within the regions displaying decreased expression. A) Analysis of all the genes contained in regions 3-2 and 7-2 by real-time quantitative polymerase chain reaction (RT-qPCR). To confirm the Affymetrix data and to determine whether decreased expression affected stretches of contiguous genes, RT-qPCR was performed on RNA from the T1207 human bladder tumor, CL1207 (a cell line derived from T1207), and normal urothelial samples (n = 4). Expression was determined for all the genes in these regions, including the genes not present on the Affymetrix array. Experiments were done in duplicate, in a TaqMan low-density array format, and expression levels were normalized relative to 18S ribosomal RNA. The histograms reflect the mean value of the duplicates. For normal samples, the error bars indicate the 95% confidence intervals for four independent samples. For additional data for all other regions, see Supplementary Figure 1, available online. B) Summary of RT-qPCR data for the nine regions containing stretches of genes with decreased expression. Experiments were done with RNA from tumor T1207 and its derived cell line, CL1207 (as shown in [A] and Supplementary Figure 1, available online). Stretches were defined by three or more consecutive genes with decreased expression in T1207 and CL1207 (ratio to average expression in normal samples <0.5). Genes that were not expressed were included in these stretches. Genes are ordered according to their chromosomal locations; their names, the orientation of transcription (shown with arrows), and the size of the regions are indicated. Genes with decreased expression are represented by a black rectangle, genes that were not expressed are indicated with a hatched rectangle and genes with no change in expression with respect to normal samples are indicated with a white rectangle. All of the CpG islands that overlap a gene promoter are shown below the genes (gray rectangles), and the numbers of CpG dinucleotides contained in the CpG islands are indicated. CpG islands that do not overlap promoters are shown for regions 2-7, 3-2, and 19-3A (hatched rectangles) because they were also used for methylation analyses.
Identification of Seven Regions Affected by Regional Epigenetic Silencing
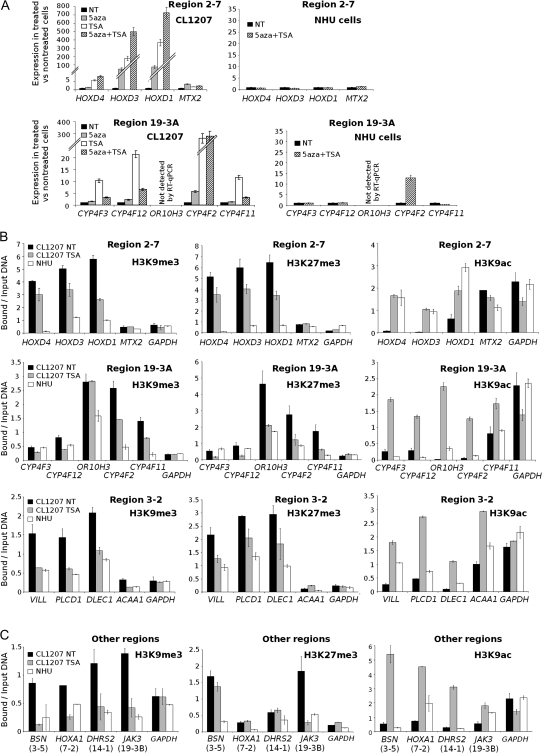
To address whether the genes within the nine gene stretches that were underexpressed in tumor vs normal samples were silenced by an epigenetic mechanism, we first treated CL1207 cells with either the DNA demethylating agent, 5-aza-deoxycytidine, and/or with the histone deacetylase inhibitor TSA, then extracted mRNA from the cells and analyzed it by RT-qPCR. Compared with mRNA from untreated cancer cells, treatment with either drug was associated with the re-expression of most genes in seven of the regions (2-7, 3-2, 3-5, 7-2, 14-1, 19-3A, and 19-3B), suggesting that these regions were silenced by DNA methylation and/or histone modifications (Figure 3, A and Supplementary Figure 2, available online). By contrast, when NHU cells were treated with 5-aza-deoxycytidine and TSA, expression was not increased for most of the genes in these regions, with the exception of some isolated genes (Figure 3, A and data not shown). Treatment of CL1207 cells with 5-aza-deoxycytidine and/or TSA led either to no re-expression or to the re-expression of only one isolated gene in each of two regions (regions 6-7 and 17-7; Supplementary Figure 2, available online). These two regions were therefore excluded from subsequent analysis.
Figure 3.
Epigenetic silencing mechanisms in regions of decreased expression. A) Effect of 5-aza-deoxycytidine (5aza) and TSA treatments on gene expression in CL1207 human bladder tumor cells vs normal human urothelial (NHU) cells. Gene expression was measured by real-time quantitative polymerase chain reaction (RT-qPCR) in RNA from CL1207 cells before (NT) and after treatment with the DNA demethylating agent 5aza, the histone deacetylase inhibitor TSA, or both and in RNA from NHU cells before (NT) and after treatment with both 5aza and TSA. Results are shown for two regions, region 2-7 (upper panels) and region 19-3A (lower panels) (for additional data for all other regions, see Supplementary Figure 2, available online). For each treatment, RT-qPCR analyses were performed in two independent experiments, with each qPCR performed in duplicate. Results are expressed as the ratio between expression in treated cells and in untreated cells. The error bars indicate the variance between the means of the two independent experiments. Treatments were scored as having an effect if there was a greater than 1.5-fold change between treated and untreated cells. B) Histone marks in regions 2-7, 19-3A, and 3-2 in chromatin from CL1207 cells in the presence or absence of TSA treatment and from untreated NHU cells. Histone marks associated with the promoters of silenced genes (H3K9me3 and H3K27me3) or expressed genes (H3K9Ac) were investigated by chromatin immunoprecipitation (ChIP) assays. ChIP assays were performed for all promoters in regions 2-7 (upper panels), 19-3A (middle panels) and 3-2 (lower panels), using antibodies against posttranslational modifications of histone H3: trimethyl lysine 9 (H3K9me3; left panels), trimethyl lysine 27 (H3K27me3; middle panels) and acetyl lysine 9 (H3K9ac; right panels). The bar graph shows the amount of immunoprecipitated target DNA, expressed as a percentage of total input DNA, measured in duplicate by qPCR. Levels of H3K9me3 and H3K27me3 in the promoter of the ubiquitously expressed GAPDH gene served as a negative control in chromatin from both cell types. The error bars indicate the variance between the means of two independent experiments. C) Histone marks in regions 3-5, 7-2, 14-1, and 19-3B in chromatin from TSA-treated or untreated CL1207 cells and from untreated NHU cells. In all other regions that presented a stretch of coordinately silenced genes that were re-expressed after TSA or 5aza treatment, experiments were done as in (B) except that ChIP assays were performed for only one gene promoter per region (eg, the promoter of BSN for region 3-5, HOXA1 for region 7-2, DHRS2 for region 14-1, and JAK3 for region 19-3B).
We then investigated the involvement of DNA methylation and histone hypoacetylation or methylation in the silencing of the seven regions that were re-expressed after 5-aza-deoxycytidine and/or TSA treatment. We assessed DNA methylation by DNA sequencing and COBRA (digestion with an enzyme targeting an original CpG site). All of the CpG islands in regions 3-2 and 19-3A were studied, whether or not they overlapped gene promoters (for their locations, see Figure 2, B). For the other regions, we studied the methylation status of all gene promoter–associated CpG islands for the genes that were re-expressed after 5-aza-deoxycytidine treatment. These methylation studies were performed on DNA from different tumors, including T1207, that had shown decreased gene expression in the regions of interest, and on DNA from the CL1207 bladder cancer cell line and from normal NHU cells. DNA methylation was rarely found in these regions and, when present, it was not necessary for silencing because some samples with the same expression profile had different methylation status (Supplementary Figure 3, available online, and data not shown).
We assessed histone modifications by ChIP followed by quantitative PCR, using chromatin from CL1207 cells with and without TSA treatment and from NHU cells. We analyzed the promoter regions of all the genes located in regions 2-7, 3-2, and 19-3A (Figure 3, B) and of one gene in each of the remaining regions (Figure 3, C). We studied two histone marks commonly associated with transcriptional inactivity (trimethylation of Lys9 of histone H3 [H3K9me3] and trimethylation of Lys27 of histone H3 [H3K27me3]) and one histone mark commonly associated with transcriptional activity (acetylation of Lys9 of histone H3 [H3K9ac]). The promoters of most genes in the silenced regions displayed high levels of methylation at the two repressive marks (H3K9me3 and H3K27me3) in chromatin from CL1207 bladder cancer cells compared with that from normal (NHU) cells. Conversely, the promoters of genes in the silenced regions lacked the histone mark associated with active genes (H3K9ac) in chromatin from CL1207 cells; however, it was present in chromatin from NHU cells (except in region 19-3A). When CL1207 cells were treated for 16 hours with TSA, the histone methylation marks associated with gene inactivity decreased and the histone acetylation mark associated with gene activity increased in most gene promoters in the regions studied. These changes were accompanied by increased expression of these genes following TSA treatment.
Together, these results show that seven regions of chromatin with contiguous inactivated genes were silenced by a regional epigenetic mechanism involving aberrant histone modifications that led to the formation of facultative heterochromatin domains in cancer cells. DNA methylation was not involved in the silencing process. We termed this mechanism “RES” for regional epigenetic silencing.
MRES Phenotype in Bladder Tumors
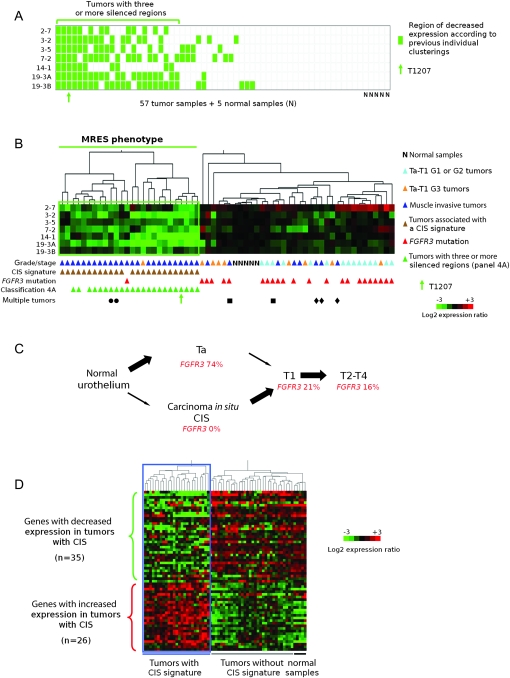
To determine whether the epigenetic silencing of these seven chromosomal regions (2-7, 3-2, 3-5, 7-2, 14-1, 19-3A, and 19-3B) occurred randomly among tumors or in particular groups of tumors, we used two different strategies. In the first approach, we used the clustering data for these seven regions (Figure 1) to assess how many of these regions were silenced in each of the 57 bladder tumors. Twenty of 57 tumors (35%) did not present any silenced regions, 14 of 57 tumors (25%) had one or two silenced regions, whereas 23 of 57 tumors (40%) had three or more silenced regions, suggesting a concomitant silencing of regions in a subgroup of tumors (Figure 4, A). In the second approach, we clustered tumors according to regional expression scores that were calculated by averaging the difference between the normalized log2-expression values for genes in these regions in the tumor sample compared with those in normal tissue samples. Using this approach, 26 of 57 tumors (46%) (including the 23 previously identified in Figure 4, A) clustered together and showed silencing of all or several of the seven regions (Figure 4, B). The simultaneous silencing of several chromosomal regions in a subgroup of tumors defines a new phenotype in cancer that we have termed the “multiple regional epigenetic silencing” (MRES) phenotype.
Figure 4.
Presence of a multiple regional epigenetic silencing (MRES) phenotype in bladder cancer and its relationship to the two pathways of bladder tumor progression. A) Determination of the number of epigenetically silenced regions for each tumor in the set of 57 bladder tumors. This panel was deduced from the cluster analysis data in Figure 1 for each of the seven epigenetically controlled regions (regions 2-7, 3-3, 3-5, 7-2, 14-1, 19-3A, and 19-3B). Each row represents a chromosomal region, and each column a tumor or normal sample. For each tumor, decreased expression of a given chromosomal region (ie, placement within the green box in Figure 1) is denoted by a green rectangle, whereas equal or increased expression is denoted by a white rectangle. Twenty-three tumors (those below the horizontal green line) displayed decreased expression of at least three regions. The position of tumor T1207 is indicated by a green arrow. B) Cluster analysis of 57 tumor samples and five normal samples according to the level of gene expression in the seven epigenetically silenced regions. Samples are clustered according to their regional expression scores, which correspond to the average levels of gene expression in each region (see “Methods”). Tumors that displayed decreased expression in several regions (below the horizontal green line) define a MRES phenotype. All samples are annotated with their stage and grade, presence or absence of a carcinoma in situ (CIS)-associated gene expression signature (Figure 4, D) and their FGFR3 mutation status. Tumors that had at least three chromosomal regions with decreased gene expression in Figure 4, A are also indicated. The tumors from the three patients with multiple tumors are indicated by three different symbols. The patient indicated by a circle had two synchronous T3-G3 tumors. The patient indicated by a square had one T2-G2 primary tumor and then a T1-G2 tumor. The patient indicated by a diamond had three synchronous T4-G3 tumors. The cluster analysis was not affected by the exclusion of regional expression scores for tumors displaying genetic loss in the corresponding region (data not shown). C) Schematic representation of the two pathways of bladder cancer progression. In bladder cancer, two different pathways can lead to invasive tumors: the superficial Ta tumor pathway, which rarely leads to progression, and the CIS pathway, in which the superficial lesions (CIS) are high grade and very often progress to lamina propria–invasive T1 and then to muscle-invasive T2-T4 tumors (29). The percentages of FGFR3 mutations at the different stages of tumor progression in the two pathways are taken from reference (30). D) Separation of the 57 bladder tumors according to the CIS-associated gene expression signature previously defined by Dyrskjøt et al. (31). Sixty-one of the 100 genes reported by Dyrskjøt et al. were represented on the Affymetrix array used (U95A). Of these 61 genes, 26 corresponded to genes with increased expression and 35 to genes with decreased expression in bladder cancers that had the CIS-associated signature compared with those that did not (31). The 62 samples (57 tumor samples and five normal urothelial samples) clustered into two groups with respect to the CIS-associated gene expression signature.
We examined the subgroup of 26 tumors with the MRES phenotype in terms of histopathologic and molecular characteristics. Strikingly, 25 of these 26 samples (96%) were from muscle-invasive tumors (≥T2), and the remaining sample was from a high-grade (G3) T1 tumor (Figure 4, B). The samples that did not display the MRES phenotype (31 tumors and five normal urothelial samples) included the eight remaining muscle-invasive tumors, eight of nine high-grade Ta and T1 tumors, all 15 low-grade (G1 and G2) Ta and T1 tumors, and all five normal samples.
We further examined whether the MRES phenotype was more associated with either of the two pathways of bladder cancer progression (Figure 4, C). It is generally thought that most muscle-invasive bladder tumors (T2–T4) develop from CIS (29). The second pathway of bladder cancer progression involves the development of stage Ta tumors, usually of low grade, which rarely progress to muscle-invasive tumors (Figure 4, C) (32). This pathway is associated with a high frequency of activating fibroblast growth factor receptor 3 (FGFR3) gene mutations, whereas CIS-associated tumors have few, if any, such mutations (30).
Dyrskjøt et al. (31) have reported a CIS-associated signature from bladder cancer transcriptome data. We looked for a possible overlap in our data between the MRES phenotype and this signature by examining expression of the 61 genes present on our Affymetrix U95A array out of the 100 genes reported by Dyrskjøt et al. (31). Twenty-five of the 57 tumors presented the CIS signature, and remarkably, all 25 displayed the MRES phenotype (Figure 4, D). The association between the MRES phenotype and the CIS signature was highly statistically significant (P < 10−14, Fisher test). Only one tumor displayed the MRES phenotype but not the CIS signature.
We also looked for the presence of FGFR3 mutation as a marker for the alternative tumor progression pathway, the Ta pathway (Figure 4, C). In our series of 57 tumors, 23 tumors had an FGFR3 mutation, and all but one of these tumors belonged to the group lacking the MRES phenotype (the FGFR3 mutated tumor with the MRES phenotype also lacked the CIS signature). The absence of FGFR3 mutation is a characteristic trait of tumors with the MRES phenotype (P < 10−6). The MRES phenotype remained associated with the CIS signature and the presence of FGFR3 mutations remained associated with non-MRES tumors if we limited the analysis to muscle-invasive tumors (P < 10−6 and P = .008, respectively).
We analyzed specimens from three patients with multiple tumors (two patients had two tumors and one patient had three tumors). All tumors from a given patient had the same MRES status and CIS signature (Figure 4, B). It should be noted that, when considering only one tumor per patient, we obtained identical results for the clustering analyses presented above and for the relationships between the MRES phenotype, the CIS signature and the absence of FGFR3 mutation (Supplementary Figure 4, available online, and data not shown).
The existence of the MRES phenotype, its association with muscle-invasive tumors with the CIS-associated signature and with a low rate of FGFR3 mutations, was also confirmed with an independent set of tumors (n = 40). These data are available online (Supplementary Figure 5, available online).
The MRES Phenotype in Bladder Cancer Cell Lines
To determine whether the MRES phenotype was also found in bladder tumor cell lines, we used a panel of established bladder cancer–derived cell lines that were representative of the diversity of bladder tumors. We used two cell lines that were derived from well-differentiated tumors (MGHU3, which has mutated FGFR3; and RT112) and four cell lines that, like CL1207, were derived from high-grade tumors (T24, TCCSUP, HT1376, and JMSU1; none with mutated FGFR3). We also included NHU cells in the analysis. We investigated the effect of TSA on the re-expression of the genes within the seven epigenetic regions that defined the MRES phenotype (Supplementary Figure 6, available online). We have provided a summary of the effects of treatment on the different cell lines (Figure 5, A). Two groups of cell lines were clearly distinguished. In the first group (NHU, MGHU3, RT112, and T24), most of the genes were not re-expressed. In the second group of cell lines (TCCSUP, HT1376, and JMSU1), as in CL1207, the genes in most of the silenced regions were re-expressed after TSA treatment, consistent with the existence of an MRES phenotype among these cell lines. Similar to what we had observed among tumors, the cell lines with the MRES phenotype were derived from high-stage and high-grade tumors, whereas all but one of the other cell lines were well differentiated (RT112) or derived from a low-grade and low-stage tumor with the FGFR3 mutation (MGHU3). The one exception, the T24 cell line, has an HRAS mutation (33), and tumors with such mutations are thought to follow the Ta pathway (34,35). ChIP experiments to assess the H3K9me3, H3K27me3, and H3K9ac histone marks were previously performed on chromatin from the CL1207 cell line and NHU cells for three regions in detail (2-7, 3-2, and 19-3A) and for one gene in each of the other regions (3-5, 7-2, 14-1, and 19-3B) (Figure 3, B and C). We studied the same marks in chromatin from two additional cell lines, one with (TCCSUP) and the other without (RT112) the MRES phenotype (Supplementary Figure 7, available online). For all seven regions, we observed higher levels of trimethylation at lysines 9 and 27 in histone 3 from the TCCSUP cells compared with histone 3 from the RT112 cells. Levels of histone 3 acetylation at lysine 9 were higher in RT112 cells for some genes. Therefore, trimethylation of lysines 9 and 27 clearly differentiated cancer cell lines with the MRES phenotype, such as TCCSUP and CL1207 cells, from normal (NHU) cells and from other cancer cell lines, such as RT112. A summary of the histone marks in the epigenetically silenced regions for all three cell lines (RT112, CL1207, and TCCSUP) compared with NHU cells is presented (Figure 5, B).
Figure 5.
The multiple regional epigenetic silencing (MRES) phenotype in bladder cancer cell lines. A) Summary of the effect of TSA treatment on gene expression in the epigenetically silenced regions in bladder cancer and normal urothelial cells in culture. A gene was considered to be re-expressed if its expression level changed 1.5-fold or more between treated and untreated cells (for original data, see Supplementary Figure 6, available online). A region was considered to be re-expressed if at least two neighboring genes were re-expressed after treatment: a yellow box indicates that a group of at least two neighboring genes were re-expressed, whereas a red box indicates that all genes were re-expressed. A cell line was considered to present the MRES phenotype if most of the regions were re-expressed following TSA treatment. B) Summary of the changes in histone marks in the epigenetically silenced regions in MRES and non-MRES tumor cell lines. This panel summarizes the changes in histone marks associated with the promoter of inactive (H3K9me3 and H3K27me3) and active (H3K9Ac) genes for the different regions in two cell lines presenting the MRES phenotype (TCCSUP and CL1207) and one cell line without the MRES phenotype (RT112) compared with NHU cells. The histone marks were assessed by chromatin immunoprecipitation, with specific antibodies directed against the different marks (for original data, see Figure 3, B and C and Supplementary Figure 7, available online). All gene promoters were studied within regions 2-7, 3-2, and 19-3A and one gene promoter per region was studied for all the other regions (the promoter of BSN for region 3-5, HOXA1 for region 7-2, DHRS2 for region 14-1 and JAK3 for region 19-3B). For each cell line, the histone marks of the promoters of the genes within the different regions were compared with the histone marks found at the same promoter in NHU cells. Within each region and for each cell line and histone mark, we have indicated the percentage of promoters with a histone mark that is modified compared with the corresponding site in NHU cells. The inactive marks (H3K9me3 and H3K27me3) were scored as being modified if we observed a twofold increase in modification at the same position in the promoter in the tumor cell line, as compared with NHU cells. For the active mark (H3K9ac), we scored the mark as modified if the mark displayed a more than twofold decrease.
Discussion
We have shown that whole chromosomal regions can be coordinately silenced by an epigenetic mechanism in a subset of bladder tumors, hence defining a MRES phenotype in cancer. The subgroup of tumors with the MRES phenotype is associated with a CIS gene expression signature, with higher tumor grade and stage, including most muscle-invasive tumors, and usually with the absence of FGFR3 mutations. The silencing occurred in association with histone H3K9 and K27 hypermethylation and H3K9 hypoacetylation of promoter regions, mimicking the formation of facultative heterochromatin domains.
Several regions of epigenetic silencing have previously been identified in cancer. Three of them overlap with regions that we identified in bladder cancer: at 3p22.3, a 1.1 Mb region in colon cancer (36), which included our 144 kb region 3-2; at 7p15.2, a region of about 100 kb of the HOXA cluster in breast cancer (12) and a region of 1.09 Mb which included the HOXA cluster in prostate cancer (14), which both overlapped with our region 7-2; and at 2q31.1, a region of 109 kb of the HOXD cluster in breast cancer (37), which overlapped with our region 2-7. The repeated identification of these sites suggests that some chromosomal regions may be prone to RES in different cancers. However, there is some cancer tissue–type specificity because the region previously identified at 2q14.2 in colorectal cancer (13) was not silenced in bladder cancer, and the 3-5, 14-1, 19-3A, and 19-3B regions identified here in bladder cancer were not silenced in breast or cervical cancers (data not shown).
In one of the silenced regions, region 14-1, one gene (NGDN) was not silenced in any of the tumors that showed silencing of this region. The reason that NGDN has “escaped” the global pattern of the chromosomal region that surrounds it is unclear, although a similar phenomenon has been reported for the inactive X chromosome in women, in which most genes are silenced, but a few can escape inactivation (38).
The different types of histone marks that we identified in the epigenetically silenced regions, especially trimethylation of lysines 27 and 9, are characteristic of silent and heterochromatic regions of the genome in normal tissue (39). Thus, it would appear that the chromatin structure of the altered chromosomal regions that we have identified has shifted from a euchromatic transcriptionally competent state in NHU cells (where such marks are not found or are found only at low levels) to a facultative heterochromatic conformation that inhibits transcription (40). There are several possible explanations for this change in chromatin structure. Among them, a condensation of chromatin and/or a modification of the localization of these regions within the nucleus (41,42) may affect their transcription status. Indeed, the HOXD cluster is regulated by such mechanisms during development (43,44) and is dysregulated in tumors with the MRES phenotype. Consequently, RES in cancer may correspond to a deregulation of the normal process of chromatin domain regulation that occurs during development. To confirm this hypothesis, it would be worth studying the status of the six other silenced regions during normal development to determine whether those genes are co-regulated, like the HOXD cluster.
Another epigenetic phenotype involving DNA hypermethylation, the CpG island methylator phenotype (CIMP), has been described in colon cancer (45). In this case, multiple isolated loci, rather than chromosomal regions, simultaneously undergo DNA methylation. This phenotype is associated with a subtype of colon tumors that harbor specific molecular and clinical characteristics such as microsatellite instability, a high frequency of BRAF V600E mutation, low frequencies of p53 mutations, proximal tumor location, poor differentiation, mucinous histology, infiltrating lymphocytes, female sex, higher age at diagnosis, and microsatellite instability (46,47).
Clinical and molecular evidence indicates that bladder cancer develops through two pathways, the Ta pathway and the CIS pathway (Figure 4, C) (29,30,48). Ta tumors, which are mostly of low grade, rarely progress to muscle-invasive disease (32). Isolated CIS is rarely observed; it is encountered predominantly with other urothelial tumors (49). The presence of concomitant CIS lesions with papillary tumors in the bladder is associated with a high risk of disease progression to a muscle-invasive stage (32), and CIS is considered to be the main precursor of muscle-invasive tumors (29). One of the strongest molecular arguments in favor of the two-pathway model is the high frequency of FGFR3-activating mutations (75%) in Ta low-grade tumors, their absence in CIS and their low frequency (10%–16%) in muscle-invasive tumors (30,50). We show here that the MRES phenotype was associated to a striking degree with muscle-invasive tumors and almost completely overlapped with the CIS-associated signature identified by Dyrskjøt et al. (31). In addition, FGFR3 mutations were found mostly in tumors without the MRES phenotype. Our data thus provide further support for the existence of two pathways of bladder tumor progression: the Ta pathway, characterized by a low propensity to progress and by a high rate of FGFR3 mutation, and the CIS pathway, characterized by an epigenetic phenotype that includes the concomitant silencing of several chromosomal regions. Further characterization of bladder cancer progression is required to determine whether the MRES is already present in CIS lesions or appears at a later stage. In support of the association between the MRES phenotype and the CIS pathway, cell lines with (or without) the MRES phenotype have the same characteristics as tumors with (or without) the MRES phenotype. Indeed, cell lines with the MRES phenotype were derived from high-stage and high-grade tumors, whereas all but one (T24) of the other cell lines were well differentiated (RT112) or derived from a low-grade and low-stage tumor with an activating FGFR3 mutation (MGHU3). However, the T24 cell line fits our model because this cell line has an HRAS mutation, and tumors with such mutations are thought to follow the Ta pathway (34,35).
In our series of bladder cancer patients, most of the muscle-invasive tumors displayed the MRES phenotype, whereas most of the Ta and T1G2 tumors did not present this phenotype, consistent with the two-pathway model of tumor progression. Most of the T1G3 tumors (eight of 11) were MRES negative. Several explanations may account for this observation: in our series, most of the T1G3 tumors may have arisen via the Ta pathway, and were therefore MRES negative; the MRES phenotype may appear principally at the transition between the T1 and T2 stages during tumor progression; or MRES-negative T1G3 tumors may reflect a third pathway of tumor progression, not derived from Ta or CIS. The detection of FGFR3 mutations, which are preferentially associated with the Ta pathway, in five of the eight T1G3 tumors without the MRES phenotype is consistent with the first hypothesis.
Our study had some limitations. Because no isolated carcinomas in situ were included in our study, we do not yet know if the MRES phenotype is already present at the CIS stage or if it appears within the CIS pathway, but at later stages. We were able to study histone marks and perform functional studies in vitro because we had a cell line derived from one tumor of our series. However, it will be important in the future to study the histone marks associated with the epigenetically silenced regions in vivo, that is, directly in tumor samples of known MRES status. In our prognostic studies, the number of bladder cancer patients in our series was relatively small, but we nonetheless found that muscle-invasive tumors with the MRES phenotype tended to have a worse outcome than muscle-invasive tumors without the MRES phenotype (P = .08). This observation requires confirmation in a larger series of tumors: too few progression events were available for non-muscle-invasive tumors for any firm conclusions to be drawn.
The identification of the MRES phenotype has potential clinical implications for the prognosis of noninvasive or muscle-invasive tumors and for treatment. The silenced regions that characterize the MRES phenotype contain known (PLCD1 [51], DLEC1 [52]) or potential (HOXA5 [53, 54]) tumor suppressor genes. We anticipate that the silencing of multiple regions produces a concomitant inactivation of tumor suppressor genes that may act in concert to drive neoplastic progression of the CIS pathway. Because epigenetic aberrations are reversible, our work suggests that treatments that target H3K9 and H3K27 methylation and/or H3K9 hypoacetylation represent a new avenue for the therapy of a particularly aggressive group of cancers.
Funding
This work was supported by the Institut Curie; the Centre National de la Recherche Scientifique; the Institut National de la Santé et de la Recherche Médicale; the Institut National Contre le Cancer (program GepiG, program Tumult); the Ligue Nationale Contre le Cancer (C.V., N.S., I.B.-P., A.H., F.R., Équipe labellisée), York Against Cancer (J.S.) and the national program Cartes d’Identité des Tumeurs (CIT), funded and developed by the Ligue Nationale Contre le Cancer.
Supplementary Material
Notes
The sponsors of this study had no role in the collection of the data, the analysis and interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript for publication.
We thank Dr Yves Denoux and Dr Anne-Catherine Baglin (Tumorothèque, Service d’Anatomie et Cytologie Pathologiques, CMC Foch, Suresnes, France), Dr M-J Terrier-Lacombe (Service de Pathologie, Institut Gustave Roussy, Villejuif, France), and Dr Karen Leroy (Plateforme de Ressources Biologiques, Pôle de Recherche Clinique, Assistance Publique Hôpitaux de Paris (APHP), Hôpital Henri Mondor, Créteil, France) for providing tumor samples. We thank Edith Heard for reading the manuscript and for her comments. We also thank Gabrielle Couchy for the TaqMan low-density array experiments.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14(Spec No 1):R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 5.Gibbons RJ. Histone modifying and chromatin remodelling enzymes in cancer and dysplastic syndromes. Hum Mol Genet. 2005;14(Spec No 1):R85–R92. doi: 10.1093/hmg/ddi106. [DOI] [PubMed] [Google Scholar]
- 6.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128(4):763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Yoo CB, Cheng JC, Jones PA. Zebularine: a new drug for epigenetic therapy. Biochem Soc Trans. 2004;32(pt 6):910–912. doi: 10.1042/BST0320910. [DOI] [PubMed] [Google Scholar]
- 8.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 9.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 10.Smith JS, Costello JF. A broad band of silence. Nat Genet. 2006;38(5):504–506. doi: 10.1038/ng0506-504. [DOI] [PubMed] [Google Scholar]
- 11.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38(12):1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 12.Novak P, Jensen T, Oshiro MM, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66(22):10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 13.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38(5):540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 14.Coolen MW, Stirzaker C, Song JZ, et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol. 2010;12(3):235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diez de Medina SG, Chopin D, El Marjou A, et al. Decreased expression of keratinocyte growth factor receptor in a subset of human transitional cell bladder carcinomas. Oncogene. 1997;14(3):323–330. doi: 10.1038/sj.onc.1200830. [DOI] [PubMed] [Google Scholar]
- 16.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80(9):1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Mostofi FK, Sobin LH, Torloni H. International Histological Classification of Tumors, No.10: Histological Typing of Urinary Bladder Tumors. Geneva, Switzerland: World Health Organization; 1973. [Google Scholar]
- 18.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 19.Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Boer WI, Houtsmuller AB, Izadifar V, Muscatelli-Groux B, Van der Kwast TH, Chopin DK. Expression and functions of EGF, FGF and TGFbeta-growth-factor family members and their receptors in invasive human transitional-cell-carcinoma cells. Int J Cancer. 1997;71(2):284–291. doi: 10.1002/(sici)1097-0215(19970410)71:2<284::aid-ijc25>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 22.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20(2):207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 23.Southgate J, Hutton KA, Thomas DF, Trejdosiewicz LK. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest. 1994;71(4):583–594. [PubMed] [Google Scholar]
- 24.Southgate J, Masters JR, Trejdosiewicz LK. Culture of Human Urothelium. In: Freshney RI, Freshney MG, editors. Culture of Epithelial Cells. New York, NY: J Wiley and Sons; 2002. pp. 381–400. [Google Scholar]
- 25.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3(10):2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Oers JM, Lurkin I, van Exsel AJ, et al. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res. 2005;11(21):7743–7748. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
- 29.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5(9):713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 30.Billerey C, Chopin D, Aubriot-Lorton MH, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158(6):1955–1959. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyrskjot L, Kruhoffer M, Thykjaer T, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64(11):4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 32.Sylvester RJ, van der Meijden AP, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49(3):466–465. doi: 10.1016/j.eururo.2005.12.031. discussion 475–477. [DOI] [PubMed] [Google Scholar]
- 33.Shih C, Weinberg RA. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell. 1982;29(1):161–169. doi: 10.1016/0092-8674(82)90100-3. [DOI] [PubMed] [Google Scholar]
- 34.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24(33):5218–5225. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 35.Zhang ZT, Pak J, Huang HY, et al. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene. 2001;20(16):1973–1980. doi: 10.1038/sj.onc.1204315. [DOI] [PubMed] [Google Scholar]
- 36.Hitchins MP, Lin VA, Buckle A, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67(19):9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- 37.Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68(20):8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 39.Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16(3):247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447(7143):413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 42.Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19(3):311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 43.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134(5):909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 44.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113(3):405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 45.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 47.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104(47):18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knowles MA. Molecular pathogenesis of bladder cancer. Int J Clin Oncol. 2008;13(4):287–297. doi: 10.1007/s10147-008-0812-0. [DOI] [PubMed] [Google Scholar]
- 49.Bostwick DG, Montironi R, Qian J, Lopez-Beltran A, Cheng L. Urothelial bladder tumor pathology. In: Lerner SP, Schoenberg MP, Sternberg CN, editors. Textbook of Bladder Cancer. Abingdon, UK: Taylor & Francis; 2006. [Google Scholar]
- 50.Zieger K, Marcussen N, Borre M, Orntoft TF, Dyrskjot L. Consistent genomic alterations in carcinoma in situ of the urinary bladder confirm the presence of two major pathways in bladder cancer development. Int J Cancer. 2009;125(9):2095–2103. doi: 10.1002/ijc.24619. [DOI] [PubMed] [Google Scholar]
- 51.Fu L, Qin YR, Xie D, et al. Characterization of a novel tumor-suppressor gene PLC delta 1 at 3p22 in esophageal squamous cell carcinoma. Cancer Res. 2007;67(22):10720–10726. doi: 10.1158/0008-5472.CAN-07-2411. [DOI] [PubMed] [Google Scholar]
- 52.Ying J, Poon FF, Yu J, et al. DLEC1 is a functional 3p22.3 tumour suppressor silenced by promoter CpG methylation in colon and gastric cancers. Br J Cancer. 2009;100(4):663–669. doi: 10.1038/sj.bjc.6604888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24(2):924–935. doi: 10.1128/MCB.24.2.924-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3(4):240–252. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.