Abstract
The opportunistic human fungal pathogen Candida glabrata is closely related to Saccharomyces cerevisiae, yet it has evolved to survive within mammalian hosts. Which traits help C. glabrata to adapt to this different environment? Which specific responses are crucial for its survival in the host? The main differences seem to include an extended repertoire of adhesin genes, high drug resistance, an enhanced ability to sustain prolonged starvation and adaptations of the transcriptional wiring of key stress response genes. Here, we discuss the properties of C. glabrata with a focus on the differences to related fungi.
Keywords: fungal pathogen, stress response, apoptosis
Introduction
Among the non-Candida albicans Candida species (Kaur et al., 2005; Marcet-Houben & Gabaldón, 2009;), Candida glabrata is the species most closely related to Saccharomyces cerevisiae (Fig. 1a). In this review, we will discuss some of the properties that allow C. glabrata to exist as a human commensal. We will briefly discuss the infectivity, genetic changes, drug and stress resistance as well as the adherence of C. glabrata. A selection of traits supporting commensalism and/or pathogenicity was proposed earlier on for fungal and bacterial pathogens. This might occur in the environment (Bliska & Casadevall, 2009), in confrontation with microbial communities or immune responses (Seider et al., 2010). A striking adaptation of C. glabrata might be its rapid growth and short generation time, which is tuned to 37 °C (Fig. 1b). Candida glabrata rarely penetrates tissue and is therefore exposed to microbial competitors on mucosal surfaces. Additionally, C. glabrata does not undergo a sexual cycle allowing the generation of resistant spores. We believe that these restrictions, combined with the host environment, favored the development of several specific traits, for example its high resistance to starvation. Compared with S. cerevisiae, C. glabrata shows only a small number of genetic adaptations. Considering its phylogenetic position (Fig. 1a), it can be assumed that C. glabrata's ability to infect humans emerged independently from that of other Candida species.
Fig. 1.
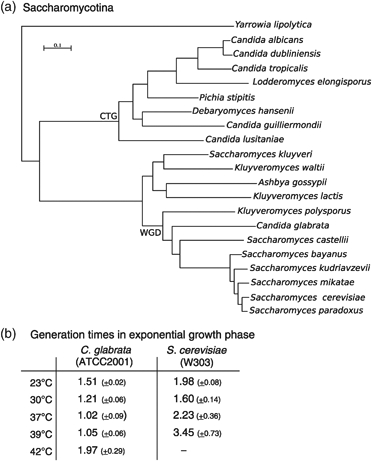
(a) Phylogenetic tree representing the evolutionary relationships between sequenced Candida and Saccharomyces species (adapted from T21 tree in Marcet-Houben & Gabaldón, 2009). The tree is based on the maximum likelihood analysis of concatenated alignments of 1137 protein families that have single-copy orthologues in the 21 species considered. Approximate likelihood ratio test support values were >0.95 for all branches, except for the one joining Debaryomyces hansenii and Candida guilliermondii (0.25). The genetic code transition in the CTG clade and the WGD events are indicated in the tree. (b) Generation time of Candida glabrata and Saccharomyces cerevisiae strains at different growth temperatures based on OD600 nm measurements during exponential growth phase in rich medium (three experiments, SDs in parentheses).
After C. albicans, C. glabrata is the most common cause of vaginal and oral candidiasis. Candida glabrata is the second most important species in terms of candidiasis in the United States (Pfaller & Diekema, 2004). In total, C. glabrata accounts for roughly 15–20% of all Candida infections, with this relative incidence increasing every year. In healthy individuals, C. glabrata is restricted because of the action of the innate immune system and the microbial communities, which counteracts dissemination by competition for nutrients and secretion of toxins (Wargo & Hogan, 2006; Pamer, 2007;). Immunocompromised persons suffering from neutropenia (e.g. cancer or transplant patients) or persons treated with antibiotics are much more susceptible to C. glabrata infections (Perlroth et al., 2007; Caston-Osorio et al., 2008; Cohen et al., 2010;). Candida glabrata is more often found in elderly patients (Hof, 2010). In murine infection models, C. glabrata infection is cleared and mice need to be rendered neutropenic to make them susceptible to pathogenic yeast infections (MacCallum et al., 2006). Together, this might indicate that C. glabrata is less virulent than C. albicans and mainly causes disease in severely ill patients.
Be creative: genetic changes
Candida glabrata shares a recent common ancestor with several Saccharomyces species, and clearly belongs to a clade different from that of other Candida species which display particular features such as the recoding of the CUG codon to Serine (Fig. 1a). As a result of this evolutionary relatedness, most S. cerevisiae genes have orthologues in C. glabrata and the chromosomal structure in terms of gene order is largely conserved between the two species (Dujon et al., 2004; Marcet-Houben & Gabaldón, 2009;). Among many others, shared genomic features include a similar stress response (Roetzer et al., 2008) and a respiratory metabolism characterized by the lack of Complex I (Marcet-Houben & Gabaldón, 2009). Despite this general resemblance, several differences in terms of gene content can be found, which might play an important role with respect to the phenotypic differences. Compared with S. cerevisiae, C. glabrata seems to have undergone an increased rate of gene loss from their common ancestor (Dujon et al., 2004). Notable absences include genes of the galactose, phosphate, nitrogen and sulfur metabolism. Similar to many other microbial pathogens (Ehrlich et al., 2008), such a reductive evolution could be related to C. glabrata's adaptation as mammalian commensal and opportunistic pathogen. Indeed, C. glabrata relies on its host to overcome important auxotrophies such as those of nicotinic acid, pyridoxine and thiamine (Kaur et al., 2005). Conversely, C. glabrata encodes genes that are absent from other Saccharomyces, including a putative racemase of bacterial origin (Marcet-Houben & Gabaldón, 2010). Other differences entail gene expansions that are specific to the C. glabrata lineage. Three of these specific expansions affect cell-wall organization and are probably related to adhesion properties of C. glabrata cells. These expansions include six copies of extracellular glycosylphosphatidylinositol-linked aspartyl proteases, eight copies of a α-1,3-mannosyltransferase involved in cell-wall biogenesis and a variable number of glycosylphosphatidylinositol-linked epithelial adhesin (EPA) genes located in subtelomeric regions. Importantly, this last family, which is required for epithelial cell adhesion and biofilm formation, seems to be a unique adaptation of C. glabrata because no significant homology can be detected in S. cerevisiae. Nevertheless, FLO genes in S. cerevisiae coding for glycosylphosphatidylinositol-linked flocculins are usually considered to be functional homologues of C. glabrata EPA genes (De Las Penas et al., 2003). Similarly to FLO genes, EPA genes are located in subtelomeric regions and are subject to SIR-mediated silencing (Kaur et al., 2007). Moreover, FLO genes have been found to be involved in cell adhesion and biofilm formation in S. cerevisiae (Reynolds & Fink, 2001). An intriguing possibility is that FLO and EPA genes are evolutionarily related but their sequences have diverged beyond recognition. Such an accelerated divergence seems to be quite frequent in yeast genes (Wolfe, 2004) and may have been favored by the extensive lineage-specific duplications undergone in the two lineages and by the acquisition of different tandemly repeated motifs.
Interestingly, despite being composed of different amino acids, the length of the tandemly repeated motifs in several FLO and EPA genes is similar (45 amino acids), perhaps reflecting a common selective pressure (Thierry et al., 2010). Finally, a recently discovered feature of the C. glabrata genome, one that is almost unique to this species, is megasatellites (Thierry et al., 2010). These long (135–417 nucleotides) repetitive sequences are enriched within genes coding for cell-wall proteins, including some EPA genes, and might also be related to the high degree of genomic plasticity observed in C. glabrata (Polakova et al., 2009). This genomic plasticity is likely to play an important role in the evolution of C. glabrata populations because a sexual cycle has not yet been observed. Remarkably, this apparent disappearance of the sexual cycle seems to be a common trait for most fungal pathogens (Butler, 2010). Thus, differences in the genome sequence and its regulation provide important hints about the way in which two closely related yeasts have adapted to radically different environments.
Be sticky: adherence and persistence
Compared with S. cerevisiae, C. glabrata has lost some of the genes needed for several metabolic pathways, for example galactose utilization. However, it has also gained certain functions necessary for its commensal lifestyle. Unlike S. cerevisiae, C. glabrata is able to adhere not only to mammalian cells but also to other surfaces (Cormack et al., 1999). An elegant screen for factors required for biofilm formation identified CgRIF1, CgSIR4 as well as the protein kinase CgYAK1 and CgEPA6 encoding an adhesin (Iraqui et al., 2005). CgEpa6 is a member of the EPA gene family of glycosylphosphatidylinositol-anchored cell-wall proteins (Castano et al., 2005; Domergue et al., 2005; Iraqui et al., 2005; Kaur et al., 2005;). Similar to the S. cerevisiae FLO lectin-like genes, the EPA genes are encoded in subtelomeric clusters and are subject to transcriptional silencing (Castano et al., 2005). Candida glabrata cells with mutations in the telomeric silencing factors Sir2, Sir3 and Sir4 are more adherent to cultured epithelial cells and show better colonization efficiency in kidneys (Castano et al., 2005; Domergue et al., 2005;). Interestingly, the lack of nicotinic acid, a precursor of NAD+, triggers adhesin (EPA6) expression (Domergue et al., 2005). Candida glabrata is an auxotroph for NAD+ and utilizes nicotinamide riboside and nicotinic acid as NAD(+) sources during disseminated infection (Ma et al., 2007). The majority of all genes transcriptionally induced by niacin limitation are also regulated by the NAD(+)-dependent histone deacetylase Hst1 (Ma et al., 2009). Therefore, nicotinic acid and nicotinamide riboside are important host-derived regulatory signals. Transcript profiling of C. glabrata cells internalized by macrophages revealed increased expression of a gene family encoding extracellular glycosylphosphatidylinositol-linked aspartyl proteases (YPS genes) (Kaur et al., 2007). YPS proteases are responsible for removing and releasing glycosylphosphatidylinositol-anchored cell-wall proteins and are thought to be necessary for cell-wall integrity and regulated adherence to mammalian cells. A recent survey identified several putative adhesin genes that had not yet been annotated (de Groot et al., 2008). Their role for C. glabrata virulence is currently unknown. It should also be noted that C. glabrata strains have different number and sequence of adhesin genes. Thus, C. glabrata uses signals from the host for growth and adherence and its adhesive repertoire is more variable and by far transcends that of S. cerevisiae.
Be adaptive: environmental stress response (ESR)
Vegetative cells of C. glabrata have high intrinsic stress tolerance. While C. albicans can survive on surfaces up to 4 months, C. glabrata can do so for even up to 5 months (Kramer et al., 2006). However, the persistence traits have not yet been explored systemically. Stress resistance might be a consequence of its natural habitat. Candida glabrata preferably grows on mucosal surfaces as biofilm. Mammalian mucosal areas cause nutrient shortage, osmotic and other stresses due to the presence of other microorganisms and protective mechanisms of the host (Garside et al., 2004; Pamer, 2007;). Furthermore, sexual reproduction as a way to generate stress-resistant spores has never been observed in C. glabrata (reviewed in Butler, 2010). So far, all clinical isolates of C. glabrata have been haploid, although it has the components of the mating machinery at its disposal (Wong et al., 2003). Candida glabrata might have lost the sexual reproduction cycle relatively recently or perhaps suppresses it in order to maximize proliferation.
Changing their transcriptional program is a major strategy of microorganisms to adapt to their immediate environment. In S. cerevisiae, the ESR comprises about 900 genes whose expression is coordinately altered to exposure of different types of stress (Gasch et al., 2000). Saccharomyces cerevisiae, C. albicans, Schizosaccharomyces pombe and C. glabrata live in different environments. Transcript analysis revealed that environmental stress (heat, hyperosmolarity, oxidative and starvation stress) induces a very similar pattern of regulated genes in these fungi (Chen et al., 2003; Enjalbert et al., 2003; Gasch, 2007;). Environmental stress activates a variety of conserved signaling mechanisms. Each of them responds to a particular cue, for example oxidative stress or carbon source starvation. Among them, mitogen-activated protein kinase (MAPK) pathways are central for relaying stress and other environmental signals. The ESR of C. albicans and S. pombe is mainly regulated by the stress-activated MAPKs CaHog1 and Sty1, respectively (Smith et al., 2004; Enjalbert et al., 2006;). CaHog1 (and most likely also C. glabrata CgHog1) is for full virulence (Alonso-Monge et al., 1999). In S. cerevisiae, the high osmolarity glycerol (HOG) MAPK pathway senses osmotic stress and, to some extent, also other stress types such as oxidative stress and acetate (Smith et al., 2010). The function of the C. glabrata HOG pathway seems to be very closely related to that of S. cerevisiae. However, unlike the S. cerevisiae HOG pathway, it also modulates resistance to longer chain weak organic acids such as sorbic acid (Gregori et al., 2007). Sorbic acid is a powerful activator of the stress transcription factors Msn2 and Msn4 in S. cerevisiae but not of the orthologous factors in C. glabrata (Schüller et al., 2004; Roetzer et al., 2008;). Sorbic acid leads to rapid localization of CgMsn2-CFP to the nucleus in S. cerevisiae but not in C. glabrata. The difference suggests an as yet unknown weak acid signaling mechanism triggering the HOG pathway in C. glabrata. In S. cerevisiae, the activation of the HOG pathway is triggered by two redundant cell surface membrane sensors, Sho1 and Sln1. These genes have orthologues in C. glabrata. Initial genetic analysis of the first completely sequenced C. glabrata strain [CBS132, ATCC 2001 (Dujon et al., 2004)] suggested that the Sln1 signaling branch was inactive because mutant strains lacking CgSHO1 displayed a high sensitivity to osmotic stress (Calcagno et al., 2005; Gregori et al., 2007;). Further analysis revealed that a component of the Sln1 branch in this particular C. glabrata strain harbors a nonsense mutation (Cgssk2-1), leading to its inactivation. Importantly, other strains (e.g. BG2) do not share this allele.
In S. cerevisiae, the partially redundant transcription factors Msn2 and Msn4 regulate many ESR genes in parallel to MAPK pathways. Msn2 and Msn4 also convey nutrient signals and are regulated by protein kinase A (Görner et al., 2002). Most Saccharomycotina species contain orthologues to MSN2. One important functional similarity of ScMsn2 orthologues and paralogues is a conserved stress-regulated nuclear export signal (NES) (Görner et al., 1998; Roetzer et al., 2008;). Msn2 orthologues from the CTG clade (Fig. 1a) do not have the stress-regulated NES and are most probably not involved in stress response. In the C. albicans, the Msn2-like protein (CaMsn4) lacks recognizable homology to the ScMsn2 NES and does not play any role in stress response (Nicholls et al., 2004). Because of selection against genetic redundancy, most gene pairs that originated during the whole-genome duplication (WGD) event lost one of the copies in subsequent lineages. Thus, retention of both Msn2 and Msn4 in many species (with the exception of Saccharomyces bajanus) suggests different roles for these factors. Like other post-WGD species, C. glabrata encodes orthologues to both Msn2 and Msn4. CgMsn2/4 are important regulators for the CgESR but seem to be less relevant for glucose signaling (Roetzer et al., 2008). So far, CgMsn2/4 have not been involved in pathogenicity and thus might play a role in persistence outside the host (Mundy & Cormack, 2009). These stress response factors might control the balance between stress resistance vs. growth required during feast and famine cycles (Berry & Gasch, 2008).
Protective response to oxidative stress is important for a pathogen facing oxidative burst attacks of innate immune cells. Compared with common S. cerevisiae laboratory strains, C. glabrata possesses high intrinsic oxidative stress resistance, which is regulated, similar to that of S. cerevisiae, by the transcription factors CgSkn7, CgYap1 and CgMsn2/4 (Cuellar-Cruz et al., 2008; Roetzer et al., 2010;). It also produces an indole-derived pigment contributing to oxidative stress resistance (Brunke et al., 2010). The loss of CgMsn2/4, CgYap1 and CgSkn7 did not affect the survival of C. glabrata when confronted with murine macrophages. Interestingly, the expression of the C. glabrata superoxide dismutase genes (CgSOD1/2) is different from that of S. cerevisiae. CgSOD1 seems to be expressed constitutively, and both CgSOD1 and 2 are induced by glucose starvation. Candida albicans is equipped with five SOD genes with highly specialized roles during host contact (Frohner et al., 2009). A slight adaptation of SOD regulation adapts C. glabrata to an environment with a higher risk of oxidative stress. In addition, many genes of the oxidative stress regulon can also be induced by other stresses such as glucose starvation (our unpublished data). Another adaptation of transcriptional regulation between C. glabrata and S. cerevisiae is the regulation of the catalase gene CgCTA1. The single catalase of C. glabrata combines both the different transcriptional regulation (carbon source vs. stress) and the different intracellular localization (cytoplasm vs. peroxisomes) of the two S. cerevisiae catalases (Roetzer et al., 2010). Thus, C. glabrata achieves a similar regulated and localized catalase activity, albeit by the regulation of one gene.
It was recently reported that certain Candida species can suppress the production of reactive oxygen species (ROS) produced by phagocytic cells in murine and human phagocytes, whereas internalization of S. cerevisiae cells enhanced ROS production (Wellington et al., 2009). Interestingly, in a cell-free system, S. cerevisiae was able to scavenge ROS in a manner similar to the one observed for C. albicans. Therefore, scavenging alone cannot account for the suppression of ROS production. We assume that an additional Candida-specific mechanism is responsible for active suppression. These results are supported by the finding that the transcription pattern of C. glabrata cells during phagocytosis did not show a prominent upregulation of oxidative stress-associated genes (Kaur et al., 2007). This might be the result of the efficient suppression and/or detoxification of ROS due to expressed protective enzymes. The role of the stress response for C. glabrata adaptation to the host environment is still being studied. So far, the available evidence points to a range of subtle changes of mechanisms known from S. cerevisiae.
Be flexible: phenotypic and morphological switching
Being largely immotile, fungi rely on passive dissemination methods and directed growth to expand into substrate. Candida glabrata primarily grows on surfaces and normally does not penetrate tissue. Under appropriate conditions, penetration into solid medium can be observed. For C. albicans, the switch between yeast and hyphae is one important contribution to virulence (Kumamoto & Vinces, 2005). In contrast, and similar to S. cerevisiae, C. glabrata can form pseudohyphae in response to nitrogen source starvation (Csank & Haynes, 2000). In S. cerevisiae, pseudohyphal growth is assumed to be a common event in search for nutrient-rich, optimal growth substrates. This morphological switch is regulated by a MAPK cascade, which has the transcription factor Ste12 as its final target (Gancedo, 2001). CgSte12 is also essential for the nitrogen starvation-induced formation of pseudohyphae in C. glabrata. In a mouse model, Cgste12Δ mutants show attenuated virulence (Calcagno et al., 2003). However, pseudohyphal forms of C. glabrata have not yet been found in clinical specimens, possibly because this is a rare event (Kaur et al., 2005).
Differently to pseudohyphal growth resulting from specific nutrient limitations, unusual cell aggregates have been observed for C. glabrata cells due to the loss of transcription factor CgAce2. These mutants have a cell separation defect and display a clumping phenotype that is similar to S. cerevisiae ace2Δ mutants (Kamran et al., 2004). Interestingly, Cgace2Δ mutants are hypervirulent in a mouse model and are able to escape from the vasculature and penetrate into tissue (MacCallum et al., 2006). Stead et al. (2010) found that lack of CgAce2 also changes the secretome of C. glabrata, which might contribute to its hypervirulence. The phenotype of the Cgace2Δ mutant supports the assumption that C. glabrata has a limited but possibly important ability to actively invade tissues despite its preferred growth on surfaces.
Phenotypic variability is important for evading the immune system. For C. albicans, switching between white and opaque cells is important for virulence (Kvaal et al., 1997, 1999). White cells are more suited for bloodstream infections, whereas opaque cells colonize skin surfaces (Lachke et al., 2003). Candida glabrata cells can undergo reversible phenotypic switching among phenotypes distinguishable by colony coloration on CuSO4-containing agar (white to very dark brown) and by the irregular wrinkle colony morphology (Lachke et al., 2002; Brockert et al., 2003;). In a mouse model, dark brown cells were reported to have an advantage in colonizing the spleen and the liver (Brockert et al., 2003; Srikantha et al., 2008;). The basis and molecular details of the reported phenotypic variability have not yet been explored in depth.
Be resistant: drug resistance
One of the reasons for the success of C. glabrata is its relatively high drug resistance, in particular toward different azole antifungals. In the last decade, C. glabrata emerged as a cause of mucosal and invasive fungal infections, in part due to its intrinsic or easily acquired resistance to azole antifungals. Therefore, echinocandins such as caspofungin are the treatment of choice. The global transcriptional response to azole treatment and the molecular mechanisms to this drug resistance are reminiscent of S. cerevisiae (Vermitsky et al., 2006). In S. cerevisiae, the zinc-cluster transcription factors ScPdr1 and ScPdr3 are central for regulating detoxification mechanisms based on transmembrane transporters such as ScPdr5 and ScSnq2. In C. glabrata, CgPdr1 activates the expression of CgCDR1 and CgCDR2, which are drug efflux pumps of the ATP-binding cassette transporter type (Sanglard et al., 2001). Gain of function mutations in CgPDR1 enhances antifungal resistance and also increases virulence (Ferrari et al., 2009). Additionally, defects in mitochondrial function causing respiratory deficiency also increase azole resistance and this, in turn, is linked to the upregulation of both CgCDR1 and CgCDR2 (Sanglard et al., 2001). In both S. cerevisiae and C. glabrata, Pdr3 – and as a consequence drug resistance – is induced by the loss of the mitochondrial genome (S. Moye-Rowley, pers. commun.; Moye-Rowley, 2005). This might be a mechanism for a dedicated retrograde signal of drug-damaged mitochondria. Because the development of intrinsic drug resistance of C. glabrata preceded antifungal drugs, we speculate that these adaptations might be due to frequent exposure to toxic substances originating from the environment of the mucosal flora.
Be aware of the source: nutrients
Like other commensal or pathogenic microorganisms, C. glabrata faces phagocytic cells of the innate immune system during dissemination or after escaping from biofilms. Transcriptome studies revealed that C. albicans, C. glabrata and the encapsulated pathogenic yeast Cryptococcus neoformans, when engulfed by macrophages, switch their expression program to genes involved in the utilization of alternative carbon sources (Lorenz et al., 2004; Fan et al., 2005; Kaur et al., 2007;). This is a default response, indicating loss of contact to the preferred carbon source glucose in the environment. Therefore, phagocytosed cells are sealed from the environment and have to rely on their endogenous resources or on those acquired from the phagocytic cell. Each successful pathogen has a unique strategy to manage this situation (Fig. 2). Unlike C. albicans, phagocytosed C. glabrata is unable to form hyphae and is trapped within the phagosome. Cryptococcus neoformans triggers phagosomal extrusions, which entails the escape of single cells without killing the phagocytes (Alvarez & Casadevall, 2006). The transcriptional response to phagocytosis also induced genes involved in autophagy, peroxisome function and lipid metabolism (Fan et al., 2005). It is thought that carbon source and thus energy shortage is a predominant challenge for engulfed pathogenic fungi.
Fig. 2.
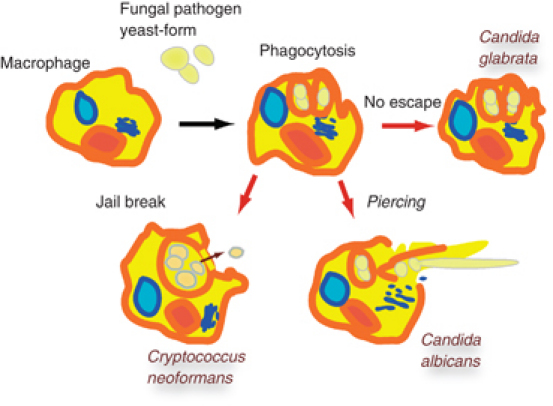
Fungal morphogenesis determines escape mechanisms of pathogenic yeasts. Upon engulfment by phagocytes, Candida albicans can form hyphae and escape from the phagosome by destroying the phagocyte. Instead of forming hyphae, Cryptococcus neoformans cells are able to escape by forming phagosomal extrusions. No specific mechanism to escape internalization is known for Candida glabrata.
An important mechanism for survival during starvation conditions is autophagy (Klionsky, 2005). Autophagy is a continuous recycling process of cellular constituents and even organelles (Xie & Klionsky, 2007). Moreover, it is important for the virulence of several fungal pathogens of plants and animals (Hu et al., 2008). In contrast, C. albicans escapes nutrient depletion by forming hyphae and killing engulfing macrophages. Consistently, mutants unable to induce autophagy survived challenge by macrophages (Palmer et al., 2007). Candida glabrata trapped in a macrophage initially induced peroxisomes, most likely as part of the switch toward utilization of alternative carbon sources (Roetzer et al., 2010). These peroxisomes were subsequently degraded via the pexophagy pathway, a specific subtype of autophagy. Both selective and bulk autophagy contribute to the survival of C. glabrata during engulfment. Candida glabrata trapped inside the phagosome requires mobilization of resources for survival. Because C. glabrata is relatively resistant to starvation, macrophages apparently have additional strategies to kill the internalized fungal cells. Macrophages are known to cause oxidative damage to microorganisms, but engulfed C. glabrata cells do not show signs of a severe oxidative stress load (Roetzer et al., 2010). However, during maturation, the phagosome fuses with lysosomes in order to decrease the pH, which initiates enzymatic digestion of engulfed microorganisms (Stuart & Ezekowitz, 2005). In vitro, only the exposure to a combination of low pH and carbon starvation caused loss of viability of C. glabrata. The autophagy-deficient atg11Δatg17Δ mutant was more sensitive to this particular combination of conditions, both in vitro and in vivo. Thus, the energy-demanding equilibration of extracellular pH with limited energy resources might be the challenging environment C. glabrata is exposed to in the phagosome.
Conclusion
Candida glabrata is an efficient human pathogen, which can infect immunocompromised and elderly persons. Its avid adherence to mammalian tissue and to other surfaces is based on the expression of a number of specific adhesins. Candida glabrata rarely penetrates tissue actively and therefore directly competes with other microbial agents on mucosal surfaces. This might have led to the selection of specific drug and oxidative stress-resistance traits. Somatic fungal cells have high resistance to starvation, a trait that might also support survival during engulfment in phagocytic cells. Prolonged survival of C. glabrata inside phagocytic cells might support the establishment of disseminated infection and thus directly relate to its success as a commensal and pathogen (Fig. 3). Robust adherence, active suppression of fungicidal drugs, stress resistance and an enhanced ability to sustain prolonged starvation render C. glabrata superior to S. cerevisiae in causing disease. Altogether, we suggest that its adaptation to the mammalian host is rather due to slight genomic fine-tuning than due to radical large-scale changes.
Fig. 3.
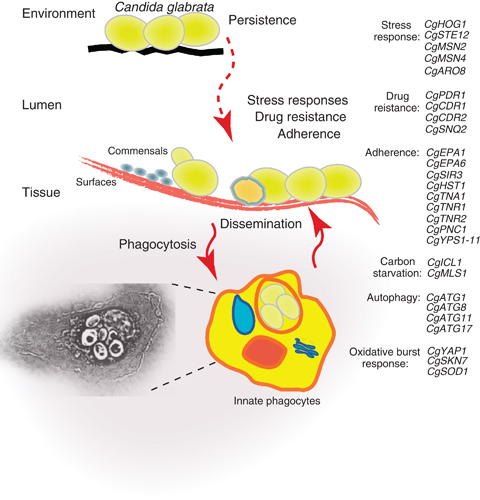
Possible habitats. Candida glabrata cells can persist on environmental surfaces for several months. As a commensal, C. glabrata is also found as part of the flora on interior mucosal areas of mammalians such as the gut. In immunocompromised persons, cells are able to disseminate into tissues and cause organ failure. Patrolling phagocytes are responsible for the extinction of invading yeast pathogens. The armory of C. glabrata: the right panel lists genes thought to be important for the opportunistic lifestyle of C. glabrata.
Acknowledgments
We regret that restrictions did not permit us to cite all important papers. We thank the anonymous referees for their very constructive comments. We are grateful to Sylvia Trnka, Wolfram Görner, Martina Dötsch and Christa Gregori for critically reading the manuscript. Work in the C.S. lab has been funded by the AMS, FWF B12-19966, the Herzfelder Foundation and Grant I031-B from the University of Vienna. T.G. is funded by grants from the Spanish Ministry of Science and Innovation (GEN2006-27784E, BFU2009-09168).
Statement
Re-use of this article is permitted in accordance with the Terms and Conditions set out at http://wileyonlinelibrary.com/onlineopen#OnlineOpen_Terms
References
- Alonso-Monge R, Navarro-Garcia F, Molero G, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska JB, Casadevall A. Intracellular pathogenic bacteria and fungi – a case of convergent evolution? Nat Rev Microbiol. 2009;7:165–171. doi: 10.1038/nrmicro2049. [DOI] [PubMed] [Google Scholar]
- Brockert PJ, Lachke SA, Srikantha T, Pujol C, Galask R, Soll DR. Phenotypic switching and mating type switching of Candida glabrata at sites of colonization. Infect Immun. 2003;71:7109–7118. doi: 10.1128/IAI.71.12.7109-7118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunke S, Seider K, Almeida RS, et al. Candida glabrata tryptophan-based pigment production via the Ehrlich pathway. Mol Microbiol. 2010;76:25–47. doi: 10.1111/j.1365-2958.2010.07052.x. [DOI] [PubMed] [Google Scholar]
- Butler G. Fungal sex and pathogenesis. Clin Microbiol Rev. 2010;23:140–159. doi: 10.1128/CMR.00053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno AM, Bignell E, Warn P, et al. Candida glabrata STE12 is required for wild-type levels of virulence and nitrogen starvation induced filamentation. Mol Microbiol. 2003;50:1309–1318. doi: 10.1046/j.1365-2958.2003.03755.x. [DOI] [PubMed] [Google Scholar]
- Calcagno AM, Bignell E, Rogers TR, Jones MD, Muhlschlegel FA, Haynes K. Candida glabrata Ste11 is involved in adaptation to hypertonic stress, maintenance of wild-type levels of filamentation and plays a role in virulence. Med Mycol. 2005;43:355–364. doi: 10.1080/13693780400006088. [DOI] [PubMed] [Google Scholar]
- Castano I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- Caston-Osorio JJ, Rivero A, Torre-Cisneros J. Epidemiology of invasive fungal infection. Int J Antimicrob Ag. 2008;32(suppl 2):S103–S109. doi: 10.1016/S0924-8579(08)70009-8. [DOI] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Karoubi P, Adrie C, Gauzit R, Marsepoil T, Zarka D, Clec'h C. Early prediction of Candida glabrata fungemia in nonneutropenic critically ill patients. Crit Care Med. 2010;38:826–830. doi: 10.1097/CCM.0b013e3181cc4734. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Ghori N, Falkow S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science. 1999;285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- Csank C, Haynes K. Candida glabrata displays pseudohyphal growth. FEMS Microbiol Lett. 2000;189:115–120. doi: 10.1111/j.1574-6968.2000.tb09216.x. [DOI] [PubMed] [Google Scholar]
- Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castano I, De Las Penas A. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell. 2008;7:814–825. doi: 10.1128/EC.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot PW, Kraneveld EA, Yin QY, et al. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot Cell. 2008;7:1951–1964. doi: 10.1128/EC.00284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Penas A, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Gene Dev. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue R, Castano I, De Las Penas A, et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Ehrlich GD, Hiller NL, Hu FZ. What makes pathogens pathogenic. Genome Biol. 2008;9:225. doi: 10.1186/gb-2008-9-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Nantel A, Whiteway M. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell. 2003;14:1460–1467. doi: 10.1091/mbc.E02-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Kraus PR, Boily MJ, Heitman J. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell. 2005;4:1420–1433. doi: 10.1128/EC.4.8.1420-1433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Ischer F, Calabrese D, et al. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 2009;5:e1000268. doi: 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol. 2009;71:240–252. doi: 10.1111/j.1365-2958.2008.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Garside P, Millington O, Smith KM. The anatomy of mucosal immune responses. Ann NY Acad Sci. 2004;1029:9–15. doi: 10.1196/annals.1309.002. [DOI] [PubMed] [Google Scholar]
- Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 2007;24:961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, et al. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Gene Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori C, Schüller C, Roetzer A, Schwarzmüller T, Ammerer G, Kuchler K. The high-osmolarity glycerol response pathway in the human fungal pathogen Candida glabrata strain ATCC 2001 lacks a signaling branch that operates in baker's yeast. Eukaryot Cell. 2007;6:1635–1645. doi: 10.1128/EC.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof H. Mycoses in the elderly. Eur J Clin Microbiol. 2010;29:5–13. doi: 10.1007/s10096-009-0822-5. [DOI] [PubMed] [Google Scholar]
- Hu G, Hacham M, Waterman SR, et al. PI3K signaling of autophagy is required for starvation tolerance and virulence of Cryptococcus neoformans. J Clin Invest. 2008;118:1186–1197. doi: 10.1172/JCI32053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui I, Garcia-Sanchez S, Aubert S, Dromer F, Ghigo JM, d'Enfert C, Janbon G. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol Microbiol. 2005;55:1259–1271. doi: 10.1111/j.1365-2958.2004.04475.x. [DOI] [PubMed] [Google Scholar]
- Kamran M, Calcagno AM, Findon H, et al. Inactivation of transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryot Cell. 2004;3:546–552. doi: 10.1128/EC.3.2.546-552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. P Natl Acad Sci USA. 2007;104:7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy. Curr Biol. 2005;15:R282–R283. doi: 10.1016/j.cub.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal CA, Srikantha T, Soll DR. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Joly S, Daniels K, Soll DR. Phenotypic switching and filamentation in Candida glabrata. Microbiology. 2002;148:2661–2674. doi: 10.1099/00221287-148-9-2661. [DOI] [PubMed] [Google Scholar]
- Lachke SA, Lockhart SR, Daniels KJ, Soll DR. Skin facilitates Candida albicans mating. Infect Immun. 2003;71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Pan SJ, Zupancic ML, Cormack BP. Assimilation of NAD(+) precursors in Candida glabrata. Mol Microbiol. 2007;66:14–25. doi: 10.1111/j.1365-2958.2007.05886.x. [DOI] [PubMed] [Google Scholar]
- Ma B, Pan SJ, Domergue R, Rigby T, Whiteway M, Johnson D, Cormack BP. High-affinity transporters for NAD+ precursors in Candida glabrata are regulated by Hst1 and induced in response to niacin limitation. Mol Cell Biol. 2009;29:4067–4079. doi: 10.1128/MCB.01461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Findon H, Kenny CC, Butler G, Haynes K, Odds FC. Different consequences of ACE2 and SWI5 gene disruptions for virulence of pathogenic and nonpathogenic yeasts. Infect Immun. 2006;74:5244–5248. doi: 10.1128/IAI.00817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome. PLoS One. 2009;4:e4357. doi: 10.1371/journal.pone.0004357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26:5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley WS. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene. 2005;354:15–21. doi: 10.1016/j.gene.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Mundy RD, Cormack B. Expression of Candida glabrata adhesins after exposure to chemical preservatives. J Infect Dis. 2009;199:1891–1898. doi: 10.1086/599120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, Straffon M, Enjalbert B, Nantel A, Macaskill S, Whiteway M, Brown AJ. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell. 2004;3:1111–1123. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer GE, Kelly MN, Sturtevant JE. Autophagy in the pathogen Candida albicans. Microbiology. 2007;153:51–58. doi: 10.1099/mic.0.2006/001610-0. [DOI] [PubMed] [Google Scholar]
- Pamer EG. Immune responses to commensal and environmental microbes. Nat Immunol. 2007;8:1173–1178. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin Microbiol Infec. 2004;10(suppl 1):11–23. doi: 10.1111/j.1470-9465.2004.t01-1-00844.x. [DOI] [PubMed] [Google Scholar]
- Polakova S, Blume C, Zarate JA, Mentel M, Jorck-Ramberg D, Stenderup J, Piskur J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. P Natl Acad Sci USA. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Fink GR. Bakers' yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Roetzer A, Gregori C, Jennings AM, et al. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol Microbiol. 2008;69:603–620. doi: 10.1111/j.1365-2958.2008.06301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A, Gratz N, Kovarik P, Schüller C. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol. 2010;12:199–216. doi: 10.1111/j.1462-5822.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Bille J. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob Agents Ch. 2001;45:1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller C, Mamnun YM, Mollapour M, et al. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seider K, Heyken A, Luttich A, Miramon P, Hube B. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol. 2010;13:392–400. doi: 10.1016/j.mib.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Morgan BA, Quinn J. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol Lett. 2010;306:1–8. doi: 10.1111/j.1574-6968.2010.01937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Daniels KJ, Wu W, et al. Dark brown is the more virulent of the switch phenotypes of Candida glabrata. Microbiology. 2008;154:3309–3318. doi: 10.1099/mic.0.2008/020578-0. [DOI] [PubMed] [Google Scholar]
- Stead DA, Walker J, Holcombe L, et al. Impact of the transcriptional regulator, Ace2, on the Candida glabrata secretome. Proteomics. 2010;10:212–223. doi: 10.1002/pmic.200800706. [DOI] [PubMed] [Google Scholar]
- Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22:539–550. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Thierry A, Dujon B, Richard GF. Megasatellites: a new class of large tandem repeats discovered in the pathogenic yeast Candida glabrata. Cell Mol Life Sci. 2010;67:671–676. doi: 10.1007/s00018-009-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol. 2006;61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, Hogan DA. Fungal–bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol. 2006;9:359–364. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Wellington M, Dolan K, Krysan DJ. Live Candida albicans suppresses production of reactive oxygen species in phagocytes. Infect Immun. 2009;77:405–413. doi: 10.1128/IAI.00860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. Evolutionary genomics: yeasts accelerate beyond BLAST. Curr Biol. 2004;14:R392–R394. doi: 10.1016/j.cub.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Wong S, Fares MA, Zimmermann W, Butler G, Wolfe KH. Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol. 2003;4:R10. doi: 10.1186/gb-2003-4-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]


