Abstract
Osteopontin (OPN) is a glycophosphoprotein with multiple intracellular and extracellular functions. In vitro, OPN enhances migration of mouse neutrophils and macrophages. In cancer, extracellular OPN facilitates migration of cancer cells via its RGD sequence. The present study was designed to investigate whether osteopontin is responsible for neutrophil and macrophage infiltration in human cancer and in particular in glioblastoma. We found that in vitro mouse neutrophil migration was RGD-dependent. In silico, we found that the OPN gene was one of the 5% most highly expressed genes in 20 out of 35 cancer microarray data sets in comparison with normal tissue in at least 30% of cancer patients. In some types of cancer, such as ovarian cancer, lung cancer and melanoma, the OPN gene was one of those with the highest expression levels in at least 90% of cancer patients. In glioblastoma, the most invasive type of brain tumours/glioma, but not in lower grades of glioma it was one of the 5% highest expressed genes in 90% of patients. In situ, we found increased protein levels of OPN in human glioblastoma versus normal human brain confirming in silico results. OPN protein expression was co-localized with neutrophils and macrophages. In conclusion, OPN in tumours not only induces migration of cancer cells but also of leucocytes.
Keywords: glioblastoma, macrophage, migration, neutrophil, osteopontin, RGD
Introduction
Osteopontin (OPN) is a multifunctional arginine-glycine-aspartate (RGD)-containing glycophosphoprotein with cytokine and chemokine properties1,2 that is expressed intra- and extracellularly and produced by cells of the immune system such as macrophages, epithelial cells, smooth muscle cells, osteoblasts, Purkinje cells and cancer cells.1,3,4 It is involved in a multitude of processes such as inflammation, angiogenesis and cancer.3,5–9 The RGD sequence of OPN binds to integrins such as αVβ3, αVβ5, αVβ1, α8β1, α9β1, α5β1 and α4β1.10 Osteopontin undergoes extensive post-translational modification such as glycosylation, phosphorylation and proteolytic cleavage by thrombin and matrix metalloproteases 3 and 75 affecting processes such as integrin binding.10 In tumours, OPN binds to cancer cells and endothelial cells in an RGD-dependent manner, which increases cell survival, cell migration and angiogenesis.5,11–13 Inhibition of OPN expression by small interfering (si) RNA in glioma cells reduces cell proliferation and motility in vitro and prevents cancer progression in vivo.14–19 Therefore, OPN is considered to be a therapeutic target in glioblastoma.15 On the other hand, OPN induces migration of neutrophils20 and macrophages,5,11 which is an activity that may counteract cancer.6,20–24
The aim of the present study was to establish (i) whether OPN gene over-expression is related to specific cancer types such as of breast,25–27 prostate,27–29 liver,5,30 head and neck31 and brain32 or whether it is a general phenomenon in cancer; (ii) OPN expression in the different grades of glioma; (iii) whether OPN expression promotes neutrophil and macrophage infiltration; and (iv) whether these effects on migrating cells are dependent on the RGD peptide sequence of OPN.24,33,34
We found in silico that elevated OPN gene expression is a general phenomenon in cancer. In some types of cancers including glioblastoma, it is one of the highest expressed genes in a large percentage of patients. In glioma, OPN gene over-expression was found only in its most malignant state, glioblastoma. Moreover, we showed that OPN protein co-localizes with neutrophils and macrophages in glioblastoma. In vitro, we observed that the RGD sequence of OPN is necessary for recruitment of neutrophils.
Materials and methods
Neutrophil migration in vitro
Animal preparation:
Eight-week-old adult OPN-null mice and their matched C57BL/6 J wild-type (WT) controls were used. The generation of these OPN-null mice has been previously described.20,35 The animals were killed by carbon dioxide exposure and cervical dislocation. All experiments were performed in duplicate and permission was granted by the Local Animal Committee.
Migration assays:
Femurs and tibias of WT and OPN-null mice (n= 6 per mouse type) were dissected and bone marrow was flushed with 2·5 ml α-minimum essential medium containing 10% fetal bovine serum and antibiotics (100 μg/ml penicillin G, 50 μg/ml gentamicin sulphate and 0·3 μg/ml fungizone). After lysis of the red blood cells with E-lyse (Cardinal, Phoenix, AZ), cells were layered on a Percoll gradient (Sigma, Oakville, ON, Canada; 3 ml of 55% Percoll, top layer; 3 ml of 65% Percoll, middle layer; 4 ml of 80% Percoll, bottom layer) and centrifuged at 1500 g for 30 min at 10°. The interface between the 65% and 80% Percoll layers, which was enriched with neutrophils (> 90%), was collected. Typical yields were 2 × 107 to 3 × 107 neutrophils/mouse, of which > 98% were viable, as indicated by Trypan blue staining.
For quantification of neutrophil migration, 24-transwell chamber plates (Corning, Acton, MA) were used. Briefly, after sampling and centrifugation of the neutrophils, a neutrophil suspension of 1 × 106 cells/100 μl phosphate buffer was placed onto polycarbonate membrane inserts (3·0-μm pore size; Sigma) coated with fetal bovine serum. The inserts were placed in the wells. Exogenous post-translationally-modified rodent macrophage OPN (mϕ-OPN; purified to homogeneity from RAW 264.7 cell-conditioned medium), full-length recombinant rat OPN (rec-OPN; produced as described previously;36 Sigma), which lacks post-translational modifications, and mutated OPN (mut-OPN; Sigma), which lacks the RGD sequence, were used in migration assays.
Neutrophils were allowed to migrate for 60 min at 37° in 5% CO2 to attach to glass cover slips (12 mm diameter). The cover slips with the neutrophils attached were gently washed with 2·0 ml PBS. The number of attached cells on each cover slip was counted (imagej software, developed by Wayne Rasband, http://rsbweb.nih.gov/ij/) in five fields of each of the two replicate wells using digital images captured with a time-lapse video microscope Eclipse E400 equipped with differential interference contrast optics and a × 40 objective (Nikon, Tokyo, Japan).
In silico analysis of OPN expression
The genome databases of Pubmed (http://www.ncbi.nlm.nih.gov/sites/entrez) were used to collect information on the human OPN gene such as Entrez Gene ID (6696), gene name (osteopontin), gene symbol (OPN) and gene synonyms (secreted phosphoprotein 1).
The Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) collections were searched to retrieve published microarray gene expression data sets comparing cancer samples with normal tissue samples. Microarray expression data were retrieved from 35 data sets (after excluding 29 data sets for various reasons such as normal tissue samples consisting of cells in culture) comparing cancer with normal human samples.37–72 For all data sets, log2-transformed signal intensity measurements were available for each probe in every sample including probes for the OPN gene. The original method of spot qualification and data normalization was maintained for each data set. To compare between platforms, an expression intensity was calculated for each Entrez Gene ID by averaging multiple probe intensities. Hence, expression parameters, namely percentage over-expression (the fraction of patients in a data set that showed over-expression of the OPN gene) and percentile of fold change (the level of over-expression of the OPN gene expressed as a percentile of expression levels of all genes in a particular data set) were used to quantify the frequency and level of OPN gene expression.37,38 A percentile of fold change larger than 0·95 was arbitrarily considered as substantial over-expression and a percentage over-expression larger than 30% was arbitrarily considered as frequent over-expression.
In situ analysis of OPN expression
Tissue samples:
We determined the expression patterns of OPN protein immunohistochemically in relation to the localization of neutrophils and macrophages in cryostat sections of samples from patients with glioblastoma. Thirteen glioblastoma samples (classified by the pathologist D.T. according to the WHO 2007 classification) and five normal brain tissue samples (normal white and grey matter post mortem samples) were selected. Tumour samples and normal samples were included only when at least 80% of the samples consisted of cancer cells, as verified by haematoxylin & eosin staining. Use of material fell under the Dutch Code of proper secondary use of human tissue.
Histochemistry:
Tissues samples were snap-frozen in liquid nitrogen in the operating room, and stored at −80° until used. Cryostat sections (8-μm thick) were cut at −25° on an HM560 cryostat (MICROM, Walldorf, Germany), picked up on glass slides, and stored at −80° until used. All staining procedures including controls were performed on serial sections of each tissue sample. Haematoxylin & eosin staining was performed to check morphology.
Immunohistochemical staining of OPN, neutrophils and macrophages was performed on cryostat sections as described.73,74 An anti-human OPN rabbit monoclonal antibody (dilution, 1 : 600 in PBS; Vector Laboratories, La Jolla, CA), mouse monoclonal anti-human macrophage CD68 (clone EBM 11, isotype IgG1, kappa; dilution, 1 : 400 in PBS) and anti-neutrophil elastase (dilution, 1 : 400 in PBS; Dako, Glostrup, Denmark) were used as primary antibodies. Immunostaining was performed manually at room temperature. After washing with PBS, three times for 5 min, endogenous peroxidase and non-specific background staining were blocked with 0·1% sodium azide, 0·3% H2O2 in PBS for 15 min. After washing with PBS, three times for 5 min, sections were incubated with the anti-OPN, anti-CD68 primary antibodies and anti-neutrophil elastase at the given dilutions for 60 min. After rinsing with PBS, three times for 5 min, sections were incubated with secondary goat anti-rabbit conjugated with peroxidase for OPN staining, with rabbit anti-mouse conjugated with peroxidase secondary for macrophages (1 : 50) and with Powervision goat anti-mouse antibodies for neutrophils (Immunologic, Duiven, the Netherlands). After rinsing with PBS, three times for 5 min, sections were incubated for 20 min with aminoethylcarbazole as peroxidase substrate. All samples were counterstained with haematoxylin for 20 seconds before mounting. Negative control incubations were performed in the absence of primary antibody.
Image analysis and semi-quantitative analysis
The OPN immunostained sections were analysed with the use of quantitative image analysis as described in detail elsewhere,75 using a Vanox-T photomicroscope with a 10 × objective (Olympus, Tokyo, Japan) and a CFW-1312M 1360 × 1024 pixel 10-bit monochrome FireWire camera (Scion, Tucson, AZ) mounted on the front port of the Vanox using adapting optics. Sections were illuminated with white light that was filtered by a monochromatic filter of 480 nm and an infrared blocking filter to correctly measure the absorbance of the coloured peroxidase reaction product, oxidized aminoethylcarbazole, without interference of the counter-stain haematoxylin. Absorbance calibration of the images was performed with the use of a calibrated 10-step density tablet (Stouffer, South Bend, IN). After measuring the step tablet, known absorbance values were related to measured grey values using the built-in calibration function of ImageJ, using the Rodbard function.75 Density calibrated images were recorded in one single run and stored on disk for analysis. The resolution used prevented distributional errors. All settings were maintained throughout the recording session and at the end of the session were verified against the step tablet values.75 Software used for capturing was swf-image, a Scion proprietary camera driver, as an extension to the image processing application ImageJ.75 ObjectJ, a plug-in for non-destructive image marking and result linking developed by Norbert Vischer and Stelian Nastase at the Department of Biology of the University of Amsterdam (http://simon.bio.uva.nl/) was used to indicate regions of interest that were measured. The regions of interest were selected on the basis of homogeneity of OPN protein levels. Using the ObjectJ plug-in, mean absorbance values within the regions of interest were collected from the test sections and control sections, allowing calculation of OPN expression as test minus control in mean absorbance units. The use of ObjectJ allows a retrospective quality control study of the areas measured.
Semi-quantitative scoring of neutrophils and macrophages in brain tissue sections was performed according to the Kellgren and Lawrence scale.76–78 The Kellgren and Lawrence scale is a general scoring system describing degrees of severity (0 = normal, 1 = doubtful, 2 = mild, 3 = moderate, 4 = severe). We have added one more degree 5 = very severe. Two observers (N.A.A. and C.J.F.V.N.) scored all sections together to obtain agreement on standards for grading in a training session. Afterwards, the two observers scored all sections again independently and in blinded fashion. Both scores were very similar; only for 10% of the sections, the score deviated by one scale at the most. These sections were then scored together to obtain a consensus score.
Data analysis
The R-program (http://www.r-project.org/) was used to determine the distribution of numbers of cells in vitro. It appeared that log10 conversion of the data resulted in normal distributions in all cases (Fig. S1), allowing the use of the Student’s t-test variant for the assumption for non-paired data with unequal variances (http://beheco.oxfordjournals.org/cgi/content/full/17/4/688) to determine significant differences (P ≤ 0·05) in the proportion of cells that had migrated towards the chemotactic gradients after the addition of different types of exogenous OPN to WT and OPN−/− neutrophils. Matlab (MatWorks, Natick, MA) was used for calculations, figures and tables. To compute and visualize in silico data (expression parameters for OPN) the R-program, Matlab and Excel 2007 were used. The same programs were used to determine semi-quantitative scoring of neutrophils and macrophages in brain tissue sections. The QQ-plot was used to determine association between normal distribution of OPN expression and presence of neutrophils and macrophages in situ. Again, the Student’s t-test was used to determine significant differences (P ≤ 0·05) between OPN expression in non-cancerous sections and glioblastoma sections. The general regression model was used in R-program to analyse the linear relation between the amount of OPN protein and presence of neutrophils and between the amount of OPN protein and presence of macrophages in non-cancerous sections and glioblastoma sections.
Results
RGD-dependent neutrophil migration in vitro
Migration of WT and OPN−/− neutrophils from mice was significantly different when stimulated with either mut-OPN lacking the RGD sequence or rec-OPN lacking post-translational modification or the post-translationally modified mϕ-OPN in vitro (Fig. 1). In the presence of mut-OPN hardly any OPN−/− neutrophil migration occurred, whereas WT neutrophils with endogenous OPN expression migrated more. The effects of exogenous mϕ-OPN were weaker on recruitment of OPN−/− neutrophils than of WT neutrophils. Exogenous rec-OPN had the strongest effect on recruitment of both WT neutrophils and OPN−/− neutrophils. Figure 1 also shows representative images of WT and OPN−/− neutrophil migration in the presence of the different types of exogenous OPN. Endogenous OPN of neutrophils had a significant effect on their migration irrespective of the type of exogenous OPN used (P < 0·05 in all cases). The major in vitro findings are that exogenous OPN-induced neutrophil migration is RGD dependent, that the endogenous OPN in neutrophils is pro-migratory and that rec-OPN without post-translational modification has the strongest effect on neutrophil migration.
Figure 1.
Quantitative analysis (log10 scale) of in vitro recruitment of wild-type (WT) and osteopontin-deficient (OPN−/−) neutrophils by three types of exogenous OPN [macrophage OPN (mϕ-OPN), recombinant OPN (rec-OPN) and mutated OPN (mut-OPN)] and representative images of recruited neutrophils by each type of OPN. Differences between the effects of the three types of OPN and between WT and OPN−/− neutrophils are significant (P < 0·05). Data are presented as log10 values because this results in a normal distribution of the data in all cases.
In silico analysis of OPN expression in cancer
The OPN mRNA levels were in general high in the 35 data sets of the various tumour types investigated. In 20 out of 35 data sets, including glioblastoma, the OPN gene was over-expressed with both percentile fold change > 0·95 and percentage over-expression > 30% (Fig. 2). In some types of cancer, such as ovarian cancer, lung cancer and melanoma, the OPN gene was one of the highest expressed genes in almost all patients. In glioblastoma, the OPN gene was one of the 5% highest expressed genes in 90% of the patients when considering the microarray data set of Bredel et al.61. Analysis of the data set of Kotliarov et al.63, also showed high OPN gene expression in glioblastoma only and not in lower-grade glioma, but the percentage of patients with high expression was much lower. Figure 3 shows that OPN gene expression in glioma was highest in glioblastoma (WHO grade 4 and histological glioblastoma) and it was lower in low-grade and anaplastic glioma and in gliomas with astrocytic or oligodendroglial elements. These data indicate that OPN gene over-expression is a general phenomenon related to cancer and is grade dependent in glioma.
Figure 2.
In silico analysis of expression of the osteopontin gene OPN in cancer data sets. Heatmap of percentile fold change and percentage over-expression of the OPN gene expression in 20 cancer data sets that have a percentile fold change > 0·95 and percentage over-expression > 30% meaning that the OPN gene is one of the 5% of genes that are most highly expressed in cancer in at least 30% of the patients. Number of normal tissue samples (N) and cancer samples (C) per data set are given.
Figure 3.
Percentile fold change and percentage over-expression of expression of osteopontin gene OPN in different WHO grades and histological grades of glioma. (a, c) Heatmap of WHO grade (WHO-2, WHO-3 and WHO-4 = glioblastoma) in the data sets of Bredel et al.61 and Kotliarov et al.63 The number of tumour samples included are nine WHO-2, 10 WHO-3 and 31 WHO-4 grades in the Bredel et al. data set and 45 WHO-2, 31 WHO-3 and 81 WHO-4 grades in the Kotliarov et al. data set. (b, d) Heatmap of histological grades (oligodenroglial, astrocytic and glioblastoma) in the data sets of Bredel et al.61 and Kotliarov et al.63 The number of tumour samples included are nine oligodenroglial, two astrocytic and 31 glioblastoma in the Bredel et al. data set61 and 50 oligodenroglial, 26 astrocytic and 81 glioblastoma in the Kotliarov et al. data set.63
Relation between in situ OPN protein and the presence of neutrophils and macrophages
Figure 4 shows localization of OPN protein, neutrophils and macrophages in normal brain tissue as detected immunohistochemically. In normal brain tissue, OPN protein expression (Fig. 4a) was low, whereas neutrophils (Fig. 4b) and macrophages (Fig. 4c) were absent. Localization of the OPN protein in normal brain was mainly intracellular in endothelial cells, microglia cells (Fig. 4a) and a few neurons (not shown).
Figure 4.
Immunohistochemical analysis of osteopontin (OPN) protein expression (a), neutrophils (b) and macrophages (c) in normal human brain tissue. OPN is present in endothelial cells (arrows) and microglial cells (arrowheads). Neutrophils and macrophages are absent. Bar = 40 μm.
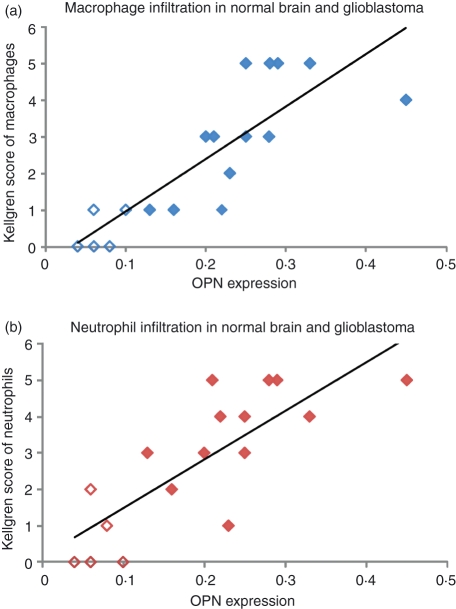
Figure 5 shows localization of OPN protein, neutrophils and macrophages in glioblastoma. The OPN protein in glioblastoma was mainly present extracellularly, and its level was highest around necrotic areas and in the vicinity of blood vessels. It was absent within necrotic areas (Fig. 5a,d,g). Non-specific staining was found only in certain necrotic areas. Localization of neutrophils (Fig. 5b,e,h) and macrophages (Fig. 5g,f,i) in adjacent sections to the OPN-immunostained sections demonstrated that glioblastoma tissue was infiltrated with neutrophils and macrophages. Image analysis showed that OPN protein expression was significantly higher in glioblastoma (mean ± SD: 0·07 ± 0·02 in areas with low expression and mean ± SD: 0·18 ± 0·06 in areas with high expression; n = 13) than in normal brain (mean ± SD: 0·03 ± 0·01 in all areas; n = 5; P < 0·05). Regression analysis of the OPN protein expression levels significantly related with the presence of neutrophils (Fig. 6a; P < 0·05)) and macrophages (Fig. 6b; P < 0·05)).
Figure 5.
Immunohistochemical analysis of osteopontin (OPN) protein expression (a, d, g), neutrophils (b, e, h) and macrophages (c, f, i) in human glioblastoma samples. The OPN is highly expressed extracellularly (CC) around necrotic areas (N) and blood vessels (BV), where neutrophils and macrophages are present. Bars represent 40 μm (a, b, c, g, h, i) and 20 μm (d, e, f).
Figure 6.
Relationship between quantitative image analysis data of osteopontin (OPN) protein expression in situ and the semi-quantative scores of macrophages (a; P < 0·05) and neutrophils (b; P < 0·05) in non-cancerous human brain tissue (n = 5) and glioblastoma (n = 13).
Discussion
Our in silico analysis enabled us to investigate 35 microarray data sets of various types of cancer, comparing OPN gene expression in cancer with that in control tissue. It appeared that the OPN gene is one of the most over-expressed genes in a wide variety of cancer types in a high percentage of patients, including glioblastoma. These data confirm previous reports that OPN gene expression is elevated in various types of human cancers including glioblastoma and its expression is malignancy related.4,27,79–81
In situ, we found that OPN protein is present intracellularly in normal brain in endothelial cells, microglia cells and some neurons. This localization pattern was similar to that of vascular endothelial growth factor A (VEGF-A; data not shown). It is known that intracellular VEGF-A, particularly in neuronal cells, has cell protective effects.82,83 It has been implied that the transcription factor hypoxia inducible factor 1α (HIF-1α), which also drives OPN expression,84 is responsible for intracellular expression of VEGF-A.85
Levels of OPN protein in glioblastoma appeared to be distinctly elevated. Moreover, OPN protein was present extracellularly in glioblastoma samples. It is possible that hypoxia induced OPN over-expression in glioblastoma via HIF-1α because OPN protein was found abundantly around necrotic areas. Elevated OPN levels have been described in an experimental model of brain tumours in rats.15,86 It has also been reported that OPN is secreted in tumours into the extracellular matrix by cancer cells where it facilitates proliferation, motility and migration of cancer cells through binding to integrins and CD44.15,87,88 It also induces secretion of matrix metalloproteases, which facilitate invasion of cancer cells.89–91 Inhibition of OPN expression by siRNA in glioma cells not only reduced cell motility and cell proliferation in vitro but also down-regulated development of tumours in vivo.14–19 Therefore, OPN in glioblastoma is considered to be a therapeutic target.15 However, this assumption has to be carefully considered because OPN may have an intracellular function in normal brain as a neuroprotectant.82,83 Furthermore, OPN is also directly related to the influx of white blood cells such as neutrophils and macrophages in glioblastoma in an RGD-dependent manner, which may have an anti-cancer effect.
We showed that intracellular OPN affects neutrophil migration in vitro. Migration of OPN−/− neutrophils in the presence of mut-OPN (lacking the RGD sequence) was almost completely absent, whereas WT neutrophils showed migration. This finding suggests that the integrin-binding peptide sequence (RGD) of OPN is necessary for neutrophil migration in vitro.
It is likely that the OPN RGD sequence interacts with integrins such as αVβ3, α4β1 and α9β1.12,92–103 The latter integrin is highly expressed by neutrophils96 and macrophages.24,33,34 It may also be possible that leucocyte influx in glioblastoma was not really altered by OPN, but rather efflux and apoptosis of leucocytes33 was reduced. Accordingly, macrophages accumulate in infected brain as a result of elevated OPN levels33. The relevance of the influx of white blood cells in glioblastoma and its down-regulating effect on carcinogenesis is unknown. It has been found previously that activity of leucocytes is down-regulated in experimental tumours because cancer cells can secrete immune-down-regulating molecules such as α2-macroglobulin, prostaglandin E2 and transforming growth factor-β.74,104 Nevertheless, the potential functioning of OPN in normal brain and the effects of OPN on migration of leucocytes into tumours and the anti-cancer effects of these cells have to be taken into consideration when using OPN as a therapeutic anti-cancer target.
It is concluded that (i) OPN gene over-expression is a general phenomenon in cancer according to the in silico microarray analysis; it is one of the highest expressed genes in various types of cancer in the majority of the patients, (ii) OPN gene over-expression is linked with malignancy in glioma, (iii) the RGD sequence in OPN is necessary for migration of neutrophils in vitro, and (iv) extracellular OPN protein expression co-localizes with neutrophils and macrophages in glioblastoma.
Acknowledgments
We thank Prof. Dr A. H. Zwinderman and Dr M. Hof for assistance with statistical analysis, Prof. Dr R. O. Schlingemann for the helpful discussions and comments on the manuscript, Ms T. M. S. Pierik and Ms M. Arendse for carefully preparing the manuscript.
Disclosures
The authors declare no competing financial interest.
Supporting Information
Additional Supporting Information may be found in the online version of the article:
Figure S1. Q-Q plots of individaul log10-transformed data against theoretical quantiles of fitted normal distribution of the data shown in Fig. 1. As all data do not show strong deviations from linearity, there is no reason to reject the assumption of normal distributions of the log10-tranformed data.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Sodek J, Ganss B, Mckee MD. Osteopontin. Crit Rev Oral Biol Med. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 2.Sodek J, Zhu B, Huynth MH, Brown TJ, Ringuette M. Novel functions of the matricellular proteins osteopontin and osteonectin/SPARC. Connect Tissue Res. 2002;43:308–19. doi: 10.1080/03008200290001050. [DOI] [PubMed] [Google Scholar]
- 3.Denhardt DT, Giachelli CM, Rittling SR. Role of osteopontin in cellular signaling and toxicant injury. Annu Rev Pharmacol Toxicol. 2001;41:723–49. doi: 10.1146/annurev.pharmtox.41.1.723. [DOI] [PubMed] [Google Scholar]
- 4.Wai PY, Kuo PC. Osteopontin: regulation in tumor metastasis. Cancer Metastasis Rev. 2008;27:103–18. doi: 10.1007/s10555-007-9104-9. [DOI] [PubMed] [Google Scholar]
- 5.Ramaiah SK, Rittling S. Pathophysiological role of osteopontin in hepatic inflammation, toxicity, and cancer. Toxicol Sci. 2008;103:4–13. doi: 10.1093/toxsci/kfm246. [DOI] [PubMed] [Google Scholar]
- 6.Alstergren P, Zhu B, Glogauer M, Mak TW, Ellen RP, Sodek J. Polarization and directed migration of murine neutrophils is dependent on cell surface expression of CD44. Cell Immunol. 2004;231:146–57. doi: 10.1016/j.cellimm.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Apte UM, Mcree R, Ramaiah SK. Osteopontin induction: possible mechanism behind hepatic neutrophil infiltration during alcoholic steatohepatitis. Hepatology. 2003;38:266A. doi: 10.1016/j.taap.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Ashkar S, Salih E, Gerstenfeld LC, Glimcher MJ. Localization and identification of a chemotactic domain on osteopontin. A possible role for osteopontin and its peptides in bone healing. J Bone Min Res. 1996;11:M442. [Google Scholar]
- 9.Bennett JS, Berger BW, Billings PC. The structure and function of platelet integrins. J Thromb Haemost. 2009;7(Suppl. 1):200–5. doi: 10.1111/j.1538-7836.2009.03378.x. [DOI] [PubMed] [Google Scholar]
- 10.Ding Q, Stewart J, Prince CW, Chang PL, Trikha M, Han XS, Grammer JR, Gladson CL. Promotion of malignant astrocytoma cell migration by osteopontin expressed in the normal brain: differences in integrin signaling during cell adhesion to osteopontin versus vitronectin. Cancer Res. 2002;62:5336–43. doi: 10.1100/tsw.2002.247. [DOI] [PubMed] [Google Scholar]
- 11.Ramaiah SK, Rittling S. Role of osteopontin in regulating hepatic inflammatory responses and toxic liver injury. Expert Opin Drug Metab Toxicol. 2007;3:519–26. doi: 10.1517/17425225.3.4.519. [DOI] [PubMed] [Google Scholar]
- 12.Ramaiah SK, Jaeschke H. Role of neutrophils in the pathogenesis of acute inflammatory liver injury. Toxicol Pathol. 2007;35:757–66. doi: 10.1080/01926230701584163. [DOI] [PubMed] [Google Scholar]
- 13.Shijubo N, Uede T, Kon S, Nagata M, Abe S. Vascular endothelial growth factor and osteopontin in tumor biology. Crit Rev Oncol. 2000;11:135–46. [PubMed] [Google Scholar]
- 14.Chakraborty G, Jain S, Patil TV, Kundu GC. Down-regulation of osteopontin attenuates breast tumour progression in vivo. J Cell Mol Med. 2008;12:2305–18. doi: 10.1111/j.1582-4934.2008.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamour V, Le MM, Lefranc F, et al. Selective osteopontin knockdown exerts anti-tumoral activity in a human glioblastoma model. Int J Cancer. 2009;126:1797–805. doi: 10.1002/ijc.24751. [DOI] [PubMed] [Google Scholar]
- 16.Shevde LA, Samant RS, Paik JC, Metge BJ, Chambers AF, Casey G, Frost AR, Welch DR. Osteopontin knockdown suppresses tumorigenicity of human metastatic breast carcinoma MDA-MB-435. Clin Exp Metastasis. 2006;23:123–33. doi: 10.1007/s10585-006-9013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wai PY, Mi Z, Guo H, et al. Osteopontin silencing by small interfering RNA suppresses in vitro and in vitro CT26 murine colon adenocarcinoma metastasis. Carcinogenesis. 2005;26:741–51. doi: 10.1093/carcin/bgi027. [DOI] [PubMed] [Google Scholar]
- 18.Zhao J, Dong L, Liu B, et al. Down-regulation of osteopontin suppresses growth and metastasis of hepatocellular carcinoma via induction of apoptosis. Gastroenterology. 2008;135:956–68. doi: 10.1053/j.gastro.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Zohar R, Zhu B, Liu P, Sodek J, McCulloch CA. Increased cell death in osteopontin-deficient cardiac fibroblasts occurs by a caspase-3-independent pathway. Am J Physiol Heart Circ Physiol. 2004;287:H1730–9. doi: 10.1152/ajpheart.00098.2004. [DOI] [PubMed] [Google Scholar]
- 20.Koh A, Da Silva AP, Bansal AK, et al. Role of osteopontin in neutrophil function. Immunology. 2007;122:466–75. doi: 10.1111/j.1365-2567.2007.02682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apte UM, Banerjee A, Mcree R, Wellberg E, Ramaiah SK. Role of osteopontin in hepatic neutrophil infiltration during alcoholic steatohepatitis. Toxicol Appl Pharmacol. 2005;207:25–38. doi: 10.1016/j.taap.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Fossati G, Ricevuti G, Edwards SW, Walker C, Dalton A, Rossi ML. Neutrophil infiltration into human gliomas. Acta Neuropathol. 1999;98:349–54. doi: 10.1007/s004010051093. [DOI] [PubMed] [Google Scholar]
- 23.McDonald B, Spicer J, Giannais B, Fallavollita L, Brodt P, Ferri LE. Systemic inflammation increases cancer cell adhesion to hepatic sinusoids by neutrophil mediated mechanisms. Int J Cancer. 2009;125:1298–305. doi: 10.1002/ijc.24409. [DOI] [PubMed] [Google Scholar]
- 24.Zhu B, Suzuki K, Goldberg HA, Rittling SR, Denhardt DT, McCulloch CA, Sodek J. Osteopontin modulates CD44-dependent chemotaxis of peritoneal macrophages through G-protein-coupled receptors: evidence of a role for an intracellular form of osteopontin. J Cell Physiol. 2004;198:155–67. doi: 10.1002/jcp.10394. [DOI] [PubMed] [Google Scholar]
- 25.Bramwell VHC, Doig GS, Tuck AB, et al. Serial plasma osteopontin levels have prognostic value in metastatic breast cancer. Clin Cancer Res. 2006;12:3337–43. doi: 10.1158/1078-0432.CCR-05-2354. [DOI] [PubMed] [Google Scholar]
- 26.Singhal H, Bautista DS, Tonkin KS, O’Malley FP, Tuck AB, Chambers AF, Harris JF. Elevated plasma osteopontin in metastatic breast cancer associated with increased tumor burden and decreased survival. Clin Cancer Res. 1997;3:605–11. [PubMed] [Google Scholar]
- 27.Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, Dvorak HF, Senger DR. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145:610–23. [PMC free article] [PubMed] [Google Scholar]
- 28.Hotte SJ, Winquist EW, Stitt L, Wilson SM, Chambers AF. Plasma osteopontin: associations with survival and metastasis to bone in men with hormone-refractory prostate carcinoma. Cancer. 2002;95:506–12. doi: 10.1002/cncr.10709. [DOI] [PubMed] [Google Scholar]
- 29.Ramankulov A, Lein M, Kristiansen G, Loening SA, Jung K. Plasma osteopontin in comparison with bone markers as indicator of bone metastasis and survival outcome in patients with prostate cancer. Prostate. 2007;67:330–40. doi: 10.1002/pros.20540. [DOI] [PubMed] [Google Scholar]
- 30.Ye QH, Qin LX, Forgues M, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–23. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 31.Le QT, Denko NC, Giaccia AJ. Hypoxic gene expression and metastasis. Cancer Metastasis Rev. 2004;23:293–310. doi: 10.1023/B:CANC.0000031768.89246.d7. [DOI] [PubMed] [Google Scholar]
- 32.Matusan-Ilijas K, Behrem S, Jonjic N, Zarkovic K, Lucin K. Osteopontin expression correlates with angiogenesis and survival in malignant astrocytoma. Pathol Oncol Res. 2008;14:293–8. doi: 10.1007/s12253-008-9058-4. [DOI] [PubMed] [Google Scholar]
- 33.Burdo TH, Wood MR, Fox HS. Osteopontin prevents monocyte recirculation and apoptosis. J Leukoc Biol. 2007;81:1504–11. doi: 10.1189/jlb.1106711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burdo TH, Ellis RJ, Fox HS. Osteopontin is increased in HIV-associated dementia. J Infect Dis. 2008;198:715–22. doi: 10.1086/590504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rittling SR, Matsumoto HN, McKee MD, et al. Mice lacking osteopontin show normal development and bone structure but display altered osteoclast formation in vitro. J Bone Miner Res. 1998;13:1101–11. doi: 10.1359/jbmr.1998.13.7.1101. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee A, Lee JH, Ramaiah SK. Interaction of osteopontin with neutrophil α4β1 and α9β1 integrins in a rodent model of alcoholic liver disease. Toxicol Appl Pharmacol. 2008;233:238–46. doi: 10.1016/j.taap.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE, Bosl GJ, Chaganti RS. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vitro differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–7. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 38.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 40.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenburg ME, Liou LS, Gerry NP, Framton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welsh JB, Zarrinkar PP, Sapinoso LM, et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2001;98:1176–81. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehan E, Ben-Dor A, Liao W, et al. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007;56:175–84. doi: 10.1016/j.lungcan.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Wachi S, Yoneda K, Wu R. Interactome–transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–8. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inamura K, Fujiwara T, Hoshida Y, et al. Two subclasses of lung squamous cell carcinoma with different gene expression profiles and prognosis identified by hierarchical clustering and non-negative matrix factorization. Oncogene. 2005;24:7105–13. doi: 10.1038/sj.onc.1208858. [DOI] [PubMed] [Google Scholar]
- 46.Kimchi ET, Posner MC, Park JO. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–54. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Higgins J, Cheung ST, et al. Novel endothelial cell markers in hepatocellular carcinoma. Mod Pathol. 2004;17:1198–210. doi: 10.1038/modpathol.3800167. [DOI] [PubMed] [Google Scholar]
- 48.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 49.Quade BJ, Wang TY, Sornberger K, Dal CP, Mutter GL, Morton CC. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosomes Cancer. 2004;40:97–108. doi: 10.1002/gcc.20018. [DOI] [PubMed] [Google Scholar]
- 50.Jones MH, Virtanen C, Honjoh D, et al. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363:775–81. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 51.Cromer A, Carles A, Millon R, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–98. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–15. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virtanen C, Ishikawa Y, Honjoh D, Kimura M, Shimane M, Miyoshi T, Nomura H, Jones MH. Integrated classification of lung tumors and cell lines by expression profiling. Proc Natl Acad Sci U S A. 2002;99:12357–62. doi: 10.1073/pnas.192240599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–39. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–30. [PubMed] [Google Scholar]
- 58.Honore S, Lascols C, Malin D, Targaouchi R, Cattoir V, Legrand P, Soussy CJ, Cambau E. Investigation of the new QNR-based mechanism of quinolone resistance among enterobacterial strains isolated in Henri-Mondor hospital 2002–2005. Pathol Biol (Paris) 2006;54:270–9. doi: 10.1016/j.patbio.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 60.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Lensen JL, Moller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–8. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 61.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–89. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 62.Bredel M, Bredel C, Juric D, et al. Tumor necrosis factor-α-induced protein 3 as a putative regulator of nuclear factor-κB-mediated resistance to O6-alkylating agents in human glioblastomas. J Clin Oncol. 2006;24:274–87. doi: 10.1200/JCO.2005.02.9405. [DOI] [PubMed] [Google Scholar]
- 63.Kotliarov Y, Steed ME, Christopher N, et al. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic inbalances. Cancer Res. 2006;66:9428–36. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson AL, Wang ZC, De NA, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 65.Miccadei S, Provenzano C, Mojzisek M, Natali PG, Civitareale D. Retinoblastoma protein acts as Pax 8 transcriptional coactivator. Oncogene. 2005;24:6993–7001. doi: 10.1038/sj.onc.1208861. [DOI] [PubMed] [Google Scholar]
- 66.Kuriakose MA, Chen WT, He ZM, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61:1372–83. doi: 10.1007/s00018-004-4069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iacobuzio-Donahue CA, Maitra A, Olsen M, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–62. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–40. [PubMed] [Google Scholar]
- 69. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL96.
- 70. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5206.
- 71. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1037.
- 72. http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS1732.
- 73.Bronckers AL, Sasaguri K, Engelse MA. Transcription and immunolocalization of Runx2/Cbfa1/Pebp2αA in developing rodent and human craniofacial tissues: further evidence suggesting osteoclasts phagocytose osteocytes. Microsc Res Tech. 2003;61:540–8. doi: 10.1002/jemt.10377. [DOI] [PubMed] [Google Scholar]
- 74.Griffini P, Smorenburg SM, Vogels IM, Tigchelaar W, Van Noorden CJ. Kupffer cells and pit cells are not effective in the defense against experimentally induced colon carcinoma metastasis in rat liver. Clin Exp Metastasis. 1996;14:367–80. doi: 10.1007/BF00123396. [DOI] [PubMed] [Google Scholar]
- 75.Chieco P, Jonker A, Van Noorden CJ. Image Cytometry. Oxford: Bios, Royal Microscopical Society Handbooks; 2001. [Google Scholar]
- 76.Kellgren JH, Lawrence JS. Radiological assessment of rheumatoid arthritis. Ann Rheum Dis. 1957;16:485–93. doi: 10.1136/ard.16.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rijk PC, de Rooy TP, Coerkamp EG, Bernoski FP, Van Noorden CJ. Radiographic evaluation of the knee joint after meniscal allograft transplantation. An experimental study in rabbits. Knee Surg Sports Traumatol Arthrosc. 2002;10:241–6. doi: 10.1007/s00167-002-0284-0. [DOI] [PubMed] [Google Scholar]
- 79.Bellahcene A, Castronovo V. Increased expression of osteonectin and osteopontin, two bone matrix proteins, in human breast cancer. Am J Pathol. 1995;146:95–100. [PMC free article] [PubMed] [Google Scholar]
- 80.Guldur ME, Kibar Y, Deniz H, Bakir K. Comparison of osteopontin, beta-catenin and hnRNP B1 expression in lung carcinomas. Pathol Oncol Res. 2010;16:55–9. doi: 10.1007/s12253-009-9187-4. [DOI] [PubMed] [Google Scholar]
- 81.Park JB, Kwak HJ, Lee SH. Role of hyaluronan in glioma invasion. Cell Adh Migr. 2008;2:202–7. doi: 10.4161/cam.2.3.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin KL, Mao XO, Greenberg DA. Vascular endothelial growth factor: direct neuroprotective effect in in vitro ischemia. Proc Natl Acad Sci U S A. 2000;97:10242–7. doi: 10.1073/pnas.97.18.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nishijima K, Ng YS, Zhong L, et al. Vascular endothelial growth factor-A is a survival factor for retinal neurons and a critical neuroprotectant during the adaptive response to ischemic injury. Am J Pathol. 2007;171:53–67. doi: 10.2353/ajpath.2007.061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Said HM, Hagemann C, Staab A, et al. Expression patterns of the hypoxia-related genes osteopontin, CA9, erythropoietin, VEGF and HIF-1α in human glioma in vitro and in vitro. Radiother Oncol. 2007;83:398–405. doi: 10.1016/j.radonc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 85.Hughes JM, Groot AJ, van der Groep P, et al. Active HIF-1 in the normal human retina. J Histochem Cytochem. 2010;58:247–54. doi: 10.1369/jhc.2009.953786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jang T, Savaresse T, Low HP, et al. Osteopontin expression in intratumoral astrocytes marks tumor progression in gliomas induced by prenatal exposure to N-ethyl-N-nitrosourea. Am J Pathol. 2006;168:1676–85. doi: 10.2353/ajpath.2006.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bellail AC, Hunter SB, Brat DJ, Tan C, van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–69. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 88.Lafrance JP, Nolin L, Senecal L, Leblanc M. Predictors and outcome of cardiopulmonary resuscitation (CPR) calls in a large haemodialysis unit over a seven-year period. Nephrol Dial Transplant. 2006;21:1006–12. doi: 10.1093/ndt/gfk007. [DOI] [PubMed] [Google Scholar]
- 89.Rangaswami HR, Bulbule A, Kundu GC. Nuclear factor inducing kinase: a key regulator in osteopontin- induced MAPK/IκB kinase dependent NF-κB-mediated promatrix metalloproteinase-9 activation. Glycoconj J. 2006;23:221–32. doi: 10.1007/s10719-006-7927-1. [DOI] [PubMed] [Google Scholar]
- 90.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 91.Tuck AB, Chambers AF, Allan AL. Osteopontin overexpression in breast cancer: knowledge gained and possible implications for clinical management. J Cell Biochem. 2007;102:859–68. doi: 10.1002/jcb.21520. [DOI] [PubMed] [Google Scholar]
- 92.Chen YJ, Wei YY, Chen HT, et al. Osteopontin increases migration and MMP-9 up-regulation via αvβ3 integrin, FAK, ERK, and NF-κB-dependent pathway in human chondrosarcoma cells. J Cell Physiol. 2009;221:98–108. doi: 10.1002/jcp.21835. [DOI] [PubMed] [Google Scholar]
- 93.Diao H, Kon S, Iwabuchi K, et al. Osteopontin as a mediator of NKT cell function in T cell-mediated liver diseases. Immunity. 2004;21:539–50. doi: 10.1016/j.immuni.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 94.Diao H, Iwabuchi K, Li L, et al. Osteopontin regulates development and function of invariant natural killer T cells. Proc Natl Acad Sci U S A. 2008;105:15884–9. doi: 10.1073/pnas.0806089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fong YC, Liu SC, Huang CY, et al. Osteopontin increases lung cancer cells migration via activation of the αvβ3 integrin/FAK/Akt and NF-κB-dependent pathway. Lung Cancer. 2009;64:263–70. doi: 10.1016/j.lungcan.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 96.Ito K, Kon S, Nakayama Y, et al. The differential amino acid requirement within osteopontin in α4 and α9 integrin-mediated cell binding and migration. Matrix Biol. 2009;28:11–9. doi: 10.1016/j.matbio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 97.Moon C, Han JR, Park HJ, Hah JS, Kang JL. Synthetic RGDS peptide attenuates lipopolysaccharide-induced pulmonary inflammation by inhibiting integrin signaled MAP kinase pathways. Respir Res. 2009;10:18. doi: 10.1186/1465-9921-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakamura I, Pilkington MF, Lakkakorpi PT, Lipfert L, Sims SM, Dixon SJ, Rodan GA, Duong LT. Role of αvβ3 integrin in osteoclast migration and formation of the sealing zone. J Cell Sci. 1999;112:3985–93. doi: 10.1242/jcs.112.22.3985. [DOI] [PubMed] [Google Scholar]
- 99.Nakamura I, Duong LT, Rodan SB, Rodan GA. Involvement of αvβ3 integrins in osteoclast function. J Bone Miner Metab. 2007;25:337–44. doi: 10.1007/s00774-007-0773-9. [DOI] [PubMed] [Google Scholar]
- 100.Nishimichi N, Higashikawa F, Kinoh HH, Tateishi Y, Matsuda H, Yokosaki Y. Polymeric osteopontin employs integrin α9β1 as a receptor and attracts neutrophils by presenting a de novo binding site. J Biol Chem. 2009;284:14769–76. doi: 10.1074/jbc.M901515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith LL, Giachelli CM. Structural requirements for α9β1-mediated adhesion and migration to thrombin-cleaved osteopontin. Exp Cell Res. 1998;242:351–60. doi: 10.1006/excr.1998.4108. [DOI] [PubMed] [Google Scholar]
- 102.Yokasaki Y, Sheppard D. Mapping of the cryptic integrin-binding site in osteopontin suggests a new mechanism by which thrombin can regulate inflammation and tissue repair. Trends Cardiovasc Med. 2000;10:155–9. doi: 10.1016/s1050-1738(00)00055-4. [DOI] [PubMed] [Google Scholar]
- 103.Yokosaki Y, Matsuura N, Sasaki T, et al. The integrin α9β1 binds to a novel recognition sequence (SVVYGLR) in the thrombin-cleaved amino-terminal fragment of osteopontin. J Biol Chem. 1999;274:36328–34. doi: 10.1074/jbc.274.51.36328. [DOI] [PubMed] [Google Scholar]
- 104.Smorenburg SM, Griffini P, Tiggelman AB, Moorman AF, Boers W, Van Noorden CJ. α2-Macroglobulin is mainly produced by cancer cells and not by hepatocytes in rats with colon carcinoma metastases in liver. Hepatology. 1996;23:560–70. doi: 10.1002/hep.510230323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.