This work reports the isolation and characterization of a novel gene of the LBD family of transcription factors, which regulates phloem development during secondary woody growth of a poplar tree. This discovery reveals a novel regulatory step that directs secondary woody growth and is likely mediated through auxin signaling and suppression of meristem and/or activation of phloem identity genes.
Abstract
Regulation of secondary (woody) growth is of substantial economic and environmental interest but is poorly understood. We identified and subsequently characterized an activation-tagged poplar (Populus tremula × Populus alba) mutant with enhanced woody growth and changes in bark texture caused primarily by increased secondary phloem production. Molecular characterization of the mutation through positioning of the tag and retransformation experiments shows that the phenotype is conditioned by activation of an uncharacterized gene that encodes a novel member of the LATERAL ORGAN BOUNDARIES DOMAIN (LBD) family of transcription factors. Homology analysis showed highest similarity to an uncharacterized LBD1 gene from Arabidopsis thaliana, and we consequently named it Populus tremula × Populus alba (Pta) LBD1. Dominant-negative suppression of Pta LBD1 via translational fusion with the repressor SRDX domain caused decreased diameter growth and suppressed and highly irregular phloem development. In wild-type plants, LBD1 was most highly expressed in the phloem and cambial zone. Two key Class I KNOTTED1-like homeobox genes that promote meristem identity in the cambium were downregulated, while an Altered Phloem Development gene that is known to promote phloem differentiation was upregulated in the mutant. A set of four LBD genes, including the LBD1 gene, was predominantly expressed in wood-forming tissues, suggesting a broader regulatory role of these transcription factors during secondary woody growth in poplar.
INTRODUCTION
Wood provides a renewable resource for the production of pulp, paper, and construction timber (Skog and Nicholson, 1998) and is growing in importance as a lignocellulosic feedstock for biofuel production (Ragauskas et al., 2006). From ecological and evolutionary perspectives, wood plays an important role in the global carbon biogeochemical cycle as it dominates terrestrial ecosystem biomass (Kirilenko and Sedjo, 2007; Bonan, 2008). In the plant, wood serves many functions in water/nutrient transport, mechanical support, and storage of organic compounds, water, and gases (Brunner et al., 2004).
Many plants, especially trees, show two distinct types of growth patterns, known as primary and secondary growth (reviewed in Baucher et al., 2007). In the stem, primary growth originates in the shoot apical meristem (SAM) and is responsible for production of leaves and stems. The vasculature develops from the procambium of the primary stem. The primary growth occurs at the top of the stem a few internodes below the SAM. In the stem of poplar (Populus spp) and many other tree species, the primary growth then gives way to a secondary (woody) growth that produces secondary xylem and secondary phloem. The stem’s primary and secondary growth zones are spatiotemporally separated, easily discernable, and develop rapidly in fast-growing species like poplar (within 1 to 2 months). In poplar, for example, primary growth occurs in the top four internodes, and at the ninth internode secondary growth is already completely established (Dharmawardhana et al., 2010). Secondary growth originates in the vascular cambium, a lateral meristem that consists of meristem cells (cambial initials) organized in a single-celled radial file that forms a continuous cylinder around the stem (Mellerowicz et al., 2001). Typically, secondary xylem is produced inwards and secondary phloem outwards of the vascular cambium, although deviations from this pattern do occur (reviewed in Spicer and Groover, 2010).
Secondary xylem is made up of fibers and vessel elements that both form thick secondary (lignified) cell walls. Secondary phloem consists of sieve elements, companion cells, and phloem parenchyma cells with thin primary cell walls, which facilitate their transport functions. In addition, secondary phloem contains phloem fiber cells with thick secondary cell walls organized in bundles, which form discontinuous concentric layers around the stem diameter (Evert, 2006). Finally, a radial system of rays comprised primarily of parenchyma cells traverses both the secondary phloem and xylem and serves to transport and store substances, including photosynthate and water (Mellerowicz et al., 2001; Chaffey et al., 2002). To date, most of the research interest has focused on secondary xylem development as it dominates wood biomass (Ko et al., 2006; Roach and Deyholos, 2007). However, secondary phloem and ray cell development is an indispensable part of wood development as it provides photoassimilates, water, and signaling molecules for the growing xylem. In addition, secondary phloem is essential for bark formation, which in trees has important functions in thermo-insulation, storage, water loss prevention, and pest protection (van Bel, 2003).
Secondary tissue formation proceeds through a distinct sequence of events. Cambial initials divide periclinally to produce xylem, phloem, and ray mother cells, which in turn undergo several rounds of division followed by differentiation to form the respective cell types (reviewed in Mellerowicz et al., 2001; Baucher et al., 2007; Jansson and Douglas, 2007; Du and Groover, 2010; Spicer and Groover, 2010). This sequence of events is the engine of wood production and thus of substantial fundamental and economic interest.
The molecular mechanisms governing the process are poorly understood, but a conceptual model is laid out based on the much better understood regulation of SAM development (Schrader et al., 2004; Groover, 2005; Baucher et al., 2007). According to this model, WUSCHEL, CLAVATA, and SHOOTMERISTEMLESS (STM) or their closely related gene family members are involved in maintaining the activity and cell identity of the cambial initials. Transcription factors like AINTEGUMENTA regulate the active proliferation of the mother cells. Finally, a set of proteins important for lateral organ outgrowth and differentiation in SAM, including KANADI, HD-ZIPIII, YABBY, and ASYMMETRIC LEAVES1, is involved in radial patterning (e.g., differentiation into xylem, phloem, or rays). Several excellent reviews describing the model are available (Nieminen et al., 2004; Groover, 2005; Baucher et al., 2007), and although recent experimental evidence supports the model (Du and Groover, 2010), many of the regulatory factors and mechanisms involved are still unknown.
The lateral organ boundaries domain (LBD) proteins (also known as ASYMMETRIC LEAVES2-Like proteins) are a family of plant-specific transcription factors (Husbands et al., 2007) with 43 members in Arabidopsis thaliana (Iwakawa et al., 2002; Shuai et al., 2002), 35 in rice (Oryza sativa; Yang et al., 2006), and 57 in poplar (Zhu et al., 2007). The founding member of this family, ASYMMETRIC LEAVES2/LBD6 (AS2/LBD6), is involved in a major regulatory loop that governs meristem maintenance through interactions with STM (Scofield and Murray, 2006). LBD genes have been found to regulate a variety of developmental processes, including differentiation of emerging leaf primordia (Iwakawa et al., 2002), determination of cell fate in flower petals (Chalfun-Junior et al., 2005), rice flower development (Li et al., 2008), maize (Zea mays) branching (Bortiri et al., 2006), and maize and rice crown and lateral root development (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007; Lee et al., 2009). In addition to developmental processes, LBDs have recently been found to play a role in metabolic regulation in response to nitrogen deficiency (Rubin et al., 2009). To our knowledge, LBDs have not been previously implicated in the regulation of secondary growth.
Here, we report the characterization of a novel member of the LBD gene family, which regulates secondary phloem development in the stems of a poplar tree. We also provide evidence for the putative regulatory context of this function and the involvement of other LBD members in secondary woody growth.
RESULTS
Discovery of an Activation-Tagged Mutant with Increased Secondary Growth
We identified a poplar activation-tagged mutant that displays increased secondary growth and changes in bark texture (Figure 1). Biometric measurements showed that mutant plants had increased stem diameter in the zone undergoing secondary growth (Figures 1A and 1B). The bark of the mutant plants was deeply furrowed (Figure 1A), and leaves were slightly smaller than those of wild type-717 (WT-717) (Figure 1E). To better understand the phenotype, we studied the anatomical structure of stems undergoing secondary growth (Figures 1C and 1D). There was an increase in the production of both xylem and phloem in the mutant, but the majority of the increase in girth was a result of significantly greater secondary phloem growth (>50% increase) (Figures 1C and 1D). We therefore concluded that the mutation predominantly affects secondary growth through increase in phloem production.
Figure 1.
Morphological Changes in the Activation-Tagged Mutant Plants.
(A), (C), and (E) The mutant.
(B), (D), and (F) WT-717.
(A) and (B) Differences in bark texture and stem diameter (numbers in parentheses show stem diameter at the 20th internode in millimeters; mean ± se, n = 6, P < 0.001).
(C) and (D) Increased secondary phloem production in mutant plants (numbers in parentheses show secondary phloem [bark] width as denoted by a double-arrowhead line at the 20th internode in micrometers; mean ± se, n = 6, P < 0.001). Sections were stained with toluidine blue O. Arrows point to the number of layers with phloem fiber bundles in the mutant (C) and WT-717 (D) plants. Xy, xylem; Ph, phloem.
(E) and (F) Leaf morphology of the mutant (E) and WT-717 (F) plants. Leaves shown on the photographs were sampled from the 15th node.
Molecular Characterization of the Tagged Gene
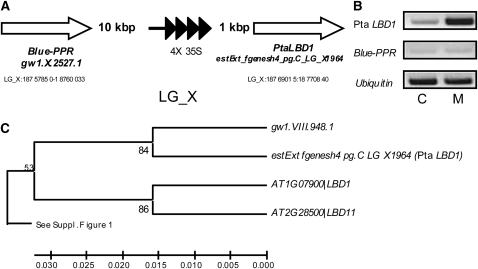
To identify the gene responsible for the observed phenotypic changes, we recovered an ~150-bp fragment of genomic sequence flanking the left border of the T-DNA insertion. Homology searches in the poplar genome positioned the sequence at LG_X:18767889-18768035. Inspection of the genomic region identified two proximal genes (Figure 2A). We compared the expression of the two genes in the mutant and WT-717 plants. The gene proximal (1063 bp from the insertion to the start ATG codon) to the enhancer corresponded to model estExt_fgenesh4_pg.C_LG_X1964 and showed clear activation (Figure 2B). The distal gene and the loading control genes showed no change (Figure 2B). Therefore, we hypothesized that estExt_fgenesh4_pg.C_LG_X1964 was likely the gene responsible for the observed phenotype.
Figure 2.
Identification of the Activation-Tagged Gene.
(A) Position of the tag in the poplar genome. White arrows represent the flanking genes and the tetrahead arrow the enhancer tetramer in the activation tagging vector.
(B) Expression of the two proximal genes in WT-717 (C) and mutant (M) plants, as determined by RT-PCR analysis (picture represents typical results from three independent biological replicates). The expression of ubiquitin was used as a loading control. Note the strong upregulation of Pta LBD1.
(C) Phylogenetic analysis of the Pta LBD1 gene in relation to close Arabidopsis putative orthologs LBD1 and LBD11. For the full phylogenetic tree, see Supplemental Figure 1 online.
We cloned and sequenced the cDNA of the activated gene, which included an open reading frame of 660 bp encoding a putative protein of 219 amino acids. The sequence shows high sequence similarity to a family of proteins containing the conserved LBD. A phylogenetic comparison of putative LBD proteins in Arabidopsis and Populus is shown in Supplemental Figure 1 online (see also Supplemental Data Set 1 online). The tagged gene shows the highest similarity (80%) to an uncharacterized gene from Arabidopsis annotated as LBD1 (Figure 2C; see Supplemental Figure 1 and Supplemental Data Sets 1 and 2 online). For consistency, we therefore named the gene Populus tremula × Populus alba (Pta) LBD1. Similar to Arabidopsis, the LBD family in Populus is comprised of two major classes that are further subdivided into nine groups (Shuai, et al., 2002; Matsumura et al., 2009) (see Supplemental Figure 1 online). Pta LBD1 and the corresponding Populus trichocarpa gene, estExt_fgenesh4_pg.C_LG_X1964, belong to Class Ia (see Supplemental Figure 1 online). Both the poplar and Arabidopsis genes are of unknown function.
Pta LBD1 Expression and Tissue Localization
We studied the expression of LBD1 in various organs and tissues, including phloem, xylem, and stems undergoing primary and secondary growth (Figure 3A). The highest levels of LBD1 transcript were found in phloem tissues and stems undergoing secondary growth (Figure 3A). Thus, the expression pattern corresponds well with the most severe phenotypic changes observed in the mutant. The gene transcript was also detectable in other tissues, but at a much lower level (Figure 3A). To better understand the localization of LBD1 expression during secondary growth, we performed in situ RT-PCR experiments in WT-717 plants. The gene transcript was localized to a narrow region that includes part of the cambial zone, which contains the cambium initials and the actively dividing mother cells (Spicer and Groover, 2010) (Figure 3B). The signal in this zone was shifted to the phloem side of the cambial zone and extended into the developing phloem (Figure 3C), which corresponded to the higher expression in the phloem (Figure 3A). To gain insight into LBD1 expression, we also examined publically available cDNA microarray databases (Sjödin et al., 2009). This analysis also pointed to phloem- and cambium-predominant expression (see Supplemental Figure 2 online), confirming the findings of our study.
Figure 3.
Native Expression and Localization of Pta LBD1.
(A) Relative expression of LBD1 in different plant organs and tissues, as determined by RT-PCR: apex (A), leaf (L), whole internodal sections of stems undergoing predominantly primary growth (PSt), whole internodal sections of stems undergoing secondary growth (SSt), phloem (Ph), xylem (Xy), and root (R). See Methods for more sampling details. Data represent relative expression (mean ± se) from three independent biological replicates and are normalized to the expression of a ubiquitin loading control gene.
(B) and (C) In situ RT-PCR localization of LBD1 in stem sections from the 10th internode. The inset in (C) represents a higher magnification of the sector positioned to the right of the double-arrowhead line.
(D) and (E) No signal was detected in the negative control (-RT). Red arrowheads and double-arrowhead line demark the cambial zone on (B) and (C), respectively.
CZ, cambium zone; PhF, phloem fiber; Ph, phloem; Xy, xylem. Bars = 100 μm in (B) and (D), 10 μm in (C) and (E), and 2 μm in the inset in (C).
Recapitulation of the Mutant Phenotype via Retransformation
To recapitulate the activation-tagged phenotype, we fused LBD1 to a strong cauliflower mosaic virus 35S promoter and transformed the construct into the WT-717 background, in which the mutant was originally discovered. We refer to these transgenic lines as LBD1-overexpressing (PtaLBD1-oe). We observed phenotypic changes early during the regeneration process (see Supplemental Figures 3 and 4 online). The shoot induction was delayed due to significant increase in callus proliferation (see Supplemental Figures 3 and 4 online). Spontaneous proliferation of callus in a few transgenic lines was observed, but only in vitro in auxin-containing media and in intact plants at the shoot/root junction (see Supplemental Figures 3A and 3B online). Most importantly, many of the regenerated shoots appeared to have thicker stems early in the regeneration process compared with control plants transformed with empty vector (see Supplemental Figures 3C and 3D online).
Multiple independent transgenic events carrying the construct were regenerated, validated for the presence and expression of the transgene, and grown in the greenhouse for ~3 months. As with the original activation-tagged mutant, we observed an increase in the stem diameter of PtaLBD1-oe plants and dramatic changes in the bark texture (Figures 4A to 4C). The differences in stem diameter were most pronounced in the base of the stem (5 cm above the soil) (Figure 4D). As in the original mutant, the leaves were slightly smaller than those of WT-717 (see Supplemental Figure 3G online). In summary, the phenotype of the PtaLBD1-oe recapitulation transgenics was similar to that of the original mutant, but in many cases accentuated, likely due to high expression from the strong constitutive promoter.
Figure 4.
Recapitulation of Mutant Phenotype via Transformation.
In all images, a representative PtaLBD1-oe transgenic plant is shown to the left and a WT-717 plant to the right.
(A) Increased stem diameter in PtaLBD1-oe plants. Inset shows stem sections displaying the differences in the stem with the red arrows delineating in the secondary phloem-bark width.
(B) and (C) Bark texture of PtaLBD1-oe plants (B) compared with that of WT-717 (C).
(D) Increased stem diameter in PtaLBD1-oe transgenic plants. Stem diameter was measured at the base of the tree and at the 10th internode in ~3-month-old greenhouse-grown plants. Values are reported as means ± se (n = 5). Asterisks indicate significance as determined by Student’s t test, with ** and * denoting P < 0.01 and P < 0.05, respectively.
Bars = 2 mm for inset in (A) and 1 cm in (B) and (C).
Wood Development in PtaLBD1-oe Transgenics
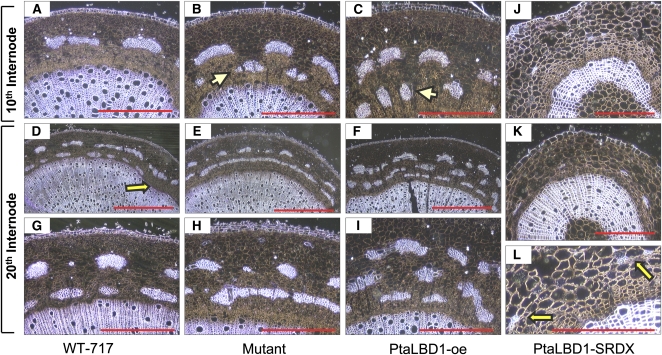
To investigate the impact of PtaLBD1 on wood formation, we sectioned stems that undergo secondary growth from WT-717, the activation-tagged mutant, and PtaLBD1-oe transgenics (Figure 5). The observed changes in PtaLBD1-oe were consistent with the abnormalities in wood development found in the original mutant. The most dramatic changes were found in secondary phloem development. In WT-717 plants, at the 10th internode, we observed only one layer of phloem fibers (Figure 5A), while the mutant and the PtaLBD1-oe plants had two layers of phloem fibers (Figures 5B and 5C). Moreover, in PtaLBD1-oe plants, the overall width of the secondary phloem was significantly increased (Figures 5C, 5F, and 5I), and in many cases this increase in PtaLBD1-oe plants was larger than in the original activation-tagged mutant (likely due to the strong 35S promoter). Similarly, at the 20th internode, PtaLBD1-oe plants had additional layers of phloem fibers and a thicker phloem/bark region (Figures 5D to 5I).
Figure 5.
Changes in Phloem Development of the Activation-Tagged Mutant, PtaLBD1-oe, and PtaLBD1-SRDX Plants.
(A), (D), and (G) Stem sections obtained from WT-717 plants. Arrow in (D) points to incomplete formation of a second phloem fiber ring at the 20th internode.
(B), (E), and (H) Increased phloem production in the mutant. Note in (E) the fully formed second phloem fiber ring.
(C), (F), and (I) Increased phloem production in PtaLBD1-oe plants. Arrows in (B) and (C) point to the secondary phloem fiber layers.
(J), (K), and (L) Suppressed phloem development in PtaLBD1-SRDX plants. Arrows point to rudimentary phloem fiber formation.
Representative sections from WT-717, mutant, and PtaLBD1-oe plants were taken from 2-month-old, greenhouse-grown plants. All plants were grown for the same period, under same conditions, and sampled at the same time at the respective internodes. Sections from PtaLBD1-SRDX plants were derived from the stems of in vitro–grown plants. The development of the WT-717 phloem in vitro has a similar pattern as in (A). Bars = 200 μm in (A) to (C), (J), and (K), 1 mm for (D) to (F), and 400 μm in (G) to (I) and (L). All images were acquired under phase contrast.
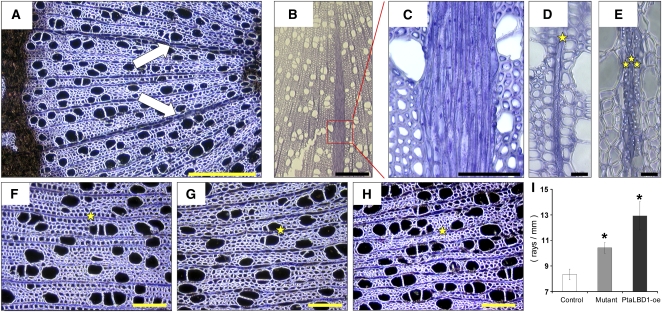
In addition to changes in secondary phloem development, we found significant changes in ray development in both mutant and PtaLBD1-oe plants (Figure 6). In poplar, rays are typically uniseriate (i.e., consist of a single cell file; Mellerowicz et al., 2001) (Figure 6D). In PtaLBD1-oe transgenics and the activation-tagged mutant, we found biseriate (two-rowed) (Figure 6A) and multiseriate (many-rowed) rays (Figures 6B, 6C, and 6E). Moreover, the density of rays was significantly higher in the mutant and PtaLBD1-oe plants than in WT-717 (Figure 6I). These results suggest that LBD1 plays a positive role in ray cell development, both at the initiation and proliferation stages.
Figure 6.
Increased Ray Production in the Activation-Tagged Mutant and PtaLBD1-oe Plants.
(A) to (C) Biseriate (A) and multiseriate ([B] and [C]) rays can be frequently observed in mutant (A) and PtaLBD1-oe plants ([B] and [C]). (C) represents a higher magnification of the boxed area shown in (B). Arrows point to biseriate rays.
(D) and (E) Typical uniseriate rays in WT-717 (D) compared with muliseriate rays in PtaLBD1-oe (E) plants. Stars indicate files of ray cells.
(F) to (H) Rays’ density in WT-717 (F), mutant (G), and PtaLBD1-oe plants (H). Stars represent typical rays that were scored to derive the data in (I).
(I) Quantification of ray density. Ray density was quantified in multiple sections, such as those shown in (F) to (H); values are reported as means ± se (n = 5). Asterisks indicate significant differences relative to WT-717 plants as determined by Student’s t test at P < 0.05.
Bars = 500 μm in (A) and (B), 100 μm in (C), 200 μm (F) to (H), and 20 μm in (D) and (E).
Generation of Loss-of-Function Phenotype via Pta LBD1-SRDX Repressor Domain Fusion
The study of LBD loss-of-function phenotypes has been hampered by significant functional redundancy in the large LBD family (Shuai et al., 2002) or by lethality of some of the knockouts (Borghi et al., 2007). Therefore, alternative approaches have often been used to investigate LBD loss-of-function phenotypes (Borghi et al., 2007; Soyano et al., 2008). These typically employ fusion of LBD with the SRDX repressor domain, a short 12–amino acid motif that converts transcription factors to dominant repressors (Hiratsu et al., 2003). Transgenic plants expressing the fusion protein display a loss-of-function phenotype (Hiratsu et al., 2003; Matsui et al., 2004; Matsui and Ohme-Takagi, 2010). We therefore transformed WT-717 with a construct expressing LBD1 fused with the transcriptional repressor domain SRDX (Hiratsu et al., 2003). Multiple transgenic lines were regenerated and validated for the presence of the transgene. PtaLBD1-SRDX transformation resulted in a very low frequency of callus production (see Supplemental Figure 4 online). This is exactly the opposite to the regeneration of PtaLBD1-oe plants, where we observed an enhanced callus production (see Supplemental Figure 4 online).
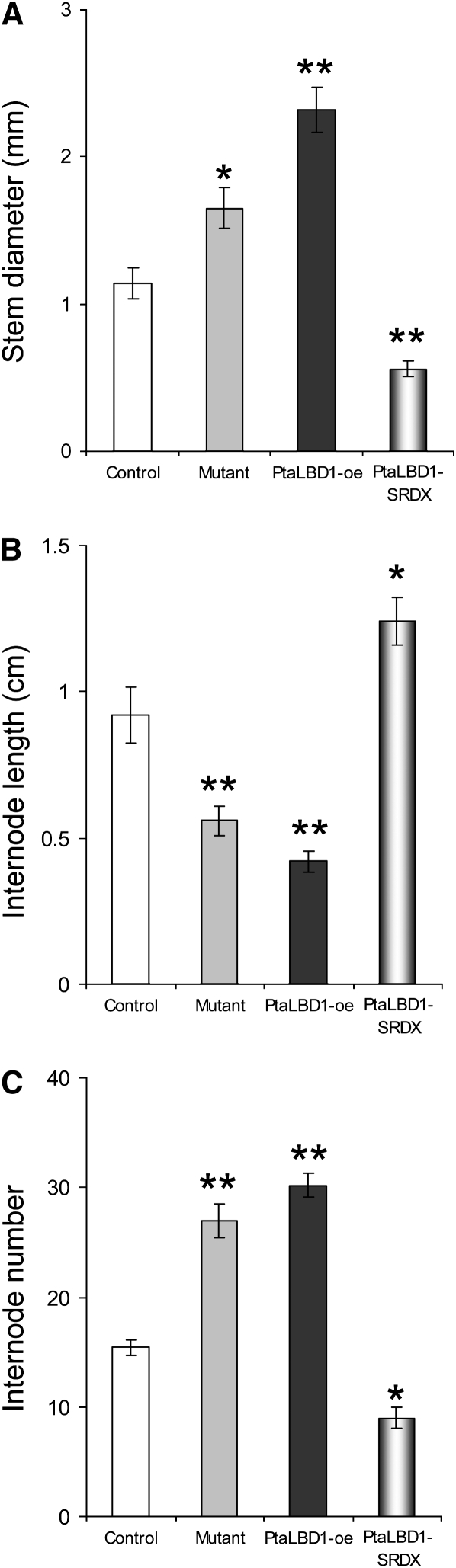
The expression of PtaLBD1-SRDX resulted in opposite phenotypes to those observed for PtaLBD1-oe plants (Figure 7). In contrast with the significantly increased stem diameter in PtaLBD1-oe and mutant plants (Figure 7A), the stem diameter of PtaLBD1-SRDX plants was significantly reduced (Figure 7A). Plants expressing the PtaLBD1-SRDX transgene had significantly increased internode lengths but decreased internode numbers (Figures 7B and 7C). Conversely, the mutant and PtaLBD1-oe plants displayed significantly shorter but a larger number of internodes (Figures 7B and 7C). Examination of stem sections of PtaLBD1-SRDX plants shows highly suppressed and irregular phloem differentiation that is in sharp contrast with the increased phloem production in the activation-tagged mutant and PtaLBD1-oe transgenics (Figures 5J to 5L).
Figure 7.
Morphological Changes in the Stems of the Activation-Tagged Mutant, PtaLBD1-oe, and PtaLBD1-SRDX Plants.
(A) Stem diameter at the base (first internodes above shoot/root junction).
(B) Internode length between 3rd and 8th node.
(C) Number of internodes measured in 2-month-old plants.
White bars represent WT-717, gray bars represent mutant, black bars represent PtaLBD1-oe, and gradient bars represent PtaLBD1-SRDX plants. Measurements were taken from ~2-month-old plants; values are reported as means ± se (n = 5). Asterisks indicate significant differences relative to WT-717 plants as determined by Student’s t test, with ** and * denoting P < 0.01 and P < 0.05, respectively.
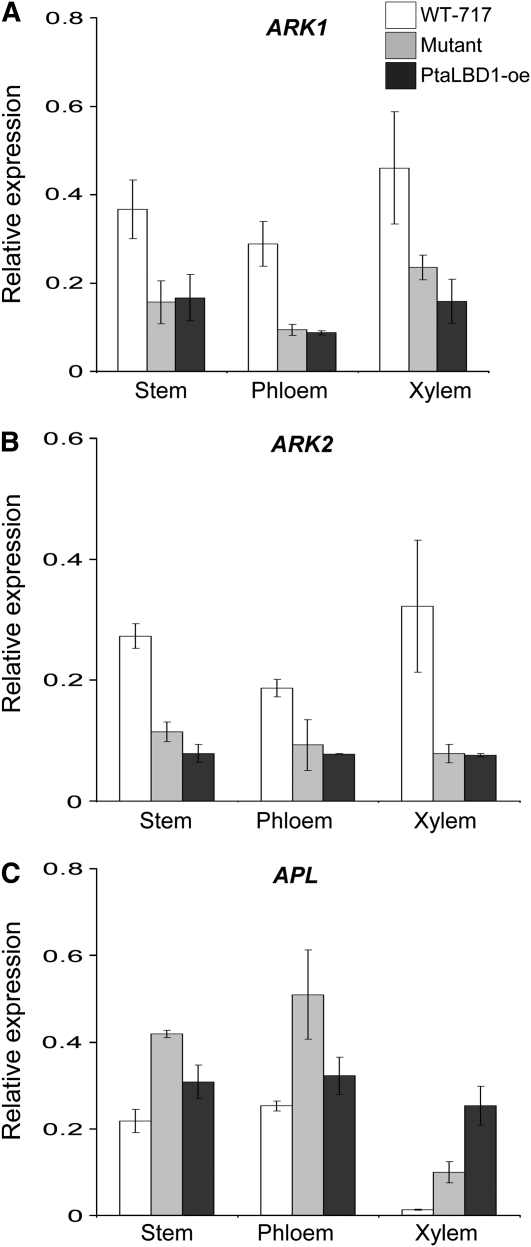
Expression of Cambium Meristem and Phloem Identity Transcription Factors in the Activation-Tagged Mutant Background
It is becoming increasingly evident that a major mechanism for LBD function in Arabidopsis SAM development is through interactions with Class I KNOTTED1-like homeobox (KNOX) transcription factors, such as STM and BREVIPEDICELLUS (BP) (Lin et al., 2003; Iwakawa et al., 2007). Recently, it has been shown that ARBORKNOX1 (ARK1) and ARK2, which are the poplar orthologs of STM and BP, respectively, play important roles as meristem identity genes in the vascular cambium of poplar (Groover et al., 2006; Du et al., 2009). We therefore investigated whether ARK1 and ARK2 expression are affected in the mutant and PtaLBD1-oe plants. Both genes were significantly downregulated to a similar extent in whole stems, phloem, and xylem tissues compared with WT-717 (Figures 8A and 8B). We thus searched the promoter regions of ARK1 and ARK2 for the presence of two cis-elements, which have been found to mediate the AS2/LBD6 repression of KNOX-I genes in Arabidopsis (Uchida et al., 2007; Guo et al., 2008; Uchida et al., 2010). Both cis-elements were present in both the ARK1 and ARK2 putative promoter regions (see Supplemental Figure 5 online).
Figure 8.
Expression of Meristem and Phloem Identity Genes Is Affected in the Activation-Tagged Mutant Plants.
(A) and (B) ARK1 (A) and ARK2 (B) are downregulated in the mutant and PtaLBD1-oe plants.
(C) The poplar ortholog of phloem identity MYB transcription factor APL is upregulated in the mutant and PtaLBD1-oe plants. Bars represent relative expression means ± se from three biological replicates. All expression estimates were normalized to the expression of a ubiquitin loading control gene.
Due to the strong evidence pointing to an involvement of Pta LBD1 in the regulation of secondary phloem development, we investigated the expression of the Populus putative ortholog of Altered Phloem Development (Pta APL), a MYB transcription factor that, in Arabidopsis, plays a role in specifying phloem identity (Bonke et al., 2003). Consistent with the role of the Arabidopsis ortholog in phloem development, Pta APL expression was found primarily in the phloem and almost completely absent in xylem of WT-717 (Figure 8C). In the activation-tagged mutant and PtaLBD1-oe plants, we found a significant increase in the expression of Pta APL in both the phloem and xylem tissue (Figure 8C).
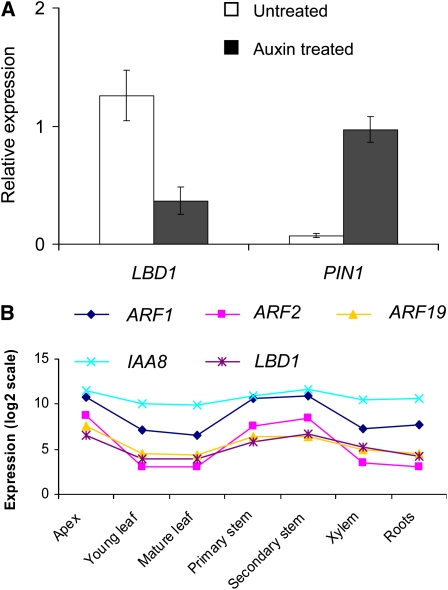
Auxin Represses Pta LBD1 Expression
PtaLBD1-oe and activation-tagged mutant poplars showed spontaneous and increased callus production compared with WT-717 plants on medium containing auxin (see Supplemental Figures 3 and 4 online). These observations suggested that PtaLBD1-oe plants may be auxin hypersensitive. We therefore studied if LBD1 expression is affected by the application of exogenous auxin. We first depleted auxin from stem sections undergoing secondary growth as previously described (Schrader et al., 2003) and then transferred the sections to auxin-containing medium (Figure 9A). Auxin caused a significant decline in LBD1 expression (Figure 9A). For a positive control, we used a poplar PIN1 homolog that is expressed in the cambium and was previously shown to be induced by auxin (Schrader et al., 2003). In contrast with the downregulation of LBD1 by auxin, PIN1 showed a clear activation after auxin application (Figure 9A) (Schrader et al., 2003). This suggests that LBD1 is negatively regulated by auxin.
Figure 9.
Pta LBD1 Is Downregulated by Auxin.
(A) LBD1 is downregulated and PIN1 upregulated by auxin in stems of WT-717 plants. Stem segments (internodes 10 to 25) were auxin depleted for 16 h, followed by incubation with (Auxin treated) and without (Untreated) 10 μM 1-naphthaleneacetic acid. See Methods for further details. Data represent relative expression (mean ± se) from three biological replicates, normalized to the expression of a ubiquitin loading control gene.
(B) Pta ARF1, ARF2, and IAA8 are coexpressed with LBD1. Expression levels (log2-transformed estimates) are based on data from microarray experiments (Pearson’s correlation, R > 0.9) (see Methods for more details).
The negative regulation of LBD1 by auxin prompted us to study if its expression coincided with the expression of known regulators of the auxin signaling cascade. Auxin response is mediated via the auxin/indole-3-acetic acid (Aux/IAA) and auxin responsive factor (ARF) signaling module (Guilfoyle and Hagen, 2001; Leyser, 2002). Auxin-mediated degradation of Aux/IAA protein allows ARF to bind to the conserved auxin response element and regulate transcription of downstream target genes (Dharmasiri and Estelle, 2004). Thus, we first searched the LBD1 putative promoter sequence and found an auxin response element 2065 bp upstream of the translation initiation codon site. Aux/IAAs and ARFs are encoded by multigene families whose expression patterns and specific interactions are thought to trigger the wealth of downstream responses and processes regulated by auxin (Guilfoyle and Hagen, 2007). We therefore studied the expression pattern of all poplar Aux/IAAs and ARFs to determine if any correlated with the expression of LBD1. Poplar homologs of Aux/IAA8, ARF1, ARF2, and ARF19 showed the most highly significant coexpression pattern with LBD1 (Figure 9B). ARFs can function as either activators or repressors of their target genes. Because LBD1 is downregulated by auxin, we investigated which ARF genes have been shown to play repressive roles. Both ARF1 and ARF2 have been previously demonstrated to be repressors (Ulmasov et al., 1999). This suggests a possible involvement of ARF1 and ARF2 in the auxin-mediated repression of Pta LBD1.
Wood-Predominant and Tissue-Specific Expression of Pta LBD Genes
One of the defining features of LBD genes is their highly specific expression pattern in the tissue/cell types that they regulate (Shuai et al., 2002; Matsumura et al., 2009). Furthermore, the different members of the LBD family show astounding functional specificities. For example, swapping the conserved LBD domain between AS2/LBD6 and the five most closely related members of the family demonstrated that none was able to complement the function of the original AS2/LBD6 (Matsumura et al., 2009). Our finding that Pta LBD1 has a regulatory role during secondary (woody) growth prompted us to investigate if other specialized LBDs were also involved in this process. We used our own and publicly available microarray data (Wilkins et al., 2009) to identify LBD members that are predominantly expressed in stems undergoing secondary growth. The GeneChip Poplar Genome Array contains probe sets that correspond to 53 of the 57 poplar LBD genes (see Supplemental Data Sets 3 and 4 online). We first applied filtering by Absence/Presence MAS5 calls and identified 34 genes/probes that are present in at least two tissue samples (see Methods for more details), indicating detectable expression beyond noise levels (see Supplemental Data Set 5 online). Analysis of variance (ANOVA) analysis of the detected genes revealed that 23 were differentially expressed in at least one of the seven tested tissue types (see Supplemental Data Set 6 online). Four LBDs, including the activation-tagged Pta LBD1, appear to be differentially upregulated (P < 0.01) (Pavlidis and Noble, 2001) in stems undergoing secondary growth (Table 1). To validate these findings, we performed expression analysis using tissues derived from WT-717 poplar plants. All four genes displayed higher transcript abundance in stems undergoing secondary growth (Figure 10A). LBD4 and LBD18 were almost exclusively expressed in secondary woody stems (Figure 10A). Additionally, LBD1 and LBD4 were mainly expressed in phloem, whereas LBD15 and LBD18 occurred in xylem tissues (Figure 10B).
Table 1.
Pta LBD Genes Predominantly Expressed in Stems Undergoing Secondary Growth
| Name | JGI Gene Model | Tissuea | AGI of Arabidopsis Putative Ortholog | Groupb | Length of the Predicted Protein (Amino Acids) |
| LBD4 | eugene3.00070807 | Phloem | AT1G31320 | Class I (Ia, Subtype B) | 146 |
| LBD1 | estExt_fgenesh4_pg.C_LG_X1964 | Phloem | AT1G07900 | Class I (Ia, Subtype B) | 219 |
| LBD18 | eugene3.00140160 | Xylem | AT2G45420 | Class I (Ia, Subtype C) | 246 |
| LBD15 | eugene3.00131258 | Xylem | AT2G40470 | Class I (Ia, Subtype B) | 223 |
AGI, Arabidopsis Genome Initiative; JGI, Joint Genome Initiative.
Based on the RT-PCR expression analysis shown in Figure 10B.
For details, see Supplemental Figure 1 online; subtypes are based on the Arabidopsis classification (Matsumura et al., 2009).
Figure 10.
Pta LBD Genes Are Predominantly Expressed in Secondary Phloem and Xylem.
(A) The predominant expression of four LBD genes (Table 1) in tissues undergoing secondary growth. PSt, stem sections derived from internodes 1 to 8; SSt, stem sections derived from internodes 10 to 20.
(B) Differential expression of the selected four LBD genes in secondary phloem and xylem.
Data in (A) and (B) represents relative expression (mean ± se) from three biological replicates normalized to the expression of a ubiquitin loading control gene.
DISCUSSION
We discovered through activation tagging an LBD gene that appears to be a positive regulator of phloem formation during secondary growth in poplar. The gene shows highest homology to LBD1 from Arabidopsis, which until now was of unknown function and was therefore named Pta LBD1. LBD genes are plant-specific transcription factors that regulate the formation of lateral vegetative and reproductive organs (Ori et al., 2000; Iwakawa et al., 2002; Shuai et al., 2002; Lin et al., 2003; Bortiri et al., 2006). Many LBD proteins are highly specialized in function and are thus expressed in defined tissue and cellular domains, typically at the boundary between the meristem and emerging lateral organ primordia (Shuai et al., 2002; Borghi et al., 2007). These highly specific expression domains are associated with the function of the respective gene members (Rast and Simon, 2008). Similarly, Pta LBD1 was expressed predominantly in stems that undergo secondary growth, and the transcript was localized in the cambial zone primarily on the phloem side (Figure 3B). Consistent with the localization experiments, LBD1 was more highly expressed in the phloem (Figure 3A), and transgenic modifications generating gain- or loss-of-function phenotypes showed predominantly phloem-related changes. Furthermore, the largest difference in phloem development resulting from LBD1 transgenic modifications was the rate of phloem production. Overexpression of LBD1 led to increased secondary phloem production, while dominant-negative transgenics showed significant suppression of phloem development. This suggests that LBD1 is a positive regulator of secondary phloem development. To our knowledge, no other genes involved in any aspect of secondary woody growth have been characterized to date by activation tagging or any other forward genetics means (Groover et al., 2010). Thus, our results show the power of the method to provide insight into the regulatory mechanisms that govern this poorly understood developmental process.
The positive regulatory role of Pta LBD1 on phloem formation could be mediated by suppression of meristem cell identity, activation of phloem differentiation, or both. We therefore tested the expression of two types of genes that are critical for these two processes. ARK1 and ARK2 are KNOX-I transcription regulators that have been shown to be essential for maintaining cambial meristem identity during secondary growth in poplar (Groover et al., 2006; Du et al., 2009). Both ARK1 and ARK2 were significantly downregulated in the activation-tagged mutant plants. Arabidopsis APL is a MYB-type transcription factor that specifies phloem-tissue identity (Bonke et al., 2003). Mutant plants showed enhanced expression of PtaAPL. Thus, it appears that activation of PtaLBD1 results in a concomitant suppression of meristem cell identity and activation of phloem differentiation. Transgenic work with ARK1 and particularly ARK2, for which gain- and loss-of function modifications are reported (Du et al., 2009), support the above conclusion. Repression of ARK2 increased, while overexpression inhibited, phloem differentiation (Du et al., 2009; Spicer and Groover, 2010), which corresponds well with the increased phloem production and ARK2 downregulation when PtaLBD1 is upregulated in transgenic plants. In summary, our data provide evidence that one of the main regulatory routes for Pta LBD1 action is through modulation of ARK1 and ARK2 expression.
In the SAM of Arabidopsis, several LBD and KNOX-1 genes have strictly delineated, and mutually exclusive, expression domains (Lin et al., 2003; Borghi et al., 2007; Rast and Simon, 2008), and the activity of LBD proteins directly or indirectly restrains the expression of the KNOX-1 genes to the SAM region (Phelps-Durr et al., 2005; Guo et al., 2008). By contrast, in the cambial region of poplar, ARK1 and ARK2 are more broadly expressed throughout the whole cambial zone, including the cambium initials (e.g., the meristem) and dividing mother cells (Groover et al., 2006; Du et al., 2009). We show here that Pta LBD1 was primarily expressed in the cambial zone and phloem. Thus, there does not appear to be mutually exclusive expression of LBD and KNOX-1 genes in the cambial region. One explanation is that, in contrast with SAM, LBD1 in the vascular cambium gradually attenuates ARK expression in the developing phloem. Such gradual attenuation would have been difficult to discern with the in situ localization techniques used to study ARK1, ARK2, and LBD1 expression. This would suggest that the transition from the cambium to differentiating tissues represents a gradual shift rather than a sharp compartmentalization. This was also suggested by the expression of a number of other genes (reviewed in Spicer and Groover, 2010). Alternatively, Pta LBD1 may modulate expression of different target gene(s) that indirectly interfere with ARK1 and ARK2 expression. Possible candidates are PINs (PIN-FORMED, auxin efflux carriers), which can be upregulated by high auxin concentrations (Schrader et al., 2003) and can self-reinforce localized auxin maxima (Zazímalová et al., 2007; Petrásek and Friml, 2009), as observed in the vascular cambium (see more discussion below). It is also possible that LBD1 directly activates APL, as suggested by the upregulation of this gene in the activation-tagged mutant plants. Absence of knowledge regarding possible regulatory interactions between these two genes in any plant species and no prior transgenic modification of APL function in poplar precludes inferences about the possible role of LBD1 in APL regulation.
The original activation-tagged mutant showed an auxin hypersensitive phenotype, and LBD1 expression was downregulated by exogenous auxin (Figure 9). Auxin concentrations in poplar and Pinus sylvestris peak at the vascular cambium and then sharply drop on the phloem side (Uggla et al., 1996; Tuominen et al., 1997). This suggests that high auxin concentrations in the cambium would prevent LBD1 expression, while lower auxin levels in the phloem would enable LBD1 expression. One possible mechanism is through interactions with the Aux/IAA and ARF regulatory module. We found putative orthologs of Aux/IAA8, ARF1, and ARF2 to be significantly coexpressed with LBD1 (Figure 9). ARF1 and ARF2 are transcription repressors in Arabidopsis (Ulmasov et al., 1999; Guilfoyle and Hagen, 2007), which would explain the downregulation of LBD1 in response to auxin treatments. IAA8 in Arabidopsis and the corresponding ortholog in Zinnia have been found to be expressed during vascular development (Groover et al., 2003), but its role in vascular differentiation is still unclear. These observations lay out a putative regulatory network that needs to be further tested by direct experimental validation.
Analysis of the activation-tagged mutant reveals that LBD1 also regulates ray formation. In the mutant and overexpressing plants, we found a higher density of rays. Furthermore, poplar species develop only uniseriate rays (Goué, et al., 2008). In the mutant and PtaLBD1-oe, we found both bi- and multiseriate ray files. These observations indicate that LBD1 is a positive regulator of both ray cells initiation and proliferation. In Populus sp, rays can comprise more than 10% of the total wood tissue volume (Mellerowicz et al., 2001), but very little is known about the regulation of their formation (van Raemdonck et al., 2005; Goué, et al., 2008). The overlapping role of LBD1 in phloem and ray cell initiation and proliferation is logical. Rays are major trafficking arteries that connect the photosynthetic assimilate stream to sink tissues, such as wood, and are also carriers of biologically active substances, like hormones, small RNAs, and proteins (van Bel, 1990). Coordination of rays and phloem proliferation may assure adequate exchange of substances between phloem, rays, and xylem.
Although the putative involvement of LBD in wood formation has thus far escaped attention, other LBD genes have been implicated in the regulation of vascular development in Arabidopsis leaves and roots (Semiarti et al., 2001; Soyano et al., 2008). One of the founding members of the family, AS2/LBD6, was shown to regulate xylem/phloem specification in leaves (Lin et al., 2003; Xu et al., 2003; Zhu et al., 2008). Plants overexpressing AS2 formed adaxialized vascular bundles with xylem surrounding the phloem tissue (Lin et al., 2003; Xu et al., 2003, 2008). The observed effect on vascular development was likely mediated via a negative feedback loop with the abaxial regulator KANADI1 (Wu et al., 2008). AS2/LBD6 was also found to regulate the level of miR165/miR166 (Ueno et al., 2007), which play an important role in xylem specification (Demura and Fukuda, 2007). In roots, two other LBD proteins were implicated in xylem tracheary element differentiation (Soyano et al., 2008). LBD18 and LBD30 were found to be expressed in immature tracheary elements (TEs). Overexpression of LBD30 and LBD18 induced cellular transdifferentiation in TE-like cells. By contrast, aberrant TEs were formed when a dominant-negative version of LBD18 was produced via fusion with the artificial repressor domain SRDX.
A defining feature of many LBD genes is their specific expression pattern at the boundary of lateral organs. For example, the LATERAL ORGAN BOUNDARIES (LOB) Arabidopsis gene was discovered via enhancer trapping by virtue of its highly specific expression in a ring at the base of all lateral organs developing from vegetative and floral meristems (Shuai et al., 2002). LOB was also expressed in a ring-like domain around the base of the emerging lateral root, at the junction with the primary root. Similarly, Pta LBD1 was found to be expressed in a ring-like pattern in the cambial zone (Figure 3). In addition to their expression specificity, LBD genes show astounding functional specificity. Swapping the conserved LOB domain between AS2/LBD6 and the five most closely related members of the family demonstrated that none was able to complement the function of the original AS2/LBD6 protein (Matsumura et al., 2009). The high functional specificity could also explain the minor pleiotropic effects observed in the PtaLBD1-oe transgenics. The leaves of these plants were only slightly smaller than those of untransformed plants, and roots developed normally. We therefore reasoned that there may be specific LBD members that regulate secondary growth and that they can be isolated based on their highly specific expression patterns. Indeed, we identified four genes (i.e., Pta LBD1, LBD4, LBD15, and LBD18) that were predominantly expressed in stems undergoing secondary (woody) growth. Two of these genes (LBD1 and LBD4) were expressed in secondary phloem and two (LBD15 and LBD18) in secondary xylem (Figure 10). To date, we have characterized only one of these genes (i.e., LBD1). Further functional analyses of all four genes in wood development will precisely delineate their functional roles and regulatory mechanisms.
From this study and the knowledge of LBD function in Arabidopsis, it appears that LBD members play an important role in radial patterning during secondary woody growth. Specific members of the family are involved in (1) differentiation of different cell/tissue types and/or (2) defining the transition between the cambium and differentiating tissues. Their function can be related to (1) regulating the expression domain and/or level of expression of KNOX-I genes involved in meristem maintenance in a manner similar to the one demonstrated in SAM. The targets of such regulation may be poplar Class I KNOX genes (ARK1 and ARK2), which have already been shown to be involved in vascular cambium maintenance (Groover et al., 2006; Du et al., 2009). (2) Their function also can be related to promoting the differentiation of secondary cell/tissue types in a manner similar to Arabidopsis LBD30 and 18 through the activation of genes, like APL, that specify tissue identity (Soyano et al., 2008).
In addition to providing fundamental insight into a poorly understood process in plants, this study reveals targets for improving the growth and physicochemical characteristics of wood, a renewable resource with growing economic and environmental importance. Several aspects of LBD transcription factor biology make them desirable targets for breeding or genetic engineering. First, in contrast with other large gene families, their high expression/functional specificity allows for precise delineation of targets. Second, the minor pleiotropic effects associated with LBD manipulations allow for specific modifications of wood characteristics. Third, economically important phenotypes associated with LBDs are typically gain-of-function dominant modifications that are relatively easy to identify and breed.
METHODS
Plant Material and Treatments
All experiments were performed in the Populus tremula × Populus alba INRA 717-IB4 genotype (WT-717). Generation of the activation tagging event was previously described (Harrison et al., 2007; Busov et al., 2010). Auxin inductive treatments were performed on stems (internodes 10 to 25) from greenhouse-grown plants after 16 h auxin depletion in half-strength Murashige and Skoog liquid medium (Schrader et al., 2003). Approximately 0.5-cm sections were floated on half-strength Murashige and Skoog solution supplemented with 10 μM 1-naphthaleneacetic acid (Sigma-Aldrich) at 23°C and gently agitated at 150 rpm for 8 h. Tissues for expression analysis were collected from three to five actively growing greenhouse plants from the same genotype (WT-717) used for the transgenic analysis and included apices, young leaves from the 2nd internode, leaves from the 8th internode, stems undergoing predominantly primary growth (internodes 1 to 8), stems undergoing secondary growth (internodes 10 to 20), and roots. Phloem was collected by peeling the stem sections from internodes 15 to 20. Exposed developing xylem was then scraped with a sharp blade (Taylor et al., 1996; Tuominen et al., 1997; Ranik et al., 2006). All sampled tissues and organs were immediately frozen in liquid nitrogen and stored at −80°C until processed.
Positioning of the Tag in the Poplar Genome Sequence
Genomic DNA flanking the insertion site was recovered via SiteFinding PCR as previously described (Tan et al., 2005). Genomic DNA was extracted using the DNeasy plant mini kit (Qiagen). The PCR mixture was as follows: 2 μL of 10× AccuTaq DNA polymerase buffer, 3 μL of mixed deoxynucleotide triphosphate solution (2 mM each of dATP, dTTP, dCTP, and dGTP), 0.5 units of AccuTaq DNA (Sigma-Aldrich), 10 pmol of SiteFinder primer 2 (5′-CACGACACGCTACTCAACACACCACCTCGCACAGCGTCCTCAAGCGGCCGCNNNNNNGCCT-3′; Tan et al., 2005), and ~100 ng of genomic DNA. A single cycle reaction was run at the following conditions: 92°C (2 min), 95°C (1 min), 25°C (1 min), ramp to 68°C over 3 min, and 68°C (10 min). Following the initial reaction, 5 μL of a reaction mixture containing 50 pmol of SFP1 primer (5′-CACGACACGCTACTCAACAC-3′; Tan et al., 2005), 10 pmol of LB1 primer (5′-AAGCCCCCATTTGGA-CGTGAATGTAGACAC-3′; Busov, et al., 2010), and 1× AccuTaq DNA polymerase buffer was added to samples on ice, the mixes were subjected to PCR, and the primary reaction was run at the following conditions: one cycle at 94°C (1 min), 30 cycles of (95°C [10 s] and 68°C [6 min]), and one cycle at 68°C (5 min). For the secondary reaction, 1 μL of the primary PCR reaction was diluted into 100 μL water, and 1 μL of the diluted products was combined with 49 μL of the secondary PCR mixture, which contained 1× AccuTaq DNA polymerase buffer, 25 μM deoxynucleotide triphosphate, 0.8 units of AccuTaq polymerase, and 0.2 μM each of LB2 (5′-TTGCTTTCGCCTATAAATACGACGGATCG-3′; Busov, et al., 2010) and SFP2 primer (5′-ACTCAACACACCACCTCGCACAGC-3′; Tan et al., 2005). For the tertiary reaction, 1 μL of the secondary PCR products was diluted into 200 μL water and processed as the secondary reaction, except that LB3 (5′-TAACGCTGCGGACATCTAC-3′; Busov et al., 2010) and SFP3 (5′-CGCACAGCGTCCTCAAGCGGCCGC-3′) primers were added. The secondary and tertiary PCR conditions were the same as the primary PCR reaction. The products from the secondary and tertiary PCR reactions were separated on 1.5% agarose gels, and the amplification specificity was evaluated by the shift in product size in the tertiary reaction compared with the secondary amplification product. Sequencing was performed directly on 1 μL of PCR reaction using the LB3 and SFP3 primers. The obtained sequence was positioned by BLASTn searches against the P. trichocarpa reference genome sequence (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). Using the genome browser, the gene prediction models closest to the insertion site were identified and further studied.
Generation of Binary Vector Constructs and Plant Transformation
The Pta LBD1 open reading frame was amplified using gene-specific primers with attB Gateway sequence attachments (PtaLBD1-B1, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATG-GATATGATTGAGCAATCTGC-3′; PtaLBD1-B2, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCATGTCCAAAGAGGTTCCCATG-3′) and AccuTaq High Fidelity polymerase (Sigma-Aldrich), following the manufacturer’s instructions. To generate the PtaLBD1-SRDX translational fusion, we used a reverse primer carrying the sequence encoding the 12 amino acids of SRDX and the attB2 recombination sites needed for the GATEWAY cloning (PtaLBD1-srdxB2, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCAAGCAAACCCTAAACGCAACTCCAAGTCTAAGTCAAGTGTCCAAAGAGGTTCCCATG-3′). The PCR products were purified from agarose gel using the QIAquick gel extraction kit (Qiagen) and inserted into pDONR221 (Invitrogen) using BP recombination reactions and following the manufacturer’s instructions (Invitrogen). All entry clones were sequence verified to guard against putative mutations being introduced by the PCR amplifications. DNA fragments were then transferred from the pDONR221 entry clone into the destination binary vector pK7WG2 (Karimi et al., 2002) using LR recombination reactions (Invitrogen). The insertions were again sequence verified and then transformed into Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991) using the freeze/thaw method (Holsters et al., 1978). Colonies growing on selection media were PCR and restriction digest verified for the presence of the binary vector. Agrobacterium-mediated transformation was performed as previously described (Han et al., 2000) into the same genetic background (i.e., clone INRA 717-IB4 P. tremula × P. alba) in which the mutation was identified. Transgenic plants were recovered and PCR verified for the presence of the transgene using the following primers: NPT_F, 5′-ATCAGGATGATCTGGACGAAGAG-3′, and NPT_R, 5′-GATACCGTAAAGCACGAGGAAG-3′.
RT-PCR Analysis
RNA was extracted as previously described (Busov et al., 2003), DNaseI treated (Qiagen), and cDNA synthesized using the RevertAid first-strand cDNA synthesis kit (Fermentas) and 1 μg of total RNA. Primers used in various experiments are shown in Supplemental Table 1 online and were designed using Primer 3 (http://frodo.wi.mit.edu/primer3). Equal amounts of cDNA were used, and 2× Green DreamTaq PCR mixture (Fermentas) was used for PCR amplification. Ubiquitin-like (UBI) gene expression was used as a loading control. The number of cycles for each gene was varied to confirm that the PCR amplification was in the linear range (see Supplemental Figure 6 online). Gel images were acquired by the UVP Gel Doc-It documentation system and quantified using ImageJ (http://rsb.info.nih.gov/ij/index.html). At least two biological and technical replicates were performed.
Microscopy and in Situ Localization
Stem sections (0.5 cm) from WT-717 and transgenic plants were sampled at the same time, immediately fixed in FAA (formaldehyde 3.7%, ethyl alcohol 50.0%, and acetic acid 5.0%), and embedded using Shandon Excelior and Histocentre 2 (Thermo Scientific). Five-micrometer-thick sections were stained with Hematoxylin-Eosin or toluidine blue O. Images were captured using a Nikon microscope coupled to a Sony 3CCD DKC-5000 camera. Measurements were made using ImageJ v1.38 (http://rsbweb.nih.gov/ij/). For in situ localization of Pta LBD1 transcript, whole-mount RT-PCR of hand-cut stem sections was used as previously described (Koltai and Bird, 2000). Sections were fixed with HISTOCHOICE MB tissue fixative (bio-WORLD) for 4 h, rinsed once in 1× PBS and once in 1× RT buffer (Fermentas). The reverse transcription was carrying out using the RevertAid first-strand cDNA synthesis kit and the PtaLBD1 reverse primer (see Supplemental Table 1 online). In-tube PCR was performed using the PtaLBD1 forward and reverse gene-specific primers (see Supplemental Table 1 online). The PCR conditions, labeling, and detection were exactly as previously described (Koltai and Bird, 2000).
Sequence and Phylogenetic Analyses
Poplar LBD1 cDNA was sequenced in both directions. Sequence homology searches and analyses were performed using the Joint Genome Initiative P. trichocarpa genome portal v. 1.1 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), Phytozome (http://www.phytozome. net/poplar.php), and the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignments were performed using the ClustalW method (Thompson et al., 1994) and phylogenetic analyses using MEGA4 (Tamura et al., 2007), using the genetic distance method with the neighbor-joining approach, and the confidences of branches of the resulting trees were statistically tested by bootstrap supporting analyses of 1000 replications. PLACE software (http://www.dna.affrc.go.jp/PLACE/) (Prestridge, 1991; Higo et al., 1999) and ClustalW were used to analyze for the presence of conserved regulatory cis-elements in the 3000 bp of putative promoter sequence upstream of the translation start site.
Microarray Data Analysis
Collection and analyses of microarray data were compliant with the MIAME standards (Brazma et al., 2001). Our own and publically available (Wilkins et al., 2009; GSE13990) microarray data were used. For each tissue, two independent biological replicates were performed. RNA was isolated as previously described using the Qiagen RNeasy plant kit (Busov et al., 2003). Prior to labeling, RNA quality was assessed using an Agilent bioanalyzer (Agilent Technologies). Total RNA was used to prepare biotinylated complementary RNA. The labeling, hybridization, and imaging procedures were performed according to Affymetrix protocols at the Center for Genomics Research and Biocomputing, Oregon State University (http://corelabs.cgrb. oregonstate.edu/affymetrix), using the Affymetrix Poplar GeneChip. Raw data were first normalized using the RMA algorithm (Bolstad et al., 2003; http://rmaexpress.bmbolstad.com/) and filtered using Present/Absent calls in the Bioconductor affy package (Liu et al., 2002; Gautier et al., 2004). Normalized and filtered data were statistically analyzed using TM4:MeV (Saeed et al., 2003; Chu et al., 2008). ANOVA (P < 0.05) adjusted by Bonferroni correction were used to isolate differentially expressed genes. For discovery of secondary stem-specific genes, the template matching algorithm was used (Pavlidis and Noble, 2001). Mean values for Pta APL (fgenesh4_pg.C_LG_VIII000710, Affymetrix probe PtpAffx.207829.1.S1_at) and Pta IRX3 (estExt_Genewise1_v1.C_LG_VI2188, Affymetrix probe Ptp.3087.1.S1_at) gene expression were used as phloem- and xylem-specific templates, respectively. To identify ARFs and Aux/IAA genes that are coexpressed with Pta LBD1, the template matching algorithm (Pearson’s correlation R > 0.9) and Pta LBD1 expression as a template (Affymetrix probe PtpAffx.20149.1.S1_a_at) were used.
Accession Numbers
The sequence flanking the left border of the activation tag insertion site corresponds to accession number HQ284166. The accession number of the Pta LBD1 cDNA sequence is HQ284165. The poplar gene models and Arabidopsis Genome Initiative numbers of the proteins and nucleotide sequences used in the comparative sequence analyses can be found in Supplemental Data Set 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of Poplar and Arabidopsis LBD Protein Families.
Supplemental Figure 2. Pta LBD1 Is Predominantly Expressed in Phloem and Cambial Tissues.
Supplemental Figure 3. Regeneration and in Vitro Development of PtaLBD1-oe Plants.
Supplemental Figure 4. Shoot Regeneration and Callus Proliferation during PtaLBD1-oe and PtaLBD1-SRDX Transformation.
Supplemental Figure 5. Localization of AS1-AS2 cis-Elements in the ARK1 and ARK2 Putative Promoter Regions.
Supplemental Figure 6. Selection of Optimal PCR Cycles for the RT-PCR Quantifications of Gene Expression.
Supplemental Table 1. Primers Used in Various RT-PCR Expression Analyses.
Supplemental Data Set 1. Text File of the Sequences Corresponding to Supplemental Figure 1.
Supplemental Data Set 2. Pairwise Distances Matrix of Populus and Arabidopsis LBD Proteins.
Supplemental Data Set 3. Gene Models Corresponding to Populus LBD Genes and AGI Numbers of Their Arabidopsis Homologs.
Supplemental Data Set 4. Normalized Expression of All LBD Genes Present on the Affymetrix Poplar GeneChip in Seven Different Tissues with Biological Replications.
Supplemental Data Set 5. Poplar LBD Genes Showing Expression Signal above Noise Level.
Supplemental Data Set 6. Differentially Expressed LBD Genes as Determined by ANOVA (Bonferoni Correction; P < 0.05).
Supplementary Material
Acknowledgments
This work was supported in part by grants from the U,S. Department of Energy (DOE), Poplar Genome Based Research for Carbon Sequestration in Terrestrial Ecosystems (DE-FG02-06ER64185 and DE-FG02-05ER64113), the Consortium for Plant Biotechnology Research (GO12026-203A), USDA Cooperative State Research, Education, and Extension Service, National Research Initiative Plant Genome (2003-04345) and USDA CSREES, Biotechnology Risk Assessment Research Grants Program (2004-35300-14687), Plant Feedstock Genomics for Bioenergy: A Joint Research Program of the USDA and the Department of Energy (2009-65504-05767), and Genome Canada. We thank Petio Kotov and Sharon Junttila from Portage Health (Houghton, MI) for help with histological analysis and Naomi Ojala (Michigan Technological University) for technical help.
References
- Baucher M., El Jaziri M., Vandeputte O. (2007). From primary to secondary growth: Origin and development of the vascular system. J. Exp. Bot. 58: 3485–3501 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bonan G.B. (2008). Forests and climate change: Forcings, feedbacks, and the climate benefits of forests. Science 320: 1444–1449 [DOI] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Borghi L., Bureau M., Simon R. (2007). Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19: 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri E., Chuck G., Vollbrecht E., Rocheford T., Martienssen R., Hake S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A., et al. (2001). Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Brunner A.M., Busov V.B., Strauss S.H. (2004). The Poplar genome sequence: Functional genomics in a keystone plant species. Trends Plant Sci. 9: 49–56 [DOI] [PubMed] [Google Scholar]
- Busov V.B., Meilan R., Pearce D.W., Ma C., Rood S.B., Strauss S.H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 132: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov V., Yordanov Y., Gou J., Meilan R., Ma C., Regan S., Strauss S. (July 8, 2010). Activation tagging is an effective gene tagging system in Populus. Tree Genet. Genomes http://dx.doi.org/10.1007/s11295-010-0317-7 [Google Scholar]
- Chaffey N., Cholewa E., Regan S., Sundberg B. (2002). Secondary xylem development in Arabidopsis: a model for wood formation. Physiol. Plant. 114: 594–600 [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior A., Franken J., Mes J.J., Marsch-Martinez N., Pereira A., Angenent G.C. (2005). ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 57: 559–575 [DOI] [PubMed] [Google Scholar]
- Chu V.T., Gottardo R., Raftery A.E., Bumgarner R.E., Yeung K.Y. (2008). MeV+R: Using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol. 9: R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T., Fukuda H. (2007). Transcriptional regulation in wood formation. Trends Plant Sci. 12: 64–70 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Estelle M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Dharmawardhana P., Brunner A.M., Strauss S.H. (March 4, 2010). Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics 11 (online), doi/10.1186/1471-2164-11-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Groover A. (2010). Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 52: 17–27 [DOI] [PubMed] [Google Scholar]
- Du J., Mansfield S.D., Groover A.T. (2009). The Populus homeobox gene ARBORKNOX2 regulates cell differentiation during secondary growth. Plant J. 60: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Evert R.F. (2006). Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function and Development, 3rd ed (Hoboken, NJ: John Wiley & Sons; ). [Google Scholar]
- Gautier L., Cope L., Bolstad B.M., Irizarry R.A. (2004). affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Goué N., Lesage-Descauses M.C., Mellerowicz E.J., Magel E., Label P., Sundberg B. (2008). Microgenomic analysis reveals cell type-specific gene expression patterns between ray and fusiform initials within the cambial meristem of Populus. New Phytol. 180: 45–56 [DOI] [PubMed] [Google Scholar]
- Groover A.T. (2005). What genes make a tree a tree? Trends Plant Sci. 10: 210–214 [DOI] [PubMed] [Google Scholar]
- Groover A.T., Mansfield S.D., DiFazio S.P., Dupper G., Fontana J.R., Millar R., Wang Y. (2006). The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 61: 917–932 [DOI] [PubMed] [Google Scholar]
- Groover A.T., Nieminen K., Helariutta Y., Mansfield S.D. (2010). Wood formation in Populus. In Genetics and Genomics of Populus, Plant Genetics and Genomics: Crops and Models, Jansson S., Rishikesh B., Andrew A., (New York: Springer; ), pp. 201–224 [Google Scholar]
- Groover A.T., Pattishall A., Jones A.M. (2003). IAA8 expression during vascular cell differentiation. Plant Mol. Biol. 51: 427–435 [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2001). Auxin response factors. J. Plant Growth Regul. 20: 281–291 [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Guo M.J., Thomas J., Collins G., Timmermans M.C.P. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K.H., Meilan R., Ma C., Strauss S.H. (2000). An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep. 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Harrison E.J., Bush M., Plett J.M., Mcphee D.P., Vitez R., O’Malley B., Sharma V., Bosnich W., Seguin A., MacKay J., Regan S. (2007). Diverse developmental mutants revealed in an activation-tagged population of poplar. Can. J. Bot. 85: 1071–1081 [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Holsters M., de Waele D., Depicker A., Messens E., van Montagu M., Schell J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Husbands A., Bell E.M., Shuai B., Smith H.M.S., Springer P.S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., Semiarti E., Machida Y., Machida C. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51: 173–184 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., Machida Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jansson S., Douglas C.J. (2007). Populus: A model system for plant biology. Annu. Rev. Plant Biol. 58: 435–458 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kirilenko A.P., Sedjo R.A. (2007). Climate change impacts on forestry. Proc. Natl. Acad. Sci. USA 104: 19697–19702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Beers E.P., Han K.H. (2006). Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol. Genet. Genomics 276: 517–531 [DOI] [PubMed] [Google Scholar]
- Koltai H., Bird D.M. (2000). High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol. 123: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo G.R., Stein P.A., Ludwig R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (NY) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Lee H.W., Kim N.Y., Lee D.J., Kim J. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. (2002). Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53: 377–398 [DOI] [PubMed] [Google Scholar]
- Li A., Zhang Y., Wu X., Tang W., Wu R., Dai Z., Liu G., Zhang H., Wu C., Chen G., Pan X. (2008). DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol. Biol. 66: 491–502 [DOI] [PubMed] [Google Scholar]
- Lin W.C., Shuai B., Springer P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.J., Wang S.F., Yu X.B., Yu J., He X.W., Zhang S.L., Shou H.X., Wu P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Liu W.M., Mei R., Di X., Ryder T.B., Hubbell E., Dee S., Webster T.A., Harrington C.A., Ho M.H., Baid J., Smeekens S.P. (2002). Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 18: 1593–1599 [DOI] [PubMed] [Google Scholar]
- Matsui K., Ohme-Takagi M. (2010). Detection of protein-protein interactions in plants using the transrepressive activity of the EAR motif repression domain. Plant J. 61: 570–578 [DOI] [PubMed] [Google Scholar]
- Matsui K., Tanaka H., Ohme-Takagi M. (2004). Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol. J. 2: 487–493 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Iwakawa H., Machida Y., Machida C. (2009). Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 58: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz E.J., Baucher M., Sundberg B., Boerjan W. (2001). Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 47: 239–274 [PubMed] [Google Scholar]
- Nieminen K.M., Kauppinen L., Helariutta Y. (2004). A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol. 135: 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Noble W.S. (2001). Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2: RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Phelps-Durr T.L., Thomas J., Vahab P., Timmermans M.C.P. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17: 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge D.S. (1991). SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput. Appl. Biosci. 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Ragauskas A.J., et al. (2006). The path forward for biofuels and biomaterials. Science 311: 484–489 [DOI] [PubMed] [Google Scholar]
- Ranik M., Creux N.M., Myburg A.A. (2006). Within-tree transcriptome profiling in wood-forming tissues of a fast-growing Eucalyptus tree. Tree Physiol. 26: 365–375 [DOI] [PubMed] [Google Scholar]
- Rast M.I., Simon R. (2008). The meristem-to-organ boundary: more than an extremity of anything. Curr. Opin. Genet. Dev. 18: 287–294 [DOI] [PubMed] [Google Scholar]
- Roach M.J., Deyholos M.K. (2007). Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol. Genet. Genomics 278: 149–165 [DOI] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Schrader J., Baba K., May S.T., Palme K., Bennett M., Bhalerao R.P., Sandberg G. (2003). Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proc. Natl. Acad. Sci. USA 100: 10096–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J., Nilsson J., Mellerowicz E., Berglund A., Nilsson P., Hertzberg M., Sandberg G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Murray J.A.H. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60: 929–946 [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Shuai B., Reynaga-Peña C.G., Springer P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A., Street N.R., Sandberg G., Gustafsson P., Jansson S. (2009). The Populus Genome Integrative Explorer (PopGenIE): A new resource for exploring the Populus genome. New Phytol. 182: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Skog K.E., Nicholson G.A. (1998). Carbon cycling through wood products: the role of wood and paper products in carbon sequestration. Forest Prod. J. 48: 75–83 [Google Scholar]
- Soyano T., Thitamadee S., Machida Y., Chua N.H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20: 3359–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer R., Groover A. (2010). Evolution of development of vascular cambia and secondary growth. New Phytol. 186: 577–592 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tan G.H., Gao Y., Shi M., Zhang X.Y., He S.P., Chen Z., An C.C. (2005). SiteFinding-PCR: A simple and efficient PCR method for chromosome walking. Nucleic Acids Res. 33: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K.C., Albrigo L.G., Chase C.D. (1996). Purification of a Zn-binding phloem protein with sequence identity to chitin-binding proteins. Plant Physiol. 110: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H., Puech L., Fink S., Sundberg B. (1997). A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 115: 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Kimura S., Koenig D., Sinha N. (2010). Coordination of leaf development via regulation of KNOX1 genes. J. Plant Res. 123: 7–14 [DOI] [PubMed] [Google Scholar]
- Uchida N., Townsley B., Chung K.H., Sinha N. (2007). Regulation of SHOOT MERISTEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proc. Natl. Acad. Sci. USA 104: 15953–15958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C., Moritz T., Sandberg G., Sundberg B. (1996). Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. USA 93: 9282–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel A.J.E. (1990). Xylem-phloem exchange via the rays: the undervalued route of transport. J. Exp. Bot. 41: 631–644 [Google Scholar]
- van Bel A.J.E. (2003). The phloem, a miracle of ingenuity. Plant Cell Environ. 26: 125–149 [Google Scholar]
- van Raemdonck D., Pesquet E., Cloquet S., Beeckman H., Boerjan W., Goffner D., El Jaziri M., Baucher M. (2005). Molecular changes associated with the setting up of secondary growth in aspen. J. Exp. Bot. 56: 2211–2227 [DOI] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N.J., Campbell M.M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lin W.C., Huang T.B., Poethig R.S., Springer P.S., Kerstetter R.A. (2008). KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Li Z., Zhu Y., Wang H., Ma H., Dong A., Huang H. (2008). Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 146: 566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A.W., Sun Y., Pi L.M., Xu Y.Q., Huang H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Yang Y., Yu X.B., Wu P. (2006). Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 39: 248–262 [DOI] [PubMed] [Google Scholar]
- Zazímalová E., Krecek P., Skůpa P., Hoyerová K., Petrásek J. (2007). Polar transport of the plant hormone auxin - the role of PIN-FORMED (PIN) proteins. Cell. Mol. Life Sci. 64: 1621–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.H., Guo A.Y., Gao G., Zhong Y.F., Xu M., Huang M., Luo J. (2007). DPTF: A database of poplar transcription factors. Bioinformatics 23: 1307–1308 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Li Z.Y., Xu B., Li H.D., Wang L.J., Dong A.W., Huang H. (2008). Subcellular localizations of AS1 and AS2 suggest their common and distinct roles in plant development. J. Integr. Plant Biol. 50: 897–905 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.