Rapid development of the male gametophyte of the water fern Marsilea vestita is posttranscriptionally regulated. This work shows that spermidine plays important roles in cell fate determination and spermatid morphogenesis by inducing the unmasking of stored mRNAs.
Abstract
Here, we show that the polyamine spermidine plays a key role as a morphogenetic determinant during spermatid development in the water fern Marsilea vestita. Spermidine levels rise first in sterile jacket cells and then increase dramatically in spermatogenous cells as the spermatids mature. RNA interference and drug treatments were employed to deplete spermidine in the gametophyte at different stages of gametogenesis. Development in spermidine-depleted gametophytes was arrested before the completion of the last round of cell divisions. In spermidine-depleted spermatogenous cells, chromatin failed to condense properly, basal body positioning was altered, and the microtubule ribbon was in disarray. When cyclohexylamine, a spermidine synthase (SPDS) inhibitor, was added at the start of spermatid differentiation, the spermatid nuclei remained round, centrin failed to localize into basal bodies, thus blocking basal body formation, and the microtubule ribbon was completely abolished. In untreated gametophytes, spermidine made in the jacket cells moves into the spermatids, where it is involved in the unmasking of stored SPDS mRNAs, leading to substantial spermidine synthesis in the spermatids. We found that treating spores directly with spermidine or other polyamines was sufficient to unmask a variety of stored mRNAs in gametophytes and arrest development. Differences in patterns of transcript distribution after these treatments suggest that specific transcripts reside in different locations in the dry spore; these differences may be linked to the timing of unmasking and translation for that mRNA during development.
INTRODUCTION
The regulation of development and cellular differentiation in multicellular eukaryotes can be triggered by a number of effectors and determinants that individually or collectively induce distinct responses in target cells. The identification and characterization of some of these determinants has provided insights on the processes involved, but many aspects of developmental regulation are obscured by complexities inherent in the organisms being studied. Because of this, we have chosen the male gametophyte of the water fern Marsilea vestita as a model system. Development of the endosporic male gametophyte of M. vestita is extremely rapid, is well defined both spatially and temporally, and is initiated by placing dry microspores into water. The microspore contains a single cell, which is a meiotic product. There are nine successive division cycles that occur at regular intervals to produce a total of 39 cells, seven of which are sterile, and 32 of which are spermatids. Each spermatid undergoes profound morphogenetic changes to differentiate into a coiled and multiciliated spermatozoid. The details of this process are well documented in the literature (Sharp, 1914; Hepler, 1976; Myles and Hepler, 1977; Klink and Wolniak, 2001). This gametophyte serves as a strikingly simple and well-ordered system for the study of mechanisms responsible for cellular morphogenesis and cell fate determination.
An important facet of spermatid development is the de novo formation of basal bodies from a particle known as a blepharoplast, which arises during the last mitotic division cycle and then differentiates to produce 140 basal bodies in each spermatid (Mizukami and Gall, 1966; Hepler, 1976). Each spermatid then forms an elaborate cytoskeleton. The anterior part of the cytoskeleton is known as a multilayered structure (MLS) and consists of a series of vanes and fins (Carothers, 1975). The top-most stratum of the MLS is the microtubule ribbon, which comprises approximately 40 cross-linked microtubules and extends along the length of the elongated and coiled nucleus (Myles and Hepler, 1977). The microtubule ribbon has long been thought to be responsible for directing the spiral elongation pattern of the cell body and the nucleus (Mizukami and Gall, 1966; Myles and Hepler, 1977). The elongation of the gamete nucleus is accompanied by the condensation of the chromatin. It has long been known that protamines replace the histones in spermatid nuclei in the liverwort Marchantia polymorpha and in M. vestita (Reynolds and Wolfe, 1978, 1984). We are interested in knowing if the extensive process of chromatin condensation underlies some of the shape change of the gamete nucleus that occurs during later stages of morphogenesis.
With regard to mechanisms that underlie cell fate, the division cycles occur at predictable times and in precise planes within the endosporic gametophyte. Since there is no cell movement, position, size, and composition define cell fate. Rapid development of the gametophyte depends mainly on large quantities of proteins and mRNAs that are stored in the dry microspore, with little or no new transcription (Hart and Wolniak, 1998, 1999; Klink and Wolniak, 2001, 2003). Thus, spatially and temporally regulated patterns of translation of stored mRNAs drive gametophyte development (Klink and Wolniak, 2001), and a key step is the release, or unmasking, of the stored transcripts. An important and unanswered question in this type of system is what cellular components trigger the unmasking of the stored mRNAs.
Spermidine is a ubiquitous polyamine (Tabor and Tabor, 1984; Kaur-Sawhney et al., 2003) that is involved in a broad range of cellular processes in plants, fungi, and animals, such as cell division (Kwak and Lee, 2002; Ackermann et al., 2003; Unal et al., 2008), rapid cell growth and differentiation (Coueé et al., 2004; Imai et al., 2004), and transcription and translation (Igarashi and Kashiwagi, 1999, 2000; Yatin, 2002; Covassin et al., 2003; Kaur-Sawhney et al., 2003; Baron and Stasolla, 2008). Intracellular levels of spermidine and other polyamines increase at specific stages of gamete development in the spermatogenous cells in a variety of animals, such as roosters (Gallus domesticus; Oliva et al., 1982), rats (Rattus norvegicus; Watts et al., 1987), sea star (Asterios forbesii; Sible et al., 1990), and humans (Homo sapiens; Quemener et al., 1992). In mice (Mus musculus; Kaipia et al., 1990), transcripts encoding proteins involved in spermidine synthesis are found in high levels in mid and late pachytene spermatocytes and in round spermatids, thus linking polyamines with early events in spermatogenesis. In rooster spermatogenesis, spermidine and other polyamines play a protective role for DNA during histone replacement by protamines (Oliva et al., 1982). Increases in the intracellular concentration of spermidine result from elevations in the enzymatic activity of spermidine synthase (SPDS). We recently isolated a SPDS cDNA from a gametophyte library, which enables us to ask whether and how the polyamine affects gametogenesis. Here, we show how changes in spermidine abundance and distribution in the gametophyte affect multiple aspects of gametophyte development and spermatid maturation through the unmasking of stored transcripts and through interactions with cytoskeletal and nuclear components in the developing spermatids.
RESULTS
We isolated a cDNA that encodes SPDS from a male gametophyte library made from M. vestita (Hart and Wolniak, 1998, 1999). This enzyme facilitates the last step in spermidine synthesis. The protein predicted to be encoded by this cDNA is aligned with other SPDSs in Supplemental Figure 1 online. At the onset of this investigation, we hypothesized that spermidine plays a role in histone replacement (Reynolds and Wolfe, 1978, 1984) and may serve as a necessary component for nuclear elongation and remodeling in the developing spermatids.
Spermidine Localization Patterns in the Developing Gametophyte
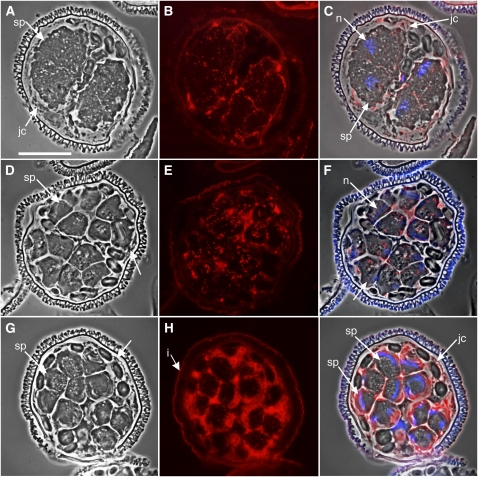
A commercially available antibody directed against spermidine was used for immunolabeling of untreated gametophytes fixed at different time points after hydration (Figure 1). Early in development, up to 4 h, spermidine was present mainly in the jacket cells and in the extracellular spaces, outside of spermatogenous cells (Figures 1A to 1C). By 6 h, low levels of spermidine were detected in the spermatogenous cells, and higher levels of spermidine were still present in the jacket cells (Figures 1D to 1F). By 8 h, the distribution was dramatically different; spermidine became abundant in the differentiating spermatids and, by comparison, was less apparent in the jacket cells (Figures 1G to 1I). Spermidine was localized in the spermatids along the anterior side of the elongated nuclei, in close proximity to the known location of the MLS and the microtubule ribbon (Myles and Hepler, 1977).
Figure 1.
Immunolocalizations in Normal Gametophytes Show That Spermidine Levels Increase Dramatically in the Spermatids after They Are Formed.
Gametophytes were allowed to develop normally for various time intervals ([A] to [C], 2 h; [D] to [F], 6 h; [G] to [I], 8 h) and then fixed, embedded in methacrylate, and sectioned. sp, spermatogenous cells; jc, jacket cells; n, nucleus. Bar = 25 μm.
(A), (D), and (G) Phase contrast images showing the morphology of the gametophytes.
(B), (E), and (H) Immunolabeling with antispermidine primary antibody and Alexa Fluor 594–conjugated secondary antibodies (red).
(C), (F), and (I) Phase contrast images (gray) were overlaid with antibody (red) and DAPI nuclear staining (blue) images to establish the relative localizations of spermidine in the gametophyte. The spore wall autofluoresces brightly (i, intine).
We were unable to obtain any antibodies directed specifically against SPDS, so we performed in situ hybridization assays on sectioned gametophytes to determine the localization pattern for SPDS transcripts (Figures 2A to 2E). In situ hybridization assays were performed because they define differences in mRNA content among gametophytes in a population, and they show whether transcripts are abundant in specific cells of a gametophyte while being absent from other cells of that same gametophyte. In a set of labeling specificity controls for the in situ assays, sense and antisense in situ hybridization probes were made and used to establish labeling patterns in untreated gametophytes, fixed after 8 h of development (see Supplemental Figure 2 online). The sense probe did not bind to cellular components. The distribution of SPDS transcripts (Figure 2) was predictive of the distribution pattern of spermidine in gametophytes fixed at different stages of development (Figure 1). SPDS mRNA was detected almost exclusively in the jacket cells until 4 h of development (Figures 2A and 2B). Between 4 and 6 h, the SPDS transcripts became detectable in the cytoplasm of spermatogenous cells initially as distinct punctae (Figure 2C, spds arrows), even in gametophytes that had been incubated in 1 to 10 μM α-amanitin, a transcriptional inhibitor, from the onset of development (Figure 2D). By 8 h, the transcripts became abundant in the cytoplasm of the spermatids, whether or not α-amanitin was added to the medium (Figure 2E). The appearance of these transcripts preceded the large increase in spermidine detectable in the spermatids in later stages of development (Figures 1H and 1I).
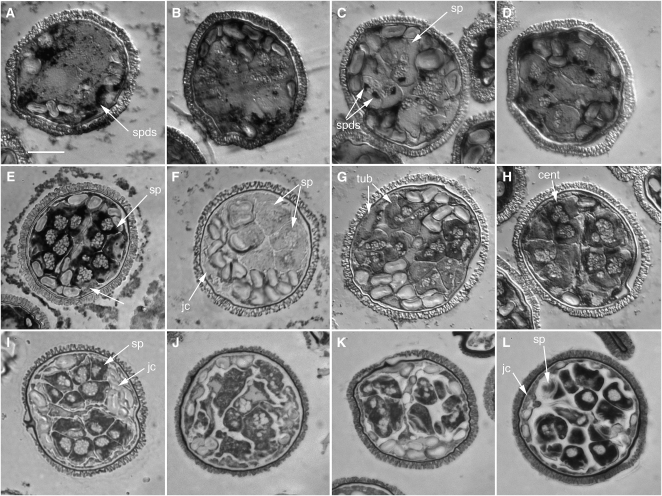
Figure 2.
SPDS in the Male Gametophyte of M. vestita.
(A) to (D) SPDS distribution and abundance in untreated gametophytes. Gametophytes were allowed to develop normally for varying time intervals ([A], 2 h; [B], 4 h; [C] and [D], 6 h; [E], 8 h), fixed in paraformaldehyde, embedded in methacrylate, and sectioned. In situ hybridization assays for SPDS mRNA were performed on semithin sections (1 to 2 μm). Images were acquired with bright-field microscopy.
(A) and (B) SPDS transcripts were detected in the jacket cells only up until 4 h of development.
(C) SPDS transcripts became detectable as distinct dots in the spermatogenous cells at 6 h (arrows).
(D) SPDS transcripts were detectable as distinct dots in the spermatogenous cells at 6 h even in gametophytes treated at the onset of development with 1 mM α-amanitin.
(E) The in situ signal for SPDS at 8 h was far more intense than at earlier time points, with the transcript abundance highest in the spermatids.
(F) to (H) SPDS silencing causes a reduction in detectable SPDS mRNA.
(F) There was no detectable SPDS mRNA label in cells fixed after 8 h of development after silencing with SPDS dsRNA.
(G) Gametophytes with silenced SPDS, fixed after 8 h of development, show intense in situ labeling in sterile and spermatogenous cells with β-tubulin probes in spite of division anomalies.
(H) Gametophytes (8 h) with silenced SPDS showing in situ labeling of centrin in sterile and spermatogenous cells in spite of division anomalies.
(I) to (L) SPDS silencing alters patterns of division in the gametophyte. Gametophytes were treated with dsRNA encoding SPDS and fixed after 8 h of development. Semithin sections (1 to 2 μm) were stained with Toluidine Blue O and observed with bright-field microscopy.
(I) Control gametophyte at 8 h. The division patterns are regular and predictable, with two clusters of intensely stained spermatids surrounded by sterile jacket cells.
(J) Severe effects of SPDS RNAi. Development was arrested at an early stage around 4 h.
(K) The majority of the treated gametophytes were arrested before the last division cycle was completed.
(L) A few of the treated gametophytes appeared to be normal with only subtle anomalies in the spermatogenous cells. Some spermatids were larger than normal cells and more elongated than control cells.
cent, centrin mRNA; jc, jacket cells; Sp, spermatogenous cells; tub, tubulin mRNA. Bar = 25 μm.
Development Is Arrested after SPDS Silencing
On the assumption that the activity of SPDS was responsible for the appearance of spermidine in the jacket cells during early development and for the large increase in spermidine in the spermatids later in development, the silencing of SPDS could reveal how the newly made copies of the enzyme (and then its product) affect gametophyte development. The SPDS gene is widely conserved in a variety of organisms and encodes a compact globular protein (Dufe et al., 2007). A number of diverse plants possess unique motifs that are attached to the 5′ end of the open reading frame that encodes the conserved enzymatic portion of the protein (Haider et al., 2005; Dufe et al., 2007). The 5′ motifs exhibit no conserved structure but are possible regulatory domains for the enzyme. The SPDS cDNA we sequenced exhibits one of these unique 5′ domains. (The amino acid sequence for the protein encoded by our cDNA is given in Supplemental Figure 1 online.) We made one long and two shorter double-stranded RNA (dsRNA) probes from our SPDS cDNA using in vitro transcription as described previously (Klink and Wolniak, 2001; van der Weele et al., 2007). Our original, long dsRNA probe was a large molecule, extending from base 47 to 1491. The SPDS open reading frame begins at base 32 of this cDNA isolate. Thus, the long dsRNA probe represents a sizable segment of the highly conserved portion of SPDS in addition to most of the unique 5′ domain. Two shorter probes (see Supplemental Figure 1 online, asterisks) were made from nonoverlapping segments of the cDNA: short probe #1 extended from base 57 to 119 and represented much of the unique 5′ domain. Short probe #2 extended from base 121 to base 162 and represented a fragment that encodes a portion of the conserved part of the protein. The longer dsRNA probe overlapped both segments of the shorter SPDS sequences. The shorter probes were made to confirm the specificity of silencing treatments that were observed in separate treatments of microspores with the long dsRNA probe. All of the dsRNA probes were introduced to the spores at the time of hydration, and the gametophytes were fixed at various intervals during development (Klink and Wolniak, 2001). No dsRNA probes were added to gametophytes that had already begun to develop because the interval when the spores are permeable to large molecules like dsRNA is brief (Klink and Wolniak, 2001; Tsai and Wolniak, 2001).
All three of the dsRNA probes elicited a similar range of phenocopies in response to SPDS silencing after 8 h of development (Figure 2; see Supplemental Figures 3 and 4 online). Each of the probes blocked gametophyte development during the cell division phase (Figure 3; see Supplemental Figure 3 online), and the timing of arrest was related to the concentration of dsRNA present in the solution at the time of spore hydration. The addition of long or short SPDS dsRNA to spores at the time of hydration resulted in a substantial reduction in SPDS mRNA staining in the gametophytes (Figure 2F; see Supplemental Figure 4 online) when compared with the untreated or α-amanitin–treated controls (Figure 2E), fixed at the same point in development. In other controls designed to test for silencing specificity, gametophytes treated to silence SPDS showed normal in situ hybridization labeling with β-tubulin (Figure 2G) or centrin (Figure 2H) mRNAs.
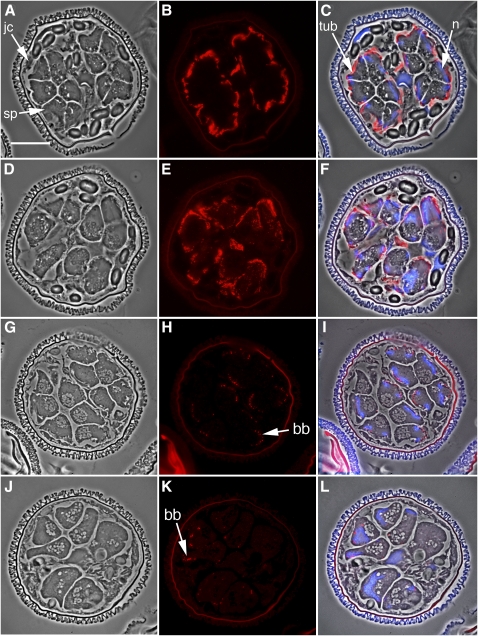
Figure 3.
The Silencing of SPDS Results in Arrested Cell Divisions, Failed Nuclear Elongation, and Disruptions in the Microtubule Ribbon and Basal Body Formation.
(A) to (F) Immunolabeling with anti-α-tubulin antibody.
(A) to (C) Untreated control gametophytes, fixed after 8 h of development.
(D) to (F) Gametophytes were treated with dsRNA encoding SPDS and fixed after 8 h of development. Gametophyte morphology was examined with phase contrast microscopy in normal (A) and RNAi-treated (D) gametophytes.
(B) and (E) The microtubule ribbon was immunolabeled with anti-α-tubulin primary antibody and an Alexa Fluor 594–conjugated secondary antibody (red).
(C) and (F) Images of phase contrast, anti-α-tubulin antibody labeling (red), and DAPI nuclear staining (blue) were overlaid to establish the relative positioning of the microtubule ribbon in the spermatids.
(G) to (L) Immunolabeling of the gametophytes with anticentrin antibody.
(G) to (I) Untreated control gametophytes, fixed after 8 h of development.
(J) to (L) Gametophytes were treated with dsRNA encoding SPDS and fixed after 8 h of development. Gametophyte morphology was examined with phase contrast microscopy in normal (G) and RNAi-treated (J) gametophytes.
(H) and (K) Immunofluorescence labeling with anticentrin primary antibodies and Alexa Fluor 594–conjugated secondary antibodies (red).
(I) and (L) Images of phase contrast, centrin immunolabeling (red), and DAPI nuclear staining (blue) were overlaid to establish the relative cellular localization of centrin in the gametophytes. The spore wall autofluoresces brightly.
bb, basal bodies; jc, jacket cells; n, nucleus; sp, spermatogenous cells. Bar = 25 μm.
The majority of the gametophytes treated to silence SPDS (>60%) were arrested after 4 to 5 h of development (Figures 2K). These gametophytes had fewer and larger spermatogenous cells when compared with controls, while the jacket cells appeared normal in both size and position. Increases in cell size and reductions in cell number occur because some of the later division cycles are slowed or blocked, and at the time of fixation, the last round of cell divisions, which produces the spermatids, has failed to occur. Since the jacket cell division cycles precede the divisions of the spermatogenous initials, and since the division planes within the gametophyte are highly predictable for all of the successive division cycles, gametophytes undergoing the arrest of later division cycles exhibit patterns of organization that are far different from gametophytes that were blocked in earlier division cycles (Klink and Wolniak, 2001; van der Weele et al., 2007). In addition to division anomalies, only a few of the spermatogenous cells in this population of gametophytes exhibited elongated nuclei. Approximately 20% of the treated gametophytes showed severe defects with developmental arrest between 2 and 4 h (Figure 2J). In this population, incomplete cell plates were sometimes present and prevented the normal partitioning of jacket cells from the spermatogenous initial cells in the gametophyte. The cells in those spermatogenous domains usually failed to divide or develop further. These gametophytes were not counted because we suspect that the inhibition is caused by excessive amounts of the dsRNA entry at the time of hydration (Klink and Wolniak, 2001; Tsai et al., 2004; van der Weele et al., 2007). Finally, fewer than 10% of the gametophytes resembled the untreated controls, though they exhibited subtle developmental anomalies (Figure 2L). In this category, gametophytes possessed the correct positioning and numbers of both spermatogenous and jacket cells, though the spermatogenous cells in this group appeared to be larger than their counterparts in the untreated gametophytes (Figure 2I). The nuclei of these cells were elongated and coiled as they are in the controls. The phenocopy with the highest percentage of occurrence (Figure 2K; >60% after this RNA interference [RNAi] treatment) and the most consistent response to the treatment was considered as the dominant effect of SPDS silencing. This phenocopy was used in the subsequent analyses performed on the samples. Immunolocalizations showed reduced antispermidine antibody labeling in gametophytes with silenced SPDS (see Supplemental Figures 5A to 5C online). Faint antispermidine antibody staining was punctate and predominantly distributed in the cytoplasmic areas of spermatogenous cells (see Supplemental Figure 5C online).
The Effect of SPDS Silencing on Spermatid Differentiation
After 8 h of normal development, anti-α-tubulin antibody staining shows microtubules arranged in one bundle on the outer (dorsal) side of the spermatogenous cells (Figures 3A to 3C). This microtubule bundle denotes the proper formation and location of the microtubule ribbon and the MLS (see Myles and Hepler, 1977). In gametophytes treated with SPDS dsRNA, anti-α-tubulin immunostaining showed microtubules (Figures 3E and 3F) that were formed in close association with the nucleus; however, they were scattered and diffuse instead of being organized in the coherent bundle observed in untreated spermatids (Figure 3C). In addition, nuclear shaping was affected in the treated gametophytes (Figure 3F). Within individual gametophytes, some spermatids exhibited elongated nuclei, while others remained large and polymorphic or spherical in shape (Figure 3F). Chromatin condensation failed to occur in the spermatids or spermatogenous cells after SPDS silencing.
In normally developing gametophytes, anticentrin antibody staining reveals the basal bodies as evenly spaced dots placed in close proximity to the nucleus on the dorsal side of the developing gametes after 8 h of development (Figures 3H and 3I) (Carothers, 1975; Myles and Hepler, 1977; Marc and Gunning, 1986; Hoffman and Vaughn, 1995). After SPDS silencing, centrin was translated and localized into basal body-like structures close to the nucleus (Figures 3K, arrow, and 3L). These basal body-like structures were sporadically positioned along the dorsal face of the cell, while in other spermatids, the basal bodies remained clustered near the center of the cell, close to the site where they were formed (Hepler, 1976).
Spermidine Levels Affect Spermatid Differentiation
Since there were pronounced changes in spermatid nuclear shaping and cytoskeletal development after the silencing of SPDS, populations of gametophytes were treated with the SPDS inhibitor cyclohexylamine (CHA) at 6 h of development, after division cycles were completed (Figure 1D). Unlike large dsRNAs, CHA and other small molecules are able to enter the cells of the gametophyte after development is well under way (Tsai and Wolniak, 2001). The addition of CHA at 6 h showed how spermidine synthesis affects spermatid differentiation, when nuclear elongation and chromatin condensation are major morphological events in gamete formation (Figure 1G), without perturbing earlier events in gametophyte development that may be dependent on the polyamine. CHA treatments at 6 h caused reductions in antispermidine antibody labeling (see Supplemental Figures 5D to 5F online): instead of diffuse staining in the cytosol, there was modest, punctate antispermidine labeling in the spermatids. It is likely that this detectable spermidine entered the spermatids from the jacket cells. The CHA treatments also altered the shape of the spermatid nuclei dramatically (Figure 4). The 4',6-diamidino-2-phenylindole (DAPI) staining of the treated gametophytes showed round or elliptical nuclei with largely uncondensed chromatin (Figures 4C and 4F), instead of the highly elongated nuclei with condensed chromatin observed in the untreated control spermatids (Figures 1C and 1I).
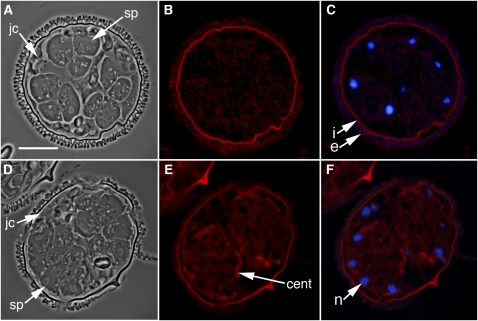
Figure 4.
The Inhibition of SPDS Enzyme by the Addition of 10 mM CHA Results in a Failure of Spermatids to Form the Microtubule Ribbon, to Make Basal Bodies, and to Undergo Nuclear Elongation with Chromatin Condensation.
(A) to (F) Gametophytes were treated with the SPDS inhibitor CHA after 6 h of normal development and then fixed after 8 h. cent, centrin; jc, jacket cells; n, nucleus; sp, spermatogenous cells. Bar = 25 μm.
(A) and (D) Phase contrast images of gametophytes, immunolabeled with anti-α-tubulin antibody ([B] and [C]) or anti-centrin antibody ([E] and [F]), respectively.
(C) and (F) Images of immunolabeling (red) and DAPI nuclear staining (blue) were overlaid to establish the relative positioning of the microtubule ribbon (B) and centrin aggregations (F). The spore wall autofluoresces brightly (i, intine; e, exine).
There was weakly detectable immunostaining with anti-α-tubulin antibody in the spermatids after the gametophytes had been treated with CHA, and this staining was uniform in the cytosol (Figure 4B); microtubule ribbons were absent in the cells. In addition, although centrin protein was translated and uniformly distributed throughout cytoplasm of the spermatids after treatment with CHA (Figure 4E), these gametophytes failed to aggregate centrin into discrete blepharoplasts. In the absence of blepharoplasts, basal bodies were undetectable in gametophytes fixed later during development (Figure 4E).
Gametophytes were treated with CHA at 6 h of development and then 10 mM spermidine was added an hour later to determine if the polyamine could offset the effects of the inhibitor on spermatid maturation (Figure 5). The spermatid nuclei (Figures 5A and 5B) remained spherical or elliptical and quite similar to the spermatid nuclei in gametophytes treated with CHA alone (Figure 4). The chromatin in the spermatogenous cells was stained intensely with DAPI, a result that suggests chromatin condensation had occurred in these nuclei (Figure 5B). Like the gametophytes treated with CHA alone (Figure 4), even after the addition of spermidine, anti-α-tubulin antibody staining was weak and diffuse (Figure 5C), indicating that the polyamine could not effectively restore cytoskeletal organization in the spermatids, at least during the time interval used in these experiments.
Figure 5.
The Addition of Spermidine to CHA-Treated Gametophytes Fails to Induce Nuclear Elongation in Spermatids of M. vestita within 60 Min.
Gametophytes were treated after 6 h of development with 10 mM CHA and an hour later with 10 mM spermidine.
(A) and (B) Round nuclei can be seen in phase contrast ([A], arrows) and with blue DAPI fluorescence (B). The polyamine did not induce nuclear elongation in the spermatids; nuclei (n) are brightly fluorescent with DAPI staining. Bar = 25 μm.
(C) Immunolabeling with anti-α-tubulin antibody showed an absence of organized microtubules in the gametophyte.
The Addition of Spermidine to Gametophytes at the Onset of Development Affects the Release of Stored mRNAs
Since a rise in spermidine concentration in the spermatogenous cells led to the apparent unmasking of SPDS mRNAs in untreated gametophytes (Figure 2C), a series of in situ hybridization assays was performed to determine if the additions of the polyamine to the gametophytes could induce the (precocious) release of stored mRNAs. As a first step, the abundance and distribution of SPDS transcripts was assayed during the first 90 min of normal gametophyte development, prior to the first mitotic division, using in situ hybridizations (Figures 6A to 6D). SPDS mRNAs are undetectable at the time the spores are placed into water (Figure 6A). After 30 min, the transcript is present in the cytoplasm near the nucleus (Figure 6B) and increases in abundance over time (Figures 6C and 6D) in the cytoplasm of the single cell of the microspore. When microspores were treated at the time of hydration with 1 mM spermidine, there was an increase in detectable SPDS mRNA in the cytoplasm that was similar to untreated microspores (Figure 6B), and development appeared to be normal, at least for the first several division cycles. With fixation at 4 h, there was a substantial accumulation of SPDS mRNA in the cytoplasm of the spermatogenous cells (Figure 6E). By contrast, the distribution of SPDS mRNA in normal gametophytes fixed at 4 h is restricted to the jacket cells (Figure 2C). The addition of 10 mM spermidine to microspores at the time of hydration resulted in an arrest of development, and by 4 h, the in situ hybridization assay for SPDS showed an intensely stained zone within the nucleus, while the cytoplasm was essentially unstained for this transcript (Figure 6F).
Figure 6.
The Addition of Polyamines to Gametophytes at the Time of Spore Hydration Results in Altered Patterns of Apparent mRNA Abundance as Detected by in Situ Hybridization Analysis.
(A) to (D) In situ hybridizations performed during early phases of development show that SPDS rapidly becomes abundant in the cytosol of the normal gametophyte.
(A) Fixation <5 min after spore hydration.
(B) Fixation at 30 min after spore hydration.
(C) Fixation 60 min after spore hydration.
(D) Fixation 90 min after spore hydration.
(E) and (F) The addition of spermidine alters the distribution and abundance of detectable SPDS mRNA in the gametophyte. In situ hybridizations were performed on gametophytes fixed at 4 h after spore hydration.
(E) The addition of 1 mM spermidine does not alter cell division frequencies, but SPDS transcripts are abundant in the cytoplasm of the antheridial initials as detected by in situ hybridization.
(F) The addition of 10 mM spermidine arrests division cycles and promotes the rapid appearance of SPDS transcripts in the nucleus of the gametophyte, where the labeling appears as an intense granule within the nucleus.
(G) The addition of 10 mM spermidine alters the pattern of apparent abundance for γ-tubulin transcripts; in situ labeling shows the mRNA to be abundant in the cytosol and present in the nucleus as an intensely staining granule. (Centrin and U620 exhibit similar patterns; see Supplemental Figures 6A and 6B online).
(H) In a pattern distinct from that of γ-tubulin, in situ analysis of PRP-19 mRNA shows that the addition of 10 mM spermidine at the time of spore hydration results in abundant PRP-19 mRNA in the cytosol and dense staining throughout the nuclei of gametophytes.
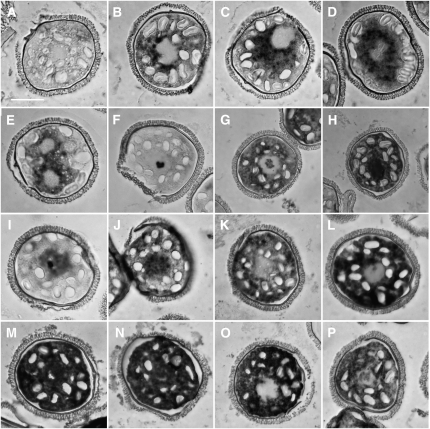
(I) to (P) The addition of different polyamines (10 mM) at the time of spore hydration causes changes in the abundance and distribution of SPDS transcripts ([I] to [L]) and PRP-19 transcripts ([M] to [P]), when analyzed by in situ hybridization after fixation at 4 h. Addition of spermidine ([I] and [M]); addition of putrecine ([J] and [N]); addition of spermine ([K] and [O]); addition of norspermidine ([L] and [P]).
Bar = 25 μm.
We next asked if the distributions of other transcripts are altered by the addition of spermidine to the gametophytes. We have previously shown that centrin mRNA becomes abundant in all cells of the gametophyte by ~4 h of development (Tsai et al., 2004) and that γ-tubulin mRNA rapidly becomes abundant after the spores are hydrated and remains abundant until ~8 h (Klink and Wolniak, 2003). Transcripts encoding U620, an RNA binding protein, are also weakly detectable within 30 min of the start of development, and like SPDS, these mRNAs become localized in the jacket cells. Later, additional U620 mRNA becomes abundant in the spermatids. PRP-19 mRNA is weakly detectable early in development but becomes localized and concentrated in the spermatogenous cells by 6 h (Tsai et al., 2004). In gametophytes treated with 10 mM spermidine and fixed at 4 h, the distribution of centrin mRNA (see Supplemental Figure 6A online) was virtually identical to that of SPDS (Figure 6F). In gametophytes treated with 10 mM spermidine at the time of hydration and fixed at 4 h, γ-tubulin mRNA was abundant in the cytoplasm and also present as a large particle in the nucleus (Figure 6G). In identically treated gametophytes, in situ labeling for U620 (see Supplemental Figure 6B online) was indistinguishable from the pattern exhibited by γ-tubulin (Figure 6G). In other gametophytes treated with 10 mM spermidine, PRP-19 mRNA was abundant in the cytoplasm, and the nuclei exhibited intense, uniform staining (Figure 6H), which is distinctive for this transcript. The elevations in transcript abundance did not result in a premature translation of the proteins they encode. Centrin immunofluorescence staining on gametophytes treated with 10 mM spermidine revealed low levels of the protein (see Supplemental Figures 6C and 6D online) at a time of development when centrin protein levels should have been high (Klink and Wolniak, 2001).
Transcript Abundance Is Affected Differently with Different Polyamines
Spermidine is a triamine and many cellular functions linked to this polyamine are related to its structure and charge (Clément et al., 1995; Geall et al., 1999). To determine whether polyamines other than spermidine can affect transcript release, putrescine, spermine, or norspermidine was added in separate experiments to microspores at the time of hydration in concentrations of 10 μm, 100 μm, 1 mM, and 10 mM. The gametophytes were fixed after 4 h of development. The majority (>90%) of the gametophytes treated with concentrations <1 mM of any of these polyamines showed normal development; transcripts encoding SPDS and PRP-19 were only weakly detectable in the cytosol within the first hour of development, and the signal became distributed in sites that would have later become the jacket cells (Figure 2B). By contrast, the vast majority of gametophytes (>90%) treated with 10 mM putrescine, spermine, or norspermidine resembled gametophytes treated with 10 mM spermidine. With 10 mM spermidine, SPDS transcripts (Figures 6F and 6I) were concentrated in the nucleus, with a prominent granule. With 10 mM putrescine (Figure 6J), SPDS transcripts showed both nuclear and cytoplasmic staining, while identical treatments using 10 mM spermine (Figure 6K) or norspermidine (Figure 6L) showed intense staining of SPDS transcripts only in the cytoplasm. A small percentage of the gametophytes (15%) showed weakly detectable nuclear staining of SPDS mRNA with spermine and norspermidine treatments. In situ hybridizations designed to detect PRP-19 mRNA showed dark nuclear and cytoplasmic staining in gametophytes treated with 10 mM spermidine (Figure 6M) or putrescine (Figure 6N) and only cytoplasmic staining in gametophytes treated with 10 mM spermine (Figure 6O). Almost 60% of the gametophytes treated with 10 mM norspermidine showed cytoplasmic staining for PRP-19 mRNA, and <50% had light staining in the nuclei (Figure 6P). Thus, the temporal and spatial pattern of transcript unmasking is dependent both on the polyamine used and on the transcript being assayed.
DISCUSSION
In this study, we show that spatial and temporal changes in spermidine concentrations in the rapidly developing gametophyte of M. vestita underlie cell fate specification, affect patterns of proliferation, and are involved in multiple aspects of spermatid maturation. Increases in spermidine concentrations follow the translation of SPDS in the jacket cells, and the accumulation of spermidine in the spermatogenous cells causes the unmasking of stored SPDS transcripts. The translation of SPDS mRNA results in the production of more spermidine in the spermatogenous cells. Clearly, spermidine affects the de novo formation of basal bodies, and it exerts profound effects on cytoskeletal elements and on the process of nuclear and cell elongation. Thus, spermidine plays multiple roles as a morphogenetic determinant in the male gametophyte of this fern. If added to the spores early in development, and at a concentration sufficient to arrest further development, the polyamine can unmask a variety of stored mRNAs, with distribution patterns that provide information on patterns of transcript localization prior to unmasking.
The silencing of SPDS mRNA prevents the early rise in spermidine concentration in the jacket cells of the gametophyte, and this results in altered division patterns. Normal increases in spermidine levels in the jacket cells coincide with a cessation of cell cycle activity, while in adjacent spermatogenous cells, with no detectable spermidine present, rapid proliferation continues. Cell cycle activity in other organisms is linked to spermidine levels, where higher concentrations of the polyamine can repress divisions (Gomurgen et al., 2004). In developing embryos of Xenopus laevis, low concentrations of spermidine in the cytosol appear to promote proliferation, whereas increasing concentrations are inhibitory to cell cycle activity (Osborne et al., 1993).
Centrin aggregation for blepharoplast, and later basal body, formation fails to occur efficiently when levels of spermidine are anomalously low as a result of SPDS silencing. The few basal bodies that formed remained clustered in the cytosol of the spermatids after SPDS silencing. Since the microtubule ribbons were anomalous (if even present) in these cells, it is likely that some of the basal body clustering observed after these treatments is a consequence of microtubule ribbon defects. Spermidine levels could directly affect the formation and elongation of the microtubule ribbon. The polyamine may alter microtubule dynamics by affecting the tubulins directly or by interacting with microtubule-associated proteins. Previous studies showed the dependence of microtubule assembly on the presence of polyamines (Pohjanpelto et al., 1981; Caruso et al., 1994; Banan et al., 1998). More recently, Needleman et al. (2004) showed that microtubules form bundles in the presence of spermidine in vitro.
Beyond the effects on basal bodies and microtubules, the silencing of SPDS resulted in a failure of the spermatid chromatin to condense. This condensation process is an essential part of nuclear remodeling in the developing motile plant gamete (Reynolds and Wolfe, 1978, 1984). Higher-order folding of chromatin requires cation-dependent charge neutralization (Pollard et al., 1999). As a small trivalent molecule, spermidine can neutralize the negative charge of condensed DNA. Our treatments of gametophytes with CHA after the spermatids had formed show that nuclear elongation is dependent on the rise in spermidine concentration. The nuclei remained round or ellipsoid in CHA-treated cells, and in the absence of nuclear elongation, the spermatid failed to undergo coiling of the cell body. The addition of spermidine after CHA treatments failed to overcome the effects of the inhibitor on nuclear elongation. In the absence of a well-organized microtubule ribbon, which is dependent on spermidine, the sudden addition of the polyamine after CHA was insufficient to promote nuclear elongation. Thus, cytoskeletal formation and elongation is essential for nuclear reshaping in the spermatid (Myles and Hepler, 1977).
Variations in Phenocopy after Silencing
Proteins involved in multiple phases of gametogenesis can produce multiple phenocopies after silencing of the transcripts that encode them (Klink and Wolniak, 2001, 2004; van der Weele et al., 2007). The changes in SPDS abundance in jacket cells and spermatogenous cells indicate that SPDS is involved at multiple stages of development, and it is not particularly surprising that SPDS silencing results in a range of responses. In our initial RNAi experiments set up to silence SPDS, we used a large fragment obtained from a gametophyte cDNA library that elicited a range of developmental anomalies in the gametophyte. To confirm that our original probe was targeting a specific mRNA, we used two additional smaller dsRNA probes that represent nonoverlapping segments of the original cDNA and encode distinct portions of the protein. There are important similarities in the outcomes of these treatments: all three of the probes produce a substantial reduction in detectable SPDS mRNA in the treated gametophytes, and all of the probes cause a similar range of anomalies in development. We cannot rule out the possibilities that our addition of a dsRNA affects multiple events in a precise developmental sequence because of variations in the concentration of dsRNA entering different spores or because of variations among spores being treated. These factors appear to contribute minimally to the response from the gametophytes because the similarities in the effects with each of the probes used to silence SPDS provide a clear indication that all three probes are acting on the same silencing target.
A Rise in Spermidine Concentration in Spermatogenous Cells Induces the Unmasking of SPDS Transcripts
In untreated gametophytes, spermidine entry into the spermatids is followed by the appearance of detectable SPDS mRNA in the spermatids (Figure 2C). This pattern of transcript appearance is also present in developing gametophytes that have been treated with 1 to 10 μM α-amanitin (Figure 2D), thereby providing a strong indication that these transcripts do not arise as a consequence of new transcription. Our earlier work suggests that the gametophyte is transcriptionally quiescent (Hart and Wolniak, 1999; Klink and Wolniak, 2003; Tsai et al., 2004), so the mRNAs that appear are masked transcripts, and they are released after the last division cycle for new translation of SPDS in the spermatids. The translation of these newly unmasked transcripts explains the rapid and dramatic rise in detectable levels of spermidine that precedes later maturation phases of the spermatids (Figure 1). In other organisms, polyamines have been implicated in the regulation of their own biosynthesis pathways (Hoyt et al., 2000). The large rise in spermidine concentration late in development can be blocked by the addition of CHA at a point when the spermatids are already present, thereby showing that the enzyme is responsible for the synthesis of the polyamine. After CHA treatment, the gametophyte exhibits arrested development with a failure of each spermatid to assemble a cytoskeleton, to elongate its nucleus, to condense its chromatin, and to develop cilia. Surprisingly, the addition of spermidine 1 h after CHA treatment could not overcome the effects of the inhibitor (Figure 5). We suspect that there are short time intervals during development when elicitors like spermidine can affect cellular components, and once that interval has passed, these cellular components fail to respond in the same way to the presence of the elicitor. The notion that chromatin condensation could serve as a force in nuclear elongation (Myles and Hepler, 1977) is likely to be partially correct, but both condensation and cytoskeletal elongation appear to be regulated by the rise in spermidine levels in the maturing spermatids. Chromatin condensation and cytoskeletal expansion work in concert to elongate the nucleus.
Spermidine and the Unmasking of Stored mRNAs
Since a rise in spermidine concentration leads to the unmasking of SPDS transcripts in the spermatids of normal gametophytes (Figures 1, 2, and 6), it was important to establish whether spermidine activity is restricted to SPDS mRNA, or alternatively, if it acts a general elicitor of transcript unmasking in the cells. The addition of 1 mM spermidine early in development is followed by normal division cycles, and SPDS unmasking occurs in the cytosol (Figure 6E). By looking at early development in gametophytes treated with higher concentrations of spermidine, a variety of stored transcripts (e.g., SPDS, PRP-19, centrin, γ-tubulin, and U620) became precociously detectable by in situ analysis. Higher spermidine concentrations were sufficient to arrest the cell division cycles, which at first glance, would be an undesirable consequence of the experimental treatment. The transcripts were detectable both in the cytosol and in the nucleus, so we conclude that they are present in both compartments early in development. High concentrations of spermidine (10 mM) cause unmasking of transcripts in their stored locations, whereas lower levels of spermidine (1 mM) allow nuclear export prior to unmasking.
To test whether the unmasking of stored transcripts was specific to spermidine additions, other polyamines were added to gametophytes and assayed for transcript abundance and distribution. Putrecine treatments resembled spermidine treatments of gametophytes for precocious unmasking of SPDS and PRP-19. The effects of spermine and norspermidine on unmasking of specific transcripts were similar to each other for SPDS and PRP-19 transcript unmasking. The differences in responses by the cells to the different polyamines are caused by differences in the affinities for each of these polyamines with RNA (Frydman et al., 1992; Lomadze et al., 2006) and perhaps by differences in binding affinity exhibited by various RNA binding proteins that are involved in masking specific species of transcripts in the cells.
Differences in spatial location of specific transcripts within the cell might determine the normal timing for unmasking and translation of these transcripts during development. We envisage early translation occurring with transcripts that are initially masked but dispersed in the cytosol of the microspore at the onset of development. Late translation occurs with mRNAs that are localized and masked within the nucleus of the microspore at the onset of development. Transcripts that are translated at multiple times or at intermediate times of development are masked and present both in the nuclear and cytoplasmic domains of the microspore at the time of hydration. Some of our future efforts will focus on these differences in spatial distribution patterns for stored transcripts, with the aim of determining whether the spatial distribution patterns influence the temporal patterns of translation for these mRNAs during rapid development of the gametophyte.
METHODS
Microspore Culture and Fixation
Marsilea vestita microspores were obtained by grinding dry sporocarps in a coffee grinder as described previously (Hepler, 1976; Klink and Wolniak, 2001; Tsai and Wolniak, 2001). Gametophytes were raised in liquid suspension culture using techniques modified from earlier studies (Hepler, 1976; Klink and Wolniak, 2001; Tsai and Wolniak, 2001). Here, cultures were started in 2-mL microcentrifuge tubes containing 4 mg of microspores/mL of commercial spring water and placed on an Orbitron shaker at 20°C, and after 5 to 10 min, individual gametophytes were separated from the spore clumps and sporangia by pipetting the liquid against the tube wall through a 1-mL pipette tip. After 1 h, spores were transferred to 50-mL flasks placed in a temperature controlled (20°C) shaking water bath as described in the earlier studies in a ratio of 4 mg of microspores/8 mL or for RNAi experiments 4 mg/10 mL water.
The gametophytes were allowed to develop for different time increments, and then fixed, dehydrated, and embedded in methacrylate, using protocols from Klink and Wolniak (2001), Tsai and Wolniak (2001), and van der Weele et al. (2007). In short, spores were filtered from the culture solution onto a polyester filter (BioDesigns Cell Microsieves) in a Buchner funnel under vacuum. The filter was rinsed with 1 mL of fixative (4% paraformaldehyde in 50 mM PIPES, pH 7.4), and the spore walls were cracked by placing the filter in a metal mortar with 50-μm-thick brass spacers using the mechanical force from one to two hammer blows onto the metal pestle (Hepler, 1976; Myles and Hepler, 1977). The gametophytes were washed from the filter into a 50-mL conical centrifuge tube with fixative solution and then placed at 4°C for 2 h. After fixation, gametophytes were rinsed, dehydrated, and embedded in methacrylate as described by Baskin et al. (1992). The gametophytes were transferred into 2-mL tubes, rinsed 3 × 15 min in 1× PBS, pH 7.4, dehydrated with 10, 25, 50, 75, and 90% ethanol for 2 h each followed by 4 × 100% ethanol for 30 min each, and then infiltrated with an ethanol-methacrylate mixture (3:1, 1:1, 1:3) for 2 h each followed by five washes of 100% methacrylate mixture of 1 h each. The gametophytes were transferred into BEEM capsules, and the samples were polymerized for 5 h in UV light at 4°C. Semithin (1 to 2 μm) sections were made using glass knives on an ultramicrotome. The sections were transferred to a microscope slide and relaxed by holding a chloroform-saturated swab in close proximity to the sections. The slides were placed on 40°C heating block to allow the sections to adhere to the glass.
RNAi Experiments
A cDNA clone encoding SPDS was isolated from our gametophyte cDNA library. The construction of our cDNA library and isolation and sequencing of the cDNA clones was described by Hart and Wolniak (1998, 1999). BLAST searches indicate that MvU185 encodes an SPDS I protein (see Supplemental Figure 1 online). Single-stranded RNA was transcribed in vitro as described by Tsai and Wolniak (2001) and van der Weele et al. (2007). To generate dsRNA, the concentration of each single-stranded RNA probe was adjusted to 1 μg/μL with RNase free water. Equal amounts of the sense and antisense RNA were added together in a microcentrifuge tube. The RNA mixture was heated to 80°C for 10 min, placed at 50°C for 5 min, followed by 37°C for 30 min. The quality of dsRNA was checked by gel electrophoresis prior to each experiment. For each RNAi treatment, one tube of 4 mg of spores was cultured with commercial spring water as control, and a second tube with 200 μg/mL dsRNA added to the commercial spring water as treatment in conditions as described above. Each of the treatments was performed at least twice with newly transcribed dsRNA and different populations of microspores. Spores were fixed, dehydrated, and embedded as described above. Semithin sections (1 to 2 μm) of the fixed gametophytes were obtained and examined with light microscopy. Since the analysis depended on the observation obtained from sectioned material, the observation of hundreds of sections was essential to construct a sense of three-dimensional representation of the spherical gametophyte so we could understand the phenocopy. The phenocopies were clustered into groups on the basis of the severity of observed effects; the percentage of each phenocopy was estimated from counts of gametophytes observed on the slides. The phenocopy with the highest percentage of occurrence, representing the majority of the affected gametophytes, was included in the analysis. Images for at least 30 different gametophytes were obtained for each treatment and assay that was performed on the treated gametophytes to ensure the accuracy of assessments of the RNAi effects. The images selected for presentation here were most representative of the effects of the different treatments and assays performed on the gametophytes. The same imaging strategy was applied for pharmacological treatments that were performed on the gametophytes.
In Situ Hybridization
Probes used for in situ hybridization were made from cDNA clones for centrin (MvU184), β-tubulin (MvU518), SPDS I (MvU185), spliceosome factor PRP-19 (MvU 89), and RNA binding protein (MvU620). A γ-tubulin transcript was isolated separately from gametophytes. In situ hybridizations were performed according to protocols described by Tsai and Wolniak (2001) and modified by van der Weele et al. (2007). In short, probes were labeled by substituting half of the dUTP with digoxigenin-11-dUTP, and semithin sections on slides were treated with acetone, proteinase K, glycine, paraformaldehyde, and triethanolamine according to procedures of Steel et al. (1998). Probes were hybridized and subsequently visualized with Nitro-blue tetrazolium and 5 bromo-4 chloro-3 indolyl-phosphate.
Cytology and Immunocytochemistry
Sections of fixed gametophytes were stained with Toluidine Blue O and viewed with bright-field microscopy (O’Brien and McCully, 1981). DAPI staining was performed as described by van der Weele et al. (2007). Immunofluorescence cytochemistry was employed to localize proteins in the gametophytes as described by Baskin and Wilson (1997) and modified from van der Weele et al. (2007) in that sections (1 to 2 μm) were etched in chloroform followed by acetone for 15 min each. Primary antibodies used were polyclonal antispermidine (1:100) (Abcam), anticentrin monoclonal antibody 20H5 directed against Chlamydomonas reinhardtii (1:100) (a kind gift from J. Salisbury, Mayo Clinic, Rochester, MN), and monoclonal anti-α-tubulin antibody (1:100) (CalBiolChem; DM1A). The secondary antibody used was an Alexa Fluor 594–conjugated goat-anti-mouse (1:1000) (Molecular Probes, Invitrogen Detection Technologies). All antibodies were diluted in PBS. Fluorescence microscopy was performed with a Zeiss Axioskop microscope equipped with a standard Texas Red filter set. Paired fluorescence and phase contrast images were made of at least 30 gametophytes from each sample.
SPDS Inhibition
Microspores were cultured using the same conditions described above except that spores were kept in 2-mL microcentrifuge tubes on the orbiting shaker during the duration of the experiment. After 6 h, CHA, an SPDS inhibitor (Unal et al., 2008) stock solution (1 M in water) was added to tubes in the appropriate volumes to bring the final concentrations to 1 μM, 10 μM, 100 μM, 1 mM, 10 mM, and 100 mM. The gametophytes were allowed to develop for two additional hours and then fixed after a total of 8 h of development and dehydrated and embedded in methacrylate for microscopy observations as described above.
Addition of Polyamines to Gametophytes
Polyamine solutions of 10 μm, 100 μm, 1 mM, 10 mM, and 100 mM were prepared in 10-mL total volumes in commercial water (Dannon). Each 1-mL polyamine solution was added to 4 mg of microspores in 2-mL microcentrifuge tubes. Gametophytes were cultured in the same way as described above except that they remained in 2-mL tubes on the orbiting shaker for the duration of the experiment. The gametophytes were fixed, dehydrated, and embedded in methacrylate mixture as described above. Thin sections (1 to 2 μm) of the fixed gametophytes were obtained and examined with a light microscope. In situ hybridization assays were performed as described above.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: β-tubulin, GI-14486017; Centrin, GI-2920834; PRP-11, GI-19569133; SPDS, HM-594947; RNA binding protein, HQ585079; and γ-tubulin, HQ585078.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of SPDSs from M. vestita, Arabidopsis thaliana, and human (Homo sapiens).
Supplemental Figure 2. In Situ Hybridization for SPDS Is Specific for Antisense SPDS Probes.
Supplemental Figure 3. Gametophytes Treated with Short dsRNA Probes Made from Segments of the SPDS cDNA Become Arrested during the Division Phase of Development.
Supplemental Figure 4. In Situ Hybridization Assays for SPDS, after SPDS Silencing with the Shortened dsRNA Probes.
Supplemental Figure 5. Spermidine Levels Are Reduced in Gametophytes after SPDS Silencing.
Supplemental Figure 6. The Addition of 10 mM Spermidine to Spores at the Time of Hydration Arrests Development and Alters Patterns of Detectable Transcripts and Proteins in Gametophytes of M. vestita.
Acknowledgments
We thank Matthew Thompson and Eliot Herman for their input and help during various phases of this project. F.D. was partially supported by a USDA Food and Agricultural Sciences National Needs Graduate and Postdoctoral Fellowship Grant (20053842015761) while this work was being performed. We gratefully acknowledge support from the Maryland Agriculture Experiment Station. We are also grateful for support for this project in the form of grants from the National Science Foundation (MCB 0720486 and DBI 0842525).
References
- Ackermann J.M., Pegg A.E., McCloskey D.E. (2003). Drugs affecting the cell cycle via actions on the polyamine metabolic pathway. Prog. Cell Cycle Res. 5: 461–468 [PubMed] [Google Scholar]
- Banan A., McCormack A., Johnson L.R. (1998). Polyamines are required for microtubule formation during gastric mucosal healing. Am. J. Physiol. Gastrointest. Liver Physiol. 274: 879–885 [DOI] [PubMed] [Google Scholar]
- Baron K., Stasolla C. (2008). The role of polyamines during in vivo and in vitro development. In Vitro Cell. Dev. Biol. Plant 44: 384–395 [Google Scholar]
- Baskin T., Busby C.H., Fowke L.C., Sammut M., Gubler F. (1992). Improvements in immunostaining samples embedded in methacrylate: Localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187: 405–413 [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Wilson J.E. (1997). Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 113: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers Z.B. (1975)Comparative studies on spermatogenesis in bryophytes. The Biology of the Male Gamete, Duckett J.G., Racey P.A., (New York: Academic Press; ), pp. 71–84 [Google Scholar]
- Caruso A., Pellati A., Bosi G., Arena N., Stabellini G. (1994). Effects of spermidine synthase inhibition on cytoskeletal organization in cultured chick embryo fibroblasts. Eur. J. Histochem. 38: 245–252 [PubMed] [Google Scholar]
- Clément S., Delcros J.G., Basu H.S., Quash G., Marton L.J., Feuerstein B.G. (1995). The structure of polyamine analogues determines haemoglobin production and cytotoxicity in murine erythroleukaemia cells. Biochem. J. 309: 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coueé I., Hummel I., Sulmon C., Gouesbet G., El Amrani A. (2004). Involvement of polyamines in root development. Plant Cell Tissue Organ Cult. 76: 1–10 [Google Scholar]
- Covassin L., Desjardins M., Soulet D., Charest-Gaudreault R., Audette M., Poulin R. (2003). Xylylated dimers of putrescine and polyamines: Influence of the polyamine backbone on spermidine transport inhibition. Bioorg. Med. Chem. Lett. 13: 3267–3271 [DOI] [PubMed] [Google Scholar]
- Dufe V.T., Qiu W., Müller I.B., Hui R., Walter R.D., Al-Karadaghi S. (2007). Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO. J. Mol. Biol. 373: 167–177 [DOI] [PubMed] [Google Scholar]
- Frydman L., Rossomando P.C., Frydman V., Fernandez C.O., Frydman B., Samejima K.S. (1992). Interactions between natural polyamines and tRNA: an 15N NMR analysis. Proc. Natl. Acad. Sci. USA 89: 9186–9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geall A.J., Eaton M.A.W., Baker T., Catterall C., Blagbrough I.S. (1999). The regiochemical distribution of positive charges along cholesterol polyamine carbamates plays significant roles in modulating DNA binding affinity and lipofection. FEBS Lett. 459: 337–342 [DOI] [PubMed] [Google Scholar]
- Gomurgen A.N., Mutlu F., Bozcuk S. (2004). Effects of polyamines (putrescine, spermidine and spermine) on root tip mitosis and chromosomes in Allium cepa L. Cytologia (Tokyo) 70: 217–224 [Google Scholar]
- Haider N., Eschbach M.-L., Dias Sde.S., Gilberger T.-W., Walter R.D., Lüersen K. (2005). The spermidine synthase of the malaria parasite Plasmodium falciparum: Molecular and biochemical characterisation of the polyamine synthesis enzyme. Mol. Biochem. Parasitol. 142: 224–236 [DOI] [PubMed] [Google Scholar]
- Hart P.E., Wolniak S.M. (1998). Spermiogenesis in Marsilea vestita: A temporal correlation between centrin expression and blepharoplast differentiation. Cell Motil. Cytoskeleton 41: 39–48 [DOI] [PubMed] [Google Scholar]
- Hart P.E., Wolniak S.M. (1999). Molecular cloning of a centrin homolog from Marsilea vestita and evidence for its translational control during spermiogenesis. Biochem. Cell Biol. 77: 101–108 [DOI] [PubMed] [Google Scholar]
- Hepler P.K. (1976). The blepharoplast of Marsilea: Its de novo formation and spindle association. J. Cell Sci. 21: 361–390 [DOI] [PubMed] [Google Scholar]
- Hoffman J.C., Vaughn K.C. (1995). Using developing spermatogenous cells of Ceratopteris to unlock the mysteries of the plant cytoskeleton. Int. J. Plant Sci. 156: 346–358 [Google Scholar]
- Hoyt M.A., Broun M., Davis R.H. (2000). Polyamine regulation of ornithine decarboxylase synthesis in Neurospora crassa. Mol. Cell. Biol. 20: 2760–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K. (1999). Polyamine transport in bacteria and yeast. Biochem. J. 344: 633–642 [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K. (2000). Polyamines: Mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271: 559–564 [DOI] [PubMed] [Google Scholar]
- Imai A., et al. (2004). Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol. 135: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaipia A., Toppari J., Mali P., Kangasniemi M., Alcivar A.A., Hecht N.B., Parvinen M. (1990). Stage- and cell-specific expression of the ornithine decarboxylase gene during rat and mouse spermatogenesis. Mol. Cell. Endocrinol. 73: 45–52 [DOI] [PubMed] [Google Scholar]
- Kaur-Sawhney R., Tiburcio A.F., Atlabells T., Galston A.W. (2003). Polyamines in plants: An overview. J. Cell. Mol. Biol. 2: 1–12 [Google Scholar]
- Klink V.P., Wolniak S.M. (2001). Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol. Biol. Cell 12: 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink V.P., Wolniak S.M. (2003). Changes in the abundance and distribution of conserved centrosomal, cytoskeletal and ciliary proteins during spermiogenesis in Marsilea vestita. Cell Motil. Cytoskeleton 56: 57–73 [DOI] [PubMed] [Google Scholar]
- Kwak S.-H., Lee S.H. (2002). The transcript-level-independent activation of ornithine decarboxylase in suspension-cultured BY2 cells entering the cell cycle. Plant Cell Physiol. 43: 1165–1170 [DOI] [PubMed] [Google Scholar]
- Lomadze N., Schneider H.-J., Albelda M.T., García-España E., Verdejo B. (2006). Dramatic selectivity differences in the association of DNA and RNA models with new ethylene- and propylene diamine derivatives and their copper complexes. Org. Biomol. Chem. 4: 1755–1759 [DOI] [PubMed] [Google Scholar]
- Marc J., Gunning B.E.S. (1986). Immunofluorescent localization of cytoskeletal tubulin and actin during spermatogenesis in Pteridium aquilinum (L.) Kuhn. Protoplasma 134: 163–177 [Google Scholar]
- Mizukami I., Gall J. (1966). Centriole replication. II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J. Cell Biol. 29: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles D.G., Hepler P.K. (1977). Spermiogenesis in the fern Marsilea microtubules, nuclear shaping, and cytomorphogenesis. J. Cell Sci. 23: 57–83 [DOI] [PubMed] [Google Scholar]
- Needleman D.J., Ojeda-Lopez M.A., Raviv U., Miller H.P., Wilson L., Safinya C.R. (2004). Higher-order assembly of microtubules by counterions: From hexagonal bundles to living necklaces. Proc. Natl. Acad. Sci. USA 101: 16099–16103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T.P., McCully M.E. 1981. The Study of Plant Structure. Principles and Selected Methods. (Melbourne, Australia: Termarcarphi Pty. Ltd.). [Google Scholar]
- Oliva R., Vidal S., Mezquita C. (1982). Cellular content and biosynthesis of polyamines during rooster spermatogenesis. Biochem. J. 208: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne H.B., Cormier P., Lorillon O., Maniey D., Bellé R. (1993). An appraisal of the developmental importance of polyamine changes in early Xenopus embryos. Int. J. Dev. Biol. 37: 615–618 [PubMed] [Google Scholar]
- Pohjanpelto P., Virtanen I., Hölttä E. (1981). Polyamine starvation causes disappearance of actin filaments and microtubules in polyamine-auxotrophic CHO cells. Nature 293: 475–477 [DOI] [PubMed] [Google Scholar]
- Pollard K.J., Samuels M.L., Crowley K.A., Hansen J.C., Peterson C.L. (1999). Functional interaction between GCN5 and polyamines: A new role for core histone acetylation. EMBO J. 18: 5622–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemener V., Blanchard Y., Lescoat D., Havouis R., Moulinoux J.P. (1992). Depletion in nuclear spermine during human spermatogenesis, a natural process of cell differentiation. Am. J. Physiol. Cell Physiol. 263: 343–347 [DOI] [PubMed] [Google Scholar]
- Reynolds W.F., Wolfe S.L. (1978). Changes in basic proteins during sperm maturation in a plant, Marchantia polymorpha. Exp. Cell Res. 116: 269–273 [DOI] [PubMed] [Google Scholar]
- Reynolds W.F., Wolfe S.L. (1984). Protamines in plant sperm. Exp. Cell Res. 152: 443–448 [DOI] [PubMed] [Google Scholar]
- Sharp L.W. (1914). Spermatogenesis in Marsilia. Bot. Gaz. 58: 419–432 [Google Scholar]
- Sible J.C., Marsh A.G., Walker C.W. (1990). Effect of extrinsic polyamines on post-spawning testes of the sea star, Asterias vulgaris (Echinodermata): Implications for the seasonal regulation of spermatogenesis. Invertebr. Reprod. Dev. 19: 257–264 [Google Scholar]
- Steel J.H., Gordon L., Polak J.M. (1998). Principles and applications of complementary RNA probes. In Situ Hybridization: Principles and Practice, Polak J., McGee J.O.D., (Oxford, UK: Oxford University Press; ), pp. 49–69 [Google Scholar]
- Tabor C.W., Tabor H. (1984). Polyamines. Annu. Rev. Biochem. 53: 749–790 [DOI] [PubMed] [Google Scholar]
- Tsai C.W., Van Der Weele C.M., Wolniak S.M. (2004). Differential segregation and modification of mRNA during spermiogenesis in Marsilea vestita. Dev. Biol. 269: 319–330 [DOI] [PubMed] [Google Scholar]
- Tsai C.W., Wolniak S.M. (2001). Cell cycle arrest allows centrin translation but not basal body formation during spermiogenesis in Marsilea. J. Cell Sci. 114: 4265–4272 [DOI] [PubMed] [Google Scholar]
- Unal M., Palavan-Unsal N., Tufekci M.A. (2008). Effects of polyamines and polyamine biosynthetic inhibitors on mitotic activity of Allium cepa root tips. Acta Biol. Hung. 59: 93–102 [DOI] [PubMed] [Google Scholar]
- van der Weele C.M., Tsai C.W., Wolniak S.M. (2007). Mago nashi is essential for spermatogenesis in Marsilea. Mol. Biol. Cell 18: 3711–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts S.A., Lee K.J., Hines G.A., Walker C.W. (1987). Determination of polyamines in digestive and reproductive tissues of adult Asterias vulgaris (Echinodermata: Asteroidea). Comp. Biochem. Physiol. B 88: 309–312 [DOI] [PubMed] [Google Scholar]
- Yatin M. (2002). Polyamines in living organisms. J. Cell Mol. Biol. 1: 57–67 [Google Scholar]