This work shows that transcription factor ABSCISIC ACID INSENSITIVE4 (ABI4) is expressed in roots and acts to repress lateral root (LR) development. ABI4 is induced by abscisic acid and cytokinin, which inhibit LR development, and is repressed by auxin, which promotes LR development. ABI4 decreases the expression of the auxin-efflux carrier PIN1 and inhibits polar auxin transport.
Abstract
Key steps in a plant’s development and adaptation to the environment are the initiation and development of lateral roots (LRs). LR development is regulated by auxin, the major plant hormone promoting LR formation, its counteracting hormones cytokinin, and abscisic acid (ABA). Here, we show that mutating ABSCISIC ACID INSENSITIVE4 (ABI4), which encodes an ABA-regulated AP2 domain transcription factor, results in an increased number of LRs. We show that ABI4 is expressed in roots and that its overexpression impairs LR development. Root expression of ABI4 is enhanced by ABA, and cytokinin and is repressed by auxin. Using hormone response promoters, we show that ABI4 also affects auxin and cytokinin profiles in the root. Furthermore, LR development in abi4 mutants is not altered or inhibited by cytokinin or ABA. Expression of the auxin-efflux carrier protein PIN1 is reduced in ABI4 overexpressors, enhanced in abi4 mutants, and is less sensitive to inhibition by cytokinin and ABA in abi4 mutants than in wild-type plants. Transport levels of exogenously applied auxin were elevated in abi4 mutants and reduced in ABI4 overexpressors. We therefore suggest that ABI4 mediates ABA and cytokinin inhibition of LR formation via reduction of polar auxin transport and that the resulting decrease in root auxin leads to a reduction in LR development.
INTRODUCTION
Plant development is unique in its ability to form new organs throughout the life of the plant. These newly developed organs include lateral organs, such as roots and shoots, as well as reproductive organs. The root system of many dicot plants has a primary root (PR) that branches to yield lateral roots (LRs), which can then undergo further orders of branching (Fukaki et al., 2007; Osmont et al., 2007; Nibau et al., 2008 and references therein). In Arabidopsis thaliana, LRs are developed in a sequence of events initiating from pericycle founder cells located opposite the xylem poles. Although pericycle cells form continuous lines adjacent to the stele, only a limited number of them become founders of LRs.
Auxin has been shown to be the key hormone in LR development (Aloni et al., 2006; Fukaki et al., 2007; Osmont et al., 2007; Nibau et al., 2008). In roots, polar auxin transport is achieved by polar localization of PIN proteins, which transport auxin. Auxin is transported toward the root tip via the stele, laterally at the base of the root tip, and backward into the cortex and epidermis. Spatial oscillation in auxin levels leads to regular spacing of LRs (De Smet et al., 2007). The polarity of PIN auxin-efflux regulatory proteins changes from anticlinal to periclinal cells during the establishment of new LR primordia (LRPs) (Benková et al., 2003). LR spacing is also influenced by root bending (Lucas et al., 2008).
The synthetic auxin-responsive promoter DR5 (Ulmasov et al., 1997) is widely used for monitoring auxin levels in planta: the auxin-responsive reporter DR5:β-glucuronidase (GUS) was used to confirm the central role of auxin during LRP development (Benková et al., 2003; Dubrovsky et al., 2008). Moreover, a large number of LR mutants have been studied (summarized in Péret et al., 2009), most of which are impaired in auxin homeostasis, signaling, and transport. In addition, most are affected at the initiation stage, whereas only a few show altered emergence or patterning.
Although auxin is the main hormone in LR development, other hormones can also influence this process (reviewed in Fukaki and Tasaka, 2009). Cytokinin is considered to act antagonistically to auxin in the development of LRs (Aloni et al., 2006; Fukaki and Tasaka, 2009). Exogenous cytokinin treatment inhibits the initiation of LRPs in pericycle founder cells (Li et al., 2006; Laplaze et al., 2007). Accordingly, mutants with reduced cytokinin levels (Werner et al., 2003), or mutated in cytokinin perception or signaling pathways (Mason et al., 2005; Riefler et al., 2006), have increased numbers of LRs.
LR formation can also be affected by the availability of macronutrients. Thus, LR density and elongation are affected by nitrogen availability: limiting nitrogen conditions enhance LR elongation, whereas high external nitrogen reduces elongation of PRs and LRs (Linkohr et al., 2002). High levels of phosphorus decrease both LR density and elongation, whereas the length of the PR is enhanced (Linkohr et al., 2002); conversely, the number of LRs is dramatically increased under severe phosphorus deficiency, and growth of the PR is inhibited (López-Bucio et al., 2002). Limiting amounts of iron and sulfate also affect LR development (Moog et al., 1995; López-Bucio et al., 2003).
Although abscisic acid (ABA) is considered the universal plant stress hormone (Verslues and Zhu, 2005; Wasilewska et al., 2008), it can also take part in non-stress-related regulatory processes (for a recent review, see Wasilewska et al., 2008). The role of ABA in root branching has recently been recognized (reviewed in De Smet et al., 2006; Wasilewska et al., 2008), and there is some evidence for ABA involvement in the repression of LR initiation and emergence. Upon prolonged water stress, Arabidopsis LRs develop as short roots, which are characterized by stubby tuberized structures (Vartanian et al., 1994). These specialized LRs enter a dormant mode and resume growth upon rehydration. This process is severely affected in ABA-insensitive mutants such as abi1-1, suggesting that ABA is involved in the signaling of LR development.
Arabidopsis ABI3 (Brady et al., 2003) and its maize (Zea mays) ortholog VP1 encode transcription factors that have been shown to affect root architecture and to mediate ABA/auxin interactions. Mutations in ENHANCED RESPONSE TO ABA1 (ERA1) resulted in increased numbers of LRs. Similar to ABA, high nitrate in the growth medium also inhibited root growth in Arabidopsis (Signora et al., 2001). The inhibitory effect of nitrate was significantly reduced in abi4 and abi5 mutants, further supporting the involvement of ABA in LR signaling. Further development of LRPs is arrested in osmotically stressed Arabidopsis plants (Deak and Malamy, 2005), a regulatory mechanism that is dependent upon ABA and LATERAL ROOT DEVELOPMENT2. These two agents have been shown to modulate root architecture, even in the absence of osmotic stress, suggesting that ABA may be a regulator of intrinsic root system development (Deak and Malamy, 2005).
In Arabidopsis, ABA can reversibly arrest LR growth by inhibiting the expression of cell cycle genes necessary for meristem activity (De Smet et al., 2003). ABI8, a plant-specific protein, is involved in signaling that also affects LR formation. In abi8 mutants, LRP formation is inhibited and the LR meristem loses division competence (Brocard-Gifford et al., 2004). The checkpoint between meristem activation and emergence of LRs can be used to modulate root architecture via ABA signaling (Signora et al., 2001; De Smet et al., 2006).
In this study, we demonstrate that abi4 mutants develop increased numbers of LRs. In addition, ABI4-overexpressing plants have a reduced number of LRs. We also show that ABI4 plays a role in mediating ABA and cytokinin repression of LR development. It also appears to affect the distributions and levels of auxin and cytokinin in the roots. ABI4 is expressed in phloem companion cells, and its expression reduces the level of the auxin-efflux carrier PIN1. ABI4 is therefore a key player in coordinating the effects on LR development determined by the balance between counteracting plant hormones.
RESULTS
abi4 Mutants Exhibit Enhanced LR Density
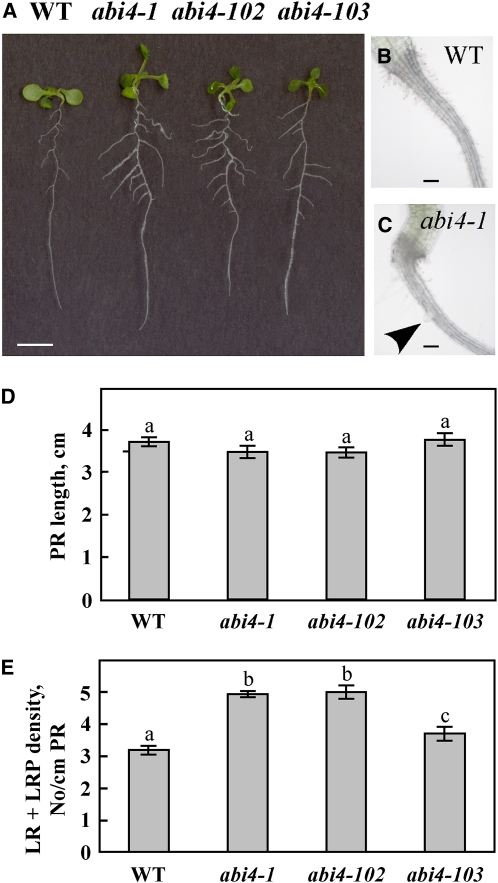
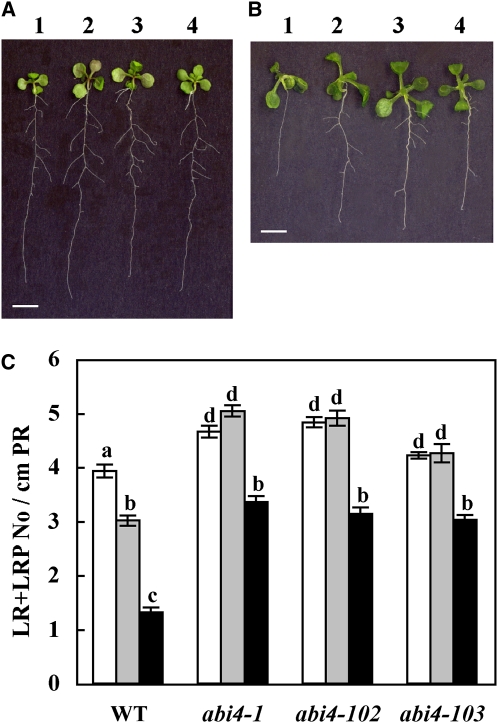
ABI4 is highly studied in germinating seedlings, and abi4 mutants have been shown to have a wild-type-like phenotype under normal growth conditions, with no glucose-induced arrest of cotyledon expansion and greening (Söderman et al., 2000; Arroyo et al., 2003; Penfield et al., 2006). Looking at Arabidopsis plants at later developmental stages, we noted that the root architecture of abi4 mutant seedlings differs from that of wild-type plants (Figure 1A). Whereas the length of the PR in seedlings mutated in three abi4 alleles (abi4-1, a deletion mutant in codon 157 [Finkelstein et al., 1998], and abi4-102 and abi4-103, nonsense point mutations at codons 80 and 39, respectively [Laby et al., 2000]) was similar to that in the wild-type plants, all three mutant lines had 125 to 135% more LRs than wild-type plants grown under the same conditions (Figure 1D). In the abi4 mutants, the first LR emerges as early as 4 d after germination, whereas in the wild type, it emerges on day 5 or 6 (Figures 1B and 1C). Moreover, LRs of abi4 mutants were longer than those of the wild-type plants (Figure 1A). This increased LR phenotype suggested a role for ABI4 in LR development at later developmental stages as well.
Figure 1.
LR Development Is Affected by Expression of ABI4: abi4 Mutants Develop More LRs.
(A) Twelve-day-old seedlings of wild-type (WT) and the specified abi4 mutants grown on solid 0.5× MS medium. Bar = 5 mm.
(B) and (C) Roots of 4-d-old wild-type and abi4-1 mutant seedlings, respectively. Arrowhead indicates emerging LR. Bars = 100 μm.
(D) Average length of PRs in 12-d-old seedlings. The data represent the mean ± se of n = 3 independent experiments each containing 50 plants per treatment. Bars with different letters represent statistically different values by Tukey’s HSD post-hoc test (P < 0.05).
(E) Density of LRs at all developmental stages (including LRP) in 12-d-old seedlings. Statistical analysis performed as in (D).
[See online article for color version of this figure.]
ABI4 Is Expressed in Roots
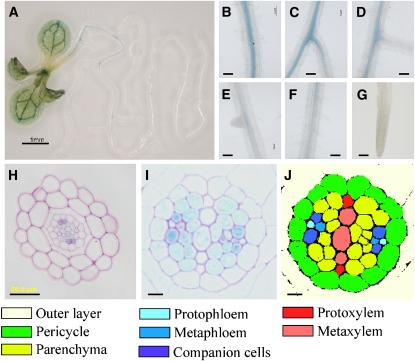
To determine whether ABI4 is indeed expressed at later developmental stages, we cloned a 2-kb promoter DNA fragment upstream of the translation start codon. This promoter sequence was subcloned into pCAMBIA 1391Z, the resulting ABI4:GUS construct was introduced into wild-type Arabidopsis plants, and the activity of the GUS reporter gene was detected histologically. Staining of germinating seedlings in the presence and absence of 7% (w/v) glucose confirmed the previously shown patterns in which ABI4 is highly expressed in cotelydons and hypocotyls of germinating seedlings (see Supplemental Figure 1 online) (Arroyo et al., 2003). As shown previously, ABI4 is also expressed in flowers and fruits (see Supplemental Figure 1 online). GUS staining was also detected in the vascular system of rosette leaves and in the petiole base and roots of Arabidopsis seedlings that were several weeks old (Figure 2; see Supplemental Figure 1 online).
Figure 2.
ABI4 Expression in Roots.
(A) to (J) Expression patterns in roots.
(A) Two-week-old Arabidopsis seedlings expressing ABI4:GUS stained histologically for GUS activity.
(B) to (G) Enlarged views of different developmental stages of the PRs and LRs (from top to bottom regions).
(B) Mature PR near the shoot-root junction.
(C) Mature LR.
(D) Newly developed LR.
(E) Emerging LR.
(F) Root hair zone. Note the expression gradient within this zone.
(G) Root tip and meristematic zone.
(H) Section of GUS-stained mid-upper zone (8 mm from the shoot-root junction) of PR counterstained with ruthenium red.
(I) Cross section of PR in the upper zone (3 mm from the shoot-root junction), treated as in (H).
(J) Colored scheme indicating the different cell types in the section depicted in (I).
Bars = 1 mm in (A), 50 μm in (B) to (H), and 10 μm in (I) and (J).
Relative quantification by quantitative RT-PCR (qRT-PCR) analysis showed that expression in the roots was almost 7 times higher than in the leaves (see Supplemental Figure 2 online). High expression levels were observed in mature regions of the PRs and LRs (Figures 2B and 2C) but not in primordia (LRPs) or young LRs (Figures 2D and 2E). Expression decreased toward the younger regions of the root. An expression gradient was also observed in the root hair zone, with low promoter activity in the upper part of the zone and no expression in the lower (younger) part (Figure 2F). No GUS staining was detected in the elongation or meristematic zones, including the root tips (Figure 2G), thereby correlating ABI4 expression levels with the root zones in which LRs develop. Root cross sections showed that ABI4 is expressed in companion cells and to a lesser extent in the metaphloem and stele parenchyma (Figures 2H to 2J). Furthermore, the extent of expression in the stele parenchyma increased with root development toward the upper parts of the root (Figures 2H and 2I).
ABI4 Expression Is Modulated by Auxin, Cytokinin, and ABA
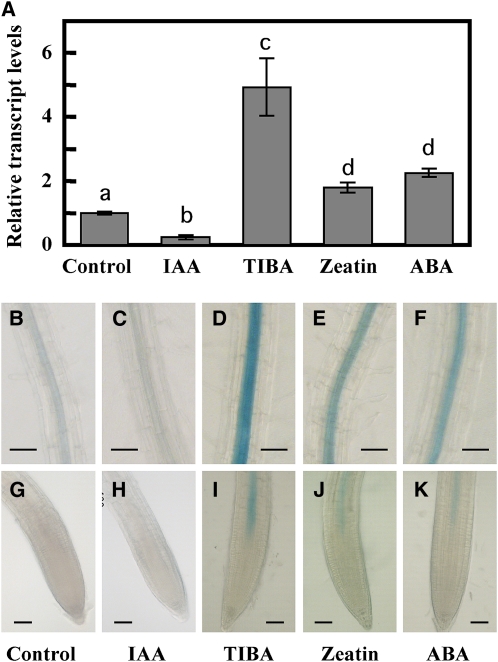
To determine whether ABI4 expression in the root system is affected by auxin and its antagonistic plant hormones, cytokinin and ABA, we used plants expressing the ABI4:GUS construct and explored the effects of these three hormones on steady state levels of ABI4 in wild-type roots using qRT-PCR analyses (Figure 3). Auxin plays a major role in promoting LR formation (Aloni et al., 2006; Fukaki et al., 2007; Osmont et al., 2007; Nibau et al., 2008). Treatment with the auxin indole-3-acetic acid (IAA) resulted in a 75% decrease in the level of ABI4 transcript in the roots. On the other hand, application of the auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA) resulted in increased levels of ABI4 transcript. LR development is known to be adversely affected by the plant hormones cytokinin and ABA (Bottger, 1974; Wightman et al., 1980), and transcript levels of ABI4 were elevated 1.8-fold by cytokinin (zeatin) treatment and 2.3-fold by ABA (Figure 3A).
Figure 3.
ABI4 Expression Is Hormone Regulated.
(A) qRT-PCR analysis of ABI4 transcripts in 2-week-old wild-type plants treated for 4 h with 20 μM of the specified agents. The data represent the mean ± se of n = 3 independent experiments. Bars with different letters represent statistically different values using Tukey’s HSD post-hoc test (P ≤ 0.05).
(B) to (K) GUS staining of roots of 2-week-old ABI4:GUS/wild-type plants treated as in (A).
(B) to (F) Upper root zone.
(G) to (K) Root tips. Bars = 50 μm.
Expression data from qRT-PCR analyses were further supported by histochemical GUS staining of ABI4:GUS Arabidopsis plants, treated with the same effectors (Figures 3B to 3K). Zeatin and ABA increased GUS staining in the upper root (Figures 3E and 3F). Furthermore, treatment with the plant hormones ABA and zeatin or the inhibitor TIBA also resulted in expansion of the ABI4 expression zone into the elongation zone of the root (Figures 3I to 3K), a region where no staining is detected in nontreated plants (Figures 2 and 3G). Thus, ABI4 expression was induced by ABA and cytokinin, plant hormones that inhibit LR formation, and repressed by auxin, the LR-promoting plant hormone. These results are in accordance with the increased number of LRs in the abi4 mutants (Figure 1).
Overexpression of ABI4 Decreases LR Development
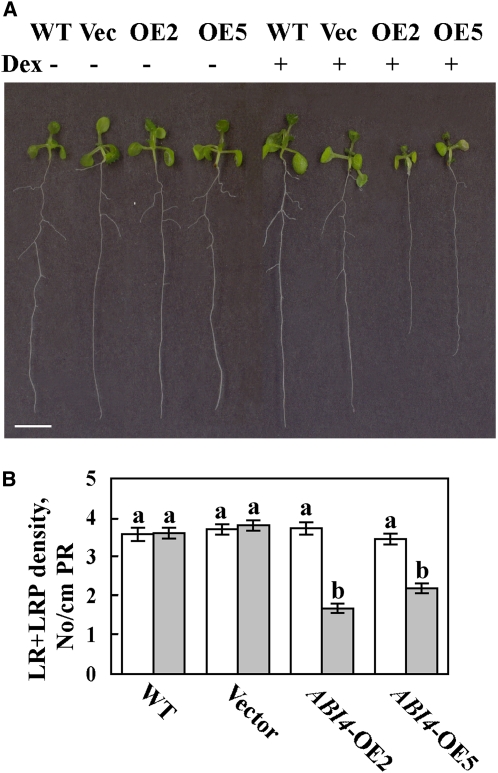
We were not able to obtain stable lines of transgenic plants constitutively expressing the ABI4 gene using 35S:ABI4 constructs. Although seeds from these transgenic lines germinated, the cotyledons senesced and died as soon as they emerged. We therefore used the inducible pOp6/LhGR expression system, which can be activated by the addition of dexamethasone (Dex) (Craft et al., 2005). When germinated on Dex-containing medium, the development of plants transformed with the pOp6:ABI4 construct in the pV-TOP vector was arrested, and the seedlings usually died at ~3 weeks of age. In the absence of Dex, the root architecture of ABI4-overexpressing plants was indistinguishable from that of wild-type plants (Figures 4A and 4B). ABI4 mRNA levels in the roots of Dex-treated ABI4-overexpressing plants were ~20,000 times the endogenous ABI4 levels expressed in the wild type and in uninduced ABI4-overexpressing plants (see Supplemental Figure 3 online). When germinated in the presence of 2 μM Dex, ABI4-overexpressing plants developed <50% of the LRPs and LRs developed in wild-type plants or plants transformed with empty vector (Figures 4A and 4B). The decrease in LRPs and LRs was similar, indicating that ABI4 inhibited LR initiation rather than subsequent LR development. The inhibitory effect of ABI4 overexpression on LR formation (Figures 4A and 4B) was in agreement with the increased number of LRs observed in the abi4 mutants (Figures 1A and 1D).
Figure 4.
Root Structure of ABI4-Overexpressing Arabidopsis.
(A) Twelve-day-old seedlings of wild-type plants (WT) , 4C-S5 plants transformed with empty vector (Vec), or two lines of ABI4 overexpressors (OE2 and OE5) germinated and grown on 0.5× MS plates with (+) or without (−) 2 μM Dex. Bar = 5 mm.
(B) Density of PRs and LRs from the LRP stage of 12-d-old seedlings. The data represent the mean ± se of n = 3 independent experiments each containing 50 plants per treatment. White bars, −Dex; gray bars, +Dex. Bars with different letters represent statistically different values by Tukey’s HSD post-hoc test (P < 0.05).
[See online article for color version of this figure.]
ABI4 Affects Formation of LRP and Elongation of Emerged LRs
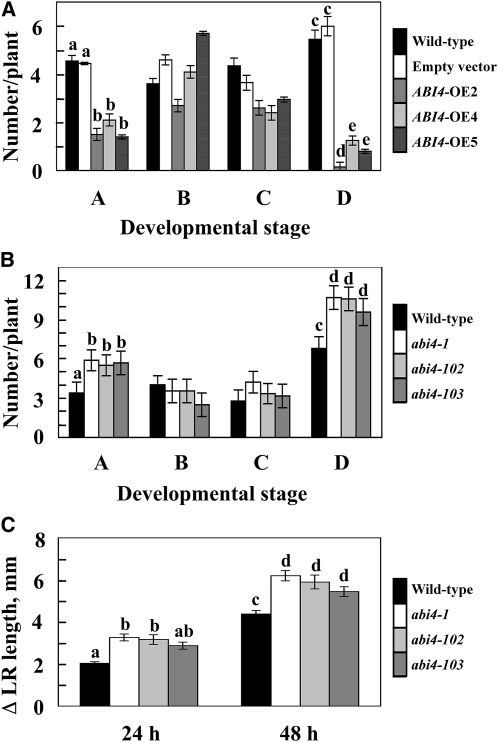
To determine the developmental checkpoints affected by ABI4, we determined the four developmental stages previously delineated by De Smet et al. (2003) and described in Methods. Overexpression of ABI4 resulted in a decrease in the number of stage A LRPs (i.e., the initial LRP developmental stage containing up to two cell layers) and in the number of LRs longer than 0.5 mm (stage D) (Figure 5A). On the other hand, the intermediate stages B and C, representing more developed LRP and short (<0.5 mm) LRs, respectively, were less affected in ABI4-overexpressing plants. The results obtained using ABI4-overexpressing plants were supported by analysis of the four developmental stages in abi4 mutants (Figure 5B). Stages A and D were enhanced with all three mutated alleles of ABI4, whereas stages B and C were not affected. The results presented in Figures 5A and 5B thus suggest that ABI4 inhibits both LR initiation and the elongation of emerged LRs.
Figure 5.
Stage-Specific Inhibitory Effects of ABI4 on LR Development.
(A) and (B) LRs were counted in roots of 12-d-old seedlings in each of four developmental stages (De Smet et al., 2003): Stage A, initials with up to two layers of cells; stage B, three or more layers of cells just prior to emergence; stage C, immediately after emergence but <0.5 mm long; stage D, LRs longer than 0.5 mm.
(A) Effects of ABI4 overexpression on LR number: wild-type plants (black bars), 4C-S5 plants transformed with empty vector (white bars), or three lines of ABI4 overexpressors (gray bars) were germinated on 0.5× MS plates with (transgenic plants) or without (wild type) 2 μM Dex.
(B) Effects on LR number in abi4 mutants: the wild type, abi4-1, abi4-102, and abi4-103 mutants were germinated on 0.5× MS plates.
(C) Initial growth rates of LRs of abi4 mutants. The lengths of the first two visible LRs of 1-week-old vertically grown seedlings were measured. The added lengths at 24 and 48 h are presented. The data represent the mean ± se of n = 3 independent experiments each containing 20 plants per treatment. Bars with different letters represent statistically different values by Tukey’s HSD post-hoc test (P ≤ 0.05).
To further confirm the effect of ABI4 on the elongation of emerged LRs, we measured the initial growth rates of the first two LRs in the abi4 mutant and wild-type plants (Figure 5C). The initial growth rates measured in the abi4 mutants 24 and 48 h postemergence were 41 to 60% and 24 to 41% higher than those determined for the wild-type seedlings at the respective time points. The growth rates, as well as the differences between the wild-type and the abi4 mutant plants, were gradually decreased with time.
Auxin's Effect on LR Formation Is Not Altered in abi4 Mutants and Is Reduced in ABI4 Overexpressors
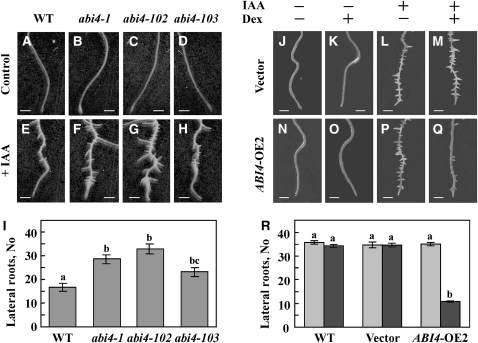
We used the abi4 mutant and ABI4-overexpressing plants to study the interaction of auxin and cytokinin with ABI4 gene activity. abi4 mutant plants were highly affected by exogenous IAA, and their root system was more branched and elongated than that of the wild-type roots when germinated and grown on auxin-containing media (see Supplemental Figure 4 online). The effect of auxin was more pronounced when 1-week-old seedlings lacking developed LRs were treated with IAA (Figures 6E to 6I; see Supplemental Figure 5 online). There was a 40 to 90% increase in LR formation in the three IAA-treated abi4 mutant lines relative to wild-type plants after the same treatment. The extent of the increase in LR number in auxin-treated abi4 mutants (Figure 6) was similar to that observed without the addition of exogenous hormone (Figure 1), suggesting that the auxin response is not altered in abi4 mutants.
Figure 6.
Interaction of ABI4 with Auxin.
(A) to (I) LR formation in control and IAA-treated wild-type (WT) and abi4 mutants. One-week-old seedlings were transferred to 0.5× MS medium ([A] to [D]) or to 0.5× MS supplemented with 20 μM IAA ([E] to [H]). Photographs were taken 1 week later. Bars = 0.5 mm.
(A) and (E) The wild type.
(B) and (F) abi4-1 mutant.
(C) and (G) abi4-102 mutant.
(D) and (H) abi4-103 mutant.
(I) Number of LRs was determined in the lower 5-mm section of the roots shown in (E) to (H).
(J) to (Q) One-week-old seedlings transformed with vector only ([J] to [M]) or pOp6:ABI4 ([N] to [Q]) were transferred to 0.5× MS containing, where indicated, 10 μM Dex and/or 20 μM IAA. Bottom sections of the roots were photographed 1 week later. Bars = 0.5 mm.
(J) and (N) Untreated.
(K) and (O) +Dex.
(L) and (P) +IAA.
(M) and (Q) +Dex and IAA.
(R) Number of LRs was determined in the lower 5-mm section of IAA treated roots. Gray bars, +IAA no Dex; black bars, +IAA +Dex. The data represent the mean ± se of n = 3 independent experiments each containing 50 plants per treatment.
Two-week-old pOp6:ABI4-transformed seedlings were transferred to medium containing Dex alone or Dex + IAA. To avoid analyzing LRs that developed prior to ABI4 induction, we counted LRs in the distal 5 mm corresponding to the root division and elongation zones. None of the tested lines developed any LRs in this region in the absence of treatment or following Dex treatment (Figures 6J, 6K, 6N, 6O, and 6R). Although IAA treatment resulted in the emergence of a large number of LRs in the distal roots of all lines (Figures 6L, 6M, and 6P to 6R), the number of LRs in Dex-induced ABI4-overexpressing plants was 30% of that in the auxin-treated wild-type and empty vector plants (both with and without added Dex) (Figures 6Q and 6R). These results suggest that the number of LR developed is inversely related to the expression levels of ABI4.
ABI4 Affects Root Auxin and Cytokinin Localization and Level
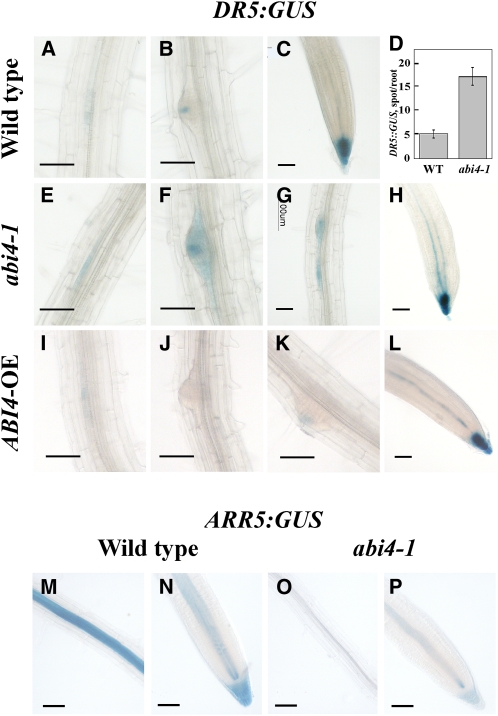
The expression activity of the synthetic DR5 promoter is widely used in studies of endogenous auxin (Ulmasov et al., 1997). This promoter contains multiple binding sites for AUXIN RESPONSIVE FACTOR transcriptional activators (Ulmasov et al., 1997). DR5:GUS-expressing transgenic plants were crossed with abi4-1-mutant plants, and the offspring were selfed to yield homozygous plants. Although DR5:GUS was expressed in LRPs and in root tips on both wild-type and abi4-1 mutant genetic backgrounds (Figure 7), the two backgrounds displayed both quantitative and qualitative differences: the number of GUS-stained loci on the abi4-1 mutant background was 350% of that observed on the wild-type background (Figure 7D; see Supplemental Figure 6 online).
Figure 7.
ABI4 Affects Auxin and Cytokinin Distribution.
(A) to (C) and (E) to (P) GUS staining.
(A) to (C) Roots of seedlings of DR5:GUS/wild type.
(E) to (H) DR5:GUS/abi4-1 mutant.
(I) to (L) DR5:GUS/ABI4 overexpressor.
(M) and (N) ARR5:GUS/wild type.
(O) and (P) ARR5:GUS/abi4-1 mutant. Bars = 50 μm.
(D) Number of stained foci in DR5:GUS/wild-type and DR5:GUS/abi4-1 mutant seedlings. The data represent the mean ± se of n = 3 independent experiments each containing 50 plants per treatment.
In the wild-type background, DR5:GUS was expressed in foci in LRPs and in root tips (Figures 7A to 7C). In preemerging wild-type LRs, DR5:GUS stained exclusively in the tip (Figure 7B). More intense GUS staining was observed for DR5:GUS in the background of the abi4-1 mutant. Staining is more dispersed in the pericycle layer along the PR (Figure 7E) and in the entire preemerging LRs (Figures 7F and 7G). This pattern might result from the increased level of GUS staining in the abi4 background. Closely spaced twin foci could be seen in the abi4-1 mutant background (Figure 7G) but seldom in wild-type plants at this developmental stage. On the other hand, DR5:GUS was markedly repressed along the PR in the ABI4-overexpressing background (Figures 7I to 7K). In the tips of emerging ABI4- overexpressing LRs, DR5:GUS expression was either undetectable (Figure 7J) or very low (Figure 7K). ABI4 overexpression did not alter DR5:GUS expression in the PR tips (Figure 7L). These results suggested that ABI4 results in inhibition of auxin localization in the root by decreasing the number of foci and the apparent concentration of the hormone within these foci.
ARR5:GUS is used as a reporter for cytokinin levels (To et al., 2004). High levels of GUS activity were observed in the stele of the PR of ARR5:GUS-expressing plants in a wild-type background (Figure 7M) all the way to the root tip, including the root cap (Figure 7N), with decreasing staining levels toward the younger regions of the root. On the other hand, expression was markedly reduced in the genetic background of the abi4-1 mutant (Figures 7O and 7P), suggesting that ABI4 might affect cytokinin levels.
LR Development in abi4 Mutants Is Insensitive to ABA and Cytokinin
Since abi4 mutants developed an increased number of LRs (Figure 1) and ABI4 expression was enhanced by ABA and cytokinin, which inhibited LR development, we tested the effects of these inhibitory hormones on LR formation in abi4 mutants. In accordance with previous studies (Bottger, 1974; Wightman et al., 1980; De Smet et al., 2003; Benková and Hejátko, 2009), treatment with ABA and zeatin reduced LR number in wild-type seedlings (Figure 8; see Supplemental Figure 7 online). By contrast, LR formation was less affected by these hormones in the three abi4 mutants (Figure 8; see Supplemental Figure 7 online), suggesting that these hormones act upstream of ABI4. Interestingly, zeatin had no effect on the frequency of LRP in abi4 mutant plants.
Figure 8.
abi4 Plants Are Insensitive to ABA and Cytokinin.
(A) and (B) One-week-old seedlings of the indicated lines were transferred to fresh 0.5× MS medium containing 10 μM ABA (A) or 10 μM zeatin (B). Plants were photographed 1 week later. 1, wild type; 2, abi4-1 mutant; 3, abi4-102 mutant; 4, abi4-103 mutant. Bars = 5 mm.
(C) LR density in abi4 mutants. White bars, no treatment control; gray bars, ABA treatment; black bars, zeatin treatment. The data represent the mean ± se of n = 3 independent experiments each containing 20 plants per treatment. Bars with different letters represent statistically different values by Tukey’s HSD post-hoc test (P ≤ 0.05). WT, wild type.
[See online article for color version of this figure.]
Distribution of Auxin-Efflux Transporter PIN1 Is Affected by ABI4
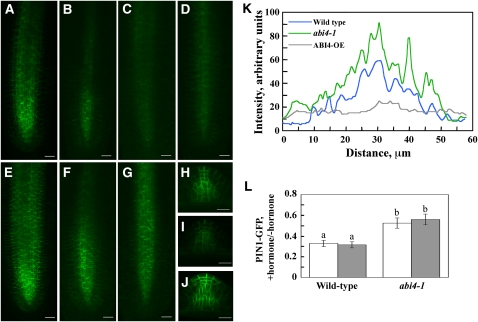
PIN1 has been shown to be very important in the polar transport of auxin from the shoot to root apices (Okada et al., 1991; Friml and Palme, 2002). PIN1 is an auxin-efflux carrier localized on the basal side of stele and endodermis cells with occasional weak expression in the quiescent center and up to the four youngest epidermis and cortex daughter cells (Gälweiler et al., 1998; Friml et al., 2002). To determine whether ABI4-mediated inhibition of LR formation involves changes in PIN1-mediated auxin transport, we crossed PIN1:PIN1-GFP–expressing (for green fluorescent protein) plants with both the abi4-1 mutants and ABI4-overexpressing Arabidopsis. In the abi4-1-mutant plants, PIN1 protein was expressed at higher levels than in the wild-type background (Figures 9A, 9E, and 9K). Moreover, in the genetic background of the abi4 mutation, PIN1 expression extended to more distal regions of the root elongation zone. In agreement with this, in the ABI4 overexpression background, PIN1 levels were reduced and confined to the stele (Figures 9D and 9K). Similarly, higher PIN1 protein levels were observed in emerging LRs of abi4 mutants, whereas low levels were seen in ABI4-overexpressing plants (Figures 9H to 9J). Application of exogenous ABA and the cytokinin zeatin markedly reduced PIN1 protein levels in the roots of wild-type plants and to a lesser extent in abi4-1 mutant plants (Figures 9B, 9C, 9F, 9G, and 9L), suggesting that these hormones affect PIN1 expression in both ABI4-dependent and ABI4-independent pathways.
Figure 9.
ABI4 Affects PIN1 Distribution.
(A) to (G) PIN1-GFP was visualized in PR tips of 12-d-old PIN1:PIN1-GFP/wild-type ([A] to [C]), PIN1:PIN1-GFP/ABI4 overexpressor (D), and PIN1:PIN1-GFP/abi4 mutant ([E] to [G]) seedlings with a fluorescent microscope.
(A), (D), and (E) Untreated seedlings.
(B) and (F) Treated with 10 μM ABA for 24 h.
(C) and (G) Treated with 10 μM zeatin for 24 h. Bars = 20 μm.
(H) to (J) PIN1-GFP expression in untreated emerging LRs.
(H) PIN1:PIN1-GFP/wild type.
(I) PIN1:PIN1-GFP/ABI4 overexpressor.
(J) PIN1:PIN1-GFP/abi4 mutant. Bars = 20 μm.
(K) and (L) Quantification of the fluorescence intensity.
(K) Images shown in (A), (D), and (E) were scanned transversally using ImageJ 1.42q software. Presented pattern was taken 60 μm above root tips. Similar patterns were obtained when other sections were analyzed.
(L) Pixel intensities of four root images were determined using ImageJ 1.42q. Ratios of the values obtained for ABA-treated (white bars) or zeatin-treated (gray bars) plants to that of nontreated plants were calculated for both wild-type and abi4-1 background. The data represent the mean ± se of n = 4. Bars with different letters represent statistically different values by Tukey’s HSD post-hoc test (P ≤ 0.05).
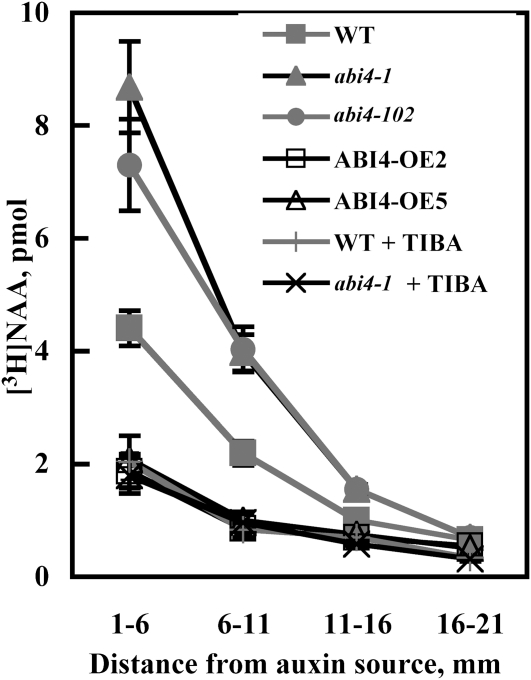
ABI4 Affects Auxin-Polar Transport
To determine if the observed effects of ABI4 on PIN1 protein expression levels alter polar auxin transport, we treated the hypocotyl-root junction with radiolabeled naphthalene acetic acid (NAA), a synthetic auxin. NAA has been shown to be an excellent tool for the study of active efflux of auxin (Delbarre et al., 1996; Petrásek et al., 2006). [3H]NAA transport was ~2-fold higher in abi4 mutants compared with wild-type plants (Figure 10). In addition, very low auxin transport was observed in roots of ABI4-overexpressing plants. These results correlate with the changes in PIN1 observed in the same genetic backgrounds. [3H]NAA appears to have been transported via the polar auxin pathway because the levels observed in abi4 and wild-type plants were almost fully diminished in the presence of the polar auxin transport inhibitor TIBA (Figure 10).
Figure 10.
Polar Auxin Transport.
Agarose blocks containing 10 μM [3H]NAA were placed on the shoot-root junction of the indicated 7-d-old seedlings. When indicated, blocks contained additional 10 μM TIBA. Roots were analyzed 10 h later as described in Methods. The data represent the mean ± se of n = 3 independent experiments each containing 15 plants per treatment. WT, wild type.
DISCUSSION
ABI4 Is Expressed in Roots and Affects LR Formation
We show here directly, both by qRT-PCR and histochemically using Arabidopsis plants expressing an ABI4:GUS reporter construct, that ABI4 is expressed in mature regions of the root and that its pattern of expression correlates with root zones in which LRs develop (Figures 2 and 3). Expression decreased toward the younger regions of the root. An expression gradient was also observed in the root hair zone, with low promoter activity in the upper part of the zone and no detectable expression in the lower (younger) part or meristematic zones, including the root tips and LRP (Figures 2E and 2G). In mature root zones, ABI4 is expressed in companion cells and to a lesser extent in the metaphloem and stele parenchyma (Figures 2H to 2J), with the extent of expression in the latter increasing with further development of the differentiated root (Figures 2H and 2I).
ABI4 has been reported to be expressed in discrete developmental windows, mainly during seed maturation and in very young seedlings postgermination (Söderman et al., 2000; Arroyo et al., 2003). At later stages, ABI4 is expressed at very low, albeit detectable, levels (Söderman et al., 2000; Arroyo et al., 2003). Other studies using ABI4:GUS expressing Arabidopsis (Arroyo et al., 2003; Bossi et al., 2009) did not see GUS staining in roots of seedlings at the postcotyledon stage. However, this may have been because the seedlings were being observed when the upper root zone had not yet reached the mature, ABI4-expressing stage or due to the use of low magnification/resolution to visualize the plants. On the other hand, a few reports have demonstrated the effects of mutations in ABI4 on later developmental stages (Signora et al., 2001; Kaliff et al., 2007), indicating that ABI4 is expressed in the roots and leaves of mature plants. Nevertheless, these studies did not directly demonstrate ABI4 expression in these tissues.
Expression of ABI4 in the root is modulated by plant hormones: it is induced by ABA and cytokinin and inhibited by auxin (Figure 3). Auxin is the key hormone affecting both initiation and development of LRs (Aloni et al., 2006; Fukaki et al., 2007; Osmont et al., 2007; Nibau et al., 2008). In agreement with the proposed inhibitory effect of ABI4 on LR formation, treatment with the LR-promoting hormone IAA decreased the expression of ABI4 (Figures 3A, 3C, and 3H). Similarly, inhibition of auxin transport by TIBA increased the expression of ABI4 (Figures 3A, 3D, and 3I). Furtheremore, cytokinin and ABA are known to counteract auxin induction of LR formation (Bottger, 1974; Wightman et al., 1980). Treatment with either of these inhibitory hormones resulted in increased ABI4 expression (Figures 3A, 3E, 3F, 3J, and 3K). It is interesting to note that, in germinating seedlings, ABI4 expression is not induced by ABA (Söderman et al., 2000), suggesting that different signaling pathways might be involved in the regulation of ABI4 expression in germinating seedlings and in mature roots.
Genetic manipulations affecting ABI4 expression result in changes in root branching. Mutating abi4 led to an increase of 25 to 35% in the number of LRs (Figure 1). In germinating seedlings, ABI4 was highly expressed at the hypocotyl-root border (see Supplemental Figure 1 online; Arroyo et al., 2003) and, thus, probably inhibits LR emergence at early developmental stages. The decrease in LRPs and LRs in abi4 mutants indicated that ABI4 inhibits the formation of LRPs. Similarly, overexpression of ABI4 decreased LR formation (Figure 4). The inhibitory effect of ABI4 overexpression on LR formation (Figures 4A and 4B) was in agreement with the increased number of LRs observed in the abi4 mutants (Figures 1A and 1D). Analysis of LR developmental stage in ABI4-overexpressing plants (Figure 5A) and in abi4 mutants (Figure 5B) confirmed that ABI4 inhibits the establishment of new LRPs (stage A) and the growth of emerged LRs (stage D), whereas the intermediate stages B and C are less affected. Moreover, ABI4 appears to decrease the initial growth rate of the newly emerged LRs (Figure 5C).
ABI4 Interactions with Auxin, Cytokinin, and ABA in Roots
Auxin is the key hormone promoting the development of LRPs and LRs (Aloni et al., 2006; Fukaki et al., 2007; Osmont et al., 2007; Nibau et al., 2008). ABA and cytokinin are known to counteract auxin activity by inhibiting LR formation (Aloni et al., 2006; De Smet et al., 2006; Wasilewska et al., 2008; Fukaki and Tasaka, 2009). Cytokinin has been suggested to inhibit LR formation by preventing establishment of the required auxin gradient via a decrease in the expression of PIN proteins (Laplaze et al., 2007; Ruzicka et al., 2009). On the other hand, the mechanism(s) governing ABA inhibition of LR formation is not understood. Our data suggest that ABI4 mediates inhibition of LR formation by ABA and cytokinin via interference with both LRP formation and further growth of emerged (>0.5 mm) LRs (Figure 5). This latter stage has also been shown to be inhibited by the application of exogenous ABA (De Smet et al., 2003). LR development in abi4 mutant plants is less sensitive to inhibition by ABA and cytokinin (Figure 8), suggesting that these hormones act upstream of ABI4. Mutants affected in the auxin influx carrier AUX1 develop a decreased number of LRs (Marchant et al., 2002). Moreover, a double pin1 pin3 mutant of the auxin-efflux transporters in Arabidopsis results in a 40 to 100% decrease in LRP number and to significant arrest in LR development (Benková et al., 2003). A triple pin1 pin3 pin4 mutant failed to develop any organized LRPs (Benková et al., 2003).
ABI4 is known to play a role in the ABA-signaling pathway in germinating seedlings (Söderman et al., 2000). Here, we show that it is also expressed in roots at later stages of plant development (Figure 2). ABI4 expression in the roots was induced by the root development inhibitory hormones ABA and cytokinin and was repressed by auxin, the LR-promoting hormone (Figure 3). Auxin treatment of abi4 mutants resulted in an increased number of LRs (Figures 6F to 6I; see Supplemental Figure 5 online). The extent of this increase was similar to that observed in the respective mutants without addition of exogenous auxin (Figure 1), suggesting that mutating abi4 does not alter the auxin response.
In addition, auxin treatment of the ABI4-overexpressing seedlings, which lack LRs, resulted in the development of some LRs (Figures 6Q and 6R). The number of LRs in Dex-induced ABI4-overexpressing plants was 30% of that in the auxin-treated wild-type and empty-vector plants (both with and without Dex) (Figures 6Q and 6R). These results suggested that abi4 mutants are not affected in auxin perception or auxin response pathways. ABI4 modulates LR development in a pathway that is distinct from that reported for ABI3 (Brady et al., 2003): the abi3 mutant displays wild-type-like root architecture, and its effects on LR formation can be seen only on the background of the ERA1 mutation or following treatment with auxin. ABI4 is expressed in upper (mature) regions of the root and not in meristematic tissues (Figure 2), whereas ABI3 is expressed in LRs. In addition ABI4 expression is reduced by auxin, while ABI3 expression is enhanced (Brady et al., 2003), and ABI3 mRNA levels in the roots were not affected in abi4 mutants.
ABI4 may inhibit the formation of LRs by interfering with auxin distribution. Using the GUS reporter gene driven by the auxin-responsive synthetic DR5 promoter, we showed a 3.5-fold increase in the number of GUS-stained foci in the genetic background of the abi4-1 mutant compared with the expression of this reporter construct in the wild-type background (Figure 7D; see Supplemental Figure 6 online). In addition, root expression of the cytokinin level reporter ARR5:GUS construct (To et al., 2004) was markedly reduced in the abi4-1 mutant background (Figures 7O and 7P) compared with the wild-type background (Figures 7M and 7N), suggesting that ABI4 might affect cytokinin levels in the roots.
ABI4 expression zone in the mature (upper) root region correlates with the root zone in which LRs develop. ABI4 is expressed mainly in phloem companion cells and to a lesser extent in the metaphloem and stele parenchyma cells (Figures 2H to 2J). ABI4 inhibits LR formation in a non-cell-autonomous fashion; its expression in the phloem and in vascular parenchyma prevents LR initiation from pericycle cells adjacent to the xylem pole. This non-cell-autonomous function is most likely mediated by auxin. Moreover, auxin (Robert and Friml, 2009), ABA (Jiang and Hartung, 2008), and cytokinin (Hirose et al., 2008) are transported in the phloem. ABA is also synthesized in companion cells, where ABI4 is highly expressed: the Arabidopsis AAO3 gene encoding an ABA biosynthetic enzyme is also localized in phloem companion and stele parenchyma cells (Koiwai et al., 2004). Auxin is believed to act as a non-cell-autonomous signal, interacting with other signaling pathways to regulate developmental processes (Swarup et al., 2002). Our data suggest that ABI4 mediates the interaction between the auxin, ABA, and cytokinin signaling pathways (Figure 11). Companion cells thus appear to be the preferred cell type for the regulatory proteins whose activities are affected or affect the balance between ABA, cytokinin, and auxin.
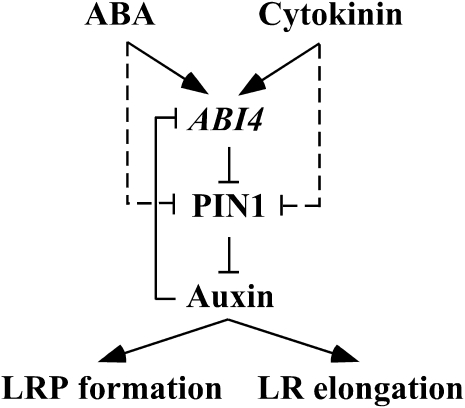
Figure 11.
A Possible Mechanism for Modulation of LR Development by ABI4.
A simplified model is shown for the hormonal regulation of LR development in Arabidopsis. ABI4 expression is enhanced by ABA and cytokinin and repressed by auxin. ABI4, in return, inhibits polar auxin transport in the root by decreasing the levels of the auxin efflux carrier PIN1, resulting in decreased auxin levels. Thus, ABI4 and auxin form a feedback-inhibition loop. In addition to ABI4-dependent pathway, ABA and cytokinin can act in an ABI4-independent way (dashed lines). Auxin is essential for LR formation and for the elongation of emerged LRs; thus, reduction in auxin levels results in inhibition of LR formation. Arrows and T-bars represent enhancement and inhibition, respectively.
Levels of the auxin-efflux transporter PIN1 were reduced in ABI4-overexpressing plants (Figures 9D, 9I, and 9K) and were enhanced in the abi4 mutants (Figures 9E to 9G, 9J, and 9K). These changes are expected to affect root auxin levels, thus affecting LR development. Indeed, direct measurements of auxin transport showed that polar auxin transport is enhanced in abi4 mutants and inhibited in ABI4-overexpressing plants (Figure 10). Since ABI4 encodes an AP2-domain transcription factor (Finkelstein, 1994), the ABI4-induced reduction of PIN1 protein levels is probably indirect. PIN1 mRNA levels were not altered in abi4 mutant or ABI4-overexpressing plants (see Supplemental Table 1 online), suggesting that ABI4 does not directly regulate PIN1 gene transcription. PIN proteins are known to be posttranscriptionally regulated (Abas et al., 2006; Malenica et al., 2007). Moreover, mutation of MODULATOR OF PIN results in a decrease in PIN1 protein levels without affecting PIN1 gene expression (Malenica et al., 2007). Thus, ABI4 may transactivate genes affecting PIN1 levels.
Although ABA is known to inhibit LR formation (De Smet et al., 2006), our results describe a direct effect of the ABA-signaling pathway on auxin transport. Exogenous cytokinin and ABA inhibited PIN1 expression in roots (Figure 9). The extent of inhibition following application of these hormones was higher in wild-type plants than in the abi4 mutants (Figure 9L), suggesting that ABA and cytokinins reduce PIN1 by both ABI4-dependent and ABI4-independent pathways. Cytokinin is known to counteract auxin activity and has been shown to negatively regulate PIN1, as reflected by a reduction in PIN1:PIN1-GFP signal in the Arabidopsis PR tip of cytokinin-treated plants (Laplaze et al., 2007; Ruzicka et al., 2009). The dampened reduction in PIN1 levels in abi4 mutants by ABA and zeatin (Figure 9) correlated with the reduced of inhibitory effects of these hormones on LR formation in these plants (Figure 8), suggesting that ABI4 activity interferes with polar auxin transport. This was supported by direct auxin transport assays (Figure 10). The increased PIN1 expression (Figure 9) and, thus, the predicated increase in auxin, is in agreement with the diffuse DR5:GUS/abi4 expression pattern (Figures 7F and 7H) resembling that observed in auxin-treated wild-type plants (Benková et al., 2003).
Our data suggest that ABI4 mediates ABA and cytokinin auxin-antagonistic activity in the modulation of root development (Figure 11). ABI4 expression is enhanced by ABA and cytokinin (Figure 3) and repressed by auxin, which is transported from the shoot to the root, or basipetally from the root tips toward the upper root sections. ABI4, in return, inhibits polar auxin transport in the root (Figure 10) by decreasing the levels of PIN1 (Figure 9), resulting in decreased auxin levels in the roots. Thus, ABI4 and auxin form a feedback inhibition loop. Reduction of PIN1 by cytokinin and ABA was also observed in the abi4-1 mutant (Figures 9F and 9G), suggesting an additional abi4-independent pathway. Moreover, the PIN1 levels in cytokinin- or ABA-treated abi4 mutants were still high enough to support the development of LRs (Figure 8). Auxin is essential for both the establishment of LRPs (Aloni et al., 2006; Fukaki et al., 2007; Osmont et al., 2007; Nibau et al., 2008) and the elongation of emerged LR (Muday and Haworth, 1994). As a result, abi4 mutant plants display an increased number of elongated LRs, whereas overexpression of ABI4 results in a decreased number of LRPs and in inhibition of elongation of emerged LRs.
METHODS
Plant Material and Growth Conditions
The following Arabidopsis thaliana (Columbia ecotype) plants were obtained from the Arabidopsis Stock Center in Columbus, OH: abi4 mutants, DR5:GUS (Ulmasov et al., 1997), ARR5:GUS (To et al., 2004), and PIN1:PIN1-GFP (Benková et al., 2003). Arabidopsis 4C-S5 and pV-TOP (Craft et al., 2005) were obtained from I. Moore (Oxford University). ABI4:GUS, pOp6:ABI4, and cauliflower mosaic virus 35S:ABI4 were constructed for this study. Seed sterilization and plant growth on solid 0.5× Murashige and Skoog (MS) + 0.5% (w/v) sucrose or in pots were as described previously (Shkolnik and Bar-Zvi, 2008). Hormone treatment was performed on Whatman No. 1 filter papers soaked with 0.5× MS medium supplemented with the indicated hormone. Each assay was performed with 50 seedlings from each line.
Constructs and Arabidopsis Plant Transformation
DNA corresponding to ABI4 promoter or coding sequences was isolated using PCR and genomic DNA as the template and primers as listed in Supplemental Table 2 online. The ABI4 promoter was subcloned upstream of the GUS coding sequence in pCAMBIA 1391Z (CAMBIA). The ABI4 coding sequence was subcloned into pCAMBIA 1200 (CAMBIA) downstream of the cauliflower mosaic virus 35S promoter or into the pV-TOP vector (obtained from I. Moore) downstream of the Dex-inducible pOp6 promoter (Craft et al., 2005). Constructs were introduced into Agrobacterium tumefaciens GV3101 and used in the transformation of wild-type Columbia or 4C-S5 Arabidopsis (Craft et al., 2005) plants, for pCAMBIA- or pV-TOP–based constructs, respectively (Clough and Bent, 1998). Transgenic plants were selected on plates containing hygromycin. Plants were selfed twice, and T3 homozygous plants were used in this study. At least five lines of independent transformants were assayed. Transgenic Arabidopsis DR5:GUS, ARR5:GUS, and PIN1:PIN1-GFP plants were used as the pollen source for crossing with emasculated wild-type or abi4-1 mutant plants. Cross-bred offspring were selected on 1 μM ABA-containing media for the abi4 trait and analyzed for GUS or GFP expression.
qRT-PCR Analysis
Total RNA was isolated using an Aurum Total RNA Mini kit (Bio-Rad Laboratories). The RNA concentration was estimated spectrally (Nano Drop ND-1000; Nano Drop Technologies). cDNA was synthesized using an ABgene Reverse-iT 1st Strand synthesis sit (ABgene). The reaction mixture contained 700 ng of total RNA and 400 ng of random decamers. Relative transcript levels were assayed by real-time PCR analysis using the 7300 real-time PCR system (Applied Biosystems). Primer sequences were designed by Primer-Express software Version 2.0 (Applied Biosystems). Wherever possible, one of the primers in each set was designed at an exon-exon border. All amplicon length ranges were between 75 and 90 bp. The reaction mixture contained 10 μL of Power SYBR Green PCR Master Mix (Applied Biosystems), 50 ng of cDNA (for endogenous control, as 18S RNA, a 1000-fold dilution was used), and 500 nM of each of the forward and reverse primers. The following default program was used: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min each, and a dissociation stage of 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. RNA relative quantification analyses were performed using 7300 System SDS software (Applied Biosystems). The list of primers used is shown in Supplemental Table 2 online. The data represent the mean ± se of n = 3 independent experiments. Each data point was determined in triplicates in each of the three biological replicates and presented as mean ± se.
GUS Staining
Plant tissues were fixed in acetone for GUS staining as described previously (Weigel and Glazebrook, 2002). Pictures were taken with a Nikon camera (DXM1200F) using a stereoscope and microscope. Each treatment was performed using three biological replicates.
PIN1:PIN1-GFP Expression
Roots of plants expressing PIN1:PIN1-GFP (Benková et al., 2003) under different generic backgrounds were imaged using Zeiss LSM510 confocal laser scanning microscope. Image quantification was performed using ImageJ 1.42q software (W.S. Rasband, ImageJ, U. S. National Institutes of Health). Images are representative of 15 roots per each treatment with three biological replicates.
Root Sections
GUS-stained seedlings were fixed, dehydrated, and directionally embedded in Technovit 7100 resin (Heraeus Kulzer) (Beeckman and Viane, 2000). Sections (4 μm) were cut with a Reichert-Jung Ultracut microtome (Leica) using a glass knife, stained with 0.05% (w/v) ruthenium red, and mounted in Entellan mounting medium (Merck) before photography.
LR Developmental Stages
Seeds were germinated on plates containing 0.5× MS supplemented with 0.5% (w/v) sucrose, 0.5% (w/v) plant agar, and where indicated also 2 μM Dex. Ten-day-old seedlings were collected, and the four defined LR developmental stages (De Smet et al., 2003) were determined using a light microscope. Dex had no effect on the number or distribution of the determined developmental stages in wild-type plants or in plants transformed with empty vector. Three biological experiments containing each 20 plants per treatment were analyzed. Error bars represent se.
LR Elongation Rate
Seedlings were grown vertically on 0.5× MS supplemented with 0.5% (w/v) sucrose and 1.5% (w/v) plant agar. The length of the first two visible LRs was marked every 24 h on the plate using color-coded markers. Root lengths at all time points were measured 9 d after emergence of the LRs. Growth rates were calculated from the difference in length after 24 and 48 h.
Root Auxin Transport Assay
[3H]NAA (Nuclear Research Center) was a gift from J. Riov (The Hebrew University, Rehovot, Israel), and NAA was obtained from Duchefa Biochemie. Seedlings were grown on vertical plates containing 0.5× MS supplemented with 0.5% (w/v) sucrose and 1.5% (w/v) plant agar. Agar blocks (2 mm × 1 mm × 1 mm) containing 10 μM [3H]NAA (4 × 104 cpm × nmol−1) were placed on top of the plants at the shoot-root junction. Plates were incubated vertically for 10 h, roots were detached 1 mm below the auxin application site, 5-mm segments were cut, placed into vials containing Quicksafe A scintillation cocktail (Zinsser Analytic), and incubated at room temperature for 2 d in the dark. Radioactivity was determined in a Tri-Carb 2100TR scintillation counter (Packard Instruments). Three biological replicates, each containing 15 plants per treatment, were examined for each line. Data are presented as mean ± se.
Statistical Analyses
Differences between groups were analyzed by Tukey’s HSD post-hoc test (PASW Statistics Data Editor of the SPSS 17.0 software package; IBM Company).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM 129580 for ABI4 mRNA, NM 106017 for PIN1 mRNA, and NM 114679 for ARR5 mRNA. The Arabidopsis Genome Initiative locus identifier for ABI4 is AT2G40220, AT1G73590 for PIN1, and AT3G48100 for ARR5.
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. GUS Staining of ABI4:GUS Plants.
Supplemental Figure 2. ABI4 Expression in Root and Leaf Tissues.
Supplemental Figure 3. Quantification of Dexamethasone-Induced ABI4 Overexpression in Transgenic Arabidopsis Plants Harboring the pOp6:ABI4 Construct.
Supplemental Figure 4. Auxin Treatment of abi4 Mutants.
Supplemental Figure 5. Lateral Root Formation in IAA-Treated abi4 Mutants.
Supplemental Figure 6. DR5:GUS Expression Pattern Is Altered on the Genetic Background of the abi4 Mutation.
Supplemental Figure 7. Lateral Root Formation in Cytokinin-Treated Wild-Type and abi4 Mutant Arabidopsis.
Supplemental Table 1. Genes Which Steady State mRNA Levels Are Not Altered in Roots of abi4 Mutants and ABI4-OE Plants.
Supplemental Table 2. Primer List.
Supplementary Material
Acknowledgments
We thank Nathan Zauberman and Abraham Minsky (Weizmann Institute of Science, Israel) for their help with the specimen sectioning, Joseph Riov (Hebrew University, Israel) for his generous gift of radiolabeled NAA, as well as Eduardo Blumwald (University of California, Davis) and Hillel Fromm and Shaul Yalovsky (Tel Aviv University, Israel) for critical reading of the manuscript.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Wirniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 Erratum. Nat. Cell Biol. 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Aloni R., Aloni E., Langhans M., Ullrich C.I. (2006). Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. (Lond.) 97: 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo A., Bossi F., Finkelstein R.R., Leon P. (2003). Three genes that affect sugar sensing (Abscisic Acid Insensitive 4, Abscisic Acid Insensitive 5, and Constitutive Triple Response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 133: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T., Viane R. (2000). Embedding thin plant specimens for oriented sectioning. Biotech. Histochem. 75: 23–26 [DOI] [PubMed] [Google Scholar]
- Benková E., Hejátko J. (2009). Hormone interactions at the root apical meristem. Plant Mol. Biol. 69: 383–396 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bossi F., Cordoba E., Dupré P., Mendoza M.S., Román C.S., León P. (2009). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Bottger M. (1974). Apical dominance in roots of Pisum sativum-L. Planta 121: 253–261 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Sarkar S.F., Bonetta D., McCourt P. (2003). The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34: 67–75 [DOI] [PubMed] [Google Scholar]
- Brocard-Gifford I., Lynch T.J., Garcia M.E., Malhotra B., Finkelstein R.R. (2004). The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16: 406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Craft J., Samalova M., Baroux C., Townley H., Martinez A., Jepson I., Tsiantis M., Moore I. (2005). New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41: 899–918 [DOI] [PubMed] [Google Scholar]
- De Smet I., Signora L., Beeckman T., Inzé D., Foyer C.H., Zhang H.M. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33: 543–555 [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I., Zhang H.M., Inzé D., Beeckman T. (2006). A novel role for abscisic acid emerges from underground. Trends Plant Sci. 11: 434–439 [DOI] [PubMed] [Google Scholar]
- Deak K.I., Malamy J. (2005). Osmotic regulation of root system architecture. Plant J. 43: 17–28 [DOI] [PubMed] [Google Scholar]
- Delbarre A., Muller P., Imhoff V., Guern J. (1996). Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benková E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R. (1994). Mutations at 2 new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5: 765–771 [Google Scholar]
- Finkelstein R.R., Wang M.L., Lynch T.J., Rao S., Goodman H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J., Palme K. (2002). Polar auxin transport—Old questions and new concepts? Plant Mol. Biol. 49: 273–284 [PubMed] [Google Scholar]
- Fukaki H., Okushima Y., Tasaka M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256: 111–137 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69: 437–449 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C.H., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H., Sakakibara H. (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Jiang F., Hartung W. (2008). Long-distance signalling of abscisic acid (ABA): The factors regulating the intensity of the ABA signal. J. Exp. Bot. 59: 37–43 [DOI] [PubMed] [Google Scholar]
- Kaliff M., Staal J., Myrenås M., Dixelius C. (2007). ABA is required for Leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol. Plant Microbe Interact. 20: 335–345 [DOI] [PubMed] [Google Scholar]
- Koiwai H., Nakaminami K., Seo M., Mitsuhashi W., Toyomasu T., Koshiba T. (2004). Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 134: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby R.J., Kincaid M.S., Kim D.G., Gibson S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Laplaze L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mo X.R., Shou H.X., Wu P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Linkohr B.I., Williamson L.C., Fitter A.H., Leyser H.M.O. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29: 751–760 [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Cruz-Ramírez A., Herrera-Estrella L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6: 280–287 [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Hernández-Abreu E., Sánchez-Calderón L., Nieto-Jacobo M.F., Simpson J., Herrera-Estrella L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Godin C., Jay-Allemand C., Laplaze L. (2008). Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J. Exp. Bot. 59: 55–66 [DOI] [PubMed] [Google Scholar]
- Malenica N., Abas L., Benjamins R., Kitakura S., Sigmund H.F., Jun K.S., Hauser M.T., Friml J., Luschnig C. (2007). MODULATOR OF PIN genes control steady-state levels of Arabidopsis PIN proteins. Plant J. 51: 537–550 [DOI] [PubMed] [Google Scholar]
- Marchant A., Bhalerao R., Casimiro I., Eklöf J., Casero P.J., Bennett M., Sandberg G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog P.R., van der Kooij T.A.W., Brüggemann W., Schiefelbein J.W., Kuiper P.J.C. (1995). Responses to iron deficiency in Arabidopsis thaliana: The Turbo iron reductase does not depend on the formation of root hairs and transfer cells. Planta 195: 505–513 [DOI] [PubMed] [Google Scholar]
- Muday G.K., Haworth P. (1994). Tomato root-growth, gravitropism, and lateral development-correlation with auxin transport. Plant Physiol. Biochem. 32: 193–203 [PubMed] [Google Scholar]
- Nibau C., Gibbs D.J., Coates J.C. (2008). Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont K.S., Sibout R., Hardtke C.S. (2007). Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Penfield S., Li Y., Gilday A.D., Graham S., Graham I.A. (2006). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. (2009). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Petrásek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Ruzicka K., Simásková M., Duclercq J., Petrásek J., Zazímalová E., Simon S., Friml J., Van Montagu M.C.E., Benková E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik D., Bar-Zvi D. (2008). Tomato ASR1 abrogates the response to abscisic acid and glucose in Arabidopsis by competing with ABI4 for DNA binding. Plant Biotechnol. J. 6: 368–378 [DOI] [PubMed] [Google Scholar]
- Signora L., De Smet I., Foyer C.H., Zhang H.M. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Söderman E.M., Brocard I.M., Lynch T.J., Finkelstein R.R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Parry G., Graham N., Allen T., Bennett M. (2002). Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol. Biol. 49: 411–426 [DOI] [PubMed] [Google Scholar]
- To J.P.C., Haberer G., Ferreira F.J., Deruère J., Mason M.G., Schaller G.E., Alonso J.M., Ecker J.R., Kieber J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian N., Marcotte L., Giraudat J. (1994). Drought rhizogenesis in Arabidopsis thaliana (Differential responses of hormonal mutants). Plant Physiol. 104: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E., Zhu J.K. (2005). Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem. Soc. Trans. 33: 375–379 [DOI] [PubMed] [Google Scholar]
- Wasilewska A., Vlad F., Sirichandra C., Redko Y., Jammes F., Valon C., Frei dit Frey N., Leung J. (2008). An update on abscisic acid signaling in plants and more. Mol. Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman F., Schneider E.A., Thimann K.V. (1980). Hormonal factors controlling the initiation and development of lateral roots. 2. Effects of exogenous growth factors on lateral root formation in pea roots. Physiol. Plant. 49: 304–314 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.