This work examines the amino acid transporter AAP2, which localizes to the phloem throughout the plant and is key for amino acid transfer from the xylem-phloem; mutants in AAP2 perturb nitrogen-carbon balance in the seed, affect leaf metabolism and development, and increase seed yield and oil content.
Abstract
Seed development and nitrogen (N) storage depend on delivery of amino acids to seed sinks. For efficient translocation to seeds, amino acids are loaded into the phloem in source leaves and along the long distance transport pathway through xylem-phloem transfer. We demonstrate that Arabidopsis thaliana AMINO ACID PERMEASE2 (AAP2) localizes to the phloem throughout the plant. AAP2 T-DNA insertion lines showed changes in source-sink translocation of amino acids and a decrease in the amount of seed total N and storage proteins, supporting AAP2 function in phloem loading and amino acid distribution to the embryo. Interestingly, in aap2 seeds, total carbon (C) levels were unchanged, while fatty acid levels were elevated. Moreover, branch and silique numbers per plant and seed yield were strongly increased. This suggests changes in N and C delivery to sinks and subsequent modulations of sink development and seed metabolism. This is supported by tracer experiments, expression studies of genes of N/C transport and metabolism in source and sink, and by phenotypic and metabolite analyses of aap2 plants. Thus, AAP2 is key for xylem to phloem transfer and sink N and C supply; moreover, modifications of N allocation can positively affect C assimilation and source-sink transport and benefit sink development and oil yield.
INTRODUCTION
Nitrogen (N) and carbon (C) are two of the most important nutrients for plant growth, and in the majority of plant species, they are transported in the phloem to sink organs in the form of amino acids and sucrose, respectively. Extensive studies have been performed on long-distance transport of sucrose. Sucrose transporters (SUC/SUT) were localized to the phloem of a number of plant species, and mutant analyses and antisense approaches demonstrated that the sucrose transporters play key roles in phloem loading of carbohydrates (for review, see Sauer, 2007; Tegeder et al., 2011). By contrast, molecular mechanisms for amino acid phloem loading remain unclear, although members of the ATF amino acid transporter family were suggested to be involved in this process (Lalonde et al., 2004; Tegeder and Rentsch, 2010).
Synthesis of amino acids mainly takes place in mature roots and source leaves, which export the N to supply sinks like flowers and seeds (Lam et al., 1996). Transport of amino acids to sinks happens mainly via the phloem, and to achieve this, the leaf-synthesized N compounds are loaded into the phloem of the minor veins. In cases where amino acid synthesis occurs in roots, they are transported via the xylem with the transpiration stream, mainly to mature leaves. Here, they are imported into the mesophyll cells for transient storage or metabolism or, alternatively, loaded into the leaf phloem of the major veins for redistribution to fruits and seeds (Rentsch et al., 2007). Transfer of root-assimilated amino acids from the xylem to the phloem also occurs along the long-distance transport pathway (e.g., root and stem) for immediate supply of rapidly growing sinks (Pate et al., 1975, 1977). Generally, all protein amino acids are transported within the phloem to the seed, but in many plant species, including Arabidopsis thaliana, Gln, Glu, Asp, Asn, Ser, or Ala seem to dominate (Riens et al., 1991; Lam et al., 1995; Lohaus and Möllers, 2000; Wilkinson and Douglas, 2003; Hunt et al., 2006). Within the seed sink, amino acids are released from the seed coat into the apoplast from where they are taken up by the embryo for development, storage product synthesis (i.e., enzymes for starch, protein, and fatty acid/oil biosynthesis), or storage protein accumulation (Tegeder et al., 2000, 2007; Miranda et al., 2001, 2003; Rolletschek et al., 2005; Tan et al., 2008; Sanders et al., 2009). Amino acid transporters in source and sink tissues are considered to be essential in partitioning of assimilates, and they are potentially rate limiting for distribution of organic N to, and into, seed sinks (Lalonde et al., 2004). Sink number, size, and storage product accumulation are dependent on organic N transport processes from source to sink (Crawford, 1995; Song et al., 1997; Ma et al., 1998; Forde, 2002a, 2002b; Schmidt et al., 2007; Hunt et al., 2010).
While the role of transporter function in phloem loading of amino acids has not been demonstrated up to now, it was shown in potato (Solanum tuberosum) that transporters play a role in long-distance transport of amino acids (Koch et al., 2003). In Arabidopsis, promoter-reporter gene and RNA localization studies have identified three potential phloem loaders for amino acids, AMINO ACID PERMEASE2 (AAP2), AAP3, and AAP8 (Hirner et al., 1998; Okumoto et al., 2002, 2004; Lee et al., 2007). Work by Okumoto et al. (2004) demonstrated that AAP3 is specifically expressed in the root phloem, but mutants of the transporter did not display any phenotype. Cellular expression of AAP8 has not been resolved, but promoter-β-glucuronidase (GUS) studies reported AAP8 expression in the veins of young flowers, peduncle, and siliques. The transporter is also expressed in very young seeds, and aap8 mutants showed a reduction in seed number per silique, suggesting a function of AAP8 in amino acid import into the seed (Schmidt et al., 2007). With respect to AAP2, its RNA was localized to the vascular system of Arabidopsis seedlings (Kwart et al., 1993), which was confirmed by promoter-GUS analyses (Hirner et al., 1998). The latter studies also indicated that AAP2 is expressed in the vascular bundle throughout mature Arabidopsis plants. While the cellular localization of AAP2 in Arabidopsis has not been demonstrated, expression of the Arabidopsis AAP2-promoter-GUS construct in tobacco (Nicotiana tabacum) showed GUS staining in the stem phloem. Analysis of the substrate selectivity of AAP2 clarified that the transporter recognizes a wide spectrum of proteogenic amino acids when expressed in yeast mutants defective in endogenous amino acid transport systems (Kwart et al., 1993; Fischer et al., 1995), and electrophysiological transport studies in Xenopus laevis oocytes demonstrated that AAP2 transports Glu and neutral amino acids with moderate or low affinity (Fischer et al., 2002).
This study addresses the significance of AAP2 in xylem-to-phloem transfer of amino acids and its role in source and sink physiology. We first show that AAP2 is localized to the plasma membrane and the phloem and then demonstrate that in aap2 mutants, source-sink translocation of amino acids is changed, affecting seed protein and oil levels as well as seed yield. Phenotypic, molecular, and biochemical analyses provide support for these changes through modulations in N and C metabolism, organ development, and partitioning processes in source and sink. The possible role of AAP2 in xylem-phloem transfer of amino acids and the implications of its function for plant growth and N/C assimilation, metabolism, and transport are discussed.
RESULTS
AAP2 Is Localized to the Phloem throughout the Arabidopsis Plant
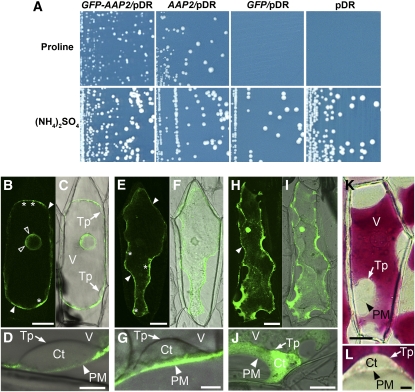
To resolve the cellular and subcellular location of AAP2 expression, green fluorescent protein (GFP)-AAP2 protein localization studies were performed. First, it was tested whether the GFP-AAP2 gene construct encoded a functional AAP2 transporter by expressing it in yeast cells (22574D) deficient in Pro transport (Figure 1A). When plated on media with Pro as sole nitrogen source, yeast cells expressing the empty vector failed to grow, while expression of GFP-AAP2 fusion protein restored cell growth.
Figure 1.
Expression of GFP-AAP2 in Yeast and Onion Cells.
(A) Yeast complementation by GFP-AAP2 fusion protein. Pro uptake-deficient yeast mutant (22574d) was transformed with GFP-AAP2 in pDR. AAP2 (positive) and GFP in pDR and empty pDR (negative) were used as controls for growth under selective conditions. Minimal media was supplemented with Pro (0.3 g/L) (top images) or ammonium sulfate (5 g/L) (bottom images) as a sole nitrogen source. GFP-AAP2 restores yeast growth on Pro containing medium similar to the AAP2 control, indicating that GFP-AAP2 is a functional protein.
(B) to (L) Subcellular localization of GFP-AAP2 fusion protein in onion cells following particle bombardment. Shown are confocal images ([B], [E], and [H]), overlaid bright-field images ([C], [D], [F], [G], [I], and [J]), and bright-field images ([K] and [L]). The cells were plasmolyzed to distinguish between plasma membrane and vacuole. asterisks, Golgi apparatus; Ct, cytosol; open arrowhead, endoplasmic reticulum; PM/closed arrowhead, plasma membrane; Tp, tonoplast; V, vacuole. Bars = 50 μm in (B), (C), (E), (F), (H), (I), and (K) and 10 μm in (D), (G), (J), and (L).
(B) to (D) GFP-AAP2 expression.
(E) to (G) Arabidopsis amino acid transporter ProT2 (Grallath et al., 2005) was used as control for membrane localization.
(H) to (J) Localization of free GFP (control).
(K) and (L) Nontransgenic cells from red onions, illustrating that the cytosol and vacuole can be easily detected due to storage of the red pigments or anthocynins in the vacuole (control).
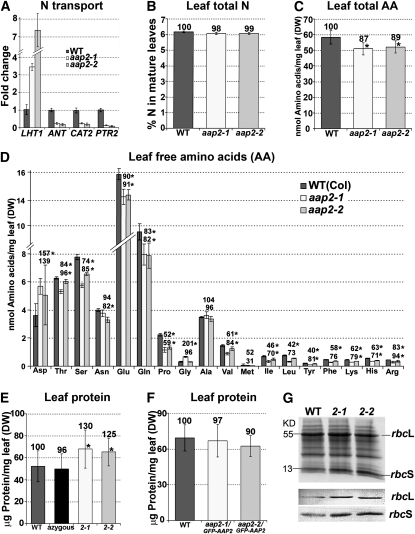
We further tested functionality of the GFP-AAP2 protein in plants by transforming two aap2 mutant lines (see below) with an AAP2 promoter-GFP-AAP2 construct. Analysis of the complementation lines showed that soluble protein levels were similar for complementation lines and wild-type plants, while protein amounts in the aap2 plants were increased, demonstrating that GFP-AAP2 was able to restore the aap2 phenotype (for details, see text below and Figures 7E and 7F).
Figure 7.
Expression of Amino acid Transporters and Analysis of Total N, Free Amino Acids, and Soluble Protein in Source Rosette Leaves of Wild-Type and aap2 Plants.
Error bars depict sd. Asterisks indicate significant differences from the wild type (WT; P < 0.05). Values above the columns indicate the percentage compared with the wild type, which was set to 100%. DW, dry weight.
(A) Real-time PCR analysis of amino acid transporter LHT1 (At5g40780; plasma membrane localized), ANT and CAT2 (At3g30390 and At1g58030, respectively; both in tonoplast), and vacuolar peptide transporter PTR2 (At2g02040; see Rentsch et al., 2007). Three technical repetitions were done for each line using Arabidopsis ubiquitin 11 (UBQ11, At4g05050) and tubulin β-9 chain (TUB9, At4g20890), respectively, as control genes. Shown is the fold change in gene expression relative to UBQ11 expression, determined from the CT values using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
(B) Total leaf N levels (n = 4).
(C) Total free amino acid levels in leaves (n = 4).
(D) Free amino acid composition and concentration in leaves (n = 4).
(E) Amounts of soluble leaf protein. Wild-type and azygous plants were used as controls (n = 5).
(F) Amounts of soluble leaf protein in wild-type and complementation lines aap2-1/GFP-AAP2 and aap2-2/GFP-AAP2 (n = 8).
(G) SDS-PAGE analysis of leaf proteins (top image) and immunoblot analysis using antibodies against small and large subunits of Rubisco (bottom image).
After confirming that GFP-AAP2 is a functional protein in planta, GFP-AAP2 under the control of cauliflower mosaic virus 35S promoter was transferred into onion (Allium cepa) epidermal cells via particle bombardment for examination of subcellular localization (Figures 1B to 1D). The cells were plasmolyzed to distinguish between plasma membrane and vacuole. Arabidopsis plasma membrane and proline transporter ProT2 (Figures 1E to 1G; Grallath et al., 2005) and free GFP (Figures 1H to 1J) were used as controls as well as nontransgenic cells from red onions, in which the cytosol and vacuole can be easily differentiated due to vacuolar storage of the red pigments or anthocyanins (Figures 1K and 1L). Analysis of onion cells expressing GFP-AAP2 showed that the transporter is primarily localized to the plasma membranes (closed arrowheads; Figures 1B and 1D). Green fluorescence was also detected in membranes surrounding the nucleus, presumably in the endoplasmic reticulum (open arrowheads; Figure 1B). Some GFP-AAP2 and GFP-ProT2 fluorescence was also seen in structures at the plasma membranes that might represent the Golgi apparatus (asterisks; Figures 1B and 1E). Since AAP2 and ProT2 travel via the different compartments prior to being incorporated into the plasma membrane, the localization to these compartments is probably due to the overexpression of the GFP transporter proteins in onion cells. High-magnification images showed no fluorescence in the tonoplast or cytosol in onion cells bombarded with GFP-AAP2 or GFP-ProT2 (Figures 1D and 1G).
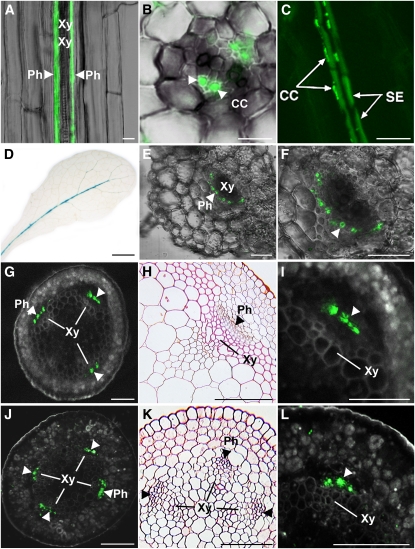
Promoter-GUS studies and RNA localization studies indicated that Arabidopsis AAP2 is expressed in the vascular tissue (Kwart et al., 1993; Hirner et al., 1998). To resolve the cellular localization of AAP2, the GFP-AAP2 fusion construct was expressed in Arabidopsis under the control of the endogenous AAP2 promoter. Analysis of the transgenic roots revealed GFP-AAP2 localization in the phloem of roots (Figure 2A). Cross and longitudinal root sections further resolved that AAP2 is present in the companion cells (Figures 2B and 2C). Some weak fluorescence was also detected in the sieve element, and future studies using anti-AAP2 antibodies are needed to clarify if the observed fluorescence is background or if AAP2 is localized to both companion cells and sieve element cells as well. Expression of AAP2 was also found in the phloem throughout the plant (Figures 2A to 2L). Using AAP2 promoter-GUS studies, transporter expression was detected in the vascular tissue of the major but not minor veins (Figure 2D), consistent with previous work by Hirner et al. (1998). Fluorescence of AAP2 protein in the leaf was weak, and we were only able to record GFP-AAP2 in the phloem of the leaf petiole by confocal imaging (Figures 2E and 2F). Localization studies in stems showed that AAP2 expression is restricted to the phloem in this tissue as well (Figures 2G to 2I). GFP-AAP2 fluorescence was also found in the phloem of other tissues along the transport path to seed sinks such as the phloem of the silique peduncle (Figures 2J to 2L). No fluorescence was observed in tissues or cells other than phloem, consistent with a role of AAP2 in phloem loading of amino acids.
Figure 2.
Localization of GFP-AAP2 in Arabidopsis Plants and AAP2 Promoter-GUS Expression Analysis in Leaves.
(A) to (C) GFP-AAP2 localization in roots under control of the AAP2 promoter. Longitudinal sections ([A] and [C]); cross section (B).
(D) AAP2 promoter-GUS studies in leaves. GUS staining was detected in the major vein.
(E) to (F) Leaf petiole cross sections.
(G) to (I) Stem cross sections.
(J) to (L) Cross sections of silique peduncle.
GFP fluorescence is confined to the phloem along the transport pathway. Resin sections stained with safranin O ([H] and [K]). CC, companion cells; Ph, phloem; SE, sieve element; Xy, xylem. Arrowheads point to the phloem. Bars = 7.5 μm in (B), 15 μm in (A), 20 μm in (E) and (F), 30 μm in (C), 100 μm in (G) to (L), and 4 mm in (D).
Identification of aap2 Mutants and AAP2 Expression Analysis
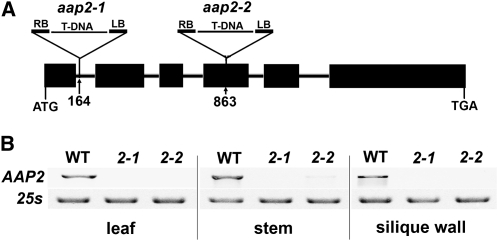
To analyze the function of AAP2 in Arabidopsis plants, a reverse genetics approach was applied. Two independent AAP2 T-DNA insertion lines (aap2-1, SALK_047237; aap2-2, SALK_143597) were obtained from the ABRC at Ohio State University (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/index.html), screened for homozygosity, and then analyzed. Allele aap2-1 contains an insertion in the 1st intron, 164 nucleotides downstream of the start codon of the AAP2 gene, and aap2-2 carries a T-DNA insertion in the 4th exon at 863 nucleotides (Figure 3A). To analyze AAP2 expression in the mutants, RT-PCR analysis was performed and revealed that AAP2 mRNA is absent in aap2-1 and aap2-2 plants (Figure 3B).
Figure 3.
Molecular Characterization of aap2 Mutants.
(A) Structure of the AAP2 gene and positions of the T-DNA insertions in aap2-1 and aap2-2 mutants. Boxes and lines represent exon and intron regions, respectively. The triangles indicate the sites of T-DNA insertions. The aap2-1 allele contains a T-DNA insertion in the first intron, 164 nucleotides downstream of the ATG. Aap2-2 carries an insertion in the fourth exon in position 863. LB, left border of T-DNA; RB, right border.
(B) RT-PCR analysis of AAP2 expression in rosette leaf, stem, and silique wall of wild-type (WT), aap2-1, and aap2-2 plants. 25S rRNA was amplified as control for equal amounts of RNA in the different samples.
Phloem Gln Levels Are Decreased and Long-Distance Transport of [14C]L-Gln Is Altered
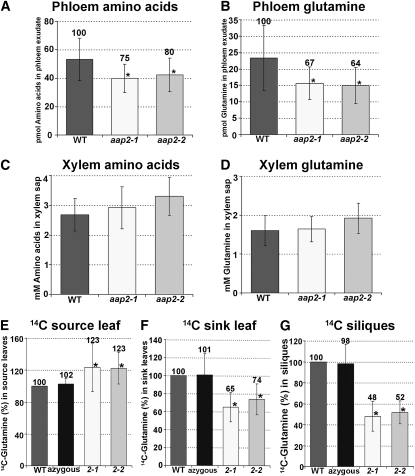
Localization of AAP2 in the leaf major veins and in the phloem throughout the plant suggests a function of the transporter in phloem loading along the transport pathway. Amino acids synthesized in the root are translocated in the xylem mainly to the leaf. For further partitioning to the sinks, they may then be transferred in the leaf major veins from the xylem to the phloem. Leaf phloem exudates from aap2-1 and aap2-2 plants were analyzed to determine if AAP2 might play a role in this process. The results showed a decrease in total phloem amino acid levels in the mutants by 20 to 25% (Figure 4A). Under the growth conditions used, Gln was found to be the main phloem amino acid transport form and presented ~45% of the total amino acids in the wild type (Figures 4A and 4B; compare with Lam et al., 1995; Hunt et al., 2006, 2010). The observed decrease in total amino acids in the mutants was solely due to a significant reduction in Gln levels. Other amino acids were present in much lower amounts than Gln (up to 25 times lower), but their phloem concentration in mutants versus the wild type was unchanged (see Supplemental Figure 1A online).
Figure 4.
Amino Acid Concentrations in Phloem Exudate and Xylem Sap of aap2 Mutants and Altered Levels of 14C-Label in Source and Sink Leaves and Siliques after Feeding of [14C]l-Gln via the aap2 Roots.
Wild-type (WT) plants and an azygous (wild-type) line were used as controls for the tracer studies. Error bars depict sd. Asterisks indicate significant differences from the wild type (P < 0.05). Values above the columns indicate the percentage compared with the wild type, which was set to 100%.
(A) Total free amino acid levels in phloem exudates from source leaves of 6-week-old plants (n = 8).
(B) Gln levels in phloem exudates from source leaves of 6-week-old plants (n = 8).
(C) Total free amino acid levels in the xylem sap of 6-week-old plants (n = 10).
(D) Gln levels in the xylem sap of 6-week-old plants (n = 10).
(E) Accumulated radioactivity in source leaves of 3-week-old plants after [14C]l-Gln feeding to the root (n = 6).
(F) Radioactivity in sink leaves of 3-week-old plants after [14C]l-Gln root feeding (n = 6).
(G) Radioactivity in siliques of 6-week-old plants after [14C]l-Gln root feeding (n = 6).
Transport studies with [14C]-amino acids were performed to further analyze if xylem to phloem transfer and source-to-sink translocation of amino acids are altered in the aap2 mutants. We first resolved which 14C-labeled amino acids should be used for these studies. Xylem sap was examined to determine which amino acids are transported in the xylem (and at what concentrations) and might be moved by AAP2 from the xylem to the phloem. The results showed that around 60% of the total amino acids in the xylem were present as Gln (Figures 4C and 4D). Levels of most other protein amino acids are relatively low (10 to 100 times lower than Gln) and range from 5 to 100 μM dependent on the amino acid (see Supplemental Figure 1B online).
Based on the xylem sap and the phloem exudate analyses (see above) and since Gln is a predicted substrate for AAP2 (Fischer et al., 1995, 2002; see also Rentsch et al., 2007), 14C-labeled Gln was used for the transport studies. Gln was also chosen because previous experiments had shown that Gln is slowly metabolized when fed to the transpiration stream (Sharkey and Pate, 1975). Roots of 3-week-old plants that were in the vegetative stage (prebolting) as well as of 6-week-old plants with fully developed branches and siliques were fed with [14C]l-Gln for 45 min followed by a 15-min chase with cold Gln (Figures 4E to 4G). Levels of 14C-label were determined in fully expanded mature leaves as well as in developing, small sink leaves of the 3-week-old plants. The results showed an increase in 14C-label in aap2 versus wild-type source leaves by 23% (Figure 4E) and a decrease in aap2 sink leaves by 26 to 35% (Figure 4F). When analyzing [14C]l-Gln translocation to siliques during the reproductive phase, results showed an ~50% reduction in label in aap2 silique sinks (Figure 4G).
We used an azygous (wild-type) line as well as wild-type plants as controls for our tracer studies (Figures 4E to 4G). The azygous plants were generated via the same procedure as transgenic plants and were the progeny from the heterozygous transgenic aap2-1 line that had lost the transgene through segregation. By contrast, wild-type plants had not gone through the transformation and tissue culture process. No significant differences were detected between wild-type and azygous lines in any of the tracer experiments, demonstrating that wild-type controls can be considered as reliable controls and that the two mutant alleles do have different properties than wild-type (and azygous) plants.
Seed Nitrogen and Carbon Contents Are Affected When AAP2 Mediated Phloem Loading of Amino Acids Is Repressed
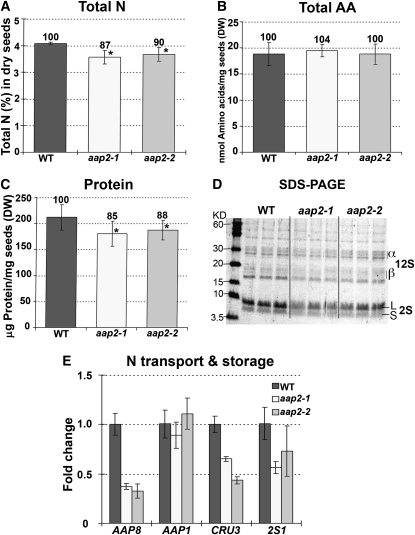
Seed development and N storage are dependent on phloem loading and delivery of amino acids to seed sinks, and feeding experiments showed a reduction in source-to-sink transport of Gln. To resolve effects on N nutrition of mutant seeds, N levels were determined in desiccated seeds of wild-type and aap2 plants (Figures 5A to 5C). While the total N amounts in aap2-1 and aap2-2 seeds were significantly reduced compared with the wild type by 13 and 10%, respectively, the concentrations and composition of free amino acids were not changed, neither in developing nor in dry seeds (Figures 5A and 5B; see Supplemental Figures 2A to 2C online). However, compared with the wild type, the amount of protein was significantly decreased in desiccated seeds of both mutants by 12 to 15% (Figure 5C). Using SDS-PAGE, we further examined if the reduction in total protein content in aap2 seeds reflects a decrease in seed storage proteins (Figure 5D). The stained protein gel revealed a decrease in storage proteins in aap2 seeds, specifically in 12S globulin and 2S albumin. This was further analyzed by expression studies showing a reduction in expression of storage protein genes 2S1 (albumin; Guerche et al., 1990) and CRU3 (globulin; Pang et al., 1988) (Figure 5E). Analysis of seed amino acid transporters demonstrated that expression of the embryo-localized AAP1 (Sanders et al., 2009; Figure 5E) was unchanged in the mutant seeds, but RNA levels of endosperm transporter AAP8 (Schmidt et al., 2007) were strongly decreased, indicating downregulation of AAP8 function when N supply is reduced (see also Sanders et al., 2009).
Figure 5.
Seed N Levels and Expression of Genes Involved in N Transport and Metabolism in Seeds of Wild-Type and aap2 Plants.
Error bars depict sd. Asterisks indicate significant differences from the wild type (WT; P < 0.05). Values above the columns indicate the percentage compared with the wild type, which was set to 100%. DW, dry weight.
(A) Total N levels (%) in dry seeds (n = 4).
(B) Total free amino acids (AA) in dry seeds (n = 4).
(C) Total amount of soluble proteins in dry seeds (n = 5).
(D) SDS-PAGE analysis of soluble proteins in dry seed. Three technical repetitions from independent extractions were done for each line as shown. α- and β-subunits of storage protein 12S globulin. S and L, small and large subunits of the 2S albumin storage protein.
(E) Real-time PCR analysis of genes of N transport and storage in developing seeds. Expression of amino acid transporter AAP8 (At1g10010) and AAP1 (At1g58360) as well as CRU3 (At4g28520) and 2S1 (At4g27140) encoding for 12S globulin and 2S albumin, respectively, were analyzed. Three technical repetitions were done for each line using Arabidopsis ubiquitin 11 (UBQ11, At4g05050) and tubulin β-9 chain (TUB9, At4g20890), respectively, as control genes. Shown is the fold change in gene expression relative to UBQ11 expression, determined from the CT values using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
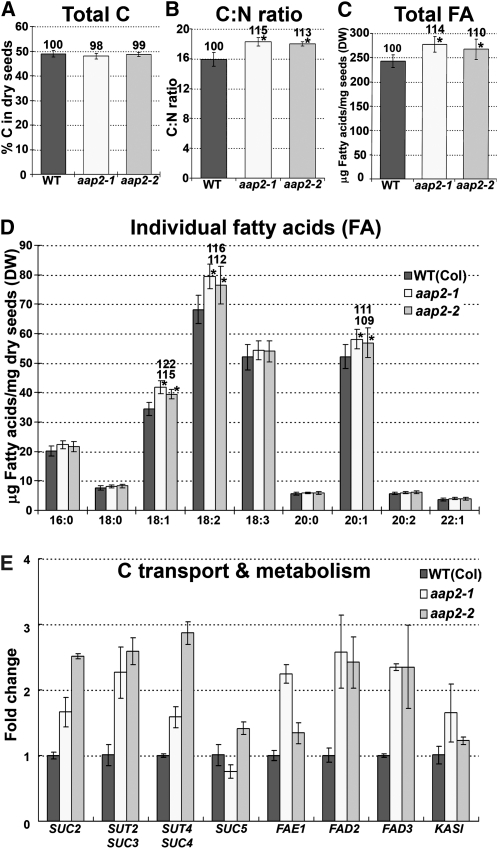
While total N and protein amounts were decreased in aap2 seeds, no differences were found in seed weight between mutants and the wild type (Figure 10E), suggesting that other seed compounds might be elevated. Based on the strong interaction of C and N metabolism (Foyer et al., 2006; Weigelt et al., 2008) total C levels were examined. No changes were detected in total elemental C levels in aap2 seeds, leading to an increased seed C:N ratio of up to ~15% depending on the mutant (Figures 6A and 6B; see also Figure 5A). Derived from these results and the fact that oils can contribute up to 40% of the storage compound content in Arabidopsis seeds (Li et al., 2006), fatty acid levels in aap2 seeds were tested. The results demonstrated that the total fatty acid content was significantly increased in mutant seeds by up to 14% (Figure 6C) and that the increase was due to elevated levels of C18:1, 18:2, and 20:1 fatty acids (Figure 6D). Analysis of transcript levels of fatty acid synthesis genes, specifically FAE1 (James et al., 1995), FAD2, (Covello and Reed, 1996), FAD3 (Browse et al., 1993), and KAS1 (Olsen et al., 2004), showed an increase in expression for all genes tested consistent with the biochemical analyses (Figure 6E). In addition, transcript levels of sucrose transporters SUC2 (Sauer and Stolz, 1994), SUT2/SUC3 (Barker et al., 2000; Meyer et al., 2004), and SUT4/SUC4 (Weise et al., 2000) were upregulated in aap2 seeds, supporting increased seed C delivery for fatty acid synthesis (Figure 6E).
Figure 10.
Yield Characteristics for aap2 and Wild-Type Plants.
Error bars depict sd. Asterisks indicate significant differences from the wild type (WT; P < 0.05). Values above the columns indicate the percentage compared with the wild type, which was set to 100%.
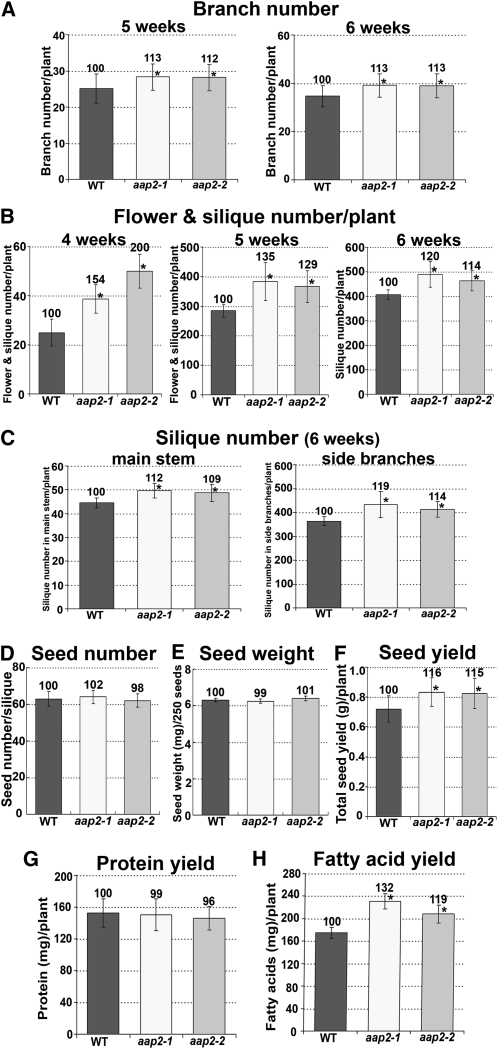
(A) Number of branches per plant after 5 and 6 weeks of plant growth (n = 22).
(B) Number of flower and siliques per plant after 4, 5, and 6 weeks of growth (n = 16).
(C) Number of siliques on the main stem or on all side branches in 6-week-old plants (n = 8).
(D) Seed number per silique (n = 60).
(E) Weight of 250 seeds (n = 5).
(F) Total seed yield (g) per plant (n = 16).
(G) Seed protein yield per plant. Calculated based on seed yield per plant (F) and total seed protein levels (see Figure 5C).
(H) Seed fatty acid yield per plant. Calculated based on seed yield per plant (F) and total seed fatty acid levels (see Figure 6C).
Figure 6.
Analysis of Seed C Levels and Expression of Genes Involved in C Transport and Metabolism in Seeds of Wild-Type and aap2 Plants.
Error bars depict sd. Asterisks indicate significant differences from the wild type (WT; P < 0.05). Values above the columns indicate the percentage compared with the wild type, which was set to 100%. DW, dry weight.
(A) Total C levels in dry seeds (n = 4).
(B) C:N ratio in dry seeds based on total amounts of C (A) and N (see Figure 5A) (n = 4).
(C) Total fatty acid (FA) levels in dry seeds (n = 4).
(D) Concentrations of individual fatty acids in dry seeds (n = 4).
(E) Real-time PCR analysis of genes of C transport and metabolism in developing seeds harvested 9 to 11 d after flowering. SUC2 (At1g22710), SUT2/SUC3 (At2g02860), SUT4/SUC4 (At1g09960), and SUC5 (At1g71890), sucrose transporters; FAE1 (At4g34520), fatty acid elongation 1; FAD2 (At3g12120), fatty acid desaturase 2; FAD3 (At2g29980), fatty acid desaturase 3; KASI (At5g46290), β-ketoacyl-acyl carrier protein synthase I. Three technical repetitions were done for each line using Arabidopsis ubiquitin 11 (UBQ11, At4g05050) and tubulin β-9 chain (TUB9, At4g20890), respectively, as control genes. Shown is the fold change in gene expression relative to UBQ11 expression, determined from the CT values using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Leaf N Metabolism and Transport Processes Are Altered in aap2 Plants
Transport studies using [14C]l-Gln indicated increased uptake of amino acids into source leaves of aap2 plants (Figure 4E). To further analyze this, we determined the leaf RNA levels of Lysine Histidine Transporter 1 (LHT1), a transporter involved in import of amino acids into mesophyll cells (Chen and Bush, 1997; Hirner et al., 2006) and found that expression of LHT1 was strongly increased in the mutant leaves (Figure 7A). However, total N amounts did not change in aap2 source leaves, but levels of most amino acids were decreased (Figures 7B to 7D). Plants store surplus amino acids in the vacuole. To analyze if transient vacuolar storage pools of organic N might be decreased in aap2 leaves, expression of Arabidopsis amino acid (ANT, At3g30390; CAT2, Su et al., 2004) and peptide (PTR2; Steiner et al., 1994) transporters that are predicted to be involved in import of the organic N into the leaf vacuole was examined (Figure 7A; Carter et al., 2004). RNA amounts of all transporters tested were decreased in aap2 leaves.
In contrast with the decreased amino acid levels, leaf protein amounts were strongly increased by up to 30% depending on the mutant (Figure 7E). This indicates that in aap2 source leaves compared with wild-type leaves, more amino acids are channeled into protein synthesis for metabolism or for transient storage. SDS-PAGE analysis and immunoblot results using antibodies against the large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) showed that this primary enzyme in CO2 assimilation was upregulated in aap2 leaves (Figure 7G). When analyzing the leaf protein levels, wild-type and azygous plants were used as controls, and no significant differences were detected between the control plants (Figure 7E). We further complemented the aap2-1 and aap2-2 lines using the AAP2 promoter-GFP-AAP2 construct and determined leaf protein levels in the resulting plants. It was found that the amounts of soluble protein were similar for complementation lines and wild-type plants, respectively (Figure 7F). Together with the tracer studies (see Figures 4E to 4G), these results further confirm that wild-type plants can be considered as true controls.
Leaf C Metabolism and Transport Processes Are Altered in aap2 Plants
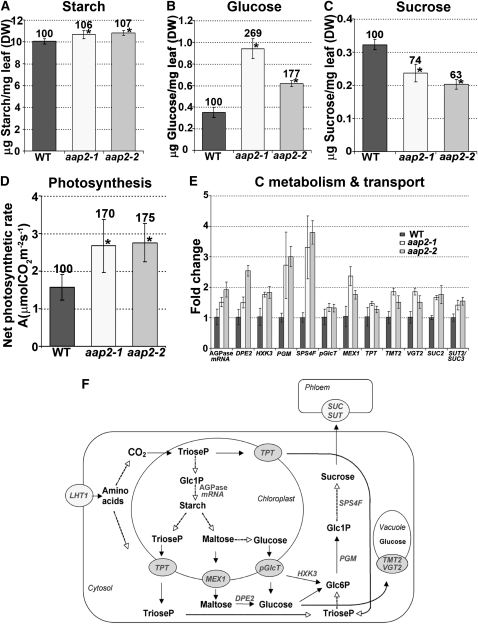
Elevated levels of Rubisco in aap2 leaves, as well as increased sucrose transporter expression and fatty acid levels in mutant seeds, point to elevated C assimilation and sucrose export out of aap2 leaves. We therefore determined the levels of products of C assimilation as well as the photosynthetic rate in aap2 leaves. It was found that the amount of starch in aap2 leaves was slightly elevated, while glucose levels were strongly increased by up to 169% (Figures 8A and 8B). Sucrose levels, on the other hand, were decreased (Figure 8C). Significant changes were also observed for the net photosynthetic rate, which was increased by 70 to 75% in the mutants (Figure 8D).
Figure 8.
Analysis of Starch, Sugar, and Gene Expression Levels of N and C Transport and C Metabolism in Source Rosette Leaves of Wild-Type and aap2 Plants.
Error bars depict sd. Asterisks indicate significant differences from the wild type (WT; P < 0.05). Values above the columns indicate the percentage compared with the wild type, which was set to 100%. DW, dry weight.
(A) Leaf starch levels (n = 5).
(B) Leaf glucose levels (n = 5).
(C) Leaf sucrose levels (n = 5).
(D) Net photosynthetic rate (n = 4). Photosynthetic rates were determined at ambient [CO2].
(E) Real-time PCR analysis of genes of C transport and metabolism in leaves. pGlcT, plastidic glucose transporter (At5g16150); MEX1, plastidic maltose transporter (At5g17520); TPT, plastidic triose phosphate/phosphate translocator (At5g46110); TMT2 (At4g35300) and VGT2 (At5g17010), vacuole hexose importer; SUC2 (At1g22710) and SUT2/SUC3 (At2g02860), plasma membrane sucrose transporters (phloem loading); RNA levels of At1g74910 encoding for AGPase (ADP glucose pyrophosphorylase); HXK3, hexokinase (At1g50460); DPE2, cytosolic transglucosidase (At2g40840); PGM, phosphoglucomutase (At1g70730); SPS4F, sucrose phosphate synthase (At4g10120). Three technical repetitions were done for each line using UBQ11 (At4g05050) and TUB9 (At4g20890), respectively, as control genes. Shown is the fold change in gene expression relative to UBQ11 expression, determined from the CT values using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
(F) Overview of carbon metabolism and transport processes in source leaves and genes analyzed in aap2 plants. For description of the specific genes and references, see Supplemental Table 1 online.
To analyze if changes in photosynthesis and metabolite levels are consistent with expression of genes of C metabolism and transport in leaf cells, RNA levels of At1g74910 encoding for ADP-glucose pyrophosphorylase/AGPase (controlling starch synthesis; Villand et al., 1993), DPE2 (releases glucose from maltose; Chia et al., 2004), HXK3 (associated with the chloroplast outer envelope; phosphorylation of glucose, probably before it enters the cytosol; Wiese et al., 1999; Karve et al., 2008), PGM (α-d-glucose 1-phosphate and α-d-glucose 6-phosphate interconversion; Fettke et al., 2008), and SPS4F (conversion of UDP-glucose + d-fructose 6-phosphate to UDP + sucrose 6-phosphate; Strand et al., 2000), as well as of C transporters were examined (Figures 8E and 8F; see Supplemental Table 1 online). Expression of the genes examined and involved in synthesis of sugars was upregulated. In addition, transcript levels of transporters exporting triose phosphate (TPT; Schneider et al., 2002), maltose (MEX1; Niittylä et al., 2004), and glucose (pGlcT; Weber et al., 2000) from the chloroplasts into the cytosol are increased, as well as expression of transporters involved in phloem loading of sucrose SUC2 (Sauer and Stolz, 1994; Truernit and Sauer, 1995) and SUC3/SUT2 (Barker et al., 2000; Meyer et al., 2004). Expression of vacuolar glucose importers (TMT2, Wormit et al., 2006; VGT2, Aluri and Büttner, 2007) were also increased in aap2 plants, indicating that the observed high glucose levels might be due to increased import and accumulation of glucose in the vacuole. Together, the results support increased C assimilation and export from the leaf for sink C supply when N partitioning within the plant is altered.
Leaf Development Is Affected in aap2 Plants
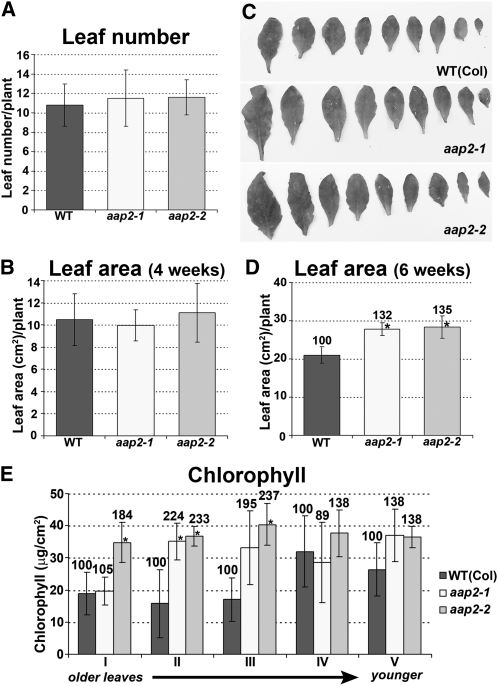
To determine if leaf development is affected in aap2 mutants, we analyzed if changes occurred in leaf number, leaf size, and leaf chlorophyll content. Leaf numbers of aap2 and wild-type plants did not significantly alter during the growth period (Figure 9A), and plants developed up to 11.6 leaves per plant, including very tiny leaves. If only sufficiently developed leaves were included, an average of nine leaves per wild-type and mutant plant was counted (Figure 9C). While no differences could be found in the leaf size of 4-week-old plants (Figure 9B), an increase was identified in aap2 after 6 weeks of growth (Figure 9C). The leaf surface was significantly increased by up to 35% dependent on the mutant (Figure 9D). When analyzing the chlorophyll content of the rosette leaves of 4- and 6-week-old plants, no differences were detected between mutants and the wild type (see Supplemental Figures 3A and 3B online). We further tested leaves of 7-week-old plants that started to senesce and found that the chlorophyll content was generally higher in the mutant compared with wild-type leaves (Figure 9E). By week 8, senescence and desiccation were strongly progressed in both wild-type and mutant plants, making further measurements impossible.
Figure 9.
Analysis of aap2 Leaf Development.
Error bars depict sd. Asterisks indicate significant differences from the wild type (WT; P < 0.05). Values above the columns indicate the percentage of each value in aap2 plants compared with the wild type, which was set to 100%.
(A) Leaf number of 6-week-old plants (n = 8).
(B) Leaf surface area of 4-week-old leaf area (n = 6).
(C) Images of rosette leaves of 6-week-old plants.
(D) Leaf surface area of 6-week-old leaf area (n = 6).
(E) Chlorophyll content of the 7-week-old plants (n = 5). Ten developed leaves in wild-type and aap2 plants were analyzed in groups of 2 along their age. Leaves 1 and 2 represent leaf group I. They were the oldest leaves of the rosette, and senescence was evident (yellowing of leaves). Leaf group V consisted of fully developed (mature) green leaves that were the youngest within the rosette.
Seed Yield Is Increased in aap2 Plants
To resolve if changes in C and N partitioning in aap2 plants affect sink set and growth, the number of side branches, flowers, and siliques that developed after 4, 5, and 6 weeks of growth was determined. Wild-type and mutants bolted after 3 weeks of growth and developed a main stem. Side branches started to grow after 4 weeks of plant growth. The branch number per aap2 plant was increased after 5 and 6 weeks by around 13% (Figure 10A). Analysis of flower and silique set on the main stem revealed an increase of 50 to 100% in the mutants after 4 weeks of growth (Figure 10B). After 5 weeks, flower and silique development was improved by up to 35% dependent on the aap2 lines. Differences in silique set up to 20% between mutant and the wild type were observed after a growth period of 6 weeks. The increased silique set was not only due to the increase in branch number in aap2 plants (Figure 10A) but also to an increase in silique number per main (9 to 12%) and side branch (14 to 19%), respectively (Figure 10C).
When analyzing yield characteristics, no effect was observed for seed set per silique, and the weight per seed was unchanged in mutants compared with the wild type (Figures 10D and 10E). Nevertheless, the improved number of siliques per plant led to an increase in total seed yield per mutant plant of 15 to 16% (Figure 10F). As a consequence, the total seed protein yield per plant was similar in wild-type and mutant plants (Figure 10G), even though the protein content per aap2 seed was decreased (Figure 5C). Even more notably, the total amount of oil in the mutants compared with the wild-type plant was significantly increased by 32 and 19% for aap2-1 and aap2-2, respectively (Figure 10H).
DISCUSSION
AAP2 Functions in Xylem-Phloem Transfer of Amino Acids
Amino acids are found in both xylem and phloem, but seeds or other sinks receive most of their organic N from the phloem, which requires efficient transfer of the N between the xylem and the phloem (Pate et al., 1975, 1977). It has been demonstrated, mainly in legumes, that movement of amino acids from xylem to phloem is quantitatively relevant in supplying fruit sinks with N (Atkins et al., 1979; for review, see van Bel, 1990; Atkins, 2000). Here, we provide evidence that AAP2 functions in xylem-phloem transfer of amino acids. It was established that AAP2 is a plasma membrane transporter that is expressed in the phloem and specifically in the companion cells (Figures 1B to 1L and 2), which is consistent with recent microarray studies identifying AAP2 as a companion cell transporter (Brady et al., 2007; Zhang et al., 2008). Our results further show that AAP2 is expressed in the phloem throughout the plant. The majority of xylem sap amino acids end up in the leaf due to high transpiration rates, and reallocation of amino acids from the xylem to the phloem occurs in the major veins, the location of AAP2 function in leaves (Figures 2D to 2F).
A more direct demonstration of AAP2 function in xylem to phloem movement and phloem loading is provided by xylem sap and leaf phloem exudate analyses (Figures 4A to 4D). Gln was the predominant amino acid in both transport streams, and in aap2 mutants, the Gln phloem concentration was strongly decreased. Furthermore, phloem and xylem levels of other amino acids, which are present at much lower concentrations than Gln, were not affected in aap2 plants. In heterologous expression systems (yeast and Xenopus oocytes), AAP2 transports a broad spectrum of amino acids (including Gln) when present in relatively high concentrations (Fischer et al., 1995, 2002; see also Rentsch et al., 2007). The observed change in only Gln in aap2 phloem exudate is consistent with the predicted role of AAP2 in moderate/low affinity but high capacity transport. It further suggests that under physiological conditions, AAP2 might preferentially transfer Gln from the xylem to phloem. A role in movement of amino acids between the different vascular systems has also recently been demonstrated for AAP6, a xylem parenchyma amino acid transporter (Okumoto et al., 2002). Differing from our studies, phloem sap analysis of the Arabidopsis aap6 mutant showed reduced levels of less (Lys, Phe, and Leu) and more (Asp) abundant amino acids (Hunt et al., 2010). However, in contrast with AAP2, AAP6 transports Asp and has a much higher affinity for neutral and acidic amino acids than AAP2 (Fischer et al., 2002).
Strong support for the predicted function of AAP2 in xylem-phloem transfer is further provided by short-term pulse-chase experiments. [14C]l-Gln fed to aap2 roots accumulated in source leaves, and allocation of label to sink leaves was decreased compared with the wild type. Similarly, flux of 14C-label to aap2 siliques was strongly reduced (Figures 4E to 4G). It cannot be excluded that some of the transported [14C]l-Gln might have been converted to other N compounds or incorporated into proteins. However, our experiments were performed over a very short time frame, and Gln seems to be slowly metabolized when fed to the transpiration stream (Sharkey and Pate, 1975).
Importance of AAP2 Function for Seed Development and Storage Reserve Accumulation
Arabidopsis seeds contain high amounts of proteins and oils (Baud et al., 2002) requiring a tight control of C/N partitioning and metabolism. Amino acids levels were unchanged in aap2 seeds, while amounts of protein were decreased (Figures 5B and 5C; see Supplemental Figures 2A to 2C online). Similar results were obtained with mutants of AAP1, an amino acid transporter responsible for amino acid import into the embryo (Sanders et al., 2009), suggesting that in seeds/embryos, a steady pool of amino acids is maintained before the N is channeled into protein synthesis. While protein amounts were decreased as a consequence of reduced N delivery, the oil content in aap2 seeds was strongly increased (Figures 5C, 6C, 6D, and 10H). This was further confirmed by an increased C:N ratio, which generally accounts for elevated oil levels in seeds (Figures 5A, 6A, and 6B; Li et al., 2006). The negative relationship between seed N levels and oil content is consistent with other reports (see Rathke et al., 2004 and references within) and has been related to competition for C skeletons during C/N metabolism (Bhatia and Rabson, 1976). Since total N amounts were decreased in aap2 seeds, more C was presumably channeled into oil synthesis. In addition, the higher C:N ratio in aap2 seeds points to increased delivery of carbohydrates, specifically sucrose, at reduced amino acid supply. This coincides with the upregulation of sucrose transporters in seeds and induction of fatty acid synthesis related gene expression (Figure 6E).
Another surprising result was that total seed yield was increased by ~15% in aap2 plants (Figures 10A to 10F), suggesting that, while xylem-phloem transfer by AAP2 is important for seed N storage (see above), abolishment of AAP2 function does not cause N deficiency leading to reduced sink development (Crawford, 1995; Ma et al., 1998; Song et al., 1997; Forde, 2002a, 2002b; Schmidt et al., 2007; Sanders et al., 2009; Hunt et al., 2010). In Arabidopsis, the majority of amino acids are synthesized in source leaves (Coruzzi, 2003), and loading of the amino N into the minor vein phloem might be essential for sink initiation and growth rather than xylem-phloem transfer. However, such phloem loaders still need to be identified and their physiological function determined (Tegeder and Rentsch, 2010).
Effects of N Reallocation on Source Leaf Metabolism
Since both organic N and C compounds are important for sink development, alterations in leaf C assimilation, metabolism, and source-sink transport might be responsible for the increased seed and oil yields in aap2 plants (Figures 10A to 10H). These changes are most probably related to modifications in N uptake into aap2 mesophyll cells, as suggested by upregulation of amino acid importer LHT1 (Figure 7A) and increased uptake of 14C-label in leaves following root feeding with [14C]l-Gln (Figure 4E). Strangely, the free amino acid levels in whole aap2 leaf extracts were decreased, while protein levels were increased (Figures 7C to 7G). Future studies need to resolve if amino acid concentrations are altered in the different cellular compartments of aap2 leaves (e.g., in the vacuole; Figure 7A) and if changes in levels of specific amino acids or other signaling compounds might trigger increased usage of amino acids for protein and enzyme synthesis.
Generally, N partitioning to leaves positively regulates photosynthesis and consequently improves allocation of carbohydrates to sink tissues for vegetative and reproductive growth (Sage, 1994; Nakano et al., 1997, Martin et al., 2002; Krapp et al., 2005; Smith and Stitt, 2007). Indeed, levels of Rubisco and rate of photosynthesis were increased in mature aap2 leaves, and expression of genes encoding for enzymes involved in C assimilation and transport were upregulated (Figures 7G, 8E, and 8F). Furthermore, starch concentrations in aap2 leaves were moderately increased and glucose concentrations strongly elevated (Figures 8A and 8B), which is in conflict with the assumption that photosynthesis and sugar metabolism are upregulated by low rather than high sugar concentrations (Rolland et al., 2006). However, many studies have described that C and N act jointly and that the C-N balance might regulate C assimilation rather than the specific C metabolite concentration (Oliveira and Coruzzi, 1999; Stitt and Krapp, 1999; Coruzzi and Zhou, 2001; Martin et al., 2002; Sato et al., 2009). On the other hand, glucose might be transiently stored in the vacuole to avoid repression of C photosynthesis as supported by upregulation of vacuolar hexose importers in aap2 leaves (Figure 8E). By contrast, sucrose levels in aap2 leaves were decreased compared with the wild type, probably caused by increased export of sucrose due to increased sink strength (Figure 8C). This coincides with upregulation of sucrose transporter expression in aap2 leaves and agrees with the increased sink numbers in aap2 plants as well as a higher C:N ratio and elevated fatty acid levels in aap2 seeds (Figures 6, 8C, and 10). Sucrose levels might also be kept low to further prevent feedback inhibition of photosynthesis (Stitt, 1991; Rolland et al., 2006), including expression of photosynthesis genes (Dijkwel et al., 1996, 1997).
Effects of N Reallocation on Source and Sink Development
When addressing leaf development, it was found that after 6 weeks of growth, the leaf surface in aap2 plants was strongly increased (Figures 9C and 9D). In addition, senescence was slightly delayed (Figure 9E). These observed effects might be due to changes in sugar levels in aap2 leaves and associated sugar-sensing mechanisms as part of a complex network regulating development and senescence of leaves (Riou-Khamlichi et al., 2000; Hanson et al., 2001; for review, see Paul and Pellny, 2003). Moreover, our data suggest that a combination of elevated photosynthesis (see above), increased leaf area for photosynthesis, and senescence (prolongation of photosynthesis) affects C partitioning from source to sink in aap2 plants, resulting in improved sink development and seed oil levels.
Decreased movement of N to seeds and increased allocation to aap2 leaves also led to increased development of side branches, flowers, and siliques in aap2 plants (Figures 10A to 10F). These differences were due to changes in the rate of sink development rather than to extended growth (Figures 10A to 10C). While timing of flowering was similar in the wild type and mutants, within the first week of inflorescence growth, flower set on the main stem was strongly increased in aap2 plants. In addition, the speed of branching was increased and more flowers and siliques developed per branch after 5 and 6 weeks of growth, respectively. These results demonstrate that alterations in N transport and subsequent changes in leaf metabolism affect the speed of progression through a developmental series finally leading to increased seed yield. While altered C levels in the phloem might provide the signal for the observed changes, modulation of N transport processes in the phloem might be equally important for the improved sink set and growth in aap2 mutants (see above) as an elevated C:N ratio in the phloem sap seems to be essential for flower induction and fruit development in Arabidopsis (Corbesier et al., 2002) in addition to the accurate amount of allocated C (Goetz et al., 2001; Zhang et al., 2001; Chen et al., 2004).
Conclusions and Perspectives
Based on the data, we can conclude that AAP2 is functioning in xylem to phloem transfer of amino acids with consequences for seed organic N and C levels and that its knockout leads to changes in C to N balance rather than causing N limitations for sink development. The aap2 plants further demonstrate the influence of N partitioning processes on C/N metabolism and transport and reinforce evidence for tight interactions of C/N metabolic pathways between source and sink. Finally, they provide a link between N allocation, source and sink growth, and the speed of development.
Several approaches have been followed with varying success to improve N use efficiency for plant growth under sufficient and limited N supply, such as manipulation of N uptake and N metabolism (for review, see Hirel et al., 2007). Our work reveals the possibility that AAP2 may control sink growth and storage compound accumulation and provides evidence that fruit development might depend upon amino acid allocation to the photosynthetic machinery and for carbohydrate synthesis. This in turn has consequences for source leaf development and C export and partitioning to sinks, affecting sink development (number and pace) and seed oil levels. Future studies might help to resolve if manipulation of N allocation and fine-tuning of N long-distance transport processes can provide a strategy for maintaining or increasing seed number and storage compounds under N limitation. Considering the complex interplay of the different pathways and organs, high-resolution analyses of the localization and concentrations of amino acids (and sugars) are needed to discover the signals that trigger the observed responses and to advance the field of amino acid transport and its importance for cell and plant physiology.
METHODS
Materials
Arabidopsis thaliana ecotype Columbia (Col-0) lines with T-DNA insertion in AAP2 (At5g09220) were obtained from the ABRC (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm). Information about these lines was acquired from the SIGnal database (Alonso et al., 2003). Homozygous plants of aap2 mutant lines (aap2-1, SALK_047237; aap2-2, SALK_143597) were identified by PCR using T-DNA left border primer SALK LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′ and gene-specific primers for aap2-1 (5′-CCGCTGCCGCCAATAACCACC-3′ and 5′-CCAAAAACAGAGTGAGAAAAACAGAG-3′) and aap2-2 (5′-GACAAAAATGCGGTTTTGCTCCAGG-3′and 5′-TGCAACTTGAACGATTCCAAGAGC-3′). The PCR products were sequenced to confirm the positions of the T-DNA insertions.
Plant Growth Conditions and Harvest
Arabidopsis plants were grown in a growth chamber in 36-well Com-Packs or single pots (T.O. Plastics) of 10 cm (xylem and phloem sap and phenotypic analyses) or 20 cm (photosynthesis measurements) in diameter. The soil was a mixture of peat (60%), pumice (20%), and sand (20%). Growth conditions were generally 16 h light at 20°C during the day and 16°C at night. Light conditions were around 200 μmol photons m−2 s−1, and humidity was 50%. Plants were given 1.75 g/L Peters 20-20-20 all-purpose plant food (Scotts-Sierra Horticultural Products) once a week. Under these growth conditions, the life cycle of the Arabidopsis plants from seeds (planting) to seeds (harvest of desiccated seeds) was ~8 weeks. Two independent sets of mutant and wild-type plants were grown for analyses and confirmation of the results.
For gene expression and most biochemical analyses, pools of source rosette leaves (first six fully developed leaves) of at least 15 plants per line were harvested after 6 weeks of growth or germination (WAG). At this time point, side branches carrying a large number of siliques (Figures 10A to 10C) were developed. Some experiments were performed with younger plants (e.g., tracer studies and phenotypic analyses). At 3 WAG, the Arabidopsis plants were in the prebolting phase and grew up to nine rosette leaves. Four of these leaves were fully expanded and were considered as source leaves, and the two smallest were used as sink leaves. Bolting and development of the main stem occurred 3 to 4 WAG, and the first side branches started to appear 4 WAG (Figure 10A). Developing seeds were collected from 6-week-old plants. Flowers were marked and seeds were collected 9 to 11 d after flowering. Alternatively, desiccated seeds were collected. For gene expression and biochemical analyses, materials were immediately frozen in liquid nitrogen and stored at −80°C or lyophilized and then stored in the dark in a desiccator. To determine the seed number per silique, fully developed siliques from the main stem (bottom 15 siliques) were transferred to 70% ethanol to remove the chlorophyll. Seeds per cleared silique were counted under the dissecting microscope (Wild Heerbrugg). For collecting dry seeds, the Arasystem was used (Lehle Seeds). Plants used for phenotypic analyses were grown without any growth restrictions.
Preparation of GFP Constructs
Constructs were prepared for GFP-AAP2 expression and localization studies in yeast cells, onion (Allium cepa) cells, and Arabidopsis, respectively. The constructs for yeast complementation experiments and particle bombardment of epidermal onion cells were made by replacing the AAP1 cDNA (NotI) in previously described constructs (Lee et al., 2007) with AAP2 cDNA, resulting in PMA1 promoter-GFP-AAP2 term/pDR196 (yeast) and cauliflower mosaic virus 35S promoter-GFP-AAP2/pDH51 (onion). For localization of GFP-AAP2 in Arabidopsis, the AAP2 promoter (Hirner et al., 1998) was amplified by PCR and cloned (HindIII, blunt) into the SmaI site of GFP-AAP2/pDH51. The AAP2 promoter-GFP-AAP2 cassette was cut with KpnI, blunted, and transferred into the binary vector pCB302 (Xiang et al., 1999). As controls, GFP constructs were used (Lee et al., 2007).
GFP-AAP2 Expression in Yeast, Onion Cells and Arabidopsis Plants
To analyze if the GFP-AAP2 encodes a functional fusion protein, functional complementation of Saccharomyces cerevisiae strain 22574d (MAT-α, ura 3-1, gap 1-1, put 4-1, uga 4-1) expressing GFP-AAP2 was performed as described by Lee et al. (2007). Selection for yeast growth was performed on nitrogen-free medium supplemented with the amino acid Pro (0.3 g/L) as sole nitrogen source. For nonselective conditions, minimal media was supplemented with ammonium sulfate (0.3 g/L).
GFP-AAP2/pDH51 and GFP/pDH51 were introduced into onion epidermal cells by particle bombardment according to Chiu et al. (1996) and the manufacturer’s protocol (Bio-Rad). Bombarded cells were incubated overnight at room temperature on moist filter paper. Epidermal onion cells were plasmolyzed using 1 M sucrose solution prior to the microscopy analysis.
Arabidopsis plants (the wild type and aap2-1 and aap2-2 lines) were transformed with AAP2 promoter-GFP-AAP2/pCB302 constructs (Clough and Bent, 1998), and transgenic plants were selected as mentioned by Lee et al. (2007). Hand sections were prepared from different organs of transgenic Arabidopsis plants as described previously (Mano et al., 2002), and the GFP-AAP2 proteins were localized using confocal microscopy (Carl Zeiss).
Analysis of Phloem Exudates and Xylem Sap, and [14C]l-Gln Transport Studies
Phloem exudates were obtained from 6-week-old Arabidopsis plants as previously described (Deeken et al., 2008). Aliquots of 100-μL phloem exudates were lyophilized and resuspended in 33 μL water, and 1/10 volume 1 n HCL was added to precipitate the EDTA. The samples were incubated on ice for 30 min, followed 30 min centrifuging at 4°C and 13,000 rpm. The supernatants were diluted 1:1 with double deionized water. Xylem sap was obtained from 6-week-old plants as previously described (Bauer et al., 1997). Briefly, the soil was saturated and plants were cut right above the root crown. Xylem sap was collected for 30 min (first drops were discarded) from 10 aap2 or wild-type plants and stored at −80°C. For HPLC analysis, xylem sap was diluted 10 times. The sap was diluted 1:50 with double deionized water. Diluted phloem and xylem sap was used for amino acid analyses by HPLC (see below).
For transport studies, 3- and 6-week-old plants grown in single pots were removed from the soil and their main root was cut ~1 cm below the stem-root transition area in 5 mM EDTA buffer (270 mosmol, pH 7.5). The plants were transferred to a microcentrifuge tube with 1 mL of a solution containing 2.5 mM MES, 5 mM EDTA, and 2 mM cold Gln and [14C]l-Gln (1 μCi, 37 kBq; Moravek Biochemical and Radiochemicals), and fed for 45 min at 300 μmol photons m−2 s−1 illumination, followed by feeding for 15 min using a chase solution (same as above but without [14C]l-Gln). The 3-week-old plants contained up to nine leaves. The first four mature source leaves and two developing sink leaves were harvested separately from a total of at least six plants and their surface was scanned. The radioactivity was determined by liquid scintillation spectrometry (Packard Instruments) and calculated per cm2 leaf area. From the 6-week-old plants, 30 siliques were harvested from the main stem of six single plants, pooled and treated as described by Schmidt et al. (2007) before the radioactivity was determined.
RNA Extraction and Gene Expression Analysis
Total RNA was extracted from seeds and rosette leaves as previously described (Pélissier and Tegeder, 2007). Following DNA digestion with TURBO DNase (Applied Biosystems/Ambion), reverse transcription was performed using 1.5 μg RNA and Moloney murine leukemia virus reverse transcriptase (MMLV-RT; Invitrogen). The resulting cDNAs were diluted 1:4 or 1:10 with water for quantitative RT-PCR according to Sanders et al. (2009). Three technical replicates were run for each primer pair using a reaction mix of Platinum Taq Buffer (Invitrogen), 200 μM deoxynucleotide triphosphate (Fisher Bio-Reagents), 2.5 mM MgCl2, 0.25× SYBR Green I, 50 nM ROX, 0.5 units Platinum Taq DNA polymerase (all Invitrogen), and 0.25 μM primers. Real-time amplification data were collected with an Applied Biosystems 7500 thermal cycler. Genes related to N or C transporters and metabolism as well as seed storage proteins were analyzed (see Supplemental Table 1 online). Gene specific primers for quantitative RT-PCR were designed in the VECTORNTI Suite, version 9, and data analysis was performed as explained earlier (Sanders et al., 2009). Primer sequences can be found in Supplemental Table 1 online.
Elemental N and C, Amino Acid and Fatty Acid Analyses
Lyophilized material of rosette leaves or dry seeds (1 to 2 mg) was used for total C and N quantification with the Costech ECS 4010 elemental analyzer as recently described (Sanders et al., 2009). To determine the amino acid content and composition, extracts were prepared from 30 mg of lyophilized seeds or rosette leaves and taken for HPLC according to Sanders et al. (2009). For oil content quantification, extracts were obtained from 250 mg of dry seeds (Sanders et al., 2009) and analyzed by gas chromatography with an Agilent 6890 series gas chromatograph outfitted with a 30 × 0.53-mm EC-WAX column and a flame ionization detector. The peak areas of all individual fatty acids were determined to establish the amount of total and single fatty acids.
Protein Extraction and Quantification
Extraction buffer (350 μL of 100 mM Tris-HCl, pH 8, 0.5% [w/v] SDS, 10% [v/v] glycerol, and 2% [v/v] 2-mercaptoethanol) was added to 1 mg lyophilized leaf or seed tissue followed by grinding and two centrifugation steps at 14,000 rpm and 4°C for 10 min. The supernatant was taken for protein quantification as well as SDS-PAGE and immunoblot analysis. The amount of protein was determined using the NanoOrange protein quantification kit according to the manufacture’s protocol (Invitrogen) and a Bio-Tek Synergy HT microplate reader (excitation, 480 nm; emission, 590 nm) equipped with KC4 v.3.4 software (http://www.biotek.com). For SDS-PAGE and immunoblot analyses, 20 μL and 10 μg of protein extracts, respectively, were added to 6× loading buffer (62.5 mM Tris-HCl, pH 6.8, 10% [v/v] glycerol, 2% [w/v] SDS, 5% [v/v] 2-mercaptoethanol, 8 M urea, and 0.002% [w/v] bromophenol blue), boiled for 10 min, loaded onto 12% (leaf extract) or 15% (seed extract) polyacrylamide gels, and separated for 2 h at 100 V using a minigel apparatus (Bio-Rad). For visualization of proteins, gels were stained with Coomassie Brilliant Blue R 250 overnight and destained in 40% (v/v) methanol and 10% (v/v) acetic acid solution. For immunoblot analysis, gels were incubated for 20 min in buffer (25 mM Tris, 192 mM glycine, 20% [v/v] methanol, and 0.02% [w/v] SDS) followed by protein transfer to a nitrocellulose membrane according the instructions (Bio-Rad). The membrane was blocked with TBST buffer (10 mM Tris-HCl, 150 nM NaCl, 0.1% Tween 20, 1% BSA, and 5% dry milk powder) for 30 min at room temperature with gentle shaking and then incubated overnight at 4°C in blocking buffer with an antibody against the large and small Rubisco subunit, respectively. Rubisco antibodies were provided by G. Edwards (anti-spinach [Spinacia oleracea] Rubisco LSU [1:10,000; courtesy of B. McFadden], anti-Amaranthus hypochondriacus Rubisco SSU [1:5000; courtesy of J. Berry]). After washing in TBST, the membrane was incubated for 2 h at room temperature in a 1:10,000 diluted secondary antibody conjugated to alkaline phosphatase (Sigma-Aldrich), washed with TBS (0.01 M Tris and 0.5 M NaCl), and equilibrated with detection buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2) for 5 min, and Rubisco was visualized using NBT/BCIP (Roche) in detection buffer for 15 to 30 min.
Carbohydrate Analyses
For analyses of glucose, sucrose, and starch content in leaves, 0.5 mL ethanol (80%) was added to 3 mg lyophilized rosette leaves, the mix was incubated at 75°C for 30 min, centrifuged at 13,000 rpm for 5 min (Eppendorf), and the supernatant was transferred to a fresh tube. Another 0.5 mL 80% ethanol was added to the pelleted plant tissue and proceeded as described. The supernatant was collected, dried under vacuum at 17,000 rpm (Savant), and dissolved in 200 μL distilled water. One hundred microliters of this solution was used for determining the amount of glucose with Amplex Red Glucose/Glucose Oxidase Assay according to the manufacture’s protocol (Invitrogen). In this assay, glucose oxidase reacts with d-glucose to form d-gluconolactone and H2O2, and the latter then reacts with horseradish peroxidase and a specific kit reagent to generate the red fluorescent oxidation product. This product was measured at wavelength 530 nm (excitation) and 590 nm (emission) with a Bio-Tek Synergy HT microplate reader equipped with KC4 v3.4 software, and the glucose concentration was determined using a standard curve. To quantify sucrose, 120 units of invertase (Sigma-Aldrich) were added to 100 μL of the sugar extract and incubated at 50°C for 10 min to hydrolyze sucrose to glucose, which then was measured. For starch quantification, the remaining pellets from the extraction described above were washed with water, dissolved in 500 μL buffer (0.2 n KOH and 0.1 n HAC, pH 4.5), and incubated at 95°C for 40 min. Three units of amylogucosidase (Sigma-Aldrich) and 0.25 units of α-amylase (Sigma-Aldrich) were added for hydrolysis of starch to glucose at 40°C overnight. The reaction mix was centrifuged (13,000 rpm, room temperature), and the supernatant was taken for determining starch-derived glucose.
Photosynthetic Gas Exchange and Chlorophyll Analysis
Gas-exchange measurements were performed using a LI-6400XT photosynthesis system (LI-COR) and six plants per aap2 mutant line and wild type. They were grown in an environmentally controlled chamber under conditions described above but at a 12-h daylength to ensure that leaves were large enough for the leaf chamber. Plants used for analysis were 8 weeks old. Two or three fully developed, mature leaves from each plant were measured. A light response curve was used to determine the PPFD at which the plants reached the highest photosynthetic rate. The PPFD used for the reported data was 300 μmol m−2 s−1 at a partial pressure (CO2) of 385 ppm.
To determine the chlorophyll content of mutant and wild-type leaves, a standard protocol for ethanol extraction was used. Leaf disks were harvested with a cork-borer from six leaves of five 4-week-old plants. For the 6- and 7-week-old plants, 10 mature leaves from five wild-type and aap2 plants, respectively, were analyzed in groups of two along their age. Leaves 1 and 2 represent leaf group I. They were the oldest leaves of the rosette, and in 7-week-old plants, senescence was evident for these leaves (yellowing of leaves). Leaf group V consisted of mature, green leaves that were the youngest within the rosette.
Statistical Analysis
Results are shown for one growth set but are representative of two independently grown sets of plants and presented as a mean standard deviation (±sd) of at least four biological repetitions, each from a pool of a minimum of 15 plants. Statistical significance was determined with a one-way analysis of variance using SigmaStat 3.0 (Systat Software).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At5g09220 (AAP2) and in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Concentrations and Composition in the Phloem and Xylem of aap2 Plants.
Supplemental Figure 2. Amino Acid Levels in Developing and Desiccated aap2 Seeds.
Supplemental Figure 3. Chlorophyll Levels in Four and 6-Week-Old aap2 Plants.
Supplemental Table 1. Overview of Primers Used for Expression Analyses of Genes of N and C Metabolism and Transport.
Supplementary Material
Acknowledgments
We acknowledge the support from our greenhouse manager Chuck Cody. We thank Ann Sanders and Gengxiang Jia for help with screening and harvesting the mutant lines and John Browse, the Washington State University Franceschi Microscopy and Imaging Center, and the Washington State University Molecular Biology Core for technical support. This work was funded by grants from the National Science Foundation (IOS 0448506 and IOS 1021286) to M.T., which are greatly appreciated.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aluri S., Büttner M. (2007). Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc. Natl. Acad. Sci. USA 104: 2537–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C.A. (2000). Biochemical aspects of assimilate transfers along the phloem path: N-solutes in lupinus. Aust. J. Plant Physiol. 27: 531–537 [Google Scholar]
- Atkins C.A., Pate J.S., Layzell D.B. (1979). Assimilation and transport of nitrogen in nonnodulated (No3-Grown) Lupinus albus L. Plant Physiol. 64: 1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L., Kühn C., Weise A., Schulz A., Gebhardt C., Hirner B., Hellmann H., Schulze W., Ward J.M., Frommer W.B. (2000). SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Boutin J.P., Miquel M., Lepiniec L., Rochat C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40: 151–160 [Google Scholar]
- Bauer A., Urquhart A.A., Joy K.W. (1997). Amino acid metabolism of pea leaves: diurnal changes and amino acid synthesis from 15N-nitrate. Plant Physiol. 59: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia C.R., Rabson R. (1976). Bioenergetic considerations in cereal breeding for protein improvement. Science 194: 1418–1421 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Browse J., McConn M., James D., Jr, Miquel M. (1993). Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268: 16345–16351 [PubMed] [Google Scholar]
- Carter C., Pan S., Zouhar J., Avila E.L., Girke T., Raikhel N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Bush D.R. (1997). LHT1, a lysine- and histidine-specific amino acid transporter in Arabidopsis. Plant Physiol. 115: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Xia X., Zheng H., Yuan Z., Huang H. (2004). The GAOLAOZHUANGREN2 gene is required for normal glucose response and development of Arabidopsis. J. Plant Res. 117: 473–476 [DOI] [PubMed] [Google Scholar]
- Chia T., Thorneycroft D., Chapple A., Messerli G., Chen J., Zeeman S.C., Smith S.M., Smith A.M. (2004). A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 37: 853–863 [DOI] [PubMed] [Google Scholar]
- Chiu W.L., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Bernier G., Périlleux C.C. (2002). C:N ratio increases in the phloem sap during floral transition of the long-day plants Sinapis alba and Arabidopsis thaliana. Plant Cell Physiol. 43: 684–688 [DOI] [PubMed] [Google Scholar]
- Coruzzi G.M. (2003). Primary N-assimilation into amino acids in Arabidopsis. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0010, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G.M., Zhou L. (2001). Carbon and nitrogen sensing and signaling in plants: Emerging ‘matrix effects’. Curr. Opin. Plant Biol. 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Covello P.S., Reed D.W. (1996). Functional expression of the extraplastidial Arabidopsis thaliana oleate desaturase gene (FAD2) in Saccharomyces cerevisiae. Plant Physiol. 111: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N.M. (1995). Nitrate: Nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G., Hedrich R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Dijkwel P.P., Huijser C., Weisbeek P.J., Chua N.H., Smeekens S.C.M. (1997). Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9: 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P.P., Kock P., Bezemer R., Weisbeek P.J., Smeekens S.C.M. (1996). Sucrose represses the developmentally controlled transient activation of the plastocyanin gene in Arabidopsis thaliana seedlings. Plant Physiol. 110: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J., Nunes-Nesi A., Alpers J., Szkop M., Fernie A.R., Steup M. (2008). Alterations in cytosolic glucose-phosphate metabolism affect structural features and biochemical properties of starch-related heteroglycans. Plant Physiol. 148: 1614–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W.N., Kwart M., Hummel S., Frommer W.B. (1995). Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J. Biol. Chem. 270: 16315–16320 [DOI] [PubMed] [Google Scholar]
- Fischer W.N., Loo D.D.F., Koch W., Ludewig U., Boorer K.J., Tegeder M., Rentsch D., Wright E.M., Frommer W.B. (2002). Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 29: 717–731 [DOI] [PubMed] [Google Scholar]
- Forde B.G. (2002a). Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Forde B.G. (2002b). The role of long-distance signalling in plant responses to nitrate and other nutrients. J. Exp. Bot. 53: 39–43 [PubMed] [Google Scholar]
- Foyer C.H., Noctor G., Verrier P. (2006). Photosynthetic carbon–nitrogen interactions: Modeling inter-pathway control and signaling. Control of Primary Metabolism in Plants, Plaxton W.C., McManus M.T., (Oxford, UK: Blackwell Publishing; ), pp. 325–347 [Google Scholar]
- Goetz M., Godt D.E., Guivarc’h A., Kahmann U., Chriqui D., Roitsch T. (2001). Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci. USA 98: 6522–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerche P., Tire C., De Sa F.G., De Clercq A., Van Montagu M., Krebbers E. (1990). Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 2: 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallath S., Weimar T., Meyer A., Gumy C., Suter-Grotemeyer M., Neuhaus J.M., Rentsch D. (2005). The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiol. 137: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J., Johannesson H., Engström P. (2001). Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol. Biol. 45: 247–262 [DOI] [PubMed] [Google Scholar]
- Hirel B., Le Gouis J., Ney B., Gallais A. (2007). The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 58: 2369–2387 [DOI] [PubMed] [Google Scholar]
- Hirner B., Fischer W.N., Rentsch D., Kwart M., Frommer W.B. (1998). Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 14: 535–544 [DOI] [PubMed] [Google Scholar]
- Hirner A., Ladwig F., Stransky H., Okumoto S., Keinath M., Harms A., Frommer W.B., Koch W. (2006). Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18: 1931–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt E., Gattolin S., Newbury H.J., Bale J.S., Tseng H.M., Barrett D.A., Pritchard J. (2010). A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J. Exp. Bot. 61: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt E.J., Pritchard J., Bennett M.J., Zhu X., Barrett D.A., Allen T., Bale J.S., Newbury H.J. (2006). The Arabidopsis thaliana/Myzus persicae model system demonstrates that a single gene can influence the interaction between a plant and a sap-feeding insect. Mol. Ecol. 15: 4203–4213 [DOI] [PubMed] [Google Scholar]
- James D.W., Jr, Lim E., Keller J., Plooy I., Ralston E., Dooner H.K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve A., Rauh B.L., Xia X., Kandasamy M., Meagher R.B., Sheen J., Moore B.D. (2008). Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Kwart M., Laubner M., Heineke D., Stransky H., Frommer W.B., Tegeder M. (2003). Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 33: 211–220 [DOI] [PubMed] [Google Scholar]
- Krapp A., Saliba-Colombani V., Daniel-Vedele F. (2005). Analysis of C and N metabolisms and of C/N interactions using quantitative genetics. Photosynth. Res. 83: 251–263 [DOI] [PubMed] [Google Scholar]
- Kwart M., Hirner B., Hummel S., Frommer W.B. (1993). Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. Plant J. 4: 993–1002 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Wipf D., Frommer W.B. (2004). Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Lam H.M., Coschigano K., Schultz C., Melo-Oliveira R., Tjaden G., Oliveira I., Ngai N., Hsieh M.H., Coruzzi G. (1995). Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.M., Coschigano K.T., Oliveira I.C., Melo-Oliveira R., Coruzzi G.M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 569–593 [DOI] [PubMed] [Google Scholar]
- Lee Y.H., Foster J., Chen J., Voll L.M., Weber A.P.M., Tegeder M. (2007). AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 50: 305–319 [DOI] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lohaus G., Möllers C. (2000). Phloem transport of amino acids in two Brassica napus L. genotypes and one B. carinata genotype in relation to their seed protein content. Planta 211: 833–840 [DOI] [PubMed] [Google Scholar]
- Ma Q., Longnecker N., Emery N., Atkins C. (1998). Growth and yield in Lupinus angustifolius are depressed by early transient nitrogen deficiency. Aust. J. Agric. Res. 49: 811–819 [Google Scholar]
- Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., Nishimura M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43: 331–341 [DOI] [PubMed] [Google Scholar]
- Martin T., Oswald O., Graham I.A. (2002). Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Lauterbach C., Niedermeier M., Barth I., Sjolund R.D., Sauer N. (2004). Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol. 134: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M., Borisjuk L., Tewes A., Dietrich D., Rentsch D., Weber H., Wobus U. (2003). Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol. 132: 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M., Borisjuk L., Tewes A., Heim U., Sauer N., Wobus U., Weber H. (2001). Amino acid permeases in developing seeds of Vicia faba L.: Expression precedes storage protein synthesis and is regulated by amino acid supply. Plant J. 28: 61–71 [DOI] [PubMed] [Google Scholar]
- Nakano H., Makino A., Mae T. (1997). The effect of elevated partial pressures of CO2 on the relationship between photosynthetic capacity and N content in rice leaves. Plant Physiol. 115: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T., Messerli G., Trevisan M., Chen J., Smith A.M., Zeeman S.C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Okumoto S., Koch W., Tegeder M., Fischer W.N., Biehl A., Leister D., Stierhof Y.D., Frommer W.B. (2004). Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. J. Exp. Bot. 55: 2155–2168 [DOI] [PubMed] [Google Scholar]
- Okumoto S., Schmidt R., Tegeder M., Fischer W.N., Rentsch D., Frommer W.B., Koch W. (2002). High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J. Biol. Chem. 277: 45338–45346 [DOI] [PubMed] [Google Scholar]
- Olsen J.G., Rasmussen A.V., von Wettstein-Knowles P., Henriksen A. (2004). Structure of the mitochondrial β-ketoacyl-[acyl carrier protein] synthase from Arabidopsis and its role in fatty acid synthesis. FEBS Lett. 577: 170–174 [DOI] [PubMed] [Google Scholar]
- Oliveira I.C., Coruzzi G.M. (1999). Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 121: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P.P., Pruitt R.E., Meyerowitz E.M. (1988). Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11: 805–820 [DOI] [PubMed] [Google Scholar]
- Pate J.S., Sharkey P.J., Atkins C.A. (1977). Nutrition of a developing legume fruit: functional economy in terms of carbon, nitrogen, water. Plant Physiol. 59: 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate J.S., Sharkey P.J., Lewis O.A.M. (1975). Xylem to phloem transfer of solutes in fruiting shoots of legumes, studied by a phloem bleeding technique. Planta 122: 11–26 [DOI] [PubMed] [Google Scholar]
- Paul M.J., Pellny T.K. (2003). Carbon metabolite feedback regulation of leaf photosynthesis and development. J. Exp. Bot. 54: 539–547 [DOI] [PubMed] [Google Scholar]
- Pélissier H.C., Tegeder M. (2007). PvUPS1 plays a role in source-sink transport of allantoin in French bean (Phaseolus vulgaris). Funct. Plant Biol. 34: 282–291 [DOI] [PubMed] [Google Scholar]
- Rathke G.W., Christen O., Diepenbrock W. (2004). Effects of nitrogen source and rate on productivity and quality of winter oilseed rape (Brassica napus L.) grown in different crop rotations. Field Crops Res. 94: 103–113 [Google Scholar]
- Rentsch D., Schmidt S., Tegeder M. (2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 581: 2281–2289 [DOI] [PubMed] [Google Scholar]
- Riens B., Lohaus G., Heineke D., Heldt H.W. (1991). Amino acid and sucrose content determined in the cytosolic, chloroplastic, and vacuolar compartments and in the phloem sap of spinach leaves. Plant Physiol. 97: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Menges M., Healy J.M.S., Murray J.A.H. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rolletschek H., Hosein F., Miranda M., Heim U., Götz K.P., Schlereth A., Borisjuk L., Saalbach I., Wobus U., Weber H. (2005). Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 137: 1236–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R.F. (1994). Acclimation of photosynthesis to increasing atmospheric CO2: The gas exchange perspective. Photosynth. Res. 39: 351–368 [DOI] [PubMed] [Google Scholar]
- Sanders A., Collier R., Trethewy A., Gould G., Sieker R., Tegeder M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 59: 540–552 [DOI] [PubMed] [Google Scholar]
- Sato T., et al. (2009). CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 60: 852–864 [DOI] [PubMed] [Google Scholar]
- Sauer N. (2007). Molecular physiology of higher plant sucrose transporters. FEBS Lett. 581: 2309–2317 [DOI] [PubMed] [Google Scholar]
- Sauer N., Stolz J. (1994). SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 6: 67–77 [DOI] [PubMed] [Google Scholar]
- Schmidt R., Stransky H., Koch W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226: 805–813 [DOI] [PubMed] [Google Scholar]
- Schneider A., Häusler R.E., Kolukisaoglu U., Kunze R., van der Graaff E., Schwacke R., Catoni E., Desimone M., Flügge U.I. (2002). An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J. 32: 685–699 [DOI] [PubMed] [Google Scholar]
- Sharkey P.J., Pate J.S. (1975). Selectivity in xylem to phloem transfer of amino acids in fruiting shoots of white lupin (Lupinus albus L.). Planta 127: 251–262 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Stitt M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Song W., Koh S., Czako M., Marton L., Drenkard E., Becker J.M., Stacey G. (1997). Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol. 114: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H.Y., Song W., Zhang L., Naider F., Becker J.M., Stacey G. (1994). An Arabidopsis peptide transporter is a member of a new class of membrane transport proteins. Plant Cell 6: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. (1991). Rising CO2 levels and their potential significance for carbon flow in photosynthetic cells. Plant Cell Environ. 14: 741–762 [Google Scholar]
- Stitt M., Krapp A. (1999). The molecular physiological basis for the interaction between elevated carbon dioxide and nutrients. Plant Cell Environ. 22: 583–622 [Google Scholar]
- Strand Å., Zrenner R., Trevanion S., Stitt M., Gustafsson P., Gardeström P. (2000). Decreased expression of two key enzymes in the sucrose biosynthesis pathway, cytosolic fructose-1,6-bisphosphatase and sucrose phosphate synthase, has remarkably different consequences for photosynthetic carbon metabolism in transgenic Arabidopsis thaliana. Plant J. 23: 759–770 [DOI] [PubMed] [Google Scholar]
- Su Y.H., Frommer W.B., Ludewig U. (2004). Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 136: 3104–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Grennan A., Rentsch D., Tegeder M. (2008). Isolation and characterization of an amino acid transporter from French bean (Phaseolus vulgaris L.). Plant Sci. 174: 348–356 [Google Scholar]
- Tegeder M., Offler C.E., Frommer W.B., Patrick J.W. (2000). Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol. 122: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M., Rentsch D. (2010). Uptake and partitioning of amino acids and peptides. Mol. Plant 3: 997–1011 [DOI] [PubMed] [Google Scholar]
- Tegeder M., Rentsch D., Patrick J.W. (2011). Organic carbon and nitrogen transporters. The Plant Plasma Membrane, Plant Cell Monographs, Murphy A., Peer W., Schulz B., (Berlin: Springer-Verlag; ), 331–352 [Google Scholar]
- Tegeder M., Tan Q., Grennan A., Patrick J.W. (2007). Amino acid transporter expression and localisation studies in pea (Pisum sativum L.). Funct. Plant Biol. 34: 1019–1028 [DOI] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- van Bel A.J.E. (1990). Xylem-phloem exchange via the rays: The undervalued route of transport. J. Exp. Bot. 41: 631–644 [Google Scholar]
- Villand P., Olsen O.A., Kleczkowski L.A. (1993). Molecular characterization of multiple cDNA clones for ADP-glucose pyrophosphorylase from Arabidopsis thaliana. Plant Mol. Biol. 23: 1279–1284 [DOI] [PubMed] [Google Scholar]
- Weber A., Servaites J.C., Geiger D.R., Kofler H., Hille D., Gröner F., Hebbeker U., Flügge U.I. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12: 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K., Küster H., Radchuk R., Müller M., Weichert H., Fait A., Fernie A.R., Saalbach I., Weber H. (2008). Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J. 55: 909–926 [DOI] [PubMed] [Google Scholar]
- Weise A., Barker L., Kühn C., Lalonde S., Buschmann H., Frommer W.B., Ward J.M. (2000). A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A., Gröner F., Sonnewald U., Deppner H., Lerchl J., Hebbeker U., Flügge U.I., Weber A. (1999). Spinach hexokinase I is located in the outer envelope membrane of plastids. FEBS Lett. 461: 13–18 [DOI] [PubMed] [Google Scholar]
- Wilkinson T.L., Douglas A.E. (2003). Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomol. Exp. Appl. 106: 103–113 [Google Scholar]
- Wormit A., Trentmann O., Feifer I., Lohr C., Tjaden J., Meyer S., Schmidt U., Martinoia E., Neuhaus H.E. (2006). Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18: 3476–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Han P., Lutziger I., Wang K., Oliver D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Zhang C., Barthelson R.A., Lambert G.M., Galbraith D.W. (2008). Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shewry P.R., Jones H., Barcelo P., Lazzeri P.A., Halford N.G. (2001). Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 28: 431–441 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.