Arabidopsis tyrosylprotein sulfotransferase (TPST) maintains the postembryonic root stem cell niche by regulating basal- and auxin-induced expression of the PLETHORA stem cell transcription factor. TPST-mediated activation of a group of sulfated peptides known as root meristem growth factors provides a link between the phytohormone auxin and PLETHORA in root stem cell niche maintenance.
Abstract
Recent identification of the Arabidopsis thaliana tyrosylprotein sulfotransferase (TPST) and a group of Tyr-sulfated peptides known as root meristem growth factors (RGFs) highlights the importance of protein Tyr sulfation in plant growth and development. Here, we report the action mechanism of TPST in maintenance of the root stem cell niche, which in the Arabidopsis root meristem is an area of four mitotically inactive quiescent cells plus the surrounding mitotically active stem cells. Mutation of TPST leads to defective maintenance of the root stem cell niche, decreased meristematic activity, and stunted root growth. We show that TPST expression is positively regulated by auxin and that mutation of this gene affects auxin distribution by reducing local expression levels of several PIN genes and auxin biosynthetic genes in the stem cell niche region. We also show that mutation of TPST impairs basal- and auxin-induced expression of the PLETHORA (PLT) stem cell transcription factor genes and that overexpression of PLT2 rescues the root meristem defects of the loss-of-function mutant of TPST. Together, these results support that TPST acts to maintain root stem cell niche by regulating basal- and auxin-induced expression of PLT1 and PLT2. TPST-dependent sulfation of RGFs provides a link between auxin and PLTs in regulating root stem cell niche maintenance.
INTRODUCTION
Plant root growth is maintained by the continuous division of cells in the meristem. Within the root meristem, the mitotically inactive quiescent center (QC) cells (four in Arabidopsis thaliana), together with the surrounding stem cells, constitute the root stem cell niche, which provides the source of cells for all tissues in roots (van den Berg et al., 1995; Scheres, 2007; Dinneny and Benfey, 2008; Kornet and Scheres, 2009). SHORT-ROOT (SHR) and SCARECROW (SCR), which encode members of the GRAS (acronym of GAI, RGA, SCR genes) family transcription factors, provide positional information along the radial axis to specify the identity of QC and to regulate the functions of the associated stem cells in the root (Di Laurenzio et al., 1996; Helariutta et al., 2000; Nakajima et al., 2001; Sabatini et al., 2003; Wildwater et al., 2005). In parallel with the SHR/SCR pathway, the phytohormone auxin, especially its gradient distribution, also plays a crucial role in the specification and maintenance of the root stem cell niche (Sabatini et al., 1999; Tanaka et al., 2006; Scheres, 2007; Benjamins and Scheres, 2008; Dinneny and Benfey, 2008). Notably, using Arabidopsis lines containing an auxin-responsive promoter (DR5) linked to β-glucuronidase (GUS), Sabatini et al. (1999) established that auxin is asymmetrically distributed in the root tip, with an apparent high concentration (auxin maximum) in the QC/columella initial region. The correlation between auxin maximum and pattern formation of the root meristem leads to a hypothesis that auxin maximum is instructive for root stem cell niche patterning (Sabatini et al., 1999; Blilou et al., 2005; Scheres, 2007; Benjamins and Scheres, 2008; Dinneny and Benfey, 2008; Ding and Friml, 2010).
The PIN-FORMED (PIN) genes, which encode key auxin efflux carriers, are essential for the formation of a proper auxin maximum and therefore regulate root patterning (Blilou et al., 2005). It was proposed that the instructive auxin gradient is somehow translated into developmental cues (Benjamins and Scheres, 2008). The auxin inducible PLETHORA1 (PLT1) and PLT2 genes, which encode members of the AP2 class of transcription factors that are essential for root stem cell niche patterning (Aida et al., 2004; Galinha et al., 2007), are potential candidates for such translations (Benjamins and Scheres, 2008). Interestingly, the expression domain of PLT genes overlaps with the auxin maximum in the root. Recent elegant work reveals an interaction network of PINs and PLTs in controlling auxin-mediated root patterning; PIN proteins restrict PLT expression in the basal embryo region to initiate root primordium. In turn, PLT genes maintain PIN transcription, which stabilizes the position of the stem cell niche (Blilou et al., 2005; Grieneisen et al., 2007; Dinneny and Benfey, 2008). In addition to PIN gene-mediated polar auxin transport, local auxin biosynthesis in the root has also been shown to be crucial for root meristem patterning (Tobeña-Santamaria et al., 2002; Ljung et al., 2005; Cheng et al., 2006, 2007; Stepanova et al., 2008; Zhao, 2010). How auxin transport and local auxin biosynthesis are coordinated remains a mystery, but it is generally believed that both processes function together to establish a state of auxin homeostasis that is required for stem cell patterning and maintenance in roots (Jiang and Feldman, 2003; Zhao, 2010).
Emerging evidence indicates that peptide ligands are also involved in root stem cell niche maintenance. CLE proteins (for CLAVATA3/ENDOSPERM SURROUNDING REGION) are small peptides with a conserved 14–amino acid CLE motif (Hobe et al., 2003). A member of the CLE protein family, CLAVATA3 (CLV3), regulates the size of the stem cell pool in the shoot meristem (Laux, 2003). Overexpression or in vitro treatment with certain CLE peptides leads to tissue layer disorganization and premature differentiation of root stem cells in Arabidopsis, suggesting a conserved mechanism in the regulation of root and shoot apical meristem (Casamitjana-Martínez et al., 2003; Hobe et al., 2003; Fiers et al., 2004, 2005, 2007; Sarkar et al., 2007). In the shoot apical meristem and as part of a feedback regulation loop, the glycosylated peptide CLV3 expressed by stem cells acts as a diffusible signal molecule to restrict the number of stem cells, while the WUSCHEL (WUS) transcription factor expressed in the stem cell organizing center acts as a stem cell–promoting factor to promote the expansion of the stem cell population (reviewed in Fiers et al., 2007; Dinneny and Benfey, 2008). Whether similar machinery is present in the root meristem is unclear. Recent work demonstrated that, similar to the CLV3-WUS feedback network in regulating shoot meristem, CLE40 and a WUS-related protein, WOX5, form a self-regulation network in roots to control the proliferation and differentiation of stem cells (Fiers et al., 2005; Sarkar et al., 2007; Stahl et al., 2009).
In addition to the CLE family of peptides, three families of Tyr-sulfated peptides, phytosulfokine (PSK), plant peptide containing sulfated tyrosine 1 (PSY1), and root meristem growth factors (RGFs), have been identified as growth factors in Arabidopsis (Matsubayashi and Sakagami, 1996; Matsubayashi et al., 2002; Amano et al., 2007; Matsuzaki et al., 2010). Protein Tyr sulfation, which is catalyzed by tyrosylprotein sulfotransferases (TPSTs), is believed to be critical for the activating of these peptides. TPSTs were first identified in mouse and human (Ouyang et al., 1998) and have been shown to play important roles in many physiological and pathological processes, including hormonal regulation, hemostasis, inflammation, and infectious diseases (Seibert and Sakmar, 2008). Interestingly, while TPST orthologs from numerous vertebrate and invertebrate species were readily identifiable based on sequence homology, no plant TPST ortholog was identified this way, suggesting that plant TPSTs have evolved in a manner distinct from their animal counterparts (Moore, 2009).
Recently, an Arabidopsis TPST was discovered by affinity purification by taking advantage of its specific interaction with the sulfation motif of the PSY1 precursor (Komori et al., 2009). Significantly, TPST is highly expressed in the root apical meristem, and the T-DNA insertion mutant tpst-1 displays stunted root growth and other developmental defects (Komori et al., 2009). The root meristem defects of tpst-1 can be restored by in vitro application of Tyr-sulfated RGF1, suggesting that TPST acts through RGF1 to maintain root stem cell niche (Matsuzaki et al., 2010). In this study, we show that mutation of the TPST gene leads to defective root stem cell niche maintenance. TPST expression is upregulated by auxin, and mutation of this gene affects the expression of PLT1/2, several PIN genes, and auxin biosynthetic genes. Our results support the conclusion that TPST acts in the auxin pathway to maintain postembryonic root stem cell niche by defining the expression of the PLT stem cell transcription factor genes.
RESULTS
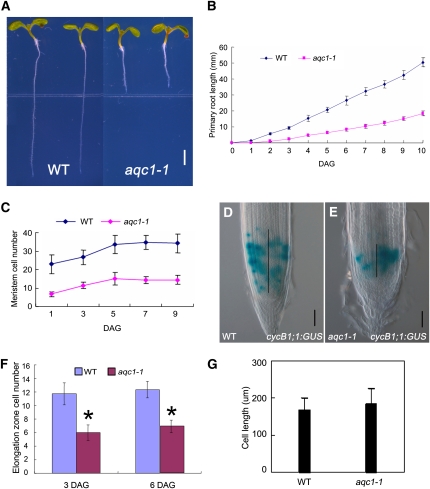
The aqc1-1 Mutant Shows Stunted Root Growth and Reduced Root Meristem Size
The active quiescent center1-1 (aqc1-1) mutant showed stunted root growth (Figures 1A and 1B) and extra QC cells (see below). As early as 2 d after germination (DAG), growth rate of the primary root was markedly reduced in the aqc1-1 mutant (Figure 1B). At 10 DAG, the primary root length of aqc1-1 was ~35% of that of the wild type (Figure 1B). We further examined the cellular defects in the meristem zone, the elongation zone, and the differentiation zone using several molecular markers. The aqc1-1 mutant showed a significantly reduced number of root meristematic cells (Figure 1C), which is defined by counting the number of cells from the cortical initial cell to the first elongated cell in the cortex layer (Casamitjana-Martínez et al., 2003; Dello Ioio et al., 2007). Consistently, the expression domain of CycB1;1:GUS, a widely used marker for the G2/M phase of the cell cycle (Colón-Carmona et al., 1999), was reduced in the mutant (Figures 1D and 1E, black bar, 177.9 ± 23.3 μm in the wild type, n = 52; 97.8 ± 12.2 μm in the mutant, n = 54; 6 DAG; t test, P < 0.01), indicating that the population of dividing cells is highly reduced in the root meristem of aqc1-1. In the elongation zone, cell number was also reduced in the aqc1-1 mutant (Figure 1F; t test, P < 0.01). Measurement of cell size in the differentiation zone indicated that the average length of the mature cortex cells in aqc1-1 was not significantly changed (Figure 1G), suggesting that the aqc1-1 mutation has little effect on final cell size. Collectively, these results indicate that AQC1 is required for maintaining the cell division activity of the root meristem.
Figure 1.
aqc1-1 Shows Reduced Root Meristem Size and Stunted Root Growth.
(A) Phenotype of wild-type Col-0 and aqc1-1 seedlings at 6 DAG. Bar = 2 mm.
(B) Primary root length of wild-type (WT) and aqc1-1 seedlings from germination to 10 DAG. Data shown are average and sd (n = 25).
(C) Root meristem cell number of the wild type and aqc1-1 from 1 to 9 DAG. The root meristem cell number is expressed as the number of cortex cells in the cortex file extending from the QC to the transition zone. Data shown are average and sd (n = 21).
(D) and (E) cyclinB1;1:GUS expression in the wild type (D) and aqc1-1 (E) at 6 DAG. Black vertical lines represent the expression area of cyclinB1;1:GUS. Bars = 50 μm.
(F) Cell numbers in elongation zone of the wild type and aqc1-1 at 3 and 6 DAG. Data shown are average and sd (n = 25). Asterisks denote Student’s t test significance compared with wild-type plants (*P < 0.01).
(G) Statistics of mature cortex cell size of the wild type and aqc1-1 at 8 DAG. Data shown are average and sd (n = 100).
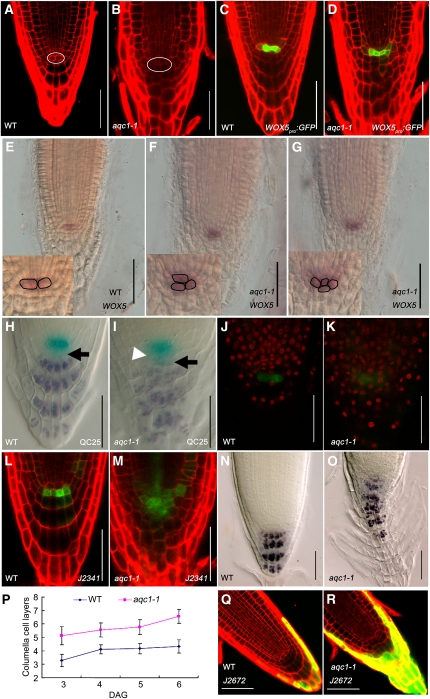
aqc1-1 Shows Defective Cellular Organization in Root Stem Cell Niche
We next examined the cellular organization of the QC and its surrounding stem cells. In Arabidopsis, the QC comprises four well-defined cells: two in the front and two in the back. Cell division is rarely observed in these cells. In aqc1-1 roots, the QC cells were characterized by morphologically aberrant cells (Figures 2A and 2B). We compared the number of QC cells between the wild type and aqc1-1 by counting QC cells in median longitudinal sections of the root meristem (Ortega-Martínez et al., 2007). Our statistics revealed that, at 4 DAG, the extra QC cell number (more than two in one focal plane) increased from 0.06 ± 0.24 in the wild type to 1.68 ± 0.30 in the mutant (n = 32; t test, P < 0.01). The aberrant QC organization of aqc1-1 was clearly visible by the altered expression domain of WOX5pro:GFP (for green fluorescent protein), a reported QC-specific marker (Blilou et al., 2005; Sarkar et al., 2007). In the wild type, WOX5pro:GFP showed specific expression in the highly localized QC cells (Figure 2C), while in aqc1-1, its expression was diffused and expanded to the adjacent cells (Figure 2D). In line with the WOX5pro:GFP results, RNA in situ hybridization also showed that WOX5 expression was expanded in aqc1-1 (Figures 2E to 2G). Similarly, aberrant cellular organization of the QC in the aqc1-1 mutant could also be visible using the promoter trap line QC25 (Figures 2H and 2I).
Figure 2.
The aqc1-1 Mutant Shows Extra QC Cells and Defective Root Stem Cell Niche Maintenance.
(A) and (B) Cellular organization of wild-type (WT) (A) and aqc1-1 (B) root tips at 4 DAG using propidium iodide staining. The white oval shows the QC cells.
(C) and (D) Expression pattern of WOX5pro:GFP in the wild type (C) and aqc1-1 (D) at 4 DAG.
(E) to (G) Whole-mount in situ hybridization with a WOX5 probe in wild-type (E) and aqc1-1 seedlings ([F] and [G], different individuals showing one or two extra QC cells) at 5 DAG. Insets show aqc1-1 roots contain supernumerary QC cells compared with the wild type.
(H) and (I) Double staining of the QC25 GUS marker (light blue) and starch granules (dark brown) in the wild type (H) and aqc1-1 (I) at 4 DAG. Black arrows, putative columella stem cell layer; white arrowhead, extra QCs containing starch granules.
(J) and (K) EdU incorporation assays in wild-type (J) and aqc1-1 (K) root meristems. Red fluorescent EdU positive nuclei in the QC cells of aqc1-1 indicate that cell division was activated in these cells in contrast with the wild type.
(L) and (M) Expression pattern of marker J2341 in the wild type (L) and aqc1-1 (M) at 4 DAG.
(N) and (O) Lugol staining of the wild type (N) and aqc1-1 (O) at 5 DAG.
(P) Number of columella cell layers of the wild type and aqc1-1 from 3 to 6 DAG. Data shown are average and sd (n = 18).
(Q) and (R) Expression pattern of marker J2672 in the wild type (Q) and aqc1-1 (R) at 4 DAG.
Bars = 50 μm.
To check whether cell cycle activities were altered in the QC of aqc1-1, we cultured 2-DAG seedlings for 24 h in the presence of 5-ethynyl-2'-deoxyuridine (EdU), a nucleoside analog of thymidine. EdU has been used to mark cell division in the root meristem because incorporation of this chemical in the nuclei is indicative for S-phase progression (Vanstraelen et al., 2009). In wild-type root meristems, most meristematic cells, including those QC-associated stem cells, incorporated EdU with red fluorescent nuclei but only occasionally in the QC cells (marked with WOX5pro:GFP), indicating their low mitotic activity (Figure 2J). In the aqc1-1 root meristem, however, the QC cells were able to incorporate EdU in a much higher frequency (13/14 in aqc1-1 versus 4/14 in the wild type, n = 14) (Figure 2K). These results suggest that the AQC1 gene is required to maintain the cellular organization of QC.
To assess the possible defects of aqc1-1 on stem cell maintenance, we analyzed the columella stem cells (CSCs), the cell layer immediately below the QC. The J2341 marker, which was specifically expressed in CSCs in the wild type (Figure 2L), expanded to more than several cell layers in aqc1-1 (Figure 2M). In the Lugol staining assay, wild-type CSCs did not contain starch granules and were distinct from the differentiated columella cells below (Figure 2H, black arrow). By contrast, aqc1-1 roots showed irregular columella cells containing starch granules directly below the aberrant QC cells, indicating the disruption of CSCs (Figure 2I, black arrow). Therefore, even though the supernumerary QC cells in aqc1-1 displayed QC identity (as evidenced by QC marker gene expression), these cells failed to execute the QC function of maintaining CSCs in an undifferentiated state. In the GUS and Logul double-stained QC marker line QC25, some GUS-staining cells also accumulated starch granules (Figure 2I, white arrowhead), further confirming that the cells derived from abnormal QC divisions were undergoing differentiation. These results suggest that AQC1 is required to maintain the cellular organization and function of QC, which in turn regulates stem cell fate of the neighboring CSCs.
Distal to the CSC layer, daughter cells of CSCs differentiate into starch-containing columella root cap cells. In the wild-type root tip, the outermost layer of root cap cells detaches after the formation of each new layer; therefore, QC remains at the same distance from the root tip (Scheres, 2007). Quantification of columella cell tiers indicated that, at 5 DAG, while wild-type roots contained 4.16 ± 0.38 tiers of regularly patterned columella cells, aqc1-1 roots contained 5.78 ± 0.55 tiers of irregularly patterned columella cells (Figures 2N to 2P; n = 18; t test, P < 0.01). We then compared the expression of the root cap cell-specific marker J2672 between the wild type and mutant. In the wild type, the J2672 maker was specifically expressed in the outermost layer of root cap cells (Figure 2Q). By contrast, several cell layers of aqc1-1 roots showed expression of the J2672 marker, indicating that the outermost columella cell tiers did not detach properly in the mutant (Figure 2R).
Interestingly, the cellular organization of the root pole in the mutant during embryo development, from the early globular to the cotyledon-stage embryo, was comparable to that of the wild type (see Supplemental Figures 1A to 1J online). In mature aqc1-1 embryo, the cellular organization of the root meristem and the QC25 expression pattern were similar to those of the wild type (see Supplemental Figures 1K and 1L online). Obvious QC defects were observed from 1 DAG onward (see Supplemental Figures 1M to 1R online), suggesting that AQC1 acts specifically to maintain root stem cell niche after germination.
AQC1 Encodes TPST, a Plant-Specific TPST
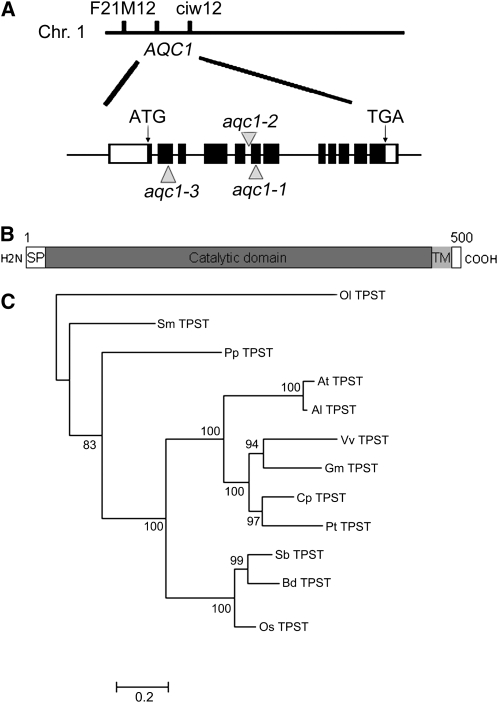
The AQC1 gene was identified using a combination of map-based cloning and thermal asymmetric interlaced-PCR (TAIL-PCR) approaches. Genetic analysis of 92 individuals showing the aqc1-1 phenotype from an F2 mapping population located the AQC1 gene on the long arm of chromosome 1 in the interval between molecular markers F21M12 and ciw12 (Figure 3A). Sequence analysis of TAIL-PCR products revealed that, in the aqc1-1 mutant, T-DNA was inserted in the sixth exon of the annotated gene At1g08030 (Figure 3A). A 6790-bp genomic DNA fragment covering the promoter and coding sequences of At1g08030 could fully complement the aqc1-1 phenotype (see Supplemental Figures 2A online), indicating that the genomic region contains the functional AQC1 gene. To identify the coding region of AQC1, we designed primers based on the sequence information and obtained a 1500-bp cDNA fragment through RT-PCR. Successful complementation of the aqc1-1 phenotype with the 1500-bp cDNA fragment driven by a 2231-bp 5′ upstream sequence or the cauliflower mosaic virus 35S promoter indicated that the cDNA fragment obtained represents the full-length coding region of AQC1 (see Supplemental Figures 2A and 2E online). Sequence comparison between cDNA and genomic DNA revealed that AQC1 contains 12 exons and 11 introns and is predicted to encode a protein of 500 amino acids (Figure 3B). Based on these results, the current annotation of At1g08030 in The Arabidopsis Information Resource needs to be corrected. RT-PCR analysis showed that the T-DNA insertion in aqc1-1 abolished the AQC1 transcript (see Supplemental Figure 2A online).
Figure 3.
AQC1 Encodes TPST.
(A) Cloning of the AQC1/TPST gene. Black boxes represent exons, lines between black boxes represent introns, and white boxes represent the 5′ and 3′ noncoding regions. Arrowheads mark the T-DNA insertion sites or the mutation site of different alleles. Chr., chromosome.
(B) Schematic representation of the AQC1/TPST protein, modified from Moore (2009). SP, signal peptide; TM, transmembrane helix.
(C) Phylogenetic analysis of AQC1/TPST and its orthologs from other plant species. The node labels indicate the proportion of trees in the posterior distribution containing the node. Sequences were from Ostreococcus lucimarinus (Ol TPST), Selaginella moellendorffii (Sm TPST), Physcomitrella patens (Pp TPST), Arabidopsis thaliana (At TPST), Arabidopsis lyrata (Al TPST), Vitis vinifera (Vv TPST), Glycine max (Gm TPST), Carica papaya (Cp TPST), Populus trichocarpa (Pt TPST), Sorghum bicolor (Sb TPST), Brachypodium distachyon (Bd TPST), and Oryza sativa (Os TPST). The sequences and alignment used are presented in Supplemental Data Set 1 online.
The authenticity of AQC1 was further confirmed by examining two independent alleles showing similar phenotypes as aqc1-1 (see Supplemental Figure 2B online). The SALK_009847 line contained a T-DNA inserting in the fifth intron of At1g08030 (Komori et al., 2009). Homozygous SALK_009847 plants, designated as aqc1-2, exhibited similar root growth defects as aqc1-1 (see Supplemental Figure 2B online). We also determined that the aqc1-3 allele contains a C-to-T transition, leading to a premature stop codon (Figures 3A; see Supplemental Figure 2B online). Genetic analyses showed that aqc1-1, aqc1-2, and aqc1-3 failed to complement each other, indicating that these mutations are allelic (see Supplemental Figure 2B online). Collectively, these results prove that AQC1 and At1g03080 are equivalent.
It was recently shown that At1g08030 encodes a tyrosylprotein sulfotransferase, named TPST, which shows a broad substrate specificity (Komori et al., 2009). We therefore refer to AQC1 as TPST hereafter. The TPST protein contains a putative signal peptide in the N terminus, a catalytic domain in the middle, and a transmembrane helix near the C terminus (Figure 3B). No obvious morphological difference was identified between wild-type and transgenic plants overexpressing TPST under the control of the cauliflower mosaic virus 35S promoter (see Supplemental Figures 2D and 2E online). Possible At TPST orthologs were identified in various plant genomes (Figure 3C) but not in yeast and animals. Significantly, the rice (Oryza sativa) ortholog of the At TPST gene, named Os TPST (Os06g06370), successfully rescued the aqc1-1 mutant phenotype (see Supplemental Figure 2C online), suggesting that the Tyr sulfation activities of the TPST proteins are conserved among some higher plants.
Auxin Induces the Expression of TPST and Several RGFs
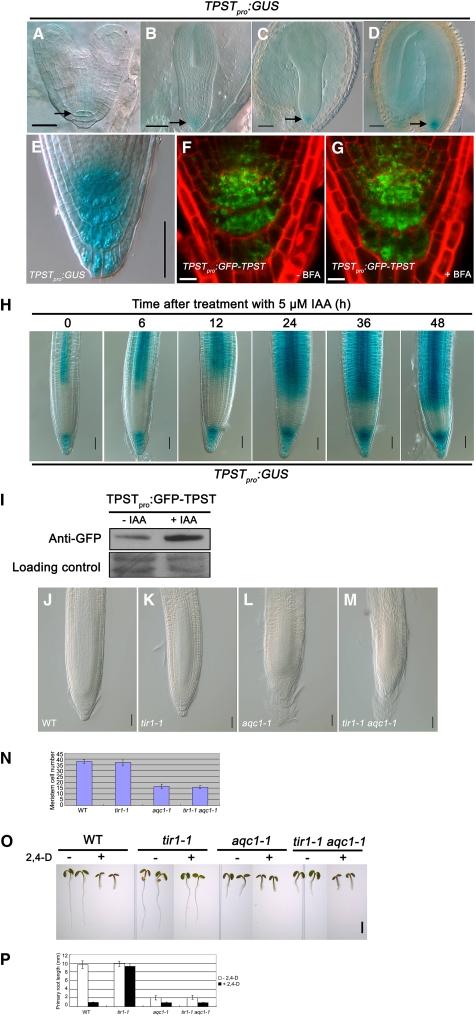
The expression pattern of TPST was investigated with transgenic plants containing a 2231-bp 5′ upstream sequence of TPST fused with a GUS reporter gene (TPSTpro:GUS). TPSTpro:GUS was first expressed in the hypophysis cells and their surrounding cells in heart-stage embryos (Figures 4A to 4D). After germination, TPST was highly expressed in the QC and surrounding stem cells, as well as the distal differentiated columella cells (Figure 4E). This expression pattern was confirmed using a GFP-TPST translational fusion construct (TPSTpro:GFP-TPST), which can complement the aqc1-1 mutant (Figures 4F and 4G; see Supplemental Figures 2C and 2E online).
Figure 4.
Expression Pattern of TPST.
(A) to (D) TPSTpro:GUS expression pattern during the embryo stage, from early heart-stage embryo (A), torpedo-stage embryo (B), early cotyledon-stage embryo (C), to cotyledon-stage embryo (D). TPSTpro:GUS is first expressed in the central cell and surrounding cells. Bars = 20 μm in (A) and 50 μm in (B) to (D).
(E) TPSTpro:GUS expression in the root tip of a representative 4 DAG seedling. Bar = 50 μm.
(F) and (G) TPST expression shown by the TPSTpro:GFP-TPST translational fusion. Seedlings (5 DAG) containing the TPSTpro:GFP-TPST construct were either not treated (F) or treated with 10 μM brefeldin A (BFA) for 2 h (G). Bars = 10 μm.
(H) Auxin-induced expression of TPSTpro:GUS. Transgenic seedlings (4 DAG) were treated with 5 μM IAA for the indicated times before GUS staining assays. Bars = 50 μm.
(I) Protein gel analysis showing that auxin upregulates the levels of the TPSTpro:GFP-TPST fusion protein. Proteins were extracted from 5-DAG transgenic seedlings treated with or without 5 μM IAA for 12 h and subjected to immunoblot using anti-GFP antibody. Ponceau S–stained membranes are shown as loading controls.
(J) to (N) Genetic analysis between aqc1-1 and tir1-1. WT, wild type.
(J) to (M) Root tips of the wild type (J), tir1-1 (K), aqc1-1 (L), and tir1-1 aqc1-1 (M) at 5 DAG. Bars = 50 μm.
(N) Meristem cell numbers of the indicated genotypes at 5 DAG. Data shown are average and sd (n = 12).
(O) Phenotypes of indicated plants without or with 30 nM 2,4-D at 3 DAG. Bar = 2 mm.
(P) Primary root lengths of the indicated plants without or with 30 nM 2,4-D at 3 DAG. Data shown are average and sd (n = 10).
In agreement with a recent study (Komori et al., 2009), transient expression of a 35Spro:GFP-TPST construct in Arabidopsis protoplasts indicated that the 35Spro:GFP-TPST fusion protein colocalized with Golgi-rk (see Supplemental Figure 3 online), a reported Golgi marker (Nelson et al., 2007). We further examined the subcellular localization of the TPST protein in planta using transgenic plants harboring the above-described functional TPSTpro:GFP-TPST construct. Confocal images of the root apex of the transgenic plants revealed that GFP fluorescence was present in a punctate pattern suggestive of organelles (Figures 4F and 4G). However, the subcellular localization of the TPSTpro:GFP-TPST fusion protein did not show an obvious difference after treatment with the vesicle trafficking inhibitor brefeldin A (Figures 4F and 4G).
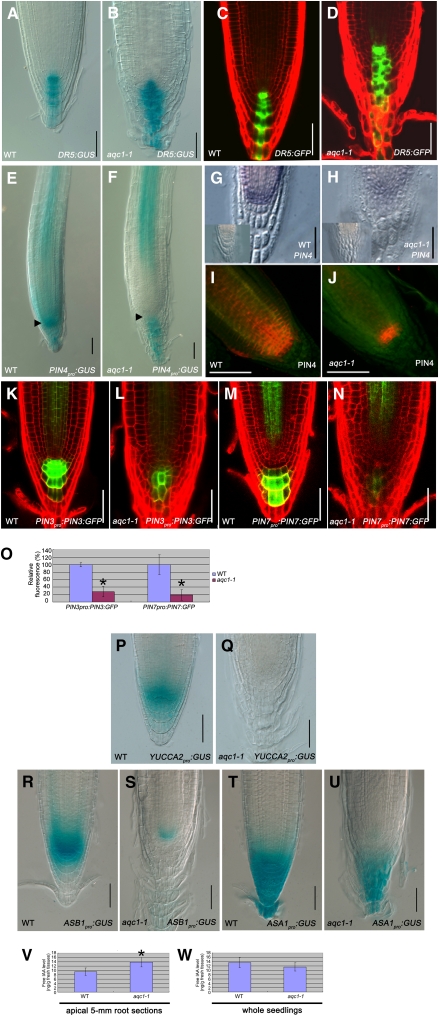
Considering that the TPSTpro:GUS and TPSTpro:GFP-TPST expression pattern in the root meristem resembles that of the DR5:GUS- and DR5:GFP-marked auxin maximum (Figures 5A and 5C), we asked whether the TPST gene expression is influenced by auxin. Without indole-3-acetic acid (IAA) treatment, TPSTpro:GUS expression was found in the distal meristem and stele of the differentiated region of the primary root (Figure 4H). Obvious induction of TPSTpro:GUS expression was detected 6 h after IAA treatment, and extended IAA treatment led to significant expansion of the TPSTpro:GUS expression domains in the root (Figure 4H). To test if auxin might regulate TPST expression at the protein level, the above-described transgenic plants containing a functional TPSTpro:GFP-TPST fusion protein were treated with IAA for 12 h, and protein immunoblot analysis was performed using an anti-GFP antibody. As shown in Figure 4I, IAA treatment markedly increased the levels of the TPSTpro:GFP-TPST fusion protein. Together, these results suggest that auxin upregulates TPST expression at both transcription and protein levels.
Figure 5.
Mutation of TPST Affects the Expression of PINs and Auxin Biosynthesis-Related Genes.
(A) to (D) Expression pattern of the DR5:GUS and DR5:GFP reporters in the wild type (WT; [A] and [C]) and aqc1-1 ([B] and [D]) at 4 DAG.
(E) and (F) PIN4pro:GUS expression in the wild type (E) and aqc1-1 (F) at 4 DAG. Black arrowheads indicate the stem cell niche.
(G) and (H) PIN4 expression in 4-DAG wild type (G) and aqc1-1 (H) revealed by RNA in situ hybridization with a PIN4 antisense probe. Insets represent the sense PIN4 probe.
(I) and (J) PIN4 expression in the wild type (I) and aqc1-1 (J) revealed by immunohistochemical localization assays using anti-PIN4 antibody in 4-DAG seedlings.
(K) and (L) PIN3pro:PIN3:GFP expression in the wild type (K) and aqc1-1 (L) at 6 DAG.
(M) and (N) PIN7pro:PIN7:GFP expression in the wild type (M) and aqc1-1 (N) at 6 DAG.
(O) Quantification of PIN3pro:PIN3:GFP and PIN7pro:PIN7:GFP GFP fluorescence in the wild type and aqc1-1 as shown in (K) to (N). Data shown are average and sd. Asterisks denote Student’s t test significant difference between wild-type and mutant plants (*P < 0.01).
(P) and (Q) YUCCA2pro:GUS expression pattern in 5-DAG wild-type (P) and aqc1-1 (Q) root tips.
(R) and (S) ASB1pro:GUS expression pattern in 5-DAG wild-type (R) and aqc1-1 (S) root tips.
(T) and (U) ASA1pro:GUS expression pattern in 5-DAG wild-type (T) and aqc1-1 (U) root tips.
Bars = 50 μm.
(V) Comparison of free IAA levels between 5-d-old wild-type and aqc1-1 root tips. IAA was extracted from apical 5-mm root sections. Data presented are mean values of nine biological repeats with sd. Asterisk denotes Student’s t test significant difference between the wild type and mutant (*P < 0.01).
(W) Comparison of free IAA levels between 5-d-old wild-type and aqc1-1 seedlings. IAA was extracted from whole seedlings. Data presented are mean values of nine biological repeats with sd. No significant difference was detected between the wild type and mutant (Student’s t test, P = 0.16).
We then generated a double mutant between aqc1-1 and the tir1-1 mutant (Ruegger et al., 1998), which harbors a mutation of the auxin receptor gene TIR1. As shown in Figures 4J to 4N, while tir1-1 did not show an obvious defect in the root meristem, root meristem phenotypes of the tir1-1 aqc1-1 double mutant were essentially similar to those of aqc1-1. In addition, the aqc1-1 mutation also suppressed the 2,4-D–insensitive phenotype of tir1-1 in root growth inhibition (Figures 4O to 4P). These data suggest that TPST acts genetically downstream of TIR1 in the auxin pathway.
Because several RGF members showed a similar expression pattern as TPST in the root stem cell region (Matsuzaki et al., 2010), we examined whether auxin regulates the expression of RGFs. Our quantitative real-time RT-PCR (qRT-PCR) assays indicated that, even though transcription of RGF1, 2, 3, and 4, which are grouped together in a phylogenetic analysis, was nonresponsive to auxin (Matsuzaki et al., 2010), transcription of RGF5, 6, 7, 8, and 9 was markedly induced by IAA treatment (see Supplemental Figure 7 online).
Mutation of TPST Affects the Local Expression of Several PINs and Auxin Biosynthetic Genes in the Stem Cell Region
We found that, in the aqc1-1 root tip, even though the expression domain of DR5:GUS or DR5:GFP showed a slight expansion compared with that of the wild type, the expression maxima of these markers was still localized in the center of the root meristem (Figures 5A to 5D). It has been shown that the auxin efflux transporter gene PIN4, whose expression pattern in the root apical meristem overlaps with that of TPST, plays an important role in stem cell niche maintenance (Friml et al., 2002; Blilou et al., 2005). The expression levels of PIN4, as shown by PIN4pro:GUS (Figures 5E and 5F), RNA in situ hybridization (Figures 5G and 5H), and immunohistological studies using the PIN4 antibody (Figures 5I and 5J), were obviously reduced in the aqc1-1 root stem cell niche. In addition, the expression levels of PIN3 and PIN7, as marked by the PIN3pro:PIN3:GFP and PIN7pro:PIN7:GFP fluorescence, were also substantially decreased in QC and columella cells of the aqc1-1 roots (Figures 5K to 5O). Interestingly, however, the expression levels of PIN1pro:PIN1:GFP and PIN2pro:PIN2:GFP were not so obviously changed by the aqc1-1 mutation (see Supplemental Figure 4 online). Taken together, the dramatic alteration in expression levels of several PIN genes in aqc1-1 suggests abnormal local auxin transport in the mutant root meristem.
In addition to PIN-mediated polar auxin transport, local auxin biosynthesis also contributes to the proper organization of the root stem cell niche (Blilou et al., 2005; Ljung et al., 2005; Zhao, 2008, 2010). Consistent with this idea, we found that the expression levels of YUCCA2pro:GUS (Figures 5P and 5Q), ASB1pro:GUS (Figures 5R and 5S), and ASA1pro:GUS (Figures 5T and 5U) were markedly reduced in the aqc1-1 root meristem compared with those in the wild type. Given the established role of YUCCA2 (Cheng et al., 2007), ASA1 (Stepanova et al., 2005; Sun et al., 2009), and ASB1 (Stepanova et al., 2005) in auxin biosynthesis, our results suggest that TPST also affects local auxin biosynthesis in the root meristem.
To test that the aqc1-1 mutation may affect local auxin concentration gradients, we measured endogenous IAA levels extracted from whole seedlings and apical 5-mm root sections. Consistent with the observation that DR5:GUS and DR5:GFP expression showed an expansion in aqc1-1 root tips (Figures 5A to 5D), free IAA levels of apical 5-mm root sections were significantly higher in aqc1-1 than those in the wild type (Figure 5V), while free IAA levels from whole seedlings did not show significant difference between aqc1-1 and the wild type (Figure 5W). These results indicated that mutation of TPST affects local expression of auxin biosynthesis- and transport-related genes and, as a consequence, leads to altered local auxin distribution in the root tip.
Mutation of TPST Affects the Expression of PLT1 and PLT2, Major Molecular Determinants of Auxin-Regulated Root Stem Cell Niche Maintenance
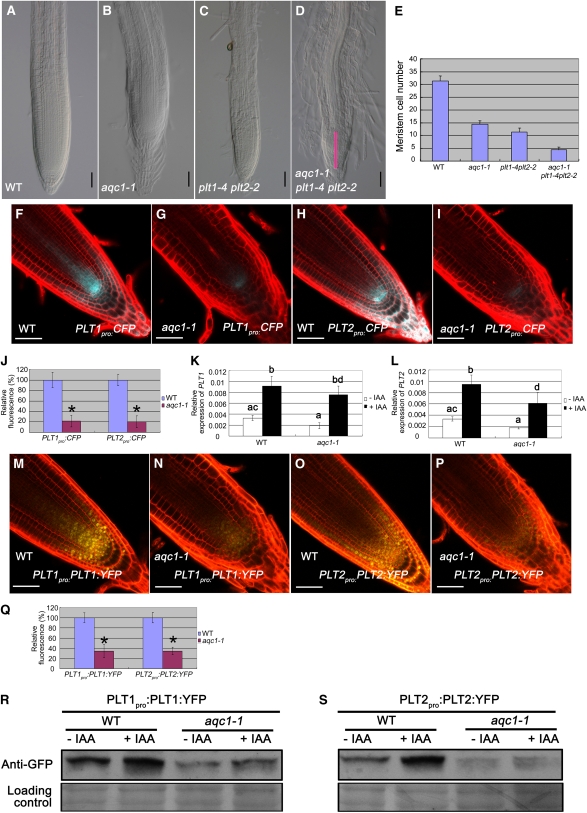
We then examined the genetic relationship between TPST and the PLT stem cell transcription factor genes. As shown in Figures 6A to 6E, even though the root growth and meristem defects of the aqc1-1 plt1-4 plt2-2 triple mutant were more severe than those of the aqc1-1 mutant or the plt1-4 plt2-2 double mutant, 95.8% (n = 24) of the aqc1-1 plt1-4 plt2-2 triple mutant seedlings contained observable root meristem at 4 DAG. By contrast, at the same stage, only 45.8% (n = 24) of the aqc1-1 scr-1 double mutant seedlings and 37.5% (n = 24) of the aqc1-1 shr-1 double mutant seedlings contained observable root meristem (see Supplemental Figures 5A to 5J online). These data, together with the finding that the aqc1-1 mutation did not affect the expression and/or the localization of SCR and SHR (see Supplemental Figures 5K to 5O online), support that TPST acts in parallel with the SHR/SCR pathway.
Figure 6.
Mutation of TPST Affects the Expression of PLT1 and PLT2.
(A) to (E) Root phenotypes of aqc1-1 and plt1-4 plt2-2 single, double, and triple mutants. WT, wild type.
(A) to (D) Root tips of the wild type (A), aqc1-1 (B), plt1-4 plt2-2 (C), and aqc1-1 plt1-4 plt2-2 (D) at 4 DAG. Pink bar indicates that the aqc1-1 plt1-4 plt2-2 triple mutant contains observable meristem at 4 DAG. Bars = 50 μm.
(E) Statistics of meristem cell number of the indicated genotypes at 4 DAG. Data shown are average and sd (n = 12).
(F) and (G) PLT1pro:CFP expression in wild-type (F) and aqc1-1 (G) root tips at 5 DAG.
(H) and (I) PLT2pro:CFP expression in wild-type (H) and aqc1-1 (I) root tips at 5 DAG.
(J) Quantification of PLT1pro:CFP and PLT2pro:CFP fluorescence as shown in (F) to (I). Data shown are average and sd (n = 15 to 20). Asterisks denote Student’s t test significant difference between wild-type and mutant plants (*P < 0.01).
(K) and (L) Analysis of auxin-regulated PLT1 (K) and PLT2 (L) expression in the wild type and aqc1-1 by qRT-PCR. Seedlings (2 DAG) were treated with or without 20 μM IAA for 24 h. Data presented are mean values of four biological repeats with sd. Samples with the different letters are significantly different at P < 0.01.
(M) and (N) PLT1pro:PLT1:YFP expression in wild-type (M) and aqc1-1 (N) root tips at 5 DAG.
(O) and (P) PLT2pro:PLT2:YFP expression in wild-type (O) and aqc1-1 (P) root tips at 5 DAG.
(Q) Quantification of PLT1pro:PLT1:YFP and PLT2pro:PLT2:YFP fluorescence as shown in (M) to (P). Data shown are average and sd (n = 15 to 20). Asterisks denote Student’s t test significant difference between wild-type and mutant plants (*P < 0.01).
(R) and (S) The TPST mutation affects accumulation of the PLT1proPLT1:YFP (R) and PLT2proPLT2:YFP (S) fusion proteins. Seedlings (5 DAG) were treated with or without 20 μM IAA for 24 h, and root tissues were harvested for protein extraction. Immunoblot was performed using anti-GFP antibody. Ponceau S–stained membranes are shown as loading controls.
Bars = 50 μm.
To test that mutation of TPST may affect the expression of PLT genes, the promoter transcriptional fusions PLT1pro:CFP (for cyan fluorescent protein) and PLT2pro:CFP were introduced into the aqc1-1 mutant through crossing. As shown in Figures 6F to 6J, CFP fluorescence levels were pronouncedly reduced in aqc1-1 compared with the wild type, suggesting that TPST affects the transcription of PLT1/2. Since it has been reported that auxin positively regulates the expression of PLT genes (Aida et al., 2004), we then quantified the PLT1/2 transcripts in the wild type and aqc1-1 with qRT-PCR assays. Without IAA treatment, levels of PLT1/2 transcripts were already markedly reduced in aqc1-1 (Figures 6K and 6L). IAA treatment led to substantial increases of PLT1 and PLT2 transcripts both in the wild type and aqc1-1. Significantly, in our repeated experiments, IAA treatment failed to increase the PLT1/2 transcripts of aqc1-1 plants to wild-type levels (Figures 6K and 6L). These results indicate that TPST affects the steady state and auxin-induced transcription of PLT1/2.
Similarly, yellow fluorescent protein (YFP) levels of the translational fusions PLT1pro:PLT1:YFP and PLT2pro:PLT2:YFP were both reduced in aqc1-1 compared with the wild type (Figures 6M to 6Q), suggesting that loss of TPST activity also affects PLT1/2 expression at the protein level. To confirm this, we conducted protein gel analyses using an anti-GFP antibody and protein extracts from transgenic plants containing the PLT1pro:PLT1:YFP or PLT2pro:PLT2:YFP translational fusion. As shown in Figures 6R and 6S, both in the absence or presence of IAA, the levels of the PLT1pro:PLT1:YFP and PLT2pro:PLT2:YFP fusion proteins were significantly reduced in aqc1-1 compared with those in the wild type. Taken together, our results reveal that the defective stem cell niche maintenance in the aqc1-1 mutant is correlated with a dramatic misregulation of PLT1/2 expression. Importantly, our findings that the aqc1-1 mutation impairs auxin-induced PLT1/2 expression at both transcription and protein levels suggest that TPST activity might be required for PLT response to auxin.
To examine that PLTs may also otherwise affect TPST expression, we transferred the TPSTpro:GUS transcriptional fusion and the TPSTpro:GFP-TPST translational fusion into the plt1-4 plt2-2 double mutant. GUS staining, qRT-PCR and GFP fluorescence assays indicated that the plt1-4 plt2-2 double mutations show little effect on the expression of TPST (see Supplemental Figure 6 online). Together, our data argue that TPST may act upstream of PLTs in regulating root stem cell niche maintenance.
Overexpression of PLT2 Partially Rescues the Root Meristem Defects of aqc1-1
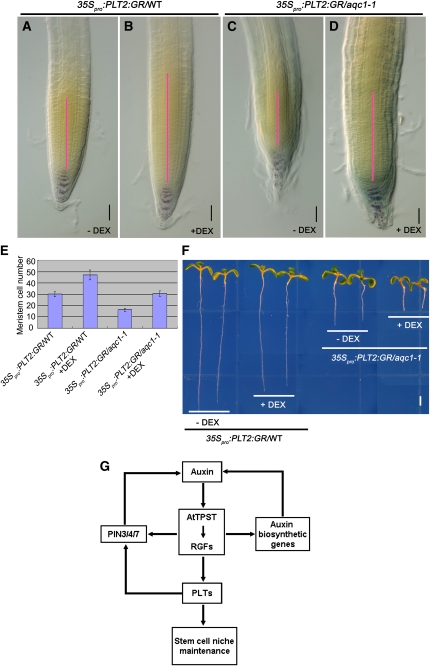
The above genetic and gene expression results prompted us to determine whether overexpression of PLT genes could rescue the aqc1-1 mutant phenotype. For these experiments, the inducible construct 35Spro:PLT2:GR (Galinha et al., 2007) was introduced into aqc1-1 through crossing. Since a short induction of PLT2 expression by adding dexamethasone (DEX) did not severely affect the growth of wild-type and mutant seedlings (Figure 7F), we examined root meristem expansion in these conditions. Consistent with previous reports (Galinha et al., 2007; Kornet and Scheres, 2009), a short induction of PLT2 expression by DEX led to substantial increase of proximal meristem cell number in wild-type roots (Figures 7A and 7B). In the aqc1-1 background, the cell number of the proximal meristem was also significantly increased after DEX induction (Figures 7C to 7E). In addition, DEX induction substantially improved the patterning of columella cells in 93.3% (n = 15) of the 35Spro:PLT2:GR/aqc1-1 seedlings (Figures 7A to 7D). Together, these data show that overexpression of PLT2 can, at least partially, bypass the root meristem defects of aqc1-1, which confirms that TPST acts in the auxin/PLT pathway.
Figure 7.
Overexpression of PLT2 Partially Rescues the Root Meristem Defects of aqc1-1.
(A) to (D) Lugol staining of 7-DAG 35Spro:PLT2:GR/wild type, and 35Spro:PLT2:GR/aqc1-1 roots without ([A] and [C]) or 2 d after 2 μM DEX treatment ([B] and [D]). Pink bars represent the root meristem of different plants extending from the QC to the transition zone. Bars = 50 μm.
(E) Cortical cell numbers in meristems of the indicated plants. Data shown are average and sd (n = 16 to 23).
(F) Phenotypes of 7-DAG 35Spro:PLT2:GR/wild type and 35Spro:PLT2:GR/aqc1-1 seedlings without or 2 d after 2 μM DEX treatment. Bar = 2 mm.
(G) A simplified model showing that TPST and the Tyr-sulfated RGF peptides maintain the root stem cell niche by defining the expression of the stem cell transcription factor PLTs. Auxin induces the expression of TPST and TPST shows a positive effect on the expression of PIN3/4/7, several auxin biosynthetic genes, and PLT1/2. Given that PLT1/2 positively regulates PIN3/4/7 expression (Blilou et al., 2005), reduced expression of PIN3/4/7 in the TPST loss-of-function mutant could be a result of downregulation of PLT1/2.
DISCUSSION
TPST-Dependent Sulfation of RGFs Is Required for Postembryonic Maintenance of the Root Stem Cell Niche
We provide evidence that the loss of function of the TPST gene, which encodes the recently identified TPST in Arabidopsis (Komori et al., 2009), led to extra QC cells, an increased number of CSCs, reduced meristem size, and defective root growth (Figures 1 and 2). It is noteworthy that, during the embryo stage, the cellular organization of aqc1-1 root pole and promeristem was comparable to that of the wild type, and obvious stem cell defects appeared during seed germination and early seedling growth (see Supplemental Figure 1 online), indicating that the TPST activity mainly acts to maintain root stem cell niche during postembryonic growth. In this regard, TPST is distinct from the SHR/SCR and the PLT transcription factors, which affect the establishment of the root stem cell niche during embryonic pattern formation. Given that the TPST protein exhibits TPST activity similar to its animal counterparts (Komori et al., 2009), our data suggest that such activity is required for postembryonic maintenance of the root stem cell niche. Considering the fact that the rice TPST ortholog readily rescues root meristem defects of the TPST loss-of-function mutant (see Supplemental Figure 2C online), it is reasonable to speculate that TPST activity and its role in root stem cell niche maintenance is conserved in the plant kingdom.
The TPST protein was shown in an in vitro assay to exhibit TPST activity on precursors of PSK and PSY1, two Tyr-sulfated peptide hormones showing growth promoting activities (Matsubayashi and Sakagami, 1996; Matsubayashi et al., 2002; Amano et al., 2007; Kutschmar et al., 2008). Tyr-sulfated PSK and PSY1, however, could not rescue stem cell defects of tpst-1, a T-DNA insertion allele of the TPST gene (Komori et al., 2009; Matsuzaki et al., 2010), suggesting the existence of other Tyr-sulfated peptide(s) that may be important for root stem cell niche maintenance. Significantly, recent elegant studies identified a group of Tyr-sulfated peptides called RGFs that could restore the meristem activity and stem cell niche defects of the tpst-1 mutant (Matsuzaki et al., 2010). Like TPST (Figures 4E to 4G), RGFs are mainly expressed in the root stem cell area and maintain the postembryonic root stem cell niche by defining the expression of stem cell transcription factor PLTs (Matsuzaki et al., 2010). Similar to tpst-1, the rgf1 rgf2 rgf3 triple mutant showed reduced meristem activity, which could be restored by external application of RGF1 (Matsuzaki et al., 2010). These data, together with our work described here, highlight the importance of TPST-dependent sulfation of RGFs in maintaining postembryonic root stem cell niche.
TPST Acts in the Auxin Pathway and Affects Local Auxin Biosynthesis and Transport in the Root Meristem
In the Arabidopsis root, two independent pathways converge to specify and maintain the identity and function of QC and the associated stem cells; the SHR/SCR pathway provides positional signal along the radial axis, whereas the auxin pathway provides the longitudinal signal (Benjamins and Scheres, 2008; Dinneny and Benfey, 2008; Petricka and Benfey, 2008). Our genetic and gene expression results suggest that TPST acts in parallel with the SHR/SCR pathway (see Supplemental Figure 5 online). We provide convincing evidence showing that TPST and RGFs act in the auxin pathway. First, auxin induces the expression of the TPST gene at both the transcriptional and protein levels (Figures 4H and 4I). Second, transcription of RGF5, 6, 7, 8, and 9 is markedly induced by IAA treatment (see Supplemental Figure 7 online). Third, a tir1-1 aqc1-1 double mutant shows similar root meristem defects as does aqc1-1, suggesting that TPST acts downstream of the auxin receptor TIR1 (Figures 4J to 4P). Fourth, the aqc1-1 mutation leads to a dramatic alteration in local expression levels of several PIN genes (Figures 5E to 5O) and auxin biosynthetic genes in the stem cell area (Figures 5P to 5U). Finally, even though mutation of TPST shows little effect on free IAA accumulation in a whole-seedling level (Figure 5W), it specifically affects local auxin accumulation in the root tip (Figure 5V). Therefore, our work revealed a regulatory loop between auxin and the TPST gene: auxin induces TPST gene expression; mutation of TPST impairs local expression levels of PIN3, 4, and 7 as well as several auxin biosynthetic genes and therefore alters local auxin accumulation in the root meristem. An important feature of the phytohormone auxin is its spatiotemporal asymmetric distribution, which provides developmental information to instruct versatile aspects of organ patterning and outgrowth (Darwin and Darwin, 1880; Tanaka et al., 2006). In this context, a significant direction of future studies is to elucidate the molecular mechanism underlying the TPST effect on local auxin biosynthesis and transport.
TPST-Mediated Sulfation of RGFs Provides a Link between Auxin and the PLT Transcription Factors in Root Stem Cell Niche Maintenance
It is generally believed that the PLT transcription factors act downstream of auxin to regulate root stem cell niche maintenance (Aida et al., 2004; Galinha et al., 2007; Benjamins and Scheres, 2008). However, relatively less is known about the molecular mechanism controlling auxin-mediated PLT expression. Here, we provide evidence showing that, in the absence or presence of exogenous IAA, the aqc1-1 mutation significantly impairs PLT1/2 expression at both the transcriptional and protein levels (Figure 6), indicating that TPST plays an important role in mediating basal- and auxin-induced expression of PLT1/2. We also show that mutations of PLT1/2 have little, if any, effect on the expression of the TPST gene (see Supplemental Figure 6 online). These data, together with our genetic analyses (Figures 6A to 6E) and the observation that overexpression of the PLT2 gene partially rescues the root meristem defects of the aqc1-1 mutant (Figures 7A to 7F) support that TPST acts in the auxin/PLT pathway. In light of the recent exciting finding that the Tyr-sulfated RGFs, but not desulfated RGFs, restore the stem cell niche defects caused by the TPST mutation (Matsuzaki et al., 2010), we proposed a simplified model showing that TPST acts in the auxin pathway to regulate root stem cell niche maintenance by affecting the expression of PLTs (Figure 7G). In this model, TPST-dependent activation of RGFs defines the expression levels and patterns of PLTs that, in turn, regulate stem cell niche maintenance. As described above, our findings that auxin upregulates the expression of TPST and several RGF members and that the aqc1-1 mutation affects local expression of PIN3/4/7 and several auxin biosynthetic genes suggest the existence of a regulation loop between local auxin accumulation and TPST expression. It is noteworthy that reduced expression of PIN3/4/7 in aqc1-1 could also be a result of downregulation of PLT1/2 because PIN3/4/7 transcription has been reported to be decreased in the plt1-4 plt2-2 double mutant (Blilou et al., 2005). Thus, TPST-mediated activation of RGFs provides a missing link between auxin and the PLT transcription factors in root stem cell niche maintenance.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes Columbia (Col), C24, Landsberg erecta, and Wassilewskija were used. Some of the plant materials used in this study were previously described: ASA1pro:GUS and ASB1pro:GUS (Stepanova et al., 2005); cycB1;1:GUS (Colón-Carmona et al., 1999); DR5:GUS (Ulmasov et al., 1997); DR5rev:GFP (Benková et al., 2003); PIN1pro:PIN1:GFP (Benková et al., 2003); PIN2pro:PIN2:GFP (Blilou et al., 2005); PIN3pro:PIN3:GFP (Blilou et al., 2005); PIN4pro:GUS (Friml et al., 2002); PIN7pro:PIN7:GFP (Blilou et al., 2005); plt1-4 plt2-2 (Aida et al., 2004); PLT1pro:CFP, PLT1pro:PLT1:YFP, PLT2pro:CFP, and PLT2pro:PLT2:YFP (Kornet and Scheres, 2009); 35Spro:PLT2:GR (Galinha et al., 2007); QC25 (Sabatini et al., 2003); scr-1 (Di Laurenzio et al., 1996); shr-1 (Benfey et al., 1993); SCRpro:GFP (Di Laurenzio et al., 1996); SHRpro:SHR:GFP (Nakajima et al., 2001); tir1-1 (Ruegger et al., 1998); WOX5pro:GFP (Blilou et al., 2005); and YUCCA2pro:GUS (Cheng et al., 2006). J2341 and J2672 are from the Haseloff enhancer trap GFP line collection (http://www.plantsci.cam.ac.uk/Haseloff). Seeds of Salk_009847 were obtained from the ABRC (Alonso et al., 2003).
Seeds were surface-sterilized for 12 min in 10% commercial kitchen bleach, washed five times with sterile water, and plated on half-strength Murashige and Skoog (MS) (Murashige and Skoog, 1962) medium with 1% sucrose and 0.8% agar. Plants were stratified at 4°C for 2 d in the darkness and then transferred to a phytotron set at 22°C with a 16-h-light/8-h-dark photoperiod (light intensity 120 μmol photons m−2 s−1) in vertically or horizontally oriented Petri dishes. Roots were examined at 1 to 12 DAG, depending on the experimental requirement.
Mutant Screening and Genetic Analysis
Seeds of a T-DNA–mutagenized population of Arabidopsis ecotype Col-0 (Zhang et al., 2005) were grown vertically on the medium described above and screened for defective root development. One seedling with significantly stunted root growth was selected, named aqc1-1, and directly transplanted to soil. The original aqc1-1 mutant was backcrossed to Col-0 for three generations, and the resulting homozygous progenies were used in this study. For T-DNA mutant genotyping, T3 segregating SALK insertion lines were plated on kanamycin-containing media for selection.
A aqc1-1 scr-1 double mutant line was identified from an F2 population generated between aqc1-1 and scr-1 (Di Laurenzio et al., 1996). A homozygous aqc1-1 mutation was identified with diagnostic PCR using gene-specific primers 5′-GGACGGAACCACTTGAGACAG-3′ and 5′-TAACCCGTATGACATGGAGGAGAT-3′. Identification of a homozygous scr-1 mutation has been described (Di Laurenzio et al., 1996). Similarly, we generated a triple mutant line between aqc1-1 and plt1-4 plt2-2 (Aida et al., 2004) and double mutant lines between aqc1-1 and shr-1 (Helariutta et al., 2000) or tir1-1 (Ruegger et al., 1998).
Measurement of the Primary Root Length
The primary root lengths of wild-type Col-0 and the aqc1-1 mutant were measured from 0 to 10 DAG using rulers (scale = 1.0 mm). Root lengths of other mutants were also measured in the same way at different DAGs. The statistical significance was evaluated by Student’s t test analysis and the Student-Newman-Keuls a.b analysis.
Measurement of IAA Levels
Five-day-old wild-type and aqc1-1 seedlings were used for IAA measurement. Approximately 60 mg (fresh weight) of whole seedlings or apical 5-mm root sections were collected for IAA extraction and measurement following a published method (Kojima et al., 2009) with minor modifications. Briefly, plant tissues were homogenized and extracted for 24 h in methanol containing 2H-IAA (CDN Isotopes) as internal standard. Purification was done with Oasis Max solid phase extract cartridge (150 mg/6 cc; Waters) after centrifugation. The entire sample was injected into a liquid chromatography–tandem mass spectrometry system consisting of an Acquity Ultra Performance Liquid Chromatograph (Acquity UPLC; Waters) and a triple quadruple tandem mass spectrometer (Quattro Premier XE; Waters).
Histology and Microscopy
Whole seedlings or different tissues were cleared in the HCG solution (chloroacetaldehyde:water:glycerol = 8:3:1) for several minutes before microscopy analysis. For Lugol staining of roots, tissues were incubated in Lugol solution (Sigma-Aldrich) for 3 to 5 min, washed in water once, and mounted in HCG solution for microscopy analysis. Histochemical staining for GUS activity in transgenic plants was performed according to the described method (Jefferson et al., 1987). Whole seedlings or different tissues were immersed in the GUS staining solution (1 mM X-glucuronide in 100 mM sodium phosphate, pH 7.2, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 0.1%Triton X-100), applied in vacuum briefly, and incubated at 37° in the dark from 5 min to overnight depending on the experimental requirement. Plants on MS medium were photographed using the Leica DFC 490 stereomicroscope and Leica DM5000B microscope. Images were processed with Adobe Photoshop CS 8.0 and Spot Flex software.
For confocal microscopy, plants were grown on MS medium from 4 to 10 d and observed using a Leica TCS SP5 confocal laser scanning microscope. Seedlings were stained with 10 μg/mL propidium iodide for 5 min and washed once in water. Propidium iodide was visualized using wavelengths of 600 to 640 nm. Wavelengths to visualize CFP, GFP, and YFP were 460 to 500, 500 to 540, and 525 to 565 nm, respectively. Fluorescence was quantified with the LAS AF Lite program on confocal sections acquired with the same microscope settings (Růžička et al., 2007). Approximately 5 to 10 images were examined, and at least three independent experiments were performed. The statistical significance was evaluated by Student’s t test analysis.
Whole-Mount in Situ Hybridization
Whole-mount in situ hybridization was performed according to the reported protocols (Weigel and Glazebrook, 2002; Hejátko et al., 2006). Antisense and sense probes were synthesized with digoxigenin-11-UTP (Roche Diagnostics) using T7 and SP6 RNA polymerases, respectively. The following primers were used to amplify the DNA template for the probe synthesis: At PIN4 primers 5′-ATAGGATCCTGTTTGGAGCTCAAGCGCTTCT-3′ and 5′-AAACTCGAGACACTTGTCGGCGGCATATGT-3′; WOX5 primers 5′-AAACTCGAGAGGCAGAAACGTCGTAAAATCT-3′ and 5′-TCAGGATCCTTAAAGAAAGCTTAATCGAAGATCTA-3′.
Sequence Analysis
The BLAST search program (Altschul et al., 1997) was used for sequence analysis. The software ClusterX and T-coffee (http://www.ebi.ac.uk/Tools/t-coffee/) were used for sequence alignment. The phylogenetic relationship of TPST in plants is inferred from protein sequences using a Bayesian approach in MrBayes (Ronquist and Huelsenbeck, 2003). The node labels are measures of support, which indicate the proportion of trees in the posterior distribution to containing the node.
TAIL-PCR and Mapping
The design of anchor primers, random primers, and the PCR reaction system were performed as described (Zhang et al., 2005). We confirmed the T-DNA insertion site by rough mapping. A segregating F2 family was constructed from a cross between the aqc1-1 in the Col-0 background and the wild type Landsberg erecta. The map position of AQC1 was obtained by analyzing DNA from 92 F2 aqc1-1 mutants of the mapping population using the simple sequence length polymorphic markers.
Plasmid Construction and Plant Transformation
A 6790-bp genomic DNA fragment from upstream 5′ noncoding region to downstream 3′ noncoding region, covering the coding sequences of At1g08030, was amplified by PCR and cloned into the KpnI site of the binary vector pCAMBIA 1300 (CAMBIA) as the complementation construct. The primers were 5′-AAGGTACCTCATCGGAACAAGTAGTAAGAGAATCAA-3′ and 5′-AAGGTACCTCTCTCTATCAACGCGTTTTACCAAAAA-3′.
The 35Spro:TPST and 35Spro:Os06g06370 (the rice [Oryza sativa] ortholog of At TPST) were prepared by inserting the PCR-amplified coding sequence of At TPST and Os06g06370 into the SpeI site and SalI/SpeI site of pCAMBIA1300-221 vector under the control of the 35S promoter, respectively. The insertion orientation was confirmed by sequencing. The primers were as follows: AtTPST-F, 5′-GGGACTAGTATGCAAATGAACTCTGTTTGGAAGC-3′; AtTPST-R, 5′-GGGACTAGTTCAAATCTTAACTTTGGAGGTTCTTCTC-3′; Os06g06370-F, 5′-ATTTGTCGACATGGCGTTCGGCGGACTC-3′; and Os06g06370-R, 5′-TGCACTAGTTCATCTGGGCATGCCTGTTAGTT-3′.
A 2231-bp genomic fragment upstream of the translation start codon was PCR amplified and inserted into the PstI/SalI sites of the binary vector pCAMBIA1391-Z (CAMBIA), resulting in the transcriptional fusion of the TPST promoter with the GUS coding region. The primers were prom-F, 5′-AAACTGCAGTCATCGGAACAAGTAGTAAGA-3′; and prom-R, 5′- AAAGTCGACTGCCTCGATTCCAAACC-3′.
TPSTpro:GFP-TPST was prepared by inserting the 2231-bp genomic fragment, putative signal peptide, enhanced GFP coding region, the rest of the coding sequence of TPST, and the NOS fragment into the pCAMBIA 1300 (CAMBIA) vector in this order. Similarly, the 35Spro:GFP-TPST was prepared by inserting the putative signal peptide, enhanced GFP coding region, the rest of the coding sequence of TPST, and the NOS fragment into the pGFP-2 transient vector. Primers were as follows: prom-sig-F, 5′-CACCTGCAGTCATCGGAACAAGTAGTAAGAGAAT-3′; prom-sig-R, 5′- AAACTGCAGTTCCGCAAAAGAGCCAATAACTGAG-3′; EGFP-F, 5′- AAACTGCAGATGGTGAGCAAGGGCGAGGAGCTGT-3′; EGFP-R, 5′- TTTGTCGACCTTGTACAGCTCGTCCATGCCGAGA-3′;. CDS-NOS-F, 5′- AATAGATCTCTTGATTTTGGCCATTGCGAAACTC-3′; and CDS-NOS-R, 5′- GCGAATTCCCGATCTAGTAACATAGATGACA-3′.
The above constructs were then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90), which was used for transformation of Arabidopsis plants by vacuum infiltration (Bechtold and Pelletier, 1998).
Marker Gene Analysis
ASA1pro:GUS, ASB1pro:GUS, cyclinB1;1:GUS, DR5:GUS, DR5:GFP, PIN1pro:PIN1:GFP, PIN2pro:PIN2:GFP, PIN3pro:PIN3:GFP, PIN4pro:PIN4:GFP, PIN7pro:PIN7:GFP, PLT1pro:CFP, PLT1pro:PLT1:YFP, PLT2pro:CFP, PLT2pro:PLT2:YFP, SCRpro:GFP, SHRpro:SHR:GFP, WOX5pro:GFP, and QC25 marker lines were crossed to aqc1-1, and plants homozygous for the aqc1-1 mutations and transgene markers were identified from the F2 population and analyzed in the next generations. In all analyses, parental transgenic lines were used to compare with those in the mutant background.
RT-PCR and qRT-PCR Assays
For RT-PCR analysis of TPST expression, 5-DAG seedlings of indicated genotypes were frozen in liquid nitrogen for RNA extraction with the RNeasy mini kit (Qiagen). First-strand cDNA was synthesized from 2 μg of total RNA using Superscript III reverse transcriptase (Invitrogen) with an oligo(dT) primer. Specific primers for RT-PCR amplification of TPST transcripts were 5′- TTGGCCATTGCGAAACTC-3′ and 5′- TGCGGCTAGACATCCTTGAC-3′. Expression levels of TPST were normalized to ACTIN8 expression levels.
For qRT-PCR analysis, whole seedlings or root tissues of the indicated genotypes were collected for RNA extraction with an RNeasy mini kit (Qiagen). First-strand cDNA was synthesized from 2 μg of total RNA using Superscript III reverse transcriptase (Invitrogen) with an oligo(dT) primer and was quantified with a cycler apparatus (Bio-Rad) with the RealMasterMix kit (SYBR Green; Tiangen) according to the manufacturer’s instructions. PCR was performed in 96-well optical reaction plates heated for 5 min at 95° to activate hot start Taq DNA polymerase, followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 58°C, and extension for 30 s at 68°C. Expression levels of target genes were normalized to those of ACTIN7. Auxin treatment of Col-0, plt1-4 plt2-2, and aqc1-1 plants was conducted according to a previously described method (Aida et al., 2004). The statistical significance was evaluated by Student’s t test analysis. For the multiple comparisons, an analysis of variance followed by Fisher’s least significant difference mean separation test (SPSS) was performed on the data. Samples with the different letters are significantly different at P < 0.01 or P < 0.05. Data presented are mean values of at least three biological repeats with sd. Gene-specific primers used for qRT-PCR amplification are listed in Supplemental Table 1 online.
Immunoblot and Immunolocalization Assays
For TPST-GFP immunoblots, 5-DAG transgenic seedlings containing the TPSTpro:GFP-TPST construct were treated with 5 μM IAA for 12 h before root tissues were harvested for protein extraction. TPSTpro:GFP-TPST fusion proteins were visualized by immunoblots using anti-GFP antibody (Abmart GFP-Tag). Ponceau S–stained membranes are shown as loading controls.
For PLT1/2-YFP immunoblots, 5-DAG wild-type and aqc1-1 seedlings containing PLT1pro:PLT1:YFP or PLT2pro:PLT2:YFP were treated with 20 μM IAA for 24 h, and total proteins were extracted from root tissues. PLT1pro:PLT1:YFP or PLT2pro:PLT2:YFP fusion proteins were visualized by immunoblots using anti-GFP antibody (Abmart GFP-Tag). Ponceau S–stained membranes are shown as loading controls.
The PIN4 immunolocalization assay was performed using the InsituPro robot (Friml et al., 2002). The following antibodies and dilutions were used: anti-PIN4 (1:400) antibody and Alexa Fluor 555 (1:500) secondary antibody (Molecular Probes). Fluorescent samples were inspected by the Zeiss Axioplan 2 confocal laser scanning microscope and Zeiss LSM 510 Image Browser software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACTIN7 (At5g09810), ACTIN8 (At1g49240), AQC1/TPST (At1g08030), ASA1 (At5g05730), ASB1 (At1g25220), CycB1;1 (At4g37490), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At1g70940), PIN4 (At2g01420), PIN7 (At1g23080), PLT1 (At3g20840), PLT2 (At1g51190), SCR (At3g54220), SHR (At4g37650), TIR1 (At3g62980), WOX5 (At3g11260), YUCCA2 (At4g13260), Os TPST (Os06g06370), Sb TPST (SORBIDRAFT_10g004010), Bd TPST (Bradi1g48760), Al TPST (ARALYDRAFT_334090), Vv TPST (LOC100255229), Gm TPST (Glyma11g20470), Cp TPST (evm.TU.supercontig_279.1), Pp TPST (PHYPADRAFT_65073), Sm TPST (SELMODRAFT_412798), Pt TPST (POPTRDRAFT_768390), Ol TPST (OSTLU_17201), and aqc1-2 (SALK_009847).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TPST Acts in the Distal Root Meristem to Maintain Stem Cell Niche after Germination.
Supplemental Figure 2. Complementation of the aqc1-1 Phenotype.
Supplemental Figure 3. Subcellular Localization of the 35Spro:GFP-TPST Fusion Protein Transiently Expressed in Arabidopsis Protoplasts.
Supplemental Figure 4. Mutation of TPST Has Little Effect on the Expression of PIN1 and PIN2.
Supplemental Figure 5. The Action of TPST on Stem Cell Niche Maintenance Is Independent of the SHR/SCR Pathway.
Supplemental Figure 6. Basal- and Auxin-Induced Expression of TPST in the Wild Type and the plt1-4 plt2-2 Double Mutant.
Supplemental Figure 7. Auxin-Induced Expression of RGFs Revealed by qRT-PCR.
Supplemental Table 1. Primers Used for qRT-PCR Analysis.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 3C.
Acknowledgments
We thank Ben Scheres, Philip N. Benfey, Malcolm J. Bennett, Klaus Palme, and Jianru Zuo for kindly providing seeds used in this study. This work was supported by grants from the Ministry of Science and Technology of China (2007CB948200 and 2006CB910604), the Chinese Academy of Sciences (KSCX2-YW-N-045), and the National Natural Science Foundation of China (90717007).
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Benfey P.N., Linstead P.J., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A. (1993). Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E., Hofhuis H.F., Xu J., Liu C.M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Darwin C., Darwin F. (1880). The Power of Movement in Plants. (London: John Murray; ). [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Ding Z., Friml J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 107: 12046–12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Benfey P.N. (2008). Plant stem cell niches: Standing the test of time. Cell 132: 553–557 [DOI] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C.M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Hause G., Boutilier K., Casamitjana-Martinez E., Weijers D., Offringa R., van der Geest L., van Lookeren Campagne M., Liu C.M. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327: 37–49 [DOI] [PubMed] [Google Scholar]
- Fiers M., Ku K.L., Liu C.-M. (2007). CLE peptide ligands and their roles in establishing meristems. Curr. Opin. Plant Biol. 10: 39–43 [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F.M., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hejátko J., Blilou I., Brewer P.B., Friml J., Scheres B., Benková E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hobe M., Müller R., Grünewald M., Brand U., Simon R. (2003). Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 213: 371–381 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Feldman L.J. (2003). Root meristem establishment and maintenance: The role of auxin. J. Plant Growth Regul. 21: 432–440 [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., Ashikari M., Ueguchi-Tanaka M., Matsuoka M., Suzuki K., Sakakibara H. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y. (2009). Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 15067–15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornet N., Scheres B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschmar A., Rzewuski G., Stührwohldt N., Beemster G.T., Inz D., Sauter M. (2008). PSK-alpha promotes root growth in Arabidopsis. New Phytol. 181: 820–831 [DOI] [PubMed] [Google Scholar]
- Laux T. (2003). The stem cell concept in plants: A matter of debate. Cell 113: 281–283 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Morita A., Sakagami Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Moore K.L. (2009). Protein tyrosine sulfation: a critical posttranslation modification in plants and animals. Proc. Natl. Acad. Sci. USA 106: 14741–14742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O., Pernas M., Carol R.J., Dolan L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510 [DOI] [PubMed] [Google Scholar]
- Ouyang Y., Lane W.S., Moore K.L. (1998). Tyrosylprotein sulfotransferase: Purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins. Proc. Natl. Acad. Sci. USA 95: 2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Benfey P.N. (2008). Root layers: Complex regulation of developmental patterning. Curr. Opin. Genet. Dev. 18: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W.M., Hobbie L., Turner J., Estelle M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Scheres B. (2007). Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8: 345–354 [DOI] [PubMed] [Google Scholar]
- Seibert C., Sakmar T.P. (2008). Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers 90: 459–477 [DOI] [PubMed] [Google Scholar]
- Stahl Y., Wink R.H., Ingram G.C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. (2005). A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Doležal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Sun J., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Dhonukshe P., Brewer P.B., Friml J. (2006). Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 63: 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobeña-Santamaria R., Bliek M., Ljung K., Sandberg G., Mol J.N., Souer E., Koes R. (2002). FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 16: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hage W., Weisbeek P., Scheres B. (1995). Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S.C., De Veylder L., Mergaert P., Kondorosi E. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2005). Generation of chemical-inducible activation tagging T-DNA insertion lines of Arabidopsis thaliana. Yi Chuan Xue Bao 32: 1082–1088 [PubMed] [Google Scholar]
- Zhao Y. (2008). The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 11: 16–22 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]