This work identifies a region of the gibberellin receptor GID1 as being involved in gibberellin-dependent GID1–DELLA protein interaction and finds that the unique GA-independent DELLA protein interaction of a GID1 present in Arabidopsis and some other dicots also depends on this loop structure.
Abstract
To investigate gibberellin (GA) signaling using the rice (Oryza sativa) GA receptor GIBBERELLIN-INSENSITIVE DWARF1 (GID1) mutant gid1-8, we isolated a suppressor mutant, Suppressor of gid1-1 (Sgd-1). Sgd-1 is an intragenic mutant containing the original gid1-8 mutation (L45F) and an additional amino acid substitution (P99S) in the loop region. GID1P99S interacts with the rice DELLA protein SLENDER RICE1 (SLR1), even in the absence of GA. Substitution of the 99th Pro with other amino acids revealed that substitution with Ala (P99A) caused the highest level of GA-independent interaction. Physicochemical analysis using surface plasmon resonance revealed that GID1P99A has smaller Ka (association) and Kd (dissociation) values for GA4 than does wild-type GID1. This suggests that the GID1P99A lid is at least partially closed, resulting in both GA-independent and GA-hypersensitive interactions with SLR1. One of the three Arabidopsis thaliana GID1s, At GID1b, can also interact with DELLA proteins in the absence of GA, so we investigated whether GA-independent interaction of At GID1b depends on a mechanism similar to that of rice GID1P99A. Substitution of the loop region or a few amino acids of At GID1b with those of At GID1a diminished its GA-independent interaction with GAI while maintaining the GA-dependent interaction. Soybean (Glycine max) and Brassica napus also have GID1s similar to At GID1b, indicating that these unique GID1s occur in various dicots and may have important functions in these plants.
INTRODUCTION
Gibberellins (GAs) are a large family of tetracyclic diterpenoid plant hormones that induce a wide range of plant growth responses, including seed germination, stem elongation, leaf expansion, flowering, and pollen maturation (Thomas et al., 2005; Aya et al., 2009). Genetic analyses using rice (Oryza sativa) and Arabidopsis thaliana mutants have provided an in-depth understanding of GA perception and signaling mechanisms. A recent breakthrough in this field is the discovery of a soluble GA receptor, GID1 (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). Interestingly, the primary structure of GID1 is similar to that of the hormone-sensitive lipase (HSL) family, which includes enzymes involved in lipid metabolism. Structural analyses of GID1 proteins of rice and Arabidopsis have revealed that its GA binding pocket corresponds to the substrate binding site of HSLs, and the movable lid located at its N-terminal portion functions to cover GA and stabilize it at the binding site (Murase et al., 2008; Shimada et al., 2008). The N-terminal lid is also involved in the GA-dependent interaction with a DELLA protein, which works as a repressor in GA signaling (Murase et al., 2008; Shimada et al., 2008). This information, together with previous information about DELLA proteins (Peng et al., 1997, 1999; Silverstone et al., 1998; Ikeda et al., 2001; Itoh et al., 2002) and GA signal–specific F-box protein (GID2 in rice and SLY1 in Arabidopsis) characteristics (McGinnis et al., 2003; Sasaki et al., 2003), leads to a model of GA perception and signaling. According to this model, the binding of GA to GID1 induces the formation of a GID1-GA-DELLA protein complex. The DELLA protein is then degraded with the aid of the F-box protein GID2/SLY1, resulting in various GA-triggered actions (Ueguchi-Tanaka et al., 2007b; Itoh et al., 2008; Harberd et al., 2009).
Although the molecular system involving GID1, DELLA, and F-box proteins is now accepted as a basic concept of GA perception, we are still far from the complete picture as to how GAs modulate plant growth, and many questions still remain to be resolved. For example, it is still unknown why GID1–DELLA protein interaction induces the interaction with an F-box protein, GID2/SLY1, and the following degradation of the DELLA protein. In this context, for example, one unique GID1 protein, Arabidopsis GID1b (At GID1b), which can interact with DELLA proteins in the absence of GA (Griffiths et al., 2006; Nakajima et al., 2006), should be interesting. To answer these remaining but important questions on the GID1–DELLA–dependent GA perception system, we attempted to isolate and characterize suppressor mutants of the gid1 phenotypes in the gid1 mutant background. We isolated five independent suppressor mutants; here, we report the characterization of one of these mutants. Through the analyses of this suppressor mutant, we identified one loop region of GID1 that is involved in GA-dependent GID1–DELLA interaction. The unique GA-independent interaction of At GID1b with DELLA proteins also depends on this loop structure. We found that soybean (Glycine max) and Brassica napus also have similar unique GID1s that have GA-independent interaction activity with DELLA, indicating that these GID1s occur in various kinds of dicot species and might have crucial roles in some developmental process.
RESULTS
Isolation and Characterization of a Suppressor Mutant of gid1
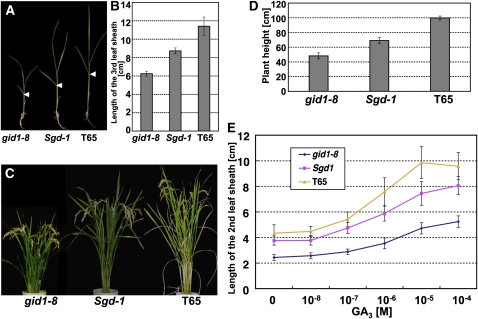
As reported previously (Ueguchi-Tanaka et al., 2007a), seven out of eight gid1 mutants cannot develop flowers and consequently are sterile. On the other hand, the mildest mutant, gid1-8, which has a reduced GA binding activity of GID1 caused by a single amino acid substitution (L45F) in the N-terminal lid region, can develop fertile flowers (Ueguchi-Tanaka et al., 2007a). To obtain suppressor mutants of gid1, we used gid1-8 as the starting material for mutagenesis. About 20,000 gid1 flowers were treated with n-nitrosourea, and plants with restored height were screened in a field at the M2 generation to identify suppressor mutants for gid1. We obtained five lines showing greater plant height than gid1-8. In this article, we focus on one mutant, Suppressor for gid1-1 (Sgd-1), which showed the clearest phenotype among the five mutants. Figures 1A and 1C show the gross morphology of Sgd-1 at the juvenile and heading stages, respectively. Sgd-1 rescued the dwarf phenotype of gid1-8 to increase height 1.5-fold at both stages, but height was not completely restored to that of the original wild-type strain, T65 (Figures 1A to 1D). The segregation of suppressive and original phenotypes in the M3 generation fitted a 3:1 ratio based on a χ2 goodness-of-fit test (Table 1), indicating that Sgd-1 is inherited in a dominant manner.
Figure 1.
Gross Morphology and GA Responsiveness of Sgd1.
(A) Gross morphology of Sgd-1 at the juvenile stage. gid1-8 (left), Sgd-1 (middle), and wild-type plants (T65; right) were grown for 4 weeks. Arrowheads represent the uppermost positions of the 3rd leaf sheath. Bar = 5 cm.
(B) Length of the 3rd leaf sheath of gid1-8 (left), Sgd-1 (middle), and T65 (right) grown for 4 weeks. Data are means ± sd, n = 14.
(C) Gross morphology of Sgd-1 at heading stage. gid1-8 (left), Sgd-1 (middle), and T65 (right) were grown for 3 months. Bar = 10 cm.
(D) Plant height of gid1-8, Sgd-1, and T65 grown for 3 months. Height was measured from the ground surface to the ear-neck node. Data are means ± sd, n = 14.
(E) Dose dependency of 2nd leaf sheath elongation of gid1-8, Sgd-1, and T65 grown on GA3 for 3 weeks. Data are means ± sd, n = 10.
Table 1.
Phenotypic Polymorphism in the M3 Generation of Sgd-1
| Phenotype of M3 Progeny |
||||
| M2 Plant Number | Tall | Short | Total | χ2 (3:1) |
| 1 | 34 (72.3%) | 13 (27.7%) | 47 | 0.177a |
| 2 | 36 (75.0%) | 12 (25.0%) | 48 | 0.000a |
| 3 | 35 (77.8%) | 10 (22.2%) | 45 | 0.185a |
Numbers in parentheses indicate the percentage relative to the total number of plants.
P ≤ 0.05, χ2 goodness of fit test.
To examine whether the phenotype of Sgd-1 is caused by rescue of the GA signaling pathway but not by other mechanisms (e.g., overproduction of active GA), we first investigated the GA sensitivity of Sgd-1 by a 2nd leaf sheath elongation experiment (Figure 1E). GA-dependent elongation of wild-type seedlings started at 10−8 M GA3 and reached a plateau at 10−5 M, while gid1-8 seedlings responded to GA3 at 10−8 to 10−7 M and reached a plateau at >10−5 M with a lower responding slope than that of the wild type, demonstrating that gid1-8 has a lower sensitivity to GA3 than the wild type. This corresponds well to the observation of lower binding activity of GID1-8 than wild-type GID1 (Ueguchi-Tanaka et al., 2007a). The GA3 response of Sgd-1 seedlings was intermediate between those of the original gid1-8 and wild-type seedlings: the incline of the Sgd-1 response curve was steeper than that of gid1-8 but more moderate than that of the wild type. This strongly suggests that the Sgd-1 phenotype depends on recovery of GA signaling but not other mechanisms.
Isolation of the SGD-1 Gene
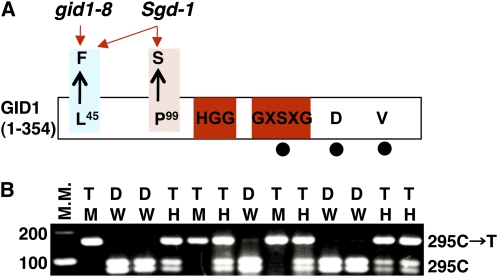
Because of the possibility that Sgd-1 might be involved in the GA signaling pathway, we attempted to isolate the SGD-1 gene. First, we crossed the suppressor genotype (gid1-8/Sgd-1) with the corresponding nonmutant genotype, T65. At the time this study was begun, we assumed that gid1-8 and Sgd-1 were at separate loci and that T65 would therefore be homozygous for wild-type alleles at two loci, G1D1 and SGD1. Based on the genetic behavior observed up to that point, we classified the mutant allele for gid1 as recessive (lowercase) and Sgd-1 as dominant (uppercase). We expected that we could classify the F2 plants into four groups based on plant height: the tallest group (GID1/Sgd-1); the second tallest, identical to T65 (GID1/SGD-1); the third tallest, identical to the original suppressor plant (gid1-8/Sgd-1); and the shortest, identical to gid1-8 (gid1-8/SGD-1). However, the plant height of the F2 plants was continuously distributed and could not be classified into groups (see Supplemental Figure 1 online). Curiously, all of the F2 plants were taller than the original gid1-8 (see Supplemental Figure 1 online). We sequenced the mutant position of gid1-8 (C133T) and confirmed that the F2 plants carrying gid1-8/gid1-8 were always taller than the original gid1-8. This led us to speculate that Sgd-1 might be an intragenic suppressor mutation of gid1-8. Thus, we sequenced the whole region of the GID1 gene in Sgd-1 and found another single nucleotide polymorphism (SNP C295T), which exchanges the 99th Pro with Ser (P99S in Figure 2A). This supports the idea that the Sgd-1 suppressor mutation for gid1-8 is caused by this SNP occurring within the GID1 locus.
Figure 2.
Sgd-1 Is Caused by an Intragenic Mutation at the GID1 Locus.
(A) Schematic structure of GID1 indicating the positions of the gid1-8 mutation (L45→F), which is present in both gid1-8 and Sgd-1, and the Sgd-1 mutation (P99→S), which is present only in the Sgd-1 suppressor mutant. Amino acid residues shared with HSL, such as HGG and GXSXG, are shown within red boxes. The residues corresponding to the catalytic triad of HSL, S, D, and V, are indicated by filled circles.
(B) Linkage analysis between the Sgd-1 phenotype and the P99S mutation in the M2 generation of Sgd-1. The P99S mutation is caused by an SNP (C295T), which introduced a cleaved amplified polymorphic sequence polymorphism after digestion with HaeIII. Plants homozygous for the wild type or mutant allele at this SNP are designated as W or M, respectively, and heterozygous plants are designated as H. Plants showing the dwarf (D) phenotype of gid1-8 are designated as D, and plants showing the taller (T) phenotype of Sgd-1 are designated as T. All plants carrying the M and H genotypes showed the Sgd-1 phenotype (T), while plants carrying the W genotype showed the gid1-8 phenotype (D), indicating complete linkage between the Sgd-1 phenotype and the P99S mutation. M.M., molecular marker.
To test this hypothesis, we performed a linkage analysis between SNP C295T and dwarfism in the M2 generation of Sgd1, which was expected to segregate into three genetically different groups, gid1-8/gid1-8, gid1-8/Sgd-1, and Sgd-1/Sgd-1. Plants not carrying the SNP for P99S (W in Figure 2B) showed dwarfism (D) typical for gid1-8, while plants carrying this SNP homozygously (M) or heterozygously (H) showed plant height similar to the original Sgd-1 suppressor plant (T) (Figure 2B). This supports the above idea and also indicates that the Sgd-1 suppressor mutation is dominant over gid1-8.
We also produced rice plants expressing Sgd-1, gid1-8, or wild-type GID1 cDNA under the control of the native GID1 promoter using a gid1 null allele, gid1-4, as the host genotype for transformation. Plants expressing the Sgd-1 cDNA were taller than the gid1-8 plants but shorter than the wild-type GID1 plants (see Supplemental Figures 2A and 2B online). Taking together all these results, we concluded that Sgd-1 is an intragenic mutation of the GID1 locus that partially suppresses the gid1-8 phenotype.
The 99th Pro Is Important for GA-Independent and GA-Dependent GID1–SLR1 Interactions
We next examined the biochemical characteristics of GID1 protein containing the P99S substitution (GID1P99S) in terms of its interaction activity with a rice DELLA protein, SLR1, using a yeast two-hybrid (Y2H) assay. Interestingly, GID1P99S showed low but detectable interaction with SLR1 even in the absence of GA4, whereas the interaction between the wild-type GID1 (GID1WT) and SLR1 was at a trace level (Figure 3A). On the other hand, the interaction between GID1P99S and SLR1 in the presence of GA was only about a quarter of that between GID1WT and SLR1. This suggests that the replacement of the 99th Pro with Ser has two effects on GID1 with respect to its interaction with SLR1: higher affinity with SLR1 compared with GID1WT under low GA conditions but lower affinity under high GA conditions. This might be the explanation for the subtle phenotype of Sgd-1.
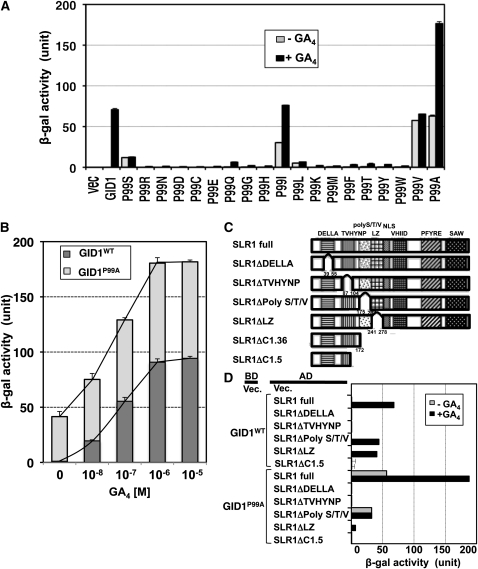
Figure 3.
Effect of Substitutions of the 99th Pro in Rice GID1 on Its GA-Dependent or GA-Independent Interactions with SLR1.
(A) SLR-interacting activity of mutated GID1s, GID1WT, and the vector control (vec) in Y2H assays. Black bars indicate activity in the presence of 10−4 M GA4; gray bars indicate activity in the absence of GA4. Data are means ± sd, n = 3. Equal expression level of mutated GID1s and GID1WT in yeast cells was confirmed by immunoblot analysis (see Supplemental Figure 6A online).
(B) Y2H assays using GID1P99A or GID1WT as bait and SLR1 as prey in the presence of various concentrations of GA4. β-Gal activity was determined by a liquid assay with yeast strain Y187 transformants (means ± sd; n = 3).
(C) The structures of various truncated SLR1 proteins.
(D) Y2H assays using GID1P99A or GID1WT as bait and the mutated SLR1s as prey, with or without 10−4 M GA4. β-Gal activity detected in a liquid assay with yeast strain Y187 transformants (means ± sd; n = 3). Black bar indicates activity in the presence of 10−4 M GA4; gray bar indicates activity in the absence of GA4. Equal expression level of mutated SLR1s in yeast cells was confirmed by immunoblot analysis (see Supplemental Figure 6E online).
We then attempted to identify other amino acids at the 99th position that would induce higher binding affinity with SLR1. For this, we exchanged the 99th Pro with all 18 remaining amino acids and examined the interaction activity with SLR1 using the Y2H assay (Figure 3A). Mutated GID1 proteins containing Ile (I), Val (V), and/or Ala (A) showed an increased interaction activity with SLR1 compared with GID1P99S, with or without GA4. In particular, mutated GID1 replaced with Ala, GID1P99A, showed the highest activity, with SLR1 binding activity in the absence of GA almost similar to that of the GID1WT–SLR1 interaction in the presence of GA. In the presence of GA, it showed almost 2.5 times higher activity than GID1WT. On the other hand, the other 15 amino acid substitutions caused not only a lack of enhancement of the GID1–SLR1 interaction in the absence of GA but also a drastic decrease in the presence of GA. The one exception was P99L, which caused a slight enhancement in the absence of GA but a dramatic decrease in the presence of GA. These results demonstrate that the 99th Pro is not only involved in the GA-independent interaction of GID1 with SLR1 but also important for its GA-dependent interaction.
Recently, Shimada et al. (2008) solved the crystal structure of GID1s from rice. Based on the analysis, the 99th Pro is located in the loop region between β2 and β3 (see Supplemental Figure 3 online). This loop region is not conserved between rice and Arabidopsis GID1s, although most other regions of the proteins are very similar (Hirano et al., 2007). To examine whether such GA-independent GID1–SLR1 interaction is due only to the 99th Pro, we performed a Y2H assay in the absence of GA, using 94 mutated GID1s in which each of the amino acids conserved among rice and the three Arabidopsis GID1 proteins (GID1a, b, and c) was replaced with Ala (see Supplemental Figure 4 online). The result indicated that P99A showed the highest GID1–SLR1 interacting activity in the absence of GA, though other mutations showed much lower activities, such as D71A, PV114, 115AA, H336A, and F347A. Again, this suggests that among the residues we tested, the 99th Pro is the most important residue for the GA-independent interaction between GID1 and SLR1.
Molecular Mechanism of GA-Independent GID1P99A–SLR1 Interaction
We further investigated the role of the 99th Pro in the GID1–SLR1 interaction. For these analyses, we used GID1P99A instead of GID1P99S because GID1P99A showed the greatest GA-independent interaction with SLR1 and also showed the highest GA-dependent interaction among the all mutagenized GID1 proteins tested (Figure 3A). First, we measured the GID1–SLR1 interacting activity at various concentrations of GA4 in a Y2H assay (Figure 3B). At 0 M GA4, the β-galactosidase (β-gal) activity was detected only in GID1P99A, but not in GID1WT, as also shown in Figure 3A. The interacting activity of GID1P99A and GID1WT with SLR1 was increased by increasing levels GA4 in both GID1P99A and GID1WT, with the responsiveness of GID1P99A being higher than that of GID1WT. This demonstrates that the P99A substitution enhances not only the GID1—SLR1 interaction in the absence of GA but also sensitivity to GA when it is present. At over 10−6 M GA4, both response curves reached a plateau; however, the maximum value for GID1P99A was almost twice that of GID1WT (~180 unit for GID1P99A versus ~90 unit for GID1WT; Figure 3B).
Next, we investigated whether the GA-independent GID1P99A–SLR1 interaction depends on a similar mechanism to that of the GA-dependent GID1WT–SLR1 interaction. We prepared various truncated SLR1 proteins (Figure 3C) and examined their interacting activity with GID1P99A with or without GA4 by Y2H (Figure 3D). GID1WT did not interact with SLR1ΔDELLA, SLR1ΔTVHYNP, or SLR1ΔC1.5 in the presence of GA4, whereas it interacted with SLR1ΔpolyS/T/V and SLR1ΔLZ as reported previously (Ueguchi-Tanaka et al., 2007a). On the other hand, GID1P99A interacted with the intact SLR1 and SLR1ΔpolyS/T/V with or without GA, whereas GID1P99A did not interact with SLR1ΔDELLA or SLR1ΔTVHYNP under either condition. The results demonstrate that both DELLA and TVHYNP domains are essential for GA-independent GID1P99A–SLR1 interaction, as in the GA-dependent GID1WT–SLR1 interaction. Furthermore, the overall trend of GA-dependent GID1P99A–SLR1 interaction was similar to that of the GA-dependent GID1WT–SLR1 interaction, suggesting that the interaction mechanisms might be similar, with the exception of a lower interaction of GID1p99A with SLR1ΔLZ compared with GID1WT. A structural change occurring via the loss of the LZ domain might affect the interaction between GID1P99A and SLR1 to a greater extent.
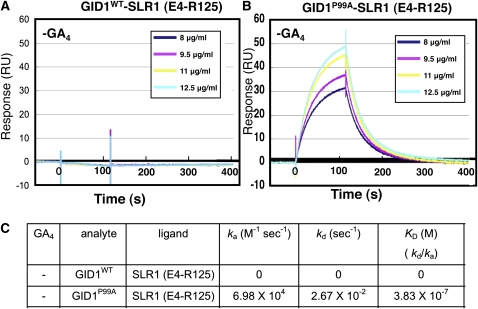
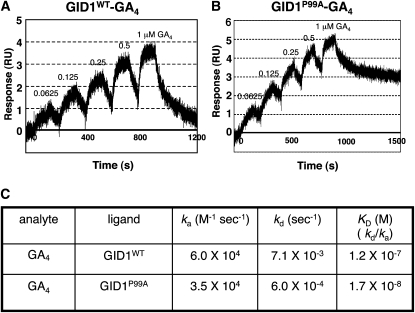
To investigate further the GA-independent GID1P99A–SLR1 interactions, we physicochemically analyzed the interaction using surface plasmon resonance (SPR). For this experiment, we used the N-terminal portion (E4-R125) of SLR1 containing DELLA and TVHYNP domains [SLR1 (E4-R125)] as a ligand. The constant amount of recombinant glutathione S-transferase (GST)-tagged SLR1 (E4-R125) was immobilized on the surface of a tip using the anti-GST antibody. Then, the solution of His-tagged GID1P99A or GID1WT at various concentrations was eluted as an analyte in the absence of GA4, and the interaction profile between SLR1 (E4-R125) and GID1s was observed (Figure 4). This in vitro assay system clearly demonstrates the interaction between GID1P99A and SLR1 (E4-R125) in the absence of GA4 (Figure 4B), whereas no interaction between GID1WT and SLR1 (E4-R125) occurred under the same conditions (Figure 4A). The Ka (association) value for the GID1P99A–SLR1 (E4-R125) interaction was estimated as 6.98 × 104 (M−1 s−1) and the Kd (dissociation) value was 2.67 × 10−2 (s−1); consequently, the KD (affinity) was calculated as 3.83 × 10−7 M (Figure 4C).
Figure 4.
Physicochemical Analysis of the Interaction between GID1P99A and SLR1 (E4-R125) without GA.
(A) GID1WT–SLR1 (E4-R125) interaction without GA4. CM5 sensor chips upon which GST-SLR1 (E4-R125) was immobilized were injected with solutions containing 8, 9.5, 11, or 12.5 μg/mL Trx·His-GID1WT. RU, resonance units.
(B) GID1P99A–SLR1 (E4-R125) interaction without GA4. CM5 sensor chips upon which GST-SLR1 (E4-R125) was immobilized were injected with solutions containing 8, 9.5, 11, or 12.5 μg/mL Trx·His-GID1P99A.
(C) Various kinetic values; ka (association rate), kd (dissociation rate), and KD (kd/ka, dissociation constant), of GID1P99A–SLR1 (E4-R125) and GID1WT–SLR1 (E4-R125) interactions in the absence of GA4.
According to a recent model of GA perception by GID1 (Murase et al., 2008; Shimada et al., 2008), when the GA molecule is trapped in its binding pocket on GID1, the GA molecule acts to close the lid region over the pocket, which induces DELLA protein interaction with the aid of the covering lid. Based on this model, we speculated that the structure of GID1P99A in the absence of GA might mimic that of GID1WT in the presence of GA, that is, a closed state of its lid region. If the structure of GID1P99A is similar to the closed state of the lid region of GID1WT, the GA molecule should enter and exit the binding pocket of GID1P99A less easily than in GID1WT. Thus, we compared the interaction kinetic values between GID1P99A–GA4 and GID1WT–GA4 by the single-cycling method of SPR (Figure 5). The Ka (association) value for GID1P99A–GA4 was estimated as 3.5 × 104, while that for GID1WT–GA4 was 6.0 × 104, indicating that binding of GA4 to GID1P99A occurs about 2 times more slowly than to GID1WT. On the other hand, the Kd (dissociation) value for GID1P99A–GA4 was 6.0 × 10−4, while that for GID1WT–GA4 was 7.1 × 10−3, indicating that dissociation of GID1P99A–GA4 occurs ~10 times more slowly than dissociation of GID1WT–GA4. The SPR results strongly support the idea that the GID1P99A lid is at least partially closed, even in the absence of GA. Furthermore, the KD value (affinity) for GID1P99A–GA4 was ~10 times smaller than that for GID1WT–GA4, indicating that GID1P99A is more sensitive for GA4 than GID1WT. Such GA hypersensitivity of GID1P99A may result in the GA-hypersensitive interaction between GID1P99A and SLR1 as shown in Y2H (Figure 3B).
Figure 5.
Physicochemical Analysis of the Interaction between GID1P99A and GA4 Using SPR.
(A) GID1WT–GA4 interaction. CM5 sensor chips upon which GST-GID1WT was immobilized were injected with solutions containing 0.0625, 0.125, 0.25, 0.5, and 1 μM GA4. RU, resonance units.
(B) GID1P99A–GA4 interaction. CM5 sensor chips upon which GST-GID1P99A was immobilized were injected with solutions containing 0.0625, 0.125, 0.25, 0.5, and 1 μM GA4.
(C) Various kinetic values; ka (association rate), kd (dissociation rate), and KD (kd/ka, dissociation constant), of GID1P99A–GA4 and GID1WT–GA4 interactions.
GA Signaling by GID1P99A in Planta
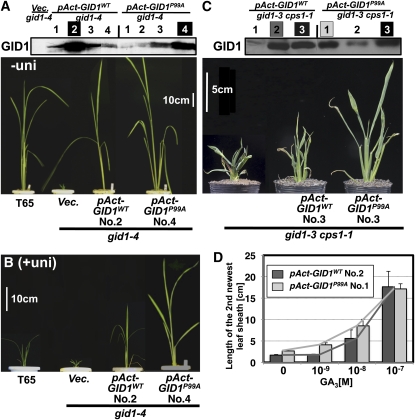
We next confirmed that GID1P99A induces GA signaling under GA-limited conditions in planta. The cDNA for GID1P99A or GID1WT was constitutively expressed under control of the rice actin promoter in plants containing gid1-4, a null allele of gid1 (Ueguchi-Tanaka et al., 2007a). To estimate precisely the effect of these GID1s, we examined the amount of transgene protein by protein immunoblot analysis using an anti-GID1 antibody (Figure 6A, top panel). Transformant No. 2 for pAct-GID1WT and transformant No. 4 for pAct-GID1P99A expressed almost same level of GID1 protein, so we compared the phenotypes of these two plants. Both plants were much taller than gid1-4 plants transformed with the control vector (Vec.) under normal conditions, whereas no clear difference in height between pAct-GID1WT and pAct-GID1P99A plants was observed (Figure 6A). This may reflect saturated GA signaling by overproduction of GID1P99A or GID1WT, as previously reported (Ueguchi-Tanaka et al., 2005). To address this possibility, we grew the plants in the presence of uniconazole, a GA synthesis inhibitor. Under such GA-limited conditions, the plants overproducing GID1P99A were much taller than plants overproducing GID1WT, whereas the vector control plant showed a dwarf phenotype caused by inhibition of GA synthesis by uniconazole (Figure 6B). To investigate further the effect of GID1P99A or GID1WT overproduction under GA-deficient conditions, we introduced the same constructs into the double mutant of cps1-1, defective in ent-copalyl diphosphate synthase (Sakamoto et al., 2004), and gid1-3 (Ueguchi-Tanaka et al., 2008). As transformant No. 3 for pAct-GID1WT and transformant No. 3 for pAct-GID1P99A expressed about same level of GID1 protein (Figure 6C, top panel), we compared these plants. The plant overproducing GID1P99A was much taller than the plant overproducing GID1WT under GA-deficient conditions in cps1-1 gid1-3 (Figure 6C, bottom panel). We also compared GA responsiveness in these plants (Figure 6D). At 0 M GA3, the GID1P99A overproducer was taller than the GID1WT overproducer. The GA-dependent elongation of the GID1P99A overproducer occurred at <10−9 M GA3, while that of the GID1WT overproducer occurred at >10−8 M GA3 with a lower responding slope compared with that of GID1P99A. At 10−7 M GA3, every leaf sheath reached to almost the same level. These results indicate that GID1P99A can more effectively activate GA signaling than GID1WT in planta under both GA-limited and GA-absent conditions.
Figure 6.
GA-Independent GA Signaling by GID1P99A in Planta.
(A) Top: Amount of GID1s produced in pAct-GID1WT (4 lines), pAct-GID1P99A (4 lines), and the vector control (Vec.) plants in gid1-4 background estimated by immunoblot analysis using an anti-GID1 antibody.
Bottom: Gross morphology of pAct-GID1WT and pAct-GID1P99A plants in gid1-4 background grown under normal conditions for 3 weeks. gid1-4 plants transformed with the vector control (Vec.) and wild-type (T65) plants are also shown. Bar = 10 cm.
(B) Same genotypes as in (A), grown with 10−6 M uniconazol for 3 weeks. Bar = 10 cm.
(C) Top: Amount of GID1s produced in pAct-GID1WT (three lines) and pAct-GID1P99A (three lines) plants in a gid1-3 cps1-1 background estimated by immunoblot analysis using an anti-GID1 antibody.
Bottom: Gross morphology of pAct-GID1WT and pAct-GID1P99A plants in a gid1-3 cps1-1 background grown under normal conditions for 3 weeks. Bar = 5 cm.
(D) Dose dependency of leaf sheath elongation of pAct-GID1WT and pAct-GID1P99A plants in a gid1-3 cps1-1 background grown in GA3 for 1 week (mean ± sd; n = 3).
GA-Independent and GA-Hypersensitive Interaction between Arabidopsis At GID1b and DELLA Proteins Depends on a Mechanism Similar to the Rice GID1P99A–SLR1 Interaction
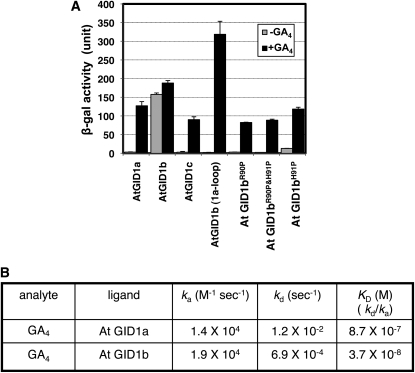
All of the above results indicate that the GID1 protein potentially has an ability to interact with DELLA proteins in the absence of GA and to interact in a GA-hypersensitive manner in the presence of GA, both of which induce GA signaling responses. This raises the interesting question of whether GID1 proteins with such unique characteristics are found in nature. There is one good candidate for this type of GID1, Arabidopsis At GID1b. Griffiths et al. (2006) and Nakajima et al. (2006) reported that At GID1b interacts with DELLA protein in the absence of GA. Interestingly, At GID1b does not contain a Pro residue at the corresponding position in the loop region containing the 99th Pro in rice GID1, whereas other At GID1s, At GID1a and At GID1c, which interact with DELLA proteins only in the presence of GA, contain Pro at that position (see Supplemental Figure 3 online). Thus, we speculated that the GA-independent At GID1b–DELLA interaction might depend on the absence of the Pro in this region. We investigated this possibility by Y2H assay. At GID1a and GID1c interacted with GAI, an Arabidopsis DELLA protein, only in the presence of GA, whereas At GID1b interacted with GAI regardless of whether GA was present, as previously described (Figure 7A; Griffiths et al., 2006; Nakajima et al., 2006). However, a modified version of At GID1b with its loop region replaced with that of At GID1a [At GID1b (1a-loop)] completely lost its GA-independent interaction with GAI, without losing the GA-dependent interaction (Figure 7A). According to the alignment of rice and Arabidopsis GID1s (see Supplemental Figure 3 online), the 99th Pro in rice GID1 corresponds to the 91st His in At GID1b. Therefore, we constructed three mutant proteins: the 91st His replaced with Pro (At GID1bH91P), the 90th Arg replaced with Pro (At GID1bR90P), and the corresponding double mutant (At GID1bR90PandH91P). Neither At GID1bR91P nor At GID1bR90PandH91P interacted with GAI in the absence of GA but both interacted with it in the presence of GA. At GID1bH91P interacted with GAI in the presence or absence of GA, showing considerably less interaction in the absence of GA. These results strongly support the above speculation that the loop region containing the 99th Pro is important for GA-independent GID1–DELLA interaction.
Figure 7.
Replacement of the At GID1b Loop with the At GID1a Loop Abolishes GA-Independent GID1–GAI Interaction.
(A) Y2H assay using mutated At GID1s as bait and GAI as prey in the presence or absence of GA4. In At GID1b (1a-loop), the entire loop region of GID1b (from S84 to T103) was replaced with the corresponding region from At GID1a. In At GID1bR90P, At GID1bR90P and H91H, and At GDI1bH91P, amino acids Arg-90 and/or His-91 were replaced with the corresponding residues from At GID1a, as indicated. β-Gal activity was determined by a liquid assay with yeast strain Y187 transformants (means ± sd; n = 3). Equal expression level of bait proteins in yeast cells was confirmed by immunoblot analysis (see Supplemental Figure 6B online).
(B) Various kinetic values; ka (association rate), kd (dissociation rate), and KD (kd/ka, dissociation constant), of At GID1a– and At GID1b–GA4 interactions by SPR.
Next, we compared the kinetic values between At GID1a–GA4 and At GID1b–GA4 by the single cycling method of SPR (Figure7B). The Ka value for At GID1a–GA4 was estimated as 1.4 × 104, whereas that for At GID1b–GA4 was 1.9 × 104, indicating that binding of GA4 to At GID1 b is almost same as that to At GID1a. On the other hand, the Kd value for GID1a–GA4 was 1.2 × 10−2, whereas that for At GID1b–GA4 was 6.9 × 10−4, indicating that dissociation of At GID1b–GA4 occurs ~17 times slower than that of At GID1a–GA4. Again, the results of SPR strongly support the idea that GA-independent and GA-hypersensitive interactions between At GID1b and DELLA proteins in Arabidopsis depend on a mechanism similar to those of the GID1P99A–SLR1 interactions in rice.
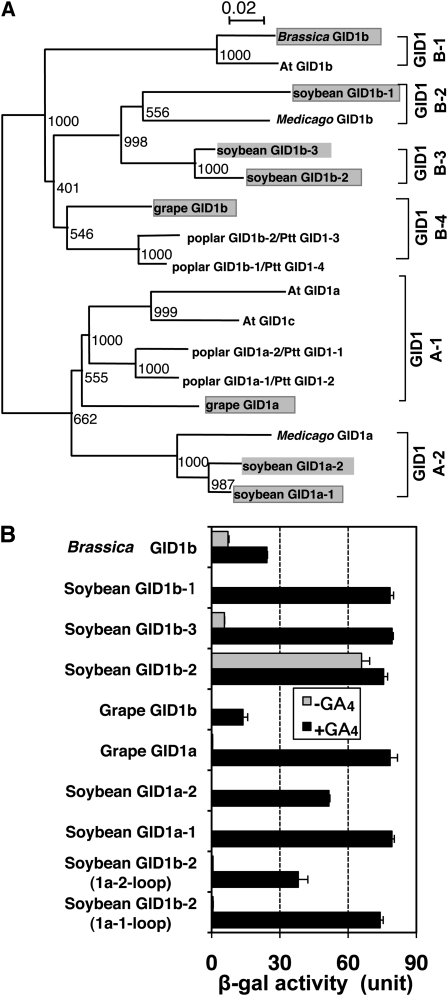
We further investigated whether the GA-independent GID1–DELLA interaction is unique to At GID1b in the natural context. According to the alignment (see Supplemental Figure 5 online) and phylogenic tree (Figure 8A) of dicot GID1s, we can divide dicot GID1s into two main groups, GID1A and GID1B, which can be further divided into several subgroups. GID1s within the GID1A-1 subgroup, including At GID1a and At GID1c, contain Pro at the corresponding position of the loop region, whereas GID1s subdivided into the GID1A-2 subgroup do not (see Supplemental Figure 5 online). Thus, we suspected that GID1s in all subgroups of GID1B (GID1B-1, 1B-2, 1B-3, and 1B-4) and in GID1A-2 might interact with DELLA proteins in the absence of GA. To test this, we selected eight GID1s, including five soybean GID1s, two grape (Vitis vinifera) GID1s, and one Brassica GID1 (indicated by shading in Figure 8A). We examined the interaction between these GID1s and GAI with or without GA using the Y2H assay. Three GID1s among the eight GID1s tested, soybean GID1b-2 and b-3 and Brassica GID1b, interacted with GAI in the absence of GA, whereas all eight GID1s interacted with GAI in the presence of GA. These results demonstrate that GA-independent GID1–DELLA protein interaction is not a unique characteristic of At GID1b; some but not all of the other GID1B proteins share this characteristic.
Figure 8.
Some Dicot GID1b-Type Proteins Show GA-Independent Interaction with Arabidopsis GAI.
(A) Phylogenetic analysis of dicot GID1s. Protein names of poplar GID1s defined by Mauriat and Moritz (2009) are shown next to each slash. A text file alignment used in this analysis is available as Supplemental Data Set 2 online.
(B) Y2H assay using various dicot GID1s and the mutated GID1s as bait and GAI as prey in the presence or absence of GA4. In soybean GID1b-2 (1a-1-loop) and soybean GID1b-2 (1a-2-loop), the entire loop region of soybean GID1b (from S82 to K102) was replaced with the corresponding region from soybean GID1a-1 and soybean GID1a-2, respectively. β-Gal activity was determined by a liquid assay of yeast strain Y187 transformants (means ± sd; n = 3). Equal expression level of bait proteins in yeast cells was confirmed by immunoblot analysis (see Supplemental Figure 6C online).
On the other hand, the GID1A-2 subgroup members, such as soybean GID1a-1 and -2, which do not contain Pro in the loop region, showed GA-dependent interaction with GAI. This indicates that Pro is not always essential for the GA-dependent interaction with DELLA protein; therefore, the GA-dependent interaction of these soybean GID1a proteins might depend on a mechanism other than that of the loop structure. Thus, we investigated whether the loop region of soybean GID1a-1 or soybean GID1a-2 can gain the GA-dependent interaction of soybean GID1b-2, which showed GA-independent interaction. Two modified versions of soybean GID1b-2 with the loop region replaced with that of soybean GID1a-1 [soybean GID1b-2 (1a-1-loop)] or GID1a-2 [soybean GID1b-2 (1a-2-loop)] completely lost their GA-independent interaction with GAI without losing the GA-dependent interaction (Figure 8B). Taken together, these results indicate that the loop structure is critical in determining the GA-dependent or -independent interaction of GID1 with DELLA proteins, but the presence or absence of Pro in the loop region is not. Pro in the loop region may be involved in the mobility of the lid (see Discussion).
DISCUSSION
In this study, we isolated an intragenic suppressor mutant for gid1, which we named Sgd-1. The mutant phenotype of Sgd-1 was inherited in a dominant manner. In addition to the SNP associating with the gid1-8 allele, the Sgd-1 allele contains a second SNP, which causes a substitution of the 99th Pro with Ser. Although the crystal structure of rice GID1 bound with GA4 and GA3 has already been analyzed, the structure of the region containing this Pro was not determined, probably because of lack of stability of the region (Shimada et al., 2008). Furthermore, comparison of amino acid sequences of GID1 proteins among various plant species revealed that this loop region is highly diverse relative to other GID1 regions (see Supplemental Figure 3 online; Hirano et al., 2007). Consequently, this region had been considered to be less significant than other regions in terms of GID1 function. In most cases, however, substitution of this Pro with other amino acids severely decreased its interaction activity with SLR1 in the presence of GA. More importantly, its substitution with several amino acids, such as Ser (Sgd-1) or Ala, conferred a new GA-independent and GA-hypersensitive SLR1-interacting activity on GID1. The particular characteristics of this Pro were also confirmed by a comprehensive Ala scanning experiment, in which 94 substitutions of conserved amino acid residues among rice and Arabidopsis GID1s did not result in this new GA-independent SLR1 interaction activity (see Supplemental Figure 4 online).
All data suggest that the molecular state of GID1P99A under GA-absent conditions mimics that of GID1WT binding the GA molecule, resulting in its GA-independent and GA-hypersensitive interaction with SLR1. For example, the GA-independent interaction of GID1P99A with some truncated versions of SLR1 is similar to that of GID1WT in the presence of GA (Figure 3D). SPR analysis also revealed lower accessibility to and slower dissociation of GA4 from GID1P99A than from GID1WT (Figure 5), suggesting that GID1P99A has a partially closed state of the lid region, similar to the GID1WT–GA complex.
Based on these observations, we propose a model for the function of this loop region (Figure 9). In GID1WT, the lid is open in the absence of GA. DELLA/TVHYNP domains of SLR1 cannot interact with GID1. When GA binds to the binding pocket, the lid closes, which allows the interaction of DELLA/TVHYNP domains of SLR1 through hydrophobic amino acid residues extruded from outside the lid region (Figure 9A; Shimada et al., 2008). In the case of GID1P99A, we propose that the lid tends to be closed even in the absence of GA, which promotes the interaction with SLR1 (Figure 9B). The closed state of the GID1P99A lid also causes slower dissociation of GA4, resulting in GA hypersensitivity and stable complex formation with SLR1. The GA-hypersensitive interaction of GID1P99A with SLR1 was confirmed by a Y2H assay (Figure 3B) as well as in planta using transgenic plants of the GA-deficient mutant cps1-1 (Figure 6D). Because the GA hypersensitivity of GID1P99A is mainly due to the lower Kd value (slower dissociation from GA), this type of receptor is expected to have a characteristic of slow response to change in GA concentration, despite its GA hypersensitivity. In other words, GID1P99A forms a stable complex with GA and SLR1 and does not easily return to its free form.
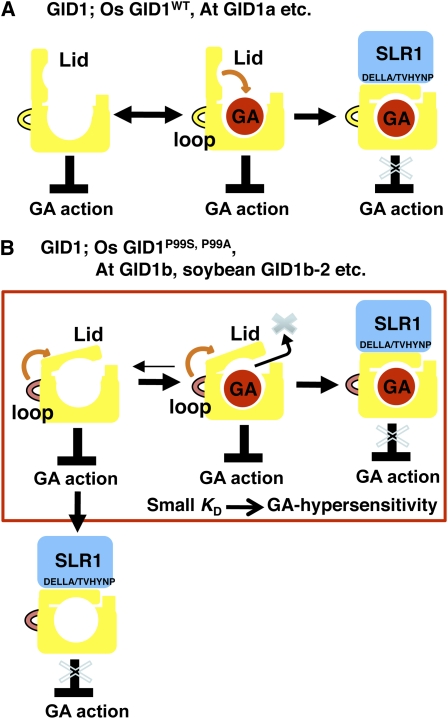
Figure 9.
Molecular Model for Formation of the GA–GID1P99A–SLR1 Complex.
(A) In typical GID1s, such as OsGID1WTand At GID1a, GID1 cannot interact with DELLA protein in the absence of GA. When GA comes into its binding pocket, the lid closes on the binding pocket, which allows the interaction between GID1 and the DELLA/TVHYNP domains of DELLA protein. This interaction between GID1 and DELLA protein leads to degradation of DELLA protein (for instance, SLR1) and allows the derepressed state of GA responses.
(B) In certain GID1s, such as Os GID1P99S and P99A, At GID1b, and soybean GID1b-2 and -3, the lid tends to be closed even in the absence of GA, which promotes the interaction with DELLA protein. The closed state of the GID1P99A lid causes lower accessibility and slower dissociation of GA4. These properties of the receptor lead to GA hypersensitivity in the formation of the GID1–GA–DELLA protein complex. The interactions between GID1 and DELLA protein both with and without GA may allow the derepressed state of GA responses.
We also demonstrated the GA-independent and GA-hypersensitive interaction of At GID1b with a DELLA protein, GAI (Figure 7A). Other GID1 proteins with this unique characteristic were also found in soybean and Brassica (Figure 8B). At GID1b and Brassica GID1b are categorized into one group with high similarity; both are Brassicaceous plants, indicating that these genes are probably derived from the same ancestral gene. However, soybean GID1b-2 and b-3, which have the same unique characteristic, are categorized into a different subgroup from At GID1b (Figure 8A). This suggests that the development and establishment of these Brassicaceous and soybean GID1bs occurred in an independent manner. Therefore, the establishment of new GID1bs may have been necessary for the evolutionary success of these plant species.
Based on the results of GID1P99A and At GID1b, we first considered that the presence of Pro at the corresponding position of P99 of rice GID1 was essential for the GA-dependent GID1–DELLA interaction. In fact, the replacement of R90 or R90/H91 of At GID1b with Pro in At GID1bR90P or At GID1bR90P and H91P resulted in the complete loss of only the GA-independent interaction with GAI, not the GA-dependent interaction, whereas At GID1bH91P maintained a slight GA-independent interaction activity (Figure 7A). However the soybean GID1a-1 and -2, lacking Pro in the loop region, showed a GA-dependent interaction with GAI (Figure 8B). This demonstrates that Pro is not always essential for the GA-dependent GID1–DELLA interaction. Even in the case of soybean GID1s, however, the loop region is essential in their GA dependency. Actually, the replacement of the loop region of soybean GID1b-2 with that of soybean GID1a-1 or -2 resulted in the complete loss of its GA-independent interaction with GAI but not in the loss of the GA-dependent interaction (Figure 8B). Taken together, these observations suggest that the loop structure is important in determining GA-dependent or -independent GID1–DELLA interaction in any GID1 protein. However, the presence of Pro in the loop region functions as an essential or sufficient residue for the GA-dependent interaction in the case of rice GID1 or At GID1s but not in the cases of soybean GID1a-1 and -2. According to the above model (Figure 9), the loop region functions like a hinge for the mobility of lid. In this context, it is possible that Pro in this loop region functions as a hinge brace itself, considering its unique structure. However, the smooth mobility of lid does not necessarily depend on Pro in the loop region; other mechanisms may be involved. Again, the fact that the loop of soybean GID1a-1 or -2 conferred GA dependency to soybean GID1b-2 supports this idea.
Why have some dicot plants acquired this new type of GID1 protein? The answer might be that dicot plants have small multigene families for GID1 and DELLA proteins, and consequently multiple interactions between GID1s and DELLA proteins occur in a tissue- or stage-dependent manner. By contrast, monocot plants have only one gene for each protein and a single pattern of GID1–DELLA interaction. The complex situation of GID1–DELLA interaction found in dicot plants may have provided leeway for the evolution of a GID1 family member with a new function. Previous expression analysis of At GID1s revealed that At GID1b is preferentially expressed in the root, whereas At GID1a and 1c are preferentially expressed in other organs (Griffiths et al., 2006). Thus, it is possible that the GID1B-type receptors function primarily in roots. As reviewed by Tanimoto (2002), there are conflicting reports of GA effects on root growth in various plants. GA at very low concentrations promotes root elongation in lettuce (Brassicaceae) and pea (Pisum sativum; Fabaceae), whereas other reports indicate that GA has no effect, or even a negative effect, on root elongation in rice, wheat (Triticum aestivum), maize (Zea mays), and tomato (Solanum lycopersicum). Interestingly, according to these results, lettuce (Lactuca sativa) and pea are grouped into the same family as Arabidopsis and soybean, having GID1B-type receptors with GA-independent and GA-hypersensitive GID1–DELLA interaction activities. Furthermore, as described above, At GID1b is preferentially expressed in root. Taken together, these results suggest that the apparently conflicting reports of GA effect on root growth might be explained by presence or absence of a GID1B-type receptor. Further studies are necessary to reveal the biological function of this type of GID1B within the roots of dicot plants.
METHODS
Screening for gid1-8 Suppressor Mutants and Growth Conditions
Sequences of primers used in this study are listed in Supplemental Data Set 1 online. Rice (Oryza sativa) plants were grown in a greenhouse at 30°C (day) and 24°C (night). gid1-8 in the background of O. sativa cv Taichung 65 was used as the original mutant strain (Ueguchi-Tanaka et al., 2007a). Twenty thousand zygotes of gid1-8 were mutagenized by N-methyl-N-nitrosourea as reported by Satoh and Omura (1979), and 12,000 M1 seeds were obtained. We screened for suppressor mutants based on rescue of the dwarf phenotype of gid1-8, and five independent mutants were isolated. To determine the presence of the sequence change in the Sgd-1 suppressor mutant, a PCR fragment was amplified with primers GID1-dCAPS/F2 and GID1-dCAPS/R2 and digested with HaeIII.
Production of Transgenic Rice Plants and Treatment with Uniconazole
pAct-GID1P99A, pAct-GID1WT, pAct vector control, pGID1-GID1WT, pGID1-Sgd-1 (L45F, P99S), pGID1-gid1-8 (L45F), and Hm12 vector control were introduced into gid1-4, which has an internal deletion between intron 1 and exon 2 of GID1 and which results in no GID1 product (Ueguchi-Tanaka et al., 2005) by Agrobacterium tumefaciens–mediated transformation (Hiei et al., 1994). gid1-4 homozygous calli were selected by PCR using GID1-4/F and GID1-4/R primers. pAct-GID1P99A and pAct-GID1WT were also introduced into the gid1-3 cps1-1 double mutant. The gid1-3 allele has a 150-bp in-frame deletion in exon2 of GID1. Genotyping of both mutations was described previously (Chhun et al., 2007). Because of the low frequency of the gid1-3 cps1-1 double mutants (Chhun et al., 2007), the double mutant calli were selected by genotyping ~2500 seeds.
For the uniconazole response test, transgenic plants were grown in the greenhouse for 3 weeks with or without 10−6 M uniconazole.
Plasmid Construction
All PCR fragments were sequenced to confirm that no mutations were induced during amplification. For the Y2H assay, pGADT7 (Clontech) and pGBKT7 (Clontech) were used as expression vectors. At GID1a, -b, and -c in pGBKT7 and GAI in pGADT7 (Nakajima et al., 2006) were kindly provided by Masatoshi Nakajima (Tokyo University). Mutagenesis and construction of Os GID1s in the pGBKT7 vector for the Ala scanning experiment and for substitution of the 99th Pro with other amino acids, the substitution of At GID1bR90 and/or H91 with Pro, and the point mutation of soybean (Glycine max) GID1 cDNAs were performed as described previously (Ueguchi-Tanaka et al., 2007a). To construct At GID1b-aloop in pGBKT7, PCR was performed against At GID1b cDNA using Eco-AtGID1b/F and aloop-GID1b/R primers to produce the 5′ region of the At GID1b-aloop and using aloop-GID1b/F and AtGID1b-Bam-Xho/R primers to produce the 3′ region of the At GID1b-aloop. Both amplified fragments were purified and mixed to produce the template for the following PCR reaction, which was performed using Eco-AtGID1b/F and AtGID1b-Bam-Xho/R primers. The PCR fragment was digested with EcoRI-BamHI and cloned into pGBKT7 at EcoRI and BamHI target sites.
GID1 cDNAs from various species for Y2H assays were produced by PCR or synthesized based on the database sequences of mRNA or predicted coding sequences. cDNAs of grape (Vitis vinifera) GID1a, grape GID1b, and Brassica napus GID1 were synthesized with EcoRI-BamHI, EcoRI-BamHI and EcoRI-SmaI sites, respectively (GenScript), and cloned into pGBKT7. cDNAs of soybean GID1a-1, a-2, b-1, b-2, and b-3 were produced as follows. Genomic DNA (gDNA) of soybean was kindly provided by Yuichi Katayose (National Institute of Agrobiological Sciences). A genomic GID1 fragment including GID1 exon 1 was amplified from gDNA of soybean using primer sets [see Supplemental Data Set 1-(8) online, colored in yellow], and GID1 exon 2 was amplified from genomic DNA of soybean using primer sets [see Supplemental Data Set 1-(8) online, colored in purple]. The amplified first and second exons of each soybean gene were joined using restriction enzyme sites and cloned into pUC18 (TAKARA) at restriction enzyme sites shown in Supplemental Data Set 1-(8) online. Inverted PCR was then performed using primer sets [see Supplemental Data Set 1-(9) online]. Each PCR product was treated with DpnI to digest the template plasmid DNA. The 5′ end of each inverted PCR fragment was phosphorylated by T4DNA kinase and self-ligated to produce cDNAs of soybean GID1a-1, -2, b-1, -2, and -3. Each cDNA was cut with appropriate restriction enzymes and cloned into pGBDT7. To construct soybean GID1b-2 (1a-2-loop) and soybean GID1b-2 (1a-1-loop) in pGBKT7, all procedures were performed as described for the construction of At GID1b-aloop using the appropriate primers described in Supplemental Data Set 1 online. The deletion series of SLR1 cDNA in the pGADT7 vector was described previously (Ueguchi-Tanaka et al., 2007a).
To construct the GST-tagged DELLA/TVHYNP fragment of SLR1 [designated GST-SLR1 (E4-R125)], a fragment for E4-R125 was produced by PCR with BamSLRE4/F and XhoSLR125R/R primers using a full-length SLR1 cDNA as the template and inserted into the BamHI-XhoI sites of the pGEX-6P-1 vector (GE Healthcare). Constructions of Trx·His-GID1WT and Trx·His-GID1P99A were described previously (Ueguchi-Tanaka et al., 2007a). To construct the GST-tagged GID1WT and GID1P99A, Trx·His-GID1WT and Trx·His-GID1P99A were digested with BamHI-EcoRI and cloned into pGEX-6P-1 vector (GE Healthcare).
To construct pAct-GID1WT and pAct-GID1P99A, the full-length wild-type GID1 gene was amplified with GID1Sma/F and GID1Sma/R primers and cloned into the SmaI site of the pBluescript vector. For GID1P99A, further mutagenesis was done using P99A/F and P99A/R primers as described previously (Ueguchi-Tanaka et al., 2007a). Each GID1WT and GID1P99A sequence was digested with SmaI and cloned into pActNos/Hm2 at the SmaI target site. To construct pGID1-GID1WT, pGID1-Sgd-1 (L45F, P99S), and pGID1-gid1-8 (L45F), PCR was performed against the 6.4-kb GID1 genomic DNA fragment, which we used previously for GID1 complementation tests (Ueguchi-Tanaka et al., 2005), to insert each corresponding mutation as described previously (Ueguchi-Tanaka et al. 2007a). Each mutated 6.4-kb GID1 genomic DNA fragment was digested with PstI, blunted by T4 DNA polymerase, and inserted into SmaI site of the Hm12 binary vector (kindly provided by Hiroyuki Hirano).
Antibody Production and Immunoblot Analysis
His-tagged GID1 protein (Shimada et al., 2008) was used for antigen to produce a rabbit polyclonal antibody. The antibody was further purified from rabbit serum using affinity column bound with GST-GID1 protein.
For immunoblot analysis of GID1 protein, crude protein extracts of rice seedlings were prepared by grinding with liquid nitrogen using a mortar and pestle in the presence of sea sand (425 to 850 mm; Wako Pure Chemical). An equal volume of 2× sample buffer was added, and samples were then boiled for 5 min. Protein samples were separated by 10% SDS-PAGE and transferred to Hybond enhanced chemiluminescence nitrocellulose membrane (GE Healthcare). For the detection of GID1 protein, the blots were treated with 5% skim milk in TBST (0.1% Tween 20 in 2 mM Tris-HCl, pH 7.6, and 13.7 mM NaCl) for 2 h and subsequently incubated with anti-Os GID1 antibody (1:2000 dilution) overnight at 4°C. Blots were washed three times with TBST for 15 min each wash. The membrane was incubated with goat anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody (Pierce; 1:10,000 dilution) for 45 min, and blots were washed following the same procedure described above. For immunoblot analyses of Myc-GID1s and HA-SLR1s, protein extraction was performed according to the Yeast Protocols Handbook (Clontech Laboratories). Myc-GID1s were detected using anti-Myc antiserum (Clontech; 1:10,000 dilution) and goat anti-mouse IgG horseradish peroxidase–conjugated secondary antibody (Pierce; 1:10,000 dilution). HA-SLR1s were detected using anti-HA antiserum (Sigma-Aldrich; 1: 10,000 dilution) and anti-mouse IgG horseradish peroxidase–conjugated secondary antibody (1:10,000 dilution).
RNA isolation and RNA Gel Blot Analysis
RNA isolation, RNA gel blot analysis, and the preparation of GID1 probe were performed as described previously (Ueguchi-Tanaka et al., 2008).
Y2H Assay
The yeast two-hybrid assay was performed as described previously (Ueguchi-Tanaka et al., 2005) using the BD Matchmaker Two-Hybrid System 3 (Clontech). The yeast strain Y187 was used as the host. GA3 dissolved in ethanol was added to the culture medium at a dilution rate of 1:1000. The yeast strain Y187 was used for detection of β-gal activity by liquid assay. Details of the methods used for the yeast assays can be found in the manufacturer’s instructions (Yeast Protocols Handbook PT3024-1; Clontech). Experiments were independently repeated at least three times.
Production of Recombinant Protein
For affinity and kinetics studies, we used GST–SLR (E4-R125), Trx·His-GID1WT, Trx·His-GID1P99A, GST-GID1WT, and GST-GID1P99A. Escherichia coli BL21 (DE3) pLysS Rosetta-gami 2 (Novagen) was used as a host strain for the production of each recombinant protein.
All procedures for producing recombinant Trx·His-GID1s were followed as described previously (Ueguchi-Tanaka et al., 2007a), with some modifications. Instead of 0.01 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 0.1 mM IPTG was used. Five hundred milliliters of Terrific Broth medium without GA4 was used for cultivation, and the cells were harvested and resuspended with buffer A containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 15 mM n-octyl-β-d-glucoside. The eluate from the affinity column was further purified by Superdex-200 gel filtration chromatography (GE Healthcare) equilibrated with buffer containing 200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 10 mM n-octyl-β-d-glucoside, and 1 mM DTT at a flow rate of 1.6 mL per min. The peak fractions corresponding to Trx·His-GID1WT and Trx·His-GID1P99A proteins were collected and used for in vitro binding experiments.
For the production of recombinant GST-SLR1 (E4-R125) protein, the cell culture and induction were performed the same as for Trx·His-GIDs except that 0.4 mM IPTG was used for induction. Cells were harvested, resuspended with buffer A containing 1 mM DTT, and disrupted by sonication (20 kHz, 5 s × 30 times). The lysate was affinity purified using 10 mL of Glutathione Sepharose 4B beads (GE Healthcare) and further purified with Superdex-200 gel filtration chromatography (GE Healthcare).
For the production of recombinant GST-GID1s, the culture, induction, and purification steps were the same as for GST-SLR1 (E4-R125) except that GA4 was added to the culture medium (0.1 mM GA4), sonication buffer (2 mM GA4), and wash buffer for glutathione affinity column chromatography (0.1 mM GA4). From the purification step using Superdex-200 gel filtration chromatography, GA4-free buffer was used.
Affinity and Kinetic Studies
The interactions between immobilized GST-SLR1 (E4-R125) protein and Trx·His-GID1 protein (either wild type or P99A) were assayed by a method based on SPR using a biosensor instrument (Biacore T100; GE Healthcare). Anti-GST antibody was initially immobilized to the CM5 sensor chip using a GST fusion capture kit (GE Healthcare). GST-SLR1 (E4-R125) protein was then immobilized to the sensor chip to level of ~2000 resonance units as ligand. Association and dissociation profiles were obtained with a continuous flow of 30 μL per min, using Trx·His-GID1WT or Trx·His-GID1P99A protein as the analyte at concentrations ranging from 8 to 12.5 μg per mL without GA4. Kinetic data were obtained using Biacore T100 evaluation software. The interactions between immobilized GST-GID1WT or GST-GID1P99A protein and GA4 were also assayed using Biacore T100. GST-GID1 was immobilized to a level over ~4000 resonance units; 0.0625, 0.125, 0.25, 0.5, and 1 μM GA4 were used as the analyte. GA4 binding was measured using single kinetic method.
Phylogenetic Analysis
Multiple sequences alignment was produced using ClustalX 2.0 (with default parameters; e.g., amino acid substitution matrix Gonnet250, slow pairwise alignments, gap opening penalty 10, gap extension penalty 0.1 for pairwise, and 0.2 for multiple alignments) (Larkin et al., 2007) and then manually adjusted to optimize alignment (available in Supplemental Data Set 2 online). A neighbor-joining tree (Saitou and Nei, 1987) was obtained with PROTDIST and NEIGHBOR in the PHYLIP version 3.69 package (http://evolution.genetics.washington.edu/phylip.html). Bootstrap analyses were performed by repeating the procedure on 1000 data sets prepared with SEQBOOT.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: Os GID1 (Q6L545), At GID1a (At3g05120), At GID1b (At3g63010), At GID1c (At5g27320), Brassica (Brassica rapa; Br) GID1 (AC155339), grape (Vitis vinifera; Vv) GID1a (AM468374), grape (Vv) GID1b (AM479851), soybean (Glycine max; Gm) GID1a-1 (Gm10g29919), soybean (Gm) GID1a-2 (Gm20g37430), soybean (Gm) GID1b-1 (Gm3g30460), soybean (Gm) GID1b-2 (Gm10g02790), soybean (Gm) GID1b-3 (Gm2g17010), poplar (Populus trichocarpa; Pt) GID1a-1/Ptt GID1-2 (POPTR_0013s02980), poplar (Pt) GID1a-2/Ptt GID1-1 (POPTR_0005s04240), poplar (Pt) GID1b-1/Ptt GID1-4 (POPTR_0002s22840), poplar (Pt) GID1b-2/Ptt GID1-3 (POPTR_0014s13170), Medicago (Medicago truncatula; Mt) GID1a (TC109095), Medicago (Mt) GID1b (TC101808), Os SLR1 (AK242577), and At GAI (At1g14920).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotype Distribution of the Segregating F2 Progeny from a Cross between Sgd-1 and the Wild Type.
Supplemental Figure 2. Transformation of gid1-4 with Sgd-1 cDNA under the Control of the Native GID1 Promoter Restored Its Height to That of Sgd-1.
Supplemental Figure 3. Sequence Alignment of Rice GID1 (Os GID1) and Its Homologs in Arabidopsis (At GID1a, b, and c).
Supplemental Figure 4. Ala Scanning Analysis of GID1 for SLR1-Interacting Activity in the Absence of GA.
Supplemental Figure 5. Sequence Alignment of Dicot GID1s.
Supplemental Figure 6. Expression Levels of the Mutated GID1 and SLR1 Proteins in Yeast Cells.
Supplemental Data Set 1. Primers Used in This Study.
Supplemental Data Set 2. Text File of the Alignment Used in the Phylogenetic Analysis Shown in Figure 8A.
Acknowledgments
We thank Rie Mitani, Hiroko Omiya, and Eriko Kouketsu (Bioscience and Biotechnology Center, Nagoya University) for expert technical assistance. We also thank Lorenzo Aleman-Sarinana (Texas Tech University), Masatoshi Nakajima (University of Tokyo), and Yuichi Katayose (National Institute of Agrobiological Sciences) for providing dicot gDNAs and cDNAs. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (M.M., M.U.-T., and H.K.), by the Target Proteins Research Program (M.M.), and by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project IP-1003; M.M.)
References
- Aya K., Ueguchi-Tanaka M., Kondo M., Hamada K., Yano K., Nishimura M., Matsuoka M. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T., Aya K., Asano K., Yamamoto E., Morinaka Y., Watanabe M., Kitano H., Ashikari M., Matsuoka M., Ueguchi-Tanaka M. (2007). Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19: 3876–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd N.P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirano K., et al. (2007). The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 19: 3058–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Matsuoka M. (2008). Molecular biology of gibberellins signaling in higher plants. Int. Rev. Cell Mol. Biol. 268: 191–221 [DOI] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Mauriat M., Moritz T. (2009). Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 58: 989–1003 [DOI] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Satoh H., Omura T. (1979). Induction of mutation by the treatment of fertilized egg cell with N-methyl-N-nitrosourea in rice. J. Fac. Agric. Kyushu Univ. 24: 165–174 [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., Kato H., Matsuoka M. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto E. (2002). Gibberellins. Plant Roots: The Hidden Half, 3rd ed, Waisel Y., Eshel A., Kafkafi U., (New York: Marcel Dekker; ), pp. 405–416 [Google Scholar]
- Thomas S.G., Rieu I., Steber C.M. (2005). Gibberellin metabolism and signaling. Vitam. Horm. 72: 289–338 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Hirano K., Hasegawa Y., Kitano H., Matsuoka M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2007a). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M. (2007b). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198 [DOI] [PubMed] [Google Scholar]