This article shows that the peculiar expression pattern of a functionally conserved floral gene (APETALA3-like) in the center of the Lacandonia schismatica flower underlies its unique inside-out arrangement (i.e., central stamens surrounded by carpels). Thus, relatively simple genetic alterations may underlie large morphological shifts fixed in extant natural populations.
Abstract
Spontaneous homeotic transformations have been described in natural populations of both plants and animals, but little is known about the molecular-genetic mechanisms underlying these processes in plants. In the ABC model of floral organ identity in Arabidopsis thaliana, the B- and C-functions are necessary for stamen morphogenesis, and C alone is required for carpel identity. We provide ABC model-based molecular-genetic evidence that explains the unique inside-out homeotic floral organ arrangement of the monocotyledonous mycoheterotroph species Lacandonia schismatica (Triuridaceae) from Mexico. Whereas a quarter million flowering plant species bear central carpels surrounded by stamens, L. schismatica stamens occur in the center of the flower and are surrounded by carpels. The simplest explanation for this is that the B-function is displaced toward the flower center. Our analyses of the spatio-temporal pattern of B- and C-function gene expression are consistent with this hypothesis. The hypothesis is further supported by conservation between the B-function genes of L. schismatica and Arabidopsis, as the former are able to rescue stamens in Arabidopsis transgenic complementation lines, and Ls-AP3 and Ls-PI are able to interact with each other and with the corresponding Arabidopsis B-function proteins in yeast. Thus, relatively simple molecular modifications may underlie important morphological shifts in natural populations of extant plant taxa.
INTRODUCTION
An ABC Model–Based Hypothesis of the Developmental Genetic Factors Underlying the Unusual Reproductive Morphology of Lacandonia schismatica
The ABC model for the specification of floral organ identity (Bowman et al., 1991; Coen and Meyerowitz, 1991; Meyerowitz et al., 1991) has played a critical role in the modern explanation of the molecular-genetic determinants of the ontogenetic development of reproductive structures in angiosperms. This combinatorial genetic model has guided diverse plant evolutionary developmental biology studies, especially during the formative years of this young field (Cronk, 2001; Cronk et al., 2002; Pruitt et al., 2003). As part of the larger discipline of evo-devo, the articulation of explanatory tools like the ABC model is considered essential for understanding the developmental mechanisms that underlie morphological innovation throughout evolutionary time (Cronk et al., 2002; Carroll et al., 2004; Gilbert, 2006; Wolpert et al., 2006).
The ABC model of flower development was based on the interpretation of floral homeotic phenotypes in Arabidopsis thaliana and Antirrhinum majus (Coen and Meyerowitz, 1991). However, the participation of homeotic transformations and other forms of heterotopy in the appearance of new organ arrangements during the evolution of angiosperms had already been considered in the botanical literature, long before the age of plant developmental genetics (e.g., Meyer, 1966; Sattler, 1984; Bowman et al., 1989; Weston, 2000). Outstanding instances of spontaneous floral homeotic phenotypes have continued to be recorded in well-characterized taxa (see, for instance, the examples presented in Bateman and DiMichele [2002] for the orchid genus Ophrys). So far, however, experimental data on the molecular mechanisms involved in the morphogenesis of plant species with naturally occurring homeotic morphologies is scarce.
In this artilce, we present molecular-genetic data collected from the Mexican endemic triurid Lacandonia schismatica (Triuridaceae), a mycoheterotrophic monocotyledonous plant with bisexual (i.e., hermaphroditic) reproductive axes, which might represent an unusual case of homeosis/heterotopy involving the male and female floral whorls. According to its original taxonomic description, L. schismatica represents the single example of a well-established angiosperm species in which the central floral whorl, composed of stamens, is surrounded by a peripheral gynoecium (Martínez and Ramos, 1989).
Observations of reproductive axes in the putative sister taxon of L. schismatica, the neotropical triurid, Triuris brevistylis, have also indicated the presence of this particular homeotic arrangement; however, such flowers are very rare and do not constitute an established floral morph in the wild (Vergara-Silva et al., 2003). To our knowledge, a peculiar teratology in the alismatid Echinodorus amazonicus (Sattler and Singh, 1978) is the only additional record of a floral homeotic/heterotopic phenotype where central stamens appear surrounded by carpels. Given that the E. amazonicus flowers described by Sattler and Singh (1978) have not been documented again, the floral organ arrangement of L. schismatica differs from the typical floral plan of thousands of known bisexual angiosperm species (72% of the ~250,000 species of extant flowering plants), all of which have central carpels.
The molecular-genetic research described in this study derives directly from our previous claim that the floral organ arrangement described by Martínez and Ramos (1989) for L. schismatica could be analyzed in the context of the ABC model of flower development (Vergara-Silva et al., 2000, 2003). The ABC model seemed to be of immediate application to the L. schismatica case, since it explicitly states that stamen identity is determined by the combined activities of two functions, labeled B and C, both of which are represented by one or more genes belonging to the MADS box multigene family (Coen and Meyerowitz, 1991). Based on this particular feature of the model, we postulated that a displacement of B-function gene expression toward the center of the floral meristem, where C-function genes are normally active, is the simplest explanation for the central position of the androecium in L. schismatica (Vergara-Silva et al., 2000).
Although our ABC model–based study of the reproductive morphology of L. schismatica could be interpreted as a test of the universality of the model in angiosperms, it depends on certain assumptions about its applicability outside Arabidopsis, Antirrhinum, and other taxa used in plant developmental genetic studies. Over the years, experimental evidence in certain nonmodel angiosperms has indicated that conserved features of floral organization are correlated with conserved ABC combinatorial gene functions (Ambrose et al., 2000; Nagasawa et al., 2003; Whipple et al., 2004, 2007). However, some divergent ABC gene expression patterns have also been documented in flowering plant species with symplesiomorphic (traits that are shared among extant taxa, as well as with their last common ancestor) phenotypic characters (within the so-called basal angiosperms), as well as in monocots or eudicots (Kramer and Irish, 1999; Kanno et al., 2003; Vandenbussche et al., 2004; Rijpkema et al., 2006; Xu et al., 2006). None of these divergent B-function gene expression patterns, however, include a case in which the B-function is restricted to stamens or is associated with a change in the relative positions of stamens and carpels in flowers.
Furthermore, B-function genes have been shown to be conserved in determining stamen identity in cases in which these genes have been functionally characterized (Ambrose et al., 2000; Nagasawa et al., 2003; Rijpkema et al., 2006). Several such studies have been performed in monocots (Ambrose et al., 2000; Nagasawa et al., 2003; Xiao et al., 2003; Xu et al., 2006). Grasses were reported to have flowers with distinctive structures in the two outer whorls (i.e., lemma and palea instead of sepals and lodicules instead of petals in the case of maize; Ambrose et al., 2000; Whipple et al., 2004). Interestingly, however, it seems that the orthologous B genes of these monocot species are also necessary for second and third whorl organ development, as in other angiosperms (Ambrose et al., 2000; Nagasawa et al., 2003; Xiao et al., 2003; Whipple et al., 2004; Xu et al., 2006). Moreover, several earlier studies of B gene expression show that APETALA3/DEFICIENS (AP3/DEF) and PISTILLATA/GLOBOSA (PI/GLO) orthologs are broadly expressed during early flower development but are restricted to second and third whorl organs later in flower development, when all organ primordial have been specified. Arabidopsis PI and its ortholog in the orchid Dendrobium crumenatum (Dc-OPI) are expressed in the fourth whorl (Goto and Meyerowitz, 1994; Xu et al., 2006), and other PI/GLO orthologs in monocots are expressed in both the fourth and the first whorls (Schwarz-Sommer et al., 1992; Chung et al., 1994; Goto and Meyerowitz, 1994; Xu et al., 2006; Ambrose et al., 2000; Lee et al., 2003). In Arabidopsis, AP3 is expressed shortly after the sepal primordia begin to form in the precursor cells of the petals and stamens, which are located in a region between the sepals and the precursor cells of the gynoecium in the center of the flower (Jack et al., 1992). However, in PI overexpression lines, the first whorl is converted into chimeric sepal/petal organs, suggesting that AP3 should be present in this whorl (Krizek and Meyerowitz, 1996; Figure 4). Furthermore, it has been reported that some AP3 orthologs in monocots are broadly expressed throughout the floral meristem (Chung et al., 1994; Moon et al., 1999; Ambrose et al., 2000; Xu et al., 2006). Regardless of differences in expression patterns, B-function is present only in the second and third whorl, where both AP3 and PI are coexpressed.
Figure 4.
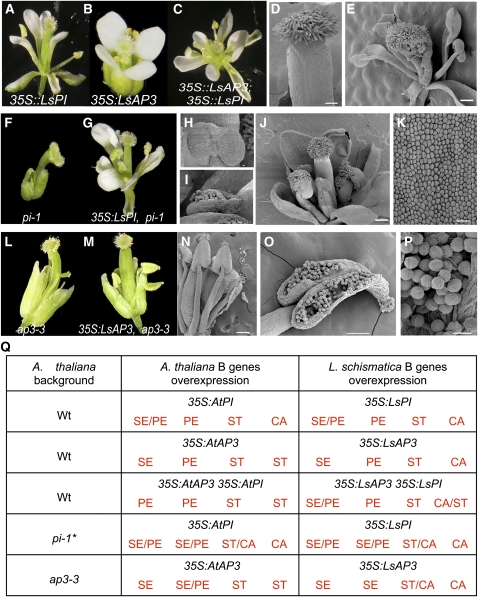
L. schismatica B Genes Are Able to Specify Petal and Stamen Cell Identity in Arabidopsis Transgenic Lines.
(A) The Ls-PI overexpression line, 35S:LsPI, in a wild-type background.
(B) The Ls-AP3 overexpression line, 35S:LsAP3, in a wild-type background.
(C) to (E) Flowers of the F2 descendants of the double overexpressor line 35S:LsAP3 35S:LsPI. The flower in (C) is similar to the single Ls-PI overexpression line in (A). Carpels do not show conversion to stamens (D). However, aberrant filamentous carpels occasionally are observed (E).
(F) pi-1 homozygous flower.
(G) 35S:LsPI in the pi-1 mutant background, showing the ability of Ls-PI overexpression to partially rescue petals and stamens of the pi-1 mutant flower.
(H) to (K) Scanning electron microscopy images of the line in (G) showing immature stamens (H) and mature stamens with viable pollen (I). Flower with a chimeric stamen/carpel organ in the third whorl (J). Second whorl petal-like cells from (G) are shown in (K).
(L) An ap3-3 homozygous flower.
(M) to (P) Examples of progeny from 35S:LsAP3 lines crossed to ap3-3 plants.
(N) Scanning electron microscopy image of the dissected perianth organs of a flower, showing at least four stamens.
(O) and (P) Scanning electron microscopy images of an opened anther.
(P) Enlargement of viable pollen. Bars = 1 mm.
(Q) Summary of floral organ phenotypes observed in overexpression lines harboring L. schismatica B genes in Arabidopsis wild-type (Wt) backgrounds and B mutant plants compared with the corresponding phenotypes in lines harboring Arabidopsis B genes (Jack et al., 1994; Krizek and Meyerowitz, 1996). Asterisk indicates the phenotype reported for the pi mutant line complemented with the Arabidopsis cDNA corresponds to data using the pi-4 allele (Piwarzyk et al., 2007). CA, carpels; PE, petals; SE, sepals; ST, stamens.
[See online article for color version of this figure.]
Thus, our assumption that the ABC model can form the basis of a molecular-genetic hypothesis for the natural occurrence of homeotic transformation in flowers remains valid. We present the results of the molecular cloning of a group of MADS box genes from L. schismatica. We found these genes to be orthologous to B- and C-function genes in various model systems, after identifying conserved domains in automated alignments and conducting Bayesian phylogenetic analyses on a matrix of angiosperm MADS box genes. Furthermore, we determined the spatio-temporal pattern of expression of these genes in several developmental stages of the L. schismatica reproductive axes by means of radioactive in situ hybridization techniques. In addition, we assessed the capacity of the L. schismatica B-function genes to complement mutations in the AP3 and PI genes in Arabidopsis using mutant transgenic lines. Finally, we tested for interactions between the proteins encoded by the cloned L. schismatica MADS box genes using yeast two- and three-hybrid systems.
Given that some of the work presented here is related to the biochemical events occurring between the transcription factors encoded by B- and C-function MADS box genes, we also considered current knowledge of the floral quartet model, which is an extension of the ABC model (Theissen and Saedler, 2001). It has been known for some time that AP3/DEF and PI/GLO carry out the B-function of the model only as obligate AP3-PI heterodimers (Jack et al., 1992, 1994; Goto and Meyerowitz, 1994; Ambrose et al., 2000; Kanno et al., 2003; Zik and Irish, 2003; Whipple et al., 2004). In eudicots, heterodimerization apparently is required to move these proteins into the nucleus (McGonigle et al., 1996) as well as for DNA binding (Schwarz-Sommer et al., 1992; Riechmann et al., 1996). Later, it was discovered that the activity of AP3-PI requires an additional protein, SEPALLATA3 (SEP3), which imparts transactivation ability to the heterodimer, stabilizes it, and facilitates its migration to the nucleus (Honma and Goto, 2001; Immink et al., 2009).
On the other hand, it has been suggested that AP3 and PI from Arabidopsis are able to form homodimers that transiently localize to the nucleus in planta (Immink et al., 2009); however, the biological significance of this homodimerization is not known. Some B-class proteins from gymnosperms and monocots can also homodimerize and bind DNA, at least in vitro (Winter et al., 2002; Kanno et al., 2003; Tzeng et al., 2004; Whipple et al., 2004; Tsai et al., 2008), while those from other monocots, such as maize (Zea mays), must heterodimerize to bind DNA (Whipple et al., 2004). Since L. schismatica is a monocotyledon, these data are relevant for the interpretation of our in vivo assays of protein interactions in a heterologous system (yeast).
Morphological Identity of Reproductive Axes of L. schismatica: True Flowers or Reduced Inflorescences?
Several interpretations for the identity of the reproductive axes of L. schismatica have been proposed (Stevens, 1991; Rudall, 2003): (1) normal flowers in which the location of stamens and carpels are secondarily inverted during early flower development, (2) compressed inflorescences or pseudanthia that resemble true flowers and have apical male flowers surrounded by female flowers at more basal positions, and (3) true homeotic flowers or euanthia in which stamens and carpels arise at inverted locations from inception. The first interpretation is refuted by observations of early development (see below).
Support for the pseudanthial hypothesis was based on morphological comparisons of mature flowers of several genera within Triuridaceae (Rudall, 2003) as well as phylogenetic analyses. Triuridaceae was placed as the sister group of Pandanaceae, a family composed of three genera (Freycinetia, Pandanus, and Sararanga), all of which have bona fide pseudanthia (Cox, 1990; Stone, 1990; Chase et al., 2000). However, additional molecular systematic analyses have placed Triuridaceae in different positions within Pandanales (e.g., Davis et al., 2004). In addition, subsequent independent studies have supported the breakdown of the Pandanaceae-Triuridaceae group, which was the starting point for the proposal of the pseudanthia interpretation (Rudall and Bateman, 2006). These new phylogenetic data led Rudall and Bateman (2006) to modify Rudall’s original pseudanthia hypothesis and to propose that carpel multiplication was involved in the origin of Triuridaceae and that this could have led to chimeric structures that exhibited a partial loss in floral determinacy, where the genetic programs underlying the development of flowers and inflorescences are mixed in a single reproductive structure (Rudall and Bateman, 2006). Even more recently, Rudall (2008) performed a comparative morphological analysis of members of the different tribes of Triuridaceae, finding a transition of centripetal to centrifugal carpel development (centrifugal carpel inception) in the tribe to which L. schismatica belongs. Assuming a euanthial understanding of triurid morphology, it was concluded that such a mode of carpel inception might indicate an ontogenetic predisposition for heterotopic change in the three central organ primordia (i.e., from carpels to stamens) of the flowers of L. schismatica (Rudall, 2008).
In parallel to the above studies, a detailed morphological analysis by Ambrose et al. (2006), in which the reproductive morphogenesis of L. schismatica was compared with that of its sister taxon, T. brevistylis, was performed. This study provided evidence in favor of the original morphological interpretation of the reproductive axes in Triuridaceae and specifically in L. schismatica. This renewed euanthial interpretation was supported by the observation that these two triurids have distinct floral meristems enclosed by a bract and that these floral buds arise from the flanks of clearly distinguishable naked inflorescence meristems. Also, the position of the protecting bract was similar to that of true flowers, namely, opposite the site where the first tepal arises. Furthermore, the temporal order of organ development recorded by Ambrose et al. (2006) was reminiscent of that commonly found in nonhomeotic flowers, where perianth organs occur in the basal-most (outermost) part of the flower, followed by stamens (in the center of the floral meristem in L. schismatica) and finally by carpels. Additionally, there was no evidence of aborted floral organs (Ambrose et al., 2006). Lastly, the fact that the organs of L. schismatica have a trimerous arrangement (i.e., three male elements in the center and six sterile organs in the periphery of the axis), as commonly happens in floral whorls of monocot species (although the latter is also true in some inflorescences), was interpreted as further evidence of the presence of true flowers in this species (Ambrose et al., 2006).
Admittedly, some of the morphological interpretations of Ambrose et al. (2006) can be contested on the grounds that not all of these characteristics are always found in true flowers and are sometimes present in inflorescences (for instance, the bracts subtending the lateral inflorescences of trimerous meristems). However, most of the data and the recently proposed alternatives to the structural interpretation of the reproductive axes in Triuridaceae support the original morphological proposal (Martínez and Ramos, 1989; Márquez-Guzmán et al., 1989), including the homeotic nature of the position of organs bearing male and female identities. Testing our ABC model–based hypothesis of the molecular-genetic basis of the reproductive phenotype in L. schismatica would thus advance our understanding of character evolution in Triuridaceae. Consequently, the experimental results presented here will be interpreted largely in euanthial terms. However, we will briefly discuss some of the implications that our results have on noneuanthial proposals of the structure of reproductive axes in Triuridaceae.
RESULTS
B- and C-Function Orthologous Genes of L. schismatica
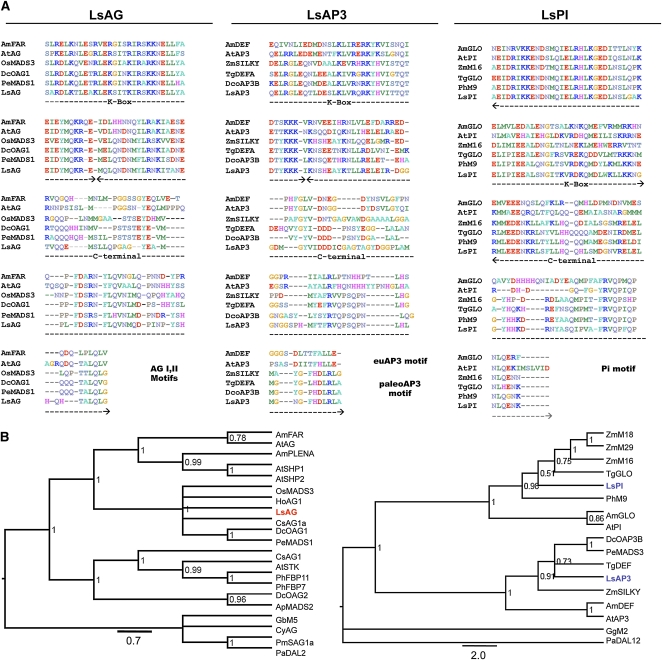
To test if the B-function of the ABC model is displaced toward the center of the reproductive meristem in L. schismatica, where male organs occur, and if the C-function genes are expressed in the male organs, we first cloned sequences putatively orthologous to the B-function (Ls-AP3 and Ls-PI) and C-function (Ls-AG) MADS box floral organ identity genes. Orthology of the cloned L. schismatica sequences was supported by visual inspection of computer-based alignments of matrices of full-length angiosperm MADS box genes and by Bayesian phylogenetic inference methods applied to such matrices. To inspect the nucleotide and amino acid matrices used in our analyses, we used previously published cladograms and phenograms of plant MADS box genes as a reference (Kramer et al., 1998, 2004). Ls-AP3 and Ls-PI display the typical MIKC domain structure of Type II MADS domain proteins (Álvarez-Buylla et al., 2000) and group within their corresponding monocot AP3 and PI lineages in the Bayesian analyses (Figure 1; see Supplemental Figure 1 and Supplemental Data Set 1 online). Furthermore, they possess the diagnostic paleoAP3 (NGFHDLRLA) and PI (IPVAFRVQPFQPNLQENK) motifs that characterize these gene families within monocotyledons (Kramer et al., 1998; Figure 1A).
Figure 1.
L. schismatica MADS Box B and C Orthologs.
(A) B and C MADS domain protein sequences of diverse angiosperms. L. schismatica orthologs possess diagnostic C-terminal motifs characteristic of each linage.
(B) Bayesian phylogenetic analysis (AG, left; B genes, right) corroborate the orthology of the L. schismatica genes (AG in red; AP3 and PI in blue) to C and B genes of Arabidopsis, respectively. Trees are supported with high posterior probabilities showed for the main branches. The L. schismatica sequences were nested within monocots. The MADS box has been excluded from this analysis to align domains with the most informative sites. Gymnosperm genes were used as outgroups. Larger alignments confirmed these results (see Supplemental Figure 1 online).
[See online article for color version of this figure.]
Ls-AG also groups within the AGAMOUS (AG) monocot clade, bearing the two diagnostic motifs (FDSRNFLQVNMQD and YSHHQHIVFQLG) characteristic of AG angiosperm orthologs (Kramer et al., 2004) in its C-terminal region and thus supporting the view that this L. schismatica gene is an ortholog of AG from Arabidopsis (Figure 1B; see Supplemental Data Set 2 online). Another C-function-like gene was cloned during our search for MADS box–containing genes from L. schismatica. An additional Bayesian analysis including a greater number of MADS box sequences further supports the orthology of the genes cloned from L. schismatica with respect to their counterparts from Arabidopsis. In the Bayesian analyses, this gene was found to be orthologous to the Arabidopsis gene SEEDSTICK (STK) and has therefore been named Ls-STK (see Supplemental Figure 1 online).
The B-Function of the ABC Model of Flower Development Is Restricted to the Central Region of the Developing Reproductive (Floral) Meristem in L. schismatica
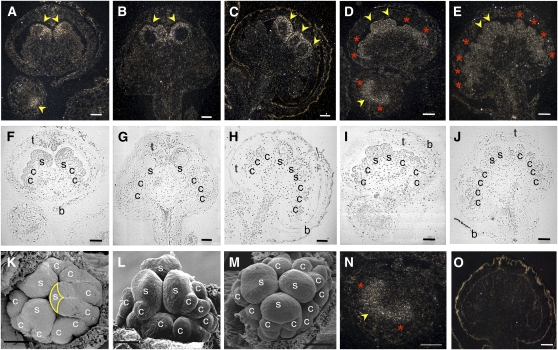
Once we tested the orthology of the cloned B and C L. schismatica genes, we determined the spatio-temporal expression patterns of Ls-AP3 and Ls-PI in different stages of floral axis development using radioactive in situ hybridization (Figure 2; see Supplemental Figure 2 online). We found that the expression of these two genes coexists only at the center of the determinate (i.e., floral) developing meristem, which is the region where stamen primordia are later formed. The restriction of Ls-AP3 to the apex of the flower meristem was observed from the early stages of flower development when stamen primordia are already apparent but no carpel primordia have differentiated (Figure 2A). Moreover, whereas Ls-AP3 continues to be restricted to the apical regions of the later-formed compound primordia, coinciding with the position where stamen primordia will, in turn, be formed (Ambrose et al., 2006; Figures 2A to 2C and 2K to 2M), Ls-PI is expressed more broadly during early flower development, but it is also expressed in such compound primordia. In contrast with Ls-AP3, Ls-PI expression was also found in the basal positions of the compound primordia, where carpels later develop to constitute the second innermost floral whorl (i.e., the gynoecium; Figures 2D, 2E, and 2N). At even later ontogenetic stages, when carpel primordia are observable, expression of Ls-AP3 remains absent in regions of the floral compound primordia that give rise to carpels, but Ls-PI expression is observed in such areas from which carpels continue to develop (Figures 2A to 2E and 2N; see Supplemental Figure 2 online). Therefore, the B-function of the ABC model of flower development, which requires the concerted expression of PI and AP3 orthologs, is restricted to the center of the flower, where stamens appear and develop in L. schismatica flowers (Figures 2A and 2D).
Figure 2.
In Situ Localization of L. schismatica B Gene (Ls-AP3 and Ls-PI) mRNA.
(A) to (E) Longitudinal sections of L. schismatica flower buds hybridized with Ls-AP3 ([A] to [C]) and Ls-PI ([D] and [E]) probes. Expression of Ls-AP3 is restricted to the apical zone (central zone in a cross section) of very early flower buds, where stamen primordia (yellow arrowhead in [A]) later arise and mature stamens will develop, while Ls-PI is expressed in a broader domain during the early stages of floral development and is then restricted to regions where both stamen (yellow arrowheads) and carpel (red asterisks) primordia develop. Ls-AP3 expression is shown in four stages of floral development in (A) to (C), and Ls-PI expression is shown for three stages in (D) and (E).
(F) to (J) Light fields of (A) to (E), respectively, for structural references.
(K) to (M) Scanning electron micrographs of three developmental stages of L. schismatica flower development for structural reference. Note that the form of the expression domain of Ls-AP3 in (A) coincides with the form of the region within the compound primordia where stamen primordia will arise (K) (yellow line).
(N) Enlarged image of Ls-PI expression in the young meristem shown in (D).
(O) In situ hybridization with an Ls-AP3 sense probe.
b, bract; c, carpel; s, stamen; t, tepal organs. Bars = 100 μm.
[See online article for color version of this figure.]
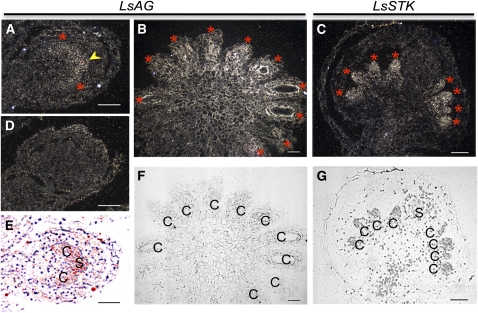
In model plant systems, stamen development requires the expression of both B and C genes. Therefore, we also performed experiments to elucidate the pattern of expression of the C-function-like genes cloned from L. schismatica, namely, Ls-AG (the putative ortholog of At-AG) and Ls-STK (the putative ortholog of At-STK; see above). In situ hybridization showed that Ls-AG was expressed in both stamen and carpel primordia, with a pattern similar to that of Ls-PI (Figures 2D,3A, and 3B), while Ls-STK was only expressed in carpels (Figure 3C).
Figure 3.
In Situ Localization of C Gene (Ls-AG and Ls-STK) mRNA in L. schismatica Flower.
(A) and (B) Longitudinal section of a young bud and of a noncentral section of an older meristem, respectively, hybridized with an antisense Ls-AG probe. Signal is observed in a pattern similar to that of Ls-PI, with a broad pattern during the early stages and in both stamen (yellow arrowhead) and carpel (red asterisks) primordia at later stages of flower development.
(C) Longitudinal section of an L. schismatica flower hybridized with the Ls-STK probe. Ls-STK mRNA is absent in stamen primordia and is strongly expressed in carpel primordia.
(D) Hybridization with a sense Ls-AG probe.
(E) Superposition of antisense Ls-AG signal (in red) shown in (A) over a bright-field view.
(F) and (G) Bright-field images of (B) and (C), respectively, for structural reference.
c, carpel; s, stamen. Bars = 100 μm.
[See online article for color version of this figure.]
Thus, both B- and C-functions are present in the center of the L. schismatica developing floral bud; however, in the second whorl, where carpels develop, only the C-function gene Ls-AG is expressed (Figures 2, 3A, and 3B; see Supplemental Figure 2 online). Except for Ls-AP3, the overall expression pattern of L. schismatica floral homeotic genes is very similar to the expression patterns that have been reported for Arabidopsis orthologs. Hence, the spatio-temporal patterns of in situ hybridization signals for the MADS box B- and C-function genes from L. schismatica are consistent with an ABC model–based explanation of its inside-out flower, provided that the B-function genes of this species are shown to be able to specify stamens.
L. schismatica B-Function Genes Are Able to Determine Stamens in Arabidopsis
L. schismatica is a mycoheterotrophic plant species, which cannot be grown in the laboratory as its survival depends on endophytic fungi (Martínez and Ramos, 1989; Vergara-Silva et al., 2003). Hence, we are unable to perform genetic experiments or induce mutations to experimentally test the roles of the B-function genes directly in this species. Therefore, as an indirect test of the ability of the coding sequences of Ls-AP3 and Ls-PI to determine petal and stamen identity, we generated ectopic expression and complementation lines for Ls-AP3 and Ls-PI in transgenic Arabidopsis plants with wild-type and mutant backgrounds, respectively, taking advantage of the known functional aspects of AP3 and PI in this dicotyledonous species.
We generated Arabidopsis plants that overexpress L. schismatica AP3 and PI orthologs (35S:LsAP3 and 35S:LsPI, respectively). Several independent lines were isolated for each genotype, and high expression 35S:LsAP3 and 35S:LsPI lines were identified by RNA gel blots and RT-PCR analyses (see Supplemental Figure 3 online). Our results show that the Arabidopsis lines bearing 35S:LsPI in a wild-type background have similar gain-of-function phenotypes to those of 35S:AtPI in Arabidopsis (Krizek and Meyerowitz, 1996; Figures 4A and 4Q). The most notable aspect of the 35S:LsPI phenotype is a partial conversion of sepals into petals in the first floral whorl. To determine if Ls-PI could rescue organ identity in the absence of endogenous PI activity, we crossed 35S:LsPI into a pi-1 mutant background. 35S:LsPI was able to rescue petal identity in the second whorl and stamen identity in the third whorl, similar to the rescue phenotype observed in pi-1 Arabidopsis plants expressing 35S:PI (Piwarzyk et al., 2007; Figures 4G to 4K and 4Q).
AP3 gain-of-function lines in Arabidopsis show a homeotic conversion of carpels to stamens (Jack et al., 1994). The 35S:LsAP3 overexpression lines did not show a conversion of carpels into stamens (Figure 4B), despite the high levels of Ls-AP3 mRNA detected in them. However, flowers of 35S:LsAP3 ap3-3 complementation lines had two outer whorls of sepals and a third whorl with stamens or staminoid organs that were able to produce functional pollen (Figures 4M to 4P and 4Q), demonstrating that Ls-AP3 is able to rescue stamen identity in Arabidopsis ap3-3 lines. However, petal identity was not rescued in these lines.
Given that MADS domain proteins corresponding to the B-function in Arabidopsis form functional heterodimers (Jack et al., 1994; Yang et al., 2003), we tested if the ectopic expression of both Ls-PI and Ls-AP3 would result in central stamens in Arabidopsis by forming a conserved heterodimer. However, we observed that the phenotype of 35S:LsPI 35S:LsAP3 double transgenic plants resembled that of plants ectopically expressing Ls-PI, where only sepals were converted into petals (Figures 4A and 4C). This indicates that the action of both Ls-PI and Ls-AP3 is not sufficient for proper stamen development (Figures 4C to 4E) in the context of Arabidopsis and that the formation of multimers might be necessary for proper Ls-AP3 function (Honma and Goto, 2001). To test for this secondary biochemical hypothesis, we performed yeast two-hybrid (Y2H) experiments with AP3 and PI orthologs of both species, as well as yeast three-hybrid (Y3H) assays that comprised all combinations of B-function proteins. In the latter class of experiments, we tested for homo- and heterodimerization with SEP3, one of the prominent components of the floral quartet model of MADS domain protein interactions (Honma and Goto, 2001; Theissen and Saedler, 2001).
L. schismatica AP3 and PI Proteins Form Heterodimers with Arabidopsis SEP3 in the Yeast Three-Hybrid System
The yeast protein hybridization results showed that L. schismatica B-function proteins were not able to heterodimerize on their own, either when both proteins from this species are used, or when used in combination with Arabidopsis proteins (Table 1). As a positive control, only truncated Arabidopsis MADS box proteins with the IKC domains were used (Yang et al., 2003).
Table 1.
Interactions between Arabidopsis AP3 and PI and L. schismatica AP3 and PI in the Y2H and Y3H Hybrid Systems
| Interactors | HIS | HIS/1 mM 3AT | HIS/3 mM 3AT | HIS/5 mM 3AT |
| Y2H (AD/BD)a | ||||
| At-AP3/At-AP3 | – | – | – | – |
| At-PI/At-PI | – | – | – | – |
| Ls-AP3/Ls-AP3 | – | – | – | – |
| Ls-PI/Ls-PI | – | – | – | – |
| Ls-AP3/At-AP3 | – | – | – | – |
| Ls-PI/At-PI | – | – | – | – |
| Ls-AP3(IKC)/At-AP3 | – | – | – | – |
| Ls-AP3(IKC)/Ls-AP3 | – | – | – | – |
| Ls-PI(IKC)/At-PI | – | – | – | – |
| Ls-PI(IKC)/Ls-PI | – | – | – | – |
| *At-AP3/At-PI | +++ | +++ | +++ | +++ |
| Ls-PI/Ls-AP3 | – | – | – | – |
| Ls-AP3(IKC)/Ls-PI | – | – | – | – |
| Ls-AP3/At-PI | – | – | – | – |
| Ls-PI/At-AP3 | – | – | – | – |
| Ls-AP3(IKC)/At-PI | – | – | – | – |
| Ls-PI(IKC)/At-AP3 | – | – | – | – |
| Ls-AP3(IKC)/Ls-PI(IKC) | – | – | – | – |
| Ls-AP3(IKC)/Ls-AP3(IKC) | – | – | – | – |
| Ls-AP3/Ls-PI(IKC) | – | – | – | – |
| Ls-PI(IKC)/Ls-PI(IKC) | – | – | – | – |
| Y3H (AD/BD/SEP3)b | ||||
| At-AP3/At-AP3 | + | +/– | – | – |
| At-PI/At-PI | + | – | – | – |
| Ls-AP3/Ls-AP3 | +++ | +++ | +++ | +++ |
| Ls-PI/Ls-PI | ++ | +/– | – | – |
| *At-AP3/Ls-AP3 | +++ | +++ | +++ | +++ |
| Ls-PI/At-PI | ++ | – | – | – |
| *At-AP3/Ls-AP3(IKC) | +++ | +++ | +++ | +++ |
| *Ls-AP3/Ls-AP3(IKC) | – | – | – | – |
| Ls-PI(IKC)/At-PI | + | +/– | – | – |
| Ls-PI(IKC)/Ls-PI | ++ | +/– | – | – |
| *PI/AP3 | +++ | +++ | +++ | +++ |
| Ls-PI/Ls-AP3 | + | + | – | – |
| Ls-AP3(IKC)/Ls-PI | ++ | – | – | – |
| Ls-AP3/At-PI | ++ | – | – | – |
| Ls-PI/At-AP3 | ++ | + | +/– | – |
| *At-PI/Ls-AP3(IKC) | +/– | – | – | – |
| Ls-PI(IKC)/At-AP3 | ++ | +/– | – | – |
Asterisk indicates the interaction took place if the genes were cloned either in the AD or BD vector.
In the Y2H assay, B-class proteins from L. schismatica and Arabidopsis were assayed for heterodimerization. Arabidopsis proteins lacked the MADS domain, and both full-length L. schismatica proteins and L. schismatica IKC domains were tested. Heterodimerization was only observed in the positive control, AP3/PI from Arabidopsis.
In the Y3H experiment, all combinations were tested between full-length AP3, PI, and SEP3 from Arabidopsis and both full-length proteins and IKC domains from L. schismatica.
When the interactions between B proteins were tested in the Y3H system using SEP3, L. schismatica B proteins were found to form heterodimers with the corresponding proteins from Arabidopsis (Ls-AP3 with At-PI and At-SEP3 and Ls-PI with At-AP3 and At-SEP3). This might explain the capacity of Ls-AP3 and Ls-PI to complement the ap3-3 and pi-1 mutant lines (Figure 4). As expected, we also confirmed the interaction of all the heterodimers when using proteins from the same species (Table 1).
DISCUSSION
L. schismatica (Lacandoniaceae = Triuridaceae; Martínez and Ramos, 1989; Maas-van de Kamer, 1995; Mabberley, 1997; Kubitzki, 1998) is a mycoheterotrophic, monocotyledonous species endemic to the Mexican Lacandon rainforest (Vergara-Silva et al., 2003), and its flowers have a cleistogamic preanthesis mode of reproduction (i.e., fertilization occurs before the flower opens; Márquez-Guzmán et al., 1993). Along with its taxonomic description (Martínez and Ramos, 1989), initial detailed studies of the reproductive morphology and anatomy of the species (Márquez-Guzmán et al., 1989) emphasized that there is an inversion in the expected position of the androecium with respect to the gynoecium. Subsequent work on the natural variation in the reproductive morphology of this rare monocot has followed the original euanthial interpretation (i.e., L. schismatica reproductive units are true bisexual flowers baring an inversion on stamen position) of its reproductive axes (Vergara-Silva et al., 2003; Ambrose et al., 2006). The aforementioned publications discuss some of the implications of alternative morphological proposals that depart from a euanthial interpretation of the reproductive axes in the entire family Triuridaceae. According to these proposals, triurid flowers should be construed instead either as pseudanthia (Rudall, 2003) or as reproductive axes where the boundary between flower and inflorescence identity is ambiguous (Rudall and Bateman, 2006; see also Rudall, 2008).
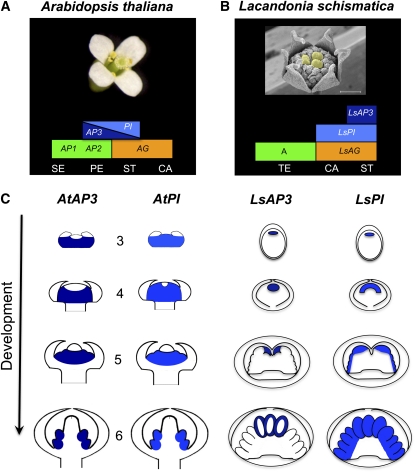
On the basis of evidence presented in Ambrose et al. (2006) and the argument set forth at the end of the Introduction in this study, we adopted a euanthial view of the reproductive axes of L. schismatica. According to this morphological perspective, our main result is describing the evolutionary shift in the spatio-temporal expression patterns of a single transcription factor (Ls-AP3) at both early and, importantly, later stages of flower development, when such patterns are compared between Arabidopsis and L. schismatica (Figures 2A to 2C and 5). In L. schismatica, we found that Ls-AP3 expression is restricted to the center (most apical zones) of the flower meristem from early stages of flower development, before stamen primordia are distinguished. As L. schismatica flower development proceeds, Ls-AP3 remains restricted to the central portion of the meristem, and at later stages it is only expressed in the area within the compound primordia, where stamens arise and develop (Figures 2A to 2C, 5B, and 5C). This pattern differs from the expression profiles of AP3 documented during the early and late developmental stages of Arabidopsis flowers (Jack et al., 1992; Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996; Figure 5A). However, as in L. schismatica, in several angiosperms (Tsai et al., 2004; Kim et al., 2005), including some model plant systems, such as Antirrhinum (Zachgo et al., 1995), the AP3 ortholog DEF is also expressed in the center during the early developmental stages. Nevertheless, expression of the PI ortholog GLO does not coincide with that of DEF in the flower center of Antirrhinum; hence, it is not associated with stamen development in the central whorl.
Figure 5.
Modified ABC Model for L. schismatica Based on Expression Data Reported Here and Compared with the ABC Model of Arabidopsis.
(A) The combinatorial ABC model of flower organ identity conserved among eudicots, with B function restricted to the petal and stamen primordia (Coen and Meyerowitz, 1991).
(B) Proposed molecular genetics hypothesis to explain the inside-out flower of L. schismatica: AP3-PI dimer function is restricted to the flower center, where stamens initiate and develop. A (green), B (AP3 dark blue and PI light blue), and C (orange) function. CA, carpels; PE, petals; SE, sepals; ST, stamens; TE, tepals. Bar = 500 μm.
(C) Changes in the in situ expression patterns of B gene mRNA along flower developmental series comparing Arabidopsis (Jack et al., 1992; Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996) and L. schismatica (this article). The differences between these two species represent two instances of the evolutionary shifts of such patterns during angiosperm evolution. Schematic representation of the developmental stages of Arabidopsis and L. schismatica flowers, starting at stage 3 of Arabidopsis flower development when sepal primordia are already visible. Sepal primordia continue to grow until they enclose the flower meristem, from stage 4 to 6. Meanwhile, at stage 5, petal and stamen primordia start to appear, and the gynoecium starts to form at stage 6. Similar stages of L. schismatica flower development were selected based on morphological traits.
[See online article for color version of this figure.]
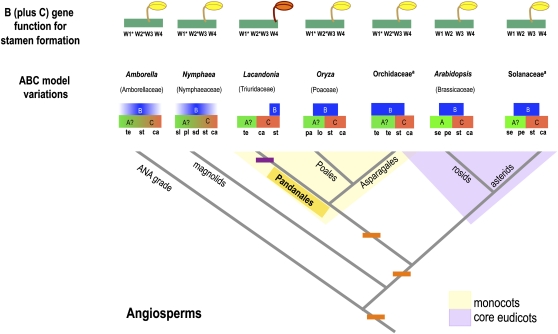
In a broader comparative framework, although the domain of expression of AP3 and PI orthologs varied over the evolution of flowering plants, it is important to note that the BC combinatory function has consistently remained restricted to the third whorl, where stamens develop in hermaphroditic flowers (Figure 6; Xiao et al., 2003; Irish and Litt, 2005; Kim et al., 2005; Soltis et al., 2007; Theissen and Melzer, 2007; Whipple et al., 2007; Mondragón-Palomino and Theissen, 2009; Álvarez-Buylla et al., 2010b). Furthermore, to the best of our knowledge, in no other case, including the one presented here, has the BC combinatory function been displaced to the flower center in bisexual flowers, resulting in central stamen development (Figure 6 and references therein).
Figure 6.
The Evolution of ABC Genes in Angiosperms.
Simplified angiosperm phylogeny (based on the Angiosperm Phylogeny Group III, 2009). Variations in the ABC model of floral organ formation for selected taxa, as well as the site where the BC combinatory function defines the development of stamens, are mapped. Representative angiosperm groups are shown in the branches of the tree with their taxonomic family in parentheses, except for Orchidaceae and Solanaceae (ª), for which the ABC model depicted stands for several species that have been experimentally analyzed (for Orchidaceae, Phalaenopsis spp [Tsai et al., 2004] and D. crumenatum [Xu et al., 2006]; for Solanaceae, Petunia hybrida [Vandenbussche et al., 2004] and tomato [Solanum lycopersicum] de Martino et al., 2006). The A function for all lineages is included for all taxa to enable comparison with Arabidopsis, although a question mark was added to emphasize its dubious role in other flowering plants, where it has not been shown to be necessary for perianth formation; rather, it is fundamental for floral meristem determination (Soltis et al., 2007). Along the phylogeny branches, the moments when the variants of the ABC genes could have evolved are indicated with orange rectangles (synapomorphies; characters shared by several lineages derived from a common ancestor). Purple rectangles indicate the appearance of traits that favored homeosis in the Triuridaceae lineage (an autapomorphy; a derived character unique to a single lineage/taxon). This autapomorphy pertains to L. schismatica AP3 expression and stamen development, which are constrained to the center of the flower during early and late developmental stages, in contrast with the early expression of AP3 orthologs in other hermaphroditic flowering plants, which can encompass several whorls, although the combined B (joint expression of AP3 and PI orthologs) and C functions are restricted to the third whorl, where stamens usually develop. ca, carpels; lo, lodicules; pe, petals; pl, petal-like tepals; ps, palea/lemma; sd, staminodes; se, sepals; sl, sepal-like tepals; st, stamens; te, petaloid tepals; sp, sepaloid petals; W1-W4, whorls within the flower primordia numbered from the outermost position to the center; asterisk, while in numerous angiosperms, W1 would stand for the sepal whorl, W2 for the petals, W3 for stamens, and W4 for the carpels, several angiosperm lineages differ from this organization either because they have a spiral arrangement (Amborella and Nimphaea), by having very modified perianth organs (Oryza), or by lacking a defined sepal/petal identity (tepals; Amborella, Lacandonia, and orchids). ABC model modified from Figure 25 of Álvarez-Buylla et al. (2010b) (data summarized from Xiao et al., 2003; Irish and Litt, 2005; Kim et al., 2005; Soltis et al., 2007; Theissen and Melzer, 2007; Whipple et al., 2007; Mondragón-Palomino and Theissen, 2009).
[See online article for color version of this figure.]
The Ls-AP3 expression pattern during flower development revealed in this study is, in turn, consistent with the euanthial molecular-genetic hypothesis previously proposed (Vergara-Silva et al., 2000) for the homeotic arrangement of male and female floral whorls in this Mexican triurid species. The additional observation that both L. schismatica B-function gene orthologs Ls-AP3 and Ls-PI are capable of rescuing stamen identity in Arabidopsis plants with AP3 or PI mutations, respectively, is also consistent with the euanthial molecular-genetic hypothesis. Furthermore, the fact that the observed expression pattern of Ls-AG is also detected in stamens, as in other flowering plants, strengthens the coherence between the molecular-genetic data and the ABC model–based molecular-genetic explanation of the homeotic floral phenotype in L. schismatica.
The data presented here could be interpreted as being discordant with the pseudanthial hypothesis to explain the reproductive structures of L. schismatica (Rudall, 2003). In our in situ hybridization analysis, we did not detect a partitioned expression pattern in the floral primordium for any of the MADS box genes assayed (Figures 2 and 3). Such a partitioned pattern of expression for ABC-like MADS box genes could correspond to minute aborted organs within multiple flowers of a pseudanthium. Such a pattern would be expected under the pseudanthial hypothesis and is supported by existing information on the early morphogenesis of reproductive structures in selected genera of Pandanaceae, where true pseudanthia occur (e.g., Freycinetia; Stone, 1990).
The in situ hybridization data presented here are less relevant to the most recent noneuanthial hypotheses proposed by Rudall and Bateman (2006) and Rudall (2008). Therefore, we suggest that additional in situ hybridization experiments should be conducted for L. schismatica homologs of other transcription factor–encoding genes that participate in flower and inflorescence development in the model systems (e.g., LEAFY [LFY]). However, in the context of the evidence presented here, the genetic alterations underlying central B-function gene expression in L. schismatica could be preliminarily understood in a phylogenetic context. Morphological analyses of the sister taxon of L. schismatica, T. brevistylis, a dioecious member of the family Triuridaceae also distributed in the Lacandon rainforest (Vergara-Silva et al., 2003), are worth mentioning. Female flowers of T. brevistylis have a strikingly similar developmental pattern to those of L. schismatica (Ambrose et al., 2006). Moreover, careful screening of the natural variation among female flowers of T. brevistylis identified the presence of central staminoids with viable pollen in a few individuals of this species (Vergara-Silva et al., 2003). Given these findings, if the common ancestor of the Mexican Triuridaceae had flowers similar to current T. brevistylis female individuals, the de novo activation of B-class genes (AP3 in particular) in the central whorl would have required a single mutational step. Alternatively, a less parsimonious scenario would involve a hermaphroditic ancestor in which the stamen identity genes underwent deactivation in an outer whorl, along with a secondary reactivation in the center of the flower meristem.
Further exploration of the implications of our data under euanthial assumptions indicates that a deeper understanding of the molecular mechanisms involved in the origin of heterotopic B-function gene expression in L. schismatica will require careful comparisons of cis-regulatory sequences of both B genes of L. schismatica and T. brevistylis, as well as analysis of the factors that regulate AP3 genes in trans in several triurids with contrasting floral arrangements. For example, transactivators of AP3 in Arabidopsis include the F-box protein UNUSUAL FLORAL ORGANS (UFO), which is an interactor of LFY. The LFY-UFO complex binds directly to the AP3 promoter and thereby induces AP3 expression specifically in floral whorls 2 and 3 (Lee et al., 1997). Moreover, it has been shown that UFO orthologs present in other angiosperms have divergent expression patterns (Souer et al., 2008), highlighting the importance of determining the expression patterns of UFO and LFY orthologs in L. schismatica.
Another MADS box gene that should be further analyzed in L. schismatica and related triurids with contrasting flower arrangements is SEP3. Our results in the Y3H analysis suggest that SEP3 is important for the formation and functioning of the AP3-PI dimer. Also, recent findings have shown that SEP3 functions not only as an overall regulator of floral meristem patterning that, besides activating B and C genes in wild-type plants (Liu et al., 2009), can induce ectopic B and C gene expression when ectopically expressed (Castillejo et al., 2005; Liu et al., 2009) but also as an element that forms ternary complexes with AP3 and PI in planta and colocalizes these proteins to the cell nucleus (Immink et al., 2009). SEP3 is also an important regulator of the transition from inflorescence to flower meristem (Liu et al., 2009). Therefore, analyses of the spatio-temporal expression patterns of putative L. schismatica SEP3 homologs, as well as of SEP3 protein complexes and regulatory interactions, should be helpful in understanding the unique expression pattern of Ls-AP3 during flower development.
As in other monocotyledons, the function of PI orthologs in L. schismatica seems to be more conserved than that of AP3-like genes (Xu et al., 2006). The 35S:LsAP3 construct was insufficient to complement a loss of petal identity in ap3-3 plants (Figure 4M) (Xu et al., 2006; Su et al., 2008). However, the relatively well-conserved MIK domains of Ls-AP3 correlate with the capacity of this protein to specify stamen identity in Arabidopsis. The so-called paleoAP3 domain of LsAP3 is also conserved (Figure 1A), although the functional role of this protein region is contentious (Lamb and Irish, 2003; Piwarzyk et al., 2007; Su et al., 2008). Conservation of the biochemical role of B-function genes/proteins and homology of petals among angiosperms is supported by functional experiments both in eudicots (Sommer et al., 1990) and in other monocots (Whipple et al., 2004, 2007). Since petal identity requires A- plus B-function in model species, the lack of B-function gene expression in the perianth organs of L. schismatica and the inability of Ls-AP3 to specify petals in Arabidopsis suggest that the perianth organs of the Mexican triurid are developmentally closer to sepals than to petals, which is in agreement with their appearance (Figure 5B). Such putative organ-type loss could be related to the presence of cleistogamy in L. schismatica, which might be associated with an absence of Darwinian selection for organs whose putative function is related to pollinator attraction.
In contrast with previous reports on Y2H AP3-PI interaction experiments for other monocots (Xu et al., 2006; Su et al., 2008), the fact that the proteins of L. schismatica form heterodimers only in the presence of At-SEP3 confirms the importance of other MADS domain proteins (AG and SEP proteins) in stabilizing the PI-AP3 interaction and specifying stamen formation (Honma and Goto, 2001). Moreover, it is also important to explain why the Ls-AP3 complementation of the Arabidopsis ap3-3 mutant lines was poorer than that of Ls-PI in Arabidopsis pi-1 mutants. Coincidently, the interaction of Ls-PI with At-AP3 or At-SEP3 was stronger than that of Ls-AP3 with At-PI/At-SEP3 (Table 1). Moreover, the Ls-AP3 protein is able to form dimers with At-AP3 as well as homodimers with itself, as found in other monocots, such as Phalaenopsis spp (Tsai et al., 2008). On the other hand, the Ls-PI/At-PI dimers and Ls-PI/Ls-PI homodimers are weaker than those of AP3 (all of these dimers were found in the presence of SEP3). These data suggest that the formation of an Ls-AP3/At-AP3/At-SEP3 complex in planta could compete with the functional Ls-AP3/At-PI/At-SEP3 heterodimeric complex, and this may explain why the Arabidopsis ap3-3 lines were not complemented as well as those of pi-1. Interestingly, the Ls-PI/Ls-AP3 dimer is weaker than the dimers formed between Ls-AP3 and either Ls-AP3 or At-AP3, and this could explain why the double overexpression line (35S:LsAP3 35S:LsPI) in a wild-type background does not present stamens in the fourth whorl as in Arabidopsis (Figures 4C and 4Q).
Finally, we propose that future studies on the expression and function of ABC model-related genes/proteins in the Mexican triurids should address the identity of the cis- or trans-acting factors underlying the expression of Ls-AP3 in the center of the flower in L. schismatica (Figure 2). If a cis-alteration is involved, and it turns out to be sufficient to explain the expression pattern of this gene, the homeotic floral organ arrangement of this species might be considered a strong candidate of morphological (evolutionary) saltation, in which a phenotypic autapomorphy (i.e., a new character state only present in a particular lineage) became fixed among natural populations through rather simple changes at the molecular level. However, additional trans-acting factors may have been involved in establishing the unique floral phenotype of this species. We suggest that research related to these questions should also be pursued in other species within families of the order Pandanales and that the use of formal modeling tools (Álvarez-Buylla et al., 2008, 2010a) might be helpful to complement such experimental investigations.
METHODS
Collections
Samples of Lacandonia schismatica were collected and processed for in situ localizations, scanning electron micrography, and DNA or RNA extraction, according to protocols previously described (Vergara-Silva et al., 2003; Ambrose et al., 2006) during Summer and Winter seasons of 1995 to 2005.
Cloning of Ls-AP3, Ls-PI, and Ls-AG
RNA was isolated from fresh or nitrogen frozen inflorescences using a standard Trizol (Invitrogen) extraction method. cDNA synthesis was followed by 3′ and 5′ rapid amplification of cDNA ends experiments, and final amplifications were based on 5′ and 3′ untranslated region sequences. Primer sequences are presented in Supplemental Table 1 online. Fragments were cloned into pGEM-T Easy vectors (Promega).
Alignment and Phylogenetic Analyses
Amino acid matrices were initially aligned using ClustalX and later aligned manually using Se-Al sequence alignment editor (http://evolve.zoo.ox.ac.uk). Bayesian phylogenetic analysis for B and C MADS box lineages was performed using unambiguously conserved domains in Mr. Bayes 3.2 (Huelsenbeck and Ronquist, 2001). In both cases, we used the mixed model option (aamodelpr=mixed) for a better estimation of the amino acid fixed rate model. Each analysis was run until the average deviation of split sequences went below 0.01 (from 1,000,000 to 1,500,000 generations). The first 10% of the total number of saved trees (1000 to 1500) was discarded to allow for the burn-in phase. Posterior probability values are indicated in each branch leading to a particular lineage. Trees were rooted using previously corroborated gymnosperm B and C genes as outgroups (Sundström et al., 1999).
In Situ Hybridization
In situ RNA hybridization was performed as previously described (Jackson, 1991). Sections (7 μm) were hybridized to 35S-labeled RNA probes obtained using the SP6/T7 transcription kit (Roche). To obtain probes, we used PCR and primers with T7 extensions to amplify fragments of ~150 bp. Primer sequences are presented in Supplemental Table 2 online. The slides were coated with NBT2 emulsion (Kodak) and exposed for 2 months. After development, the sections were stained in 20% Gills hematoxylin (British Drug House). The sections were photographed using a Leica microscope coupled to a Leica DFC 490 camera, and the contrast of the photomicrographs was adjusted using Adobe Photoshop 8.0 software.
Complementation Lines
Both Ls-PI and Ls-AP3 full-length cDNAs were amplified with primers (see Supplemental Table 3 online) that conferred XhoI and BamHI restriction sites to the 5′ and 3′ ends, respectively, to allow directional cloning. PCR products were cut with the same enzymes and inserted into pART7, a shuttle vector carrying a single copy of the cauliflower mosaic virus 35S promoter and unique NotI sites flanking the promoter and OCS terminator. These constructs were cut with NotI and subcloned into a NotI-digested cut pART27, a binary vector that confers kanamycin resistance to plants (Gleave, 1992). Wild-type Arabidopsis thaliana plants (Columbia ecotype) were transformed with the 35S:LsAP3 or 35S:LsPI constructs using the standard floral dipping procedure and selected on kanamycin plates (Clough and Bent, 1998). RNA gel blot hybridization was performed using 5 μg of total RNA from leaves and gene-specific 3′ probes (see Supplemental Table 4 online) to verify mRNA expression of B genes (see Supplemental Figure 3 online). At least four independent lines showed consistent phenotypes for 35S:LsPI, while nine independent lines were obtained for 35S:LsAP3. 35S:LsPI (ABA30) and 35S:LsAP3 (ABA13.2) F2 lines were crossed with pi-1/pi-1 and ap3-3/ap3-3 mutants, respectively. The F2 of such crosses [ABA 64-4(X) for 35S:LsPI;pi-1/pi-1 and ABA101-1(X), ABA101-3(X), and ABA102-3(X) for 35S:LsAP3;ap3-3/ap3-3] segregated individuals with wild-type, mutant, ectopic expression, and complementation phenotypes according to Mendelian ratios (see ratios in Supplemental Table 5 online). Double transgenic crosses 35S:LsAP3 × 35S:LsPI were performed, and the F2 plants [71-2(X)] were analyzed. RT-PCR was performed using specific primers (see Supplemental Table 4 online) and cDNA from leaf tissue (see Supplemental Figure 3 online). The phenotype of plants was analyzed using light microscopy and scanning electron microscopy in a Jeol microscope model JSM-5310LV, as previously described (Ambrose et al., 2006).
Y2H and Y3H Assays
All of the genes that were used in Y2H assays were cloned both into the AD vector (pDEST22) and into the BD vector (pDEST32). We transformed yeast by the Li-acetate method, as described initially by Ito et al. (1983) and modified by Lauermann (1991). All the genes in the pDEST22 vector were transformed into yeast strain Y187 (MATα; Harper et al., 1993), and all of the inserts in pDEST32 into strain PJ69-4 (MATa; James et al., 1996) and selected on SD plates lacking Trp and Leu, respectively. Cells were routinely grown on YPD propagation medium (1% yeast extract [Difco], 2% bacto-peptone [Difco], and 2% glucose) or SD selection medium (2% glucose, 0.7% yeast nitrogen base without amino acids [Difco], and 0.05 M MES [Sigma-Aldrich], pH 6) supplemented with different compounds: adenine (Ade; 30 mg/mL), uracil (Ura; 30 mg/mL), His (30 mg/mL), Trp (100 mg/mL), and Leu (100 mg/mL) at 30°C. Solid medium contained 2% agar (Difco).
Each transformant was grown on SD medium. Transformants were mated with each other by sowing each colony over the other on SD medium containing all the requirements and incubated at 30°C. The resulting diploid yeast colony was transferred to SD plates lacking both Leu and Trp and, after 3 d of growth, yeast cells were transferred to different selection plates containing SD medium lacking His, Leu, and Trp (HIS) and SD medium lacking His, Leu, and Trp, supplemented with 1 (HIS/1 mM 3AT), 3 (HIS/3 mM 3AT), and 5 mM (HIS/5 mM 3AT) 3-amino-1,2,4-triazole (3AT). These plates were incubated at 28°C and scored for growth over a 10-d period to detect protein–protein interactions.
For the Y3H experiment, all of the AD clones were crossed with the TFT vector and selected on SD medium lacking Trp and Ade and then crossed with each of the yeast lines containing the BD constructs. After this point, the same SD selection medium supplemented with various compounds was used as for Y2H, but without Ade. All of the combinations generated in the Y2H experiment were assayed, and almost all of the diploids were assayed as both AD and BD in the Y3H assay, except for LsPI(IKC)/AtSEP3/LsPI(IKC), LsAP3FL/AtSEP3/LsPI(IKC), LsPI(IKC)/AtSEP3/LsAP3(IKC), and LsAP3(IKC)/AtSEP3/LsAP3(IKC), where the first construct was in the AD vector and the third construct was in the BD vector. The results of the Y2H and Y3H assays (Table 1) were reported with the first gene in the AD vector and the second gene in the BD vector.
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information/GenBank/nucleotide data libraries under accession numbers GQ214161 (Ls-AP3), GQ214162 (Ls-PI), GQ214163 (Ls-AG), and GQ214164 (Ls-STK).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Bayesian Analysis of Members of the AG Lineage.
Supplemental Figure 2. Ls-PI in Situ Localization.
Supplemental Figure 3. Overexpression of Ls-PI and Ls-AP3 in F2 Segregating Lines in Their Respective Mutant Backgrounds.
Supplemental Table 1. Primers Sequences Used for 5′and 3′ RACE.
Supplemental Table 2. Primer Sequences for 3′ Specific in Situ Hybridization Probes.
Supplemental Table 3. Primers Sequences Used for Amplified Ls-AP3 and Ls-PI Full-Length cDNAs.
Supplemental Table 4. Specific Primers for Ls-AP3 and Ls-PI RT-PCR and RNA Gel Blot Probes.
Supplemental Table 5. Phenotypic Frequencies of an F2 Segregating Population of a Cross between the Ectopic Expression Line of L. schismatica B Genes and an Endogenous B Gene Mutant in Arabidopsis.
Supplemental Data Set 1. Gene Alignments of L. schismatica PI and AP3.
Supplemental Data Set 2. Gene Alignments of L. schismatica AG and STK.
Acknowledgments
This article is dedicated to the indigenous and peasant communities settled in the Mexican Lacandon rainforest, on whose well-being the conservation of this unique and diverse ecosystem depends. We hope this article calls attention to the importance of supporting these poor communities and the conservation of the Lacandon rainforest. The Frontera Corozal Cho'l community, as well as the Lacandon people, are acknowledged for authorizations and help during L. schismatica collection trips over the last 13 years. Special thanks to Ambrosio and his family and to Don Pascual for all their help and guidance while searching for L. schismatica populations. We thank John L. Bowman for discussions and help in the lab and field work and Stefan de Folter for advice and materials for the yeast assays. Rigoberto Vicencio Pérez-Ruiz helped with various laboratory tasks and Diana Romo with logistical support. Judith Márquez and Sonia Vázquez Santana were helpful during the early stages of this project. Financial support was from a Human Frontiers Science Program grant to E.M. and E.R.A.-B., from “Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica,” Universidad Nacional Autónoma de México (UNAM; IN230002, IN223607, IN229009-3, and IX207104 to E.R.A.-B., IN210408 to B.G.-P., and IN226510-3 to A.G.-A.), University of California-MEXUS ECO IE 271, Consejo Nacional de Ciencia y Tecnología (CONACYT) CO1.41848/A-1, 81542, and CO1.0435.B-1, and Redes temáticas de CONACYT de investigación en la red: Complejidad, Ciencia y Sociedad Grants to E.R.A.-B., 81433 to B.G.-P., and 90565 to A.G.-A. The CONACYT granted PhD scholarships to F.V.-S., A.P.-N., and E.F.-S. and an undergraduate scholarship to E.T.-B. A.P.-N.’s PhD research is done at the Posgrado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (Instituto de Ecología). B.A.A. was supported by National Science Foundation and UNAM fellowships.
References
- Álvarez-Buylla E.R., Azpeitia E., Barrio R., Benítez M., Padilla-Longoria P. (2010a). From ABC genes to regulatory networks, epigenetic landscapes and flower morphogenesis: making biological sense of theoretical approaches. Semin. Cell Dev. Biol. 21: 108–117 [DOI] [PubMed] [Google Scholar]
- Álvarez-Buylla E.R., Benítez M., Corvera-Poiré A., Chaos-Cador A., de Folter S., Gamboa de Buen A., Garay-Arroyo A., García-Ponce B., Jaimes-Miranda F., Pérez-Ruiz R.V., Piñeyro-Nelson A., Sánchez-Corrales Y.E. (March 23, 2010b). Flower development. In The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0127, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Buylla E.R., Chaos A., Aldana M., Benítez M., Cortés-Poza Y., Espinosa-Soto C., Hartasánchez D.A., Lotto R.B., Malkin D., Escalera Santos G.J., Padilla-Longoria P. (2008). Floral morphogenesis: Stochastic explorations of a gene network epigenetic landscape. PLoS ONE 3: e3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martínez-Castilla L., Yanofsky M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose B.A., Espinosa-Matías S., Vázquez-Santana S., Vergara-Silva F., Martínez E., Márquez-Guzmán J., Álvarez-Buylla E.R. (2006). Comparative developmental series of the Mexican triurids support a euanthial interpretation for the unusual reproductive axes of Lacandonia schismatica (Triuridaceae). Am. J. Bot. 93: 15–35 [Google Scholar]
- Ambrose B.A., Lerner D.R., Ciceri P., Padilla C.M., Yanofsky M.F., Schmidt R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579 [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group III (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161: 105–121 [Google Scholar]
- Bateman R.M., DiMichele W.A. (2002). Generating and filtering major phenotypic novelties: neoGoldschmidtian saltation revisited. In Developmental Genetics and Plant Evolution, Cronk Q.C.B., Bateman R.M., Hawkins J.A., (London: Taylor & Francis; ), pp. 109–159 [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Carroll S.B., Grenier J.K., Weatherbee S.D. (2004). From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design, 2nd ed (Malden, MA: Blackwell Scientific; ). [Google Scholar]
- Castillejo C., Romera-Branchat M., Pelaz S. (2005). A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 43: 586–596 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Chase M.W., Soltis D.E., Soltis P.S., Rudall P.J., Fay M.F., Hahn W.H., Sullivan S., Joseph J., Givnish T., Sytsma K.J., Pires J.C. (2000). Higher-level systematics of the monocotyledons: An assessment of current knowledge and a new classification. In Systematics and Evolution of Monocots: Proceedings of the 2nd International Monocot Symposium, Wilson K.L., Morrison D.A., (Melbourne, Australia: MCSIRO; ), pp. 3–16 [Google Scholar]
- Chung Y.Y., Kim S.R., Finkel D., Yanofsky M.F., An G. (1994). Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol. Biol. 26: 657–665 [DOI] [PubMed] [Google Scholar]
- Cox P.A. (1990). Pollination and the evolution of breeding systems in Pandanaceae. Ann. Mo. Bot. Gard. 77: 816–840 [Google Scholar]
- Cronk Q.C.B. (2001). Plant evolution and development in a post-genomic context. Nat. Rev. Genet. 2: 607–619 [DOI] [PubMed] [Google Scholar]
- Cronk Q.C.B., Bateman R.M., Hawkins J.A. (2002). Developmental Genetics and Plant Evolution. (London: Taylor and Francis; ). [Google Scholar]
- Davis J.I., et al. (2004). A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values. Syst. Bot. 29: 467–510 [Google Scholar]
- de Martino G., Pan I., Emmanuel E., Levy A., Irish V. (2006). Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.F. (2006). The generation of novelty: The province of developmental biology. Biol. Theory 1: 209–212 [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Immink R.G.H., Tonaco I.A.N., de Folter S., Shchennikova A., van Dijk A.D.J., Busscher-Lange J., Borst J.W., Angenent G.C. (2009). SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V.F., Litt A. (2005). Flower development and evolution: Gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15: 454–460 [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T., Brockman L.L., Meyerowitz E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G.L., Meyerowitz E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Jackson D. (1991). In situ hybridization in plants. In Molecular Plant Pathology, A Practical Approach, Vol. 1. Gurr S., McPherson M., Bowles D., (Oxford, UK: IRL Press; ), pp. 63–174 [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A., Saeki H., Kameya T., Saedler H., Theissen G. (2003). Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52: 831–841 [DOI] [PubMed] [Google Scholar]
- Kim S., Koh J., Yoo M.J., Kong H., Hu Y., Ma H., Soltis P.S., Soltis D.E. (2005). Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 43: 724–744 [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Dorit R.L., Irish V.F. (1998). Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149: 765–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M., Irish V.F. (1999). Evolution of genetic mechanisms controlling petal development. Nature 399: 144–148 [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Jaramillo M.A., Di Stilio V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22 [DOI] [PubMed] [Google Scholar]
- Kubitzki K. (1998). The Families and Genera of Vascular Plants, Volume 3: Flowering Plants: Monocotyledons: Lilianae (except Orchidaceae). (Berlin: Springer; ). [Google Scholar]
- Lamb R.S., Irish V.F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 100: 6558–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauermann V. (1991). Ethanol improves the transformation efficiency of intact yeast cells. Curr. Genet. 20: 1–3 [DOI] [PubMed] [Google Scholar]
- Lee I., Wolfe D.S., Nilsson O., Weigel D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7: 95–104 [DOI] [PubMed] [Google Scholar]
- Lee S., Jeon J.-S., An K., Moon Y.-H., Lee S., Chung Y.-Y., An G. (2003). Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217: 904–911 [DOI] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Maas-van de Kamer H. (1995). Triuridiflorae—Gardner’s delight? In Monocotyledons: Systematics and Evolution, Rudall P.J., Cribb P.J., Cutler D.F., Humphreys C.J., (London: Kew-Royal Botanic Gardens; ), pp. 287–301 [Google Scholar]
- Mabberley D.J. (1997). The Plant Book, 2nd ed (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Márquez-Guzmán E., Engleman E.M., Martínez-Mena A., Martínez E., Ramos C. (1989). Anatomía reproductiva de Lacandonia schismatica (Lacandoniaceae). Ann. Mo. Bot. Gard. 76: 124–127 [Google Scholar]
- Márquez-Guzmán J., Vázquez-Santana S., Engleman E.M., Martínez-Mena A., Martínez E. (1993). Pollen development and fertilization in Lacandonia schismatica (Lacandoniaceae). Ann. Mo. Bot. Gard. 80: 891–897 [Google Scholar]
- Martínez E., Ramos C.H. (1989). Lacandoniaceae (Triuridales), a new family of Mexican plants. Ann. Mo. Bot. Gard. 76: 128–135 [Google Scholar]
- McGonigle B., Bouhidel K., Irish V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10: 1812–1821 [DOI] [PubMed] [Google Scholar]
- Meyer V.G. (1966). ‘Floral Abnormalities’. Bot. Rev. 32: 165–195 [Google Scholar]
- Meyerowitz E.M., Bowman J.L., Brockman L.L., Drews G.N., Jack T., Sieburth L.E., Weigel D. (1991). A genetic and molecular model for flower development in Arabidopsis thaliana. Dev. Suppl. 1(suppl. 1): 157–167 [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theissen G. (2009). Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann. Bot. (Lond.) 104: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y.H., Jung J.Y., Kang H.G., An G. (1999). Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol. Biol. 40: 167–177 [DOI] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., Nagato Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Piwarzyk E., Yang Y.Z., Jack T. (2007). Conserved C-terminal motifs of the Arabidopsis proteins APETALA3 and PISTILLATA are dispensable for floral organ identity function. Plant Physiol. 145: 1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R.E., Bowman J.L., Grossniklaus U. (2003). Plant genetics: A decade of integration. Nat. Genet. 33(Suppl): 294–304 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema A.S., Royaert S., Zethof J., van der Weerden G., Gerats T., Vandenbussche M. (2006). Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P.J. (2003). Monocot Pseudanthia revisited: Floral structure of the mycoheterotrophic family Triuridaceae. Int. J. Plant Sci. 164: S307–S320 [Google Scholar]
- Rudall P.J. (2008). Fascicles and filamentous structures: Comparative ontogeny of morphological novelties in Triuridaceae. Int. J. Plant Sci. 169: 1023–1037 [Google Scholar]
- Rudall P.J., Bateman R.M. (2006). Morphological phylogenetic analysis of pandanales: Testing contrasting hypotheses of floral evolution. Syst. Bot. 31: 223–238 [Google Scholar]
- Sattler R. (1984). Homology – A continuing challenge. Syst. Bot. 9: 382–394 [Google Scholar]
- Sattler R., Singh V. (1978). Floral organogenesis of Echinodorus amazonicus Rataj and floral construction of the Alismatales. Bot. J. Linn. Soc. 77: 141–156 [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P.J., Hansen R., Tetens F., Lönnig W.E., Saedler H., Sommer H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Chanderbali A.S., Kim S., Buzgo M., Soltis P.S. (2007). The ABC model and its applicability to basal angiosperms. Ann. Bot. (Lond.) 100: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H., Beltrán J.P., Huijser P., Pape H., Lönnig W.E., Saedler H., Schwarz-Sommer Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E., Rebocho A.B., Bliek M., Kusters E., de Bruin R.A.M., Koes R. (2008). Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell 20: 2033–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens P.F. (1991). Lacandonia schismatica—A challenge to some recent theories of floral morphogenesis? Flower Newsletter 12: 32–33 [Google Scholar]
- Stone B.C. (1990). New evidence for the reconciliation of floral organization in Pandanaceae with normal angiosperm patterns. In Plant Diversity of Malesia, Bass P., Kalkman K., Geesink R., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 33–55 [Google Scholar]
- Su K.M., Zhao S.H., Shan H.Y., Kong H.Z., Lu W.L., Theissen G., Chen Z.D., Meng Z. (2008). The MIK region rather than the C-terminal domain of AP3-like class B floral homeotic proteins determines functional specificity in the development and evolution of petals. New Phytol. 178: 544–558 [DOI] [PubMed] [Google Scholar]
- Sundström J., Carlsbecker A., Svensson M.E., Svenson M., Johanson U., Theissen G., Engström P. (1999). MADS-box genes active in developing pollen cones of Norway spruce (Picea abies) are homologous to the B-class floral homeotic genes in angiosperms. Dev. Genet. 25: 253–266 [DOI] [PubMed] [Google Scholar]
- Theissen G., Melzer R. (2007). Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 100: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Tsai W.C., Kuoh C.S., Chuang M.H., Chen W.H., Chen H.H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 45: 831–844 [DOI] [PubMed] [Google Scholar]
- Tsai W.C., Pan Z.J., Hsiao Y.Y., Jeng M.F., Wu T.F., Chen W.H., Chen H.H. (2008). Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol. 49: 814–824 [DOI] [PubMed] [Google Scholar]
- Tzeng T.Y., Liu H.C., Yang C.H. (2004). The C-terminal sequence of LMADS1 is essential for the formation of homodimers for B function proteins. J. Biol. Chem. 279: 10747–10755 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Zethof J., Royaert S., Weterings K., Gerats T. (2004). The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara-Silva F., Espinosa-Matias S., Ambrose B.A., Vazquez-Santana S., Martinez-Mena A., Marquez-Guzman J., Martinez E., Meyerowitz E.M., Alvarez-Buylla E.R. (2003). Inside-out flowers characteristic of Lacandonia schismatica evolved at least before its divergence from a closely related taxon, Triuris brevistylis. Int. J. Plant Sci. 164: 345–357 [Google Scholar]
- Vergara-Silva F., Martinez-Castilla L., Alvarez-Buylla E.R. (2000). MADS-box genes: Development and evolution of plant body plans. J. Phycol. 36: 803–812 [Google Scholar]
- Weston P.H. (2000). Process morphology from a cladistic perspective. In Homology and Systematics: Coding Characters for Phylogenetic Analysis. Scotland R., Pennington R.T., (London: Taylor & Francis; ), pp. 124–143 [Google Scholar]
- Whipple C.J., Ciceri P., Padilla C.M., Ambrose B.A., Bandong S.L., Schmidt R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131: 6083–6091 [DOI] [PubMed] [Google Scholar]
- Whipple C.J., Zanis M.J., Kellogg E.A., Schmidt R.J. (2007). Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. USA 104: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K.U., Weiser C., Kaufmann K., Bohne A., Kirchner C., Kanno A., Saedler H., Theissen G. (2002). Evolution of class B floral homeotic proteins: Obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19: 587–596 [DOI] [PubMed] [Google Scholar]
- Wolpert L., Jessell T., Lawrence P., Meyerowitz E., Robertson E., Smith J. (2006). Principles of Development, 3rd ed (New York: Oxford University Press; ). [Google Scholar]
- Xiao H., Wang Y., Liu D., Wang W., Li X., Zhao X., Xu J., Zhai W., Zhu L. (2003). Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 52: 957–966 [DOI] [PubMed] [Google Scholar]
- Xu Y.F., Teo L.L., Zhou J., Kumar P.P., Yu H. (2006). Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46: 54–68 [DOI] [PubMed] [Google Scholar]
- Yang Y.Z., Fanning L., Jack T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59 [DOI] [PubMed] [Google Scholar]
- Zachgo S., Silva Ede , A., Motte P., Tröbner W., Saedler H., Schwarz-Sommer Z. (1995). Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121: 2861–2875 [DOI] [PubMed] [Google Scholar]
- Zik M., Irish V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]