Meiotic chromosome segregation requires extensive remodeling of cell cycle machinery. This work analyzes how TDM1, SMG7, and the cyclin TAM, which are essential for completion of meiosis, genetically interact to regulate meiotic progression in Arabidopsis pollen mother cells.
Abstract
Meiosis is a modified cell division that produces four haploid nuclei from a single diploid cell in two rounds of chromosome segregation. Here, we analyze the role of Arabidopsis thaliana SUPPRESSOR WITH MORPHOGENETIC EFFECTS ON GENITALIA7 (SMG7), THREE DIVISION MUTANT1 (TDM1), and TARDY ASYNCHRONOUS MEIOSIS (TAM) in meiotic progression. SMG7 is a conserved nonsense-mediated mRNA decay factor that is also, in Arabidopsis, essential for completion of meiosis. Examination of activating CYCLIN DEPENDENT KINASE A;1 phosophorylation at Thr-161 suggests that the meiotic arrest observed in smg7 mutants is likely caused by a failure to downregulate cyclin-dependent kinase (CDK) activity at the end of the second meiotic division. Genetic analysis indicates that SMG7 and TDM1 act in the same pathway to facilitate exit from meiosis. We further demonstrate that the cyclin TAM is specifically expressed in meiosis I and has both stimulatory and inhibitory effects on progression to meiosis II. TAM knockouts skip the second meiotic division producing unreduced gametes, but inactivation of SMG7 or TDM1 alleviates TAM’s requirement for entry into meiosis II. We propose a model that meiotic progression in Arabidopsis pollen mother cells is driven by a yet to be identified cyclin-CDK activity that is modulated by regulatory interactions between TDM1, SMG7, and TAM.
INTRODUCTION
Cell division is a carefully orchestrated process in which DNA duplication is followed by chromosome segregation and formation of two new daughter cells. The cell cycle is driven by cyclin-dependent kinases (CDKs), the activity and specificity of which is determined by its association with regulatory cyclin subunits. CDK activity is further controlled by a sophisticated network of inhibiting and activating mechanisms that fine-tune cell cycle progression according to developmental and environmental cues. Entry and progression through mitosis requires high CDK activity that, in higher eukaryotes, primarily relies on A- and B-type cyclins. CDK activity peaks at metaphase when all chromosomes attach to the spindle and align at the metaphase plate. At this point, activation of the anaphase promoting complex (APC) initiates chromosome segregation through proteolytic destruction of securin and B-type cyclins. While degradation of securin leads to the activation of separase, the protease that cleaves cohesin and triggers entry into anaphase, the destruction of cyclins results in downregulation of CDK activity, which is important for chromosome decondensation, cytokinesis, and transition to G1 (de Gramont and Cohen-Fix, 2005). Thus, activation of APC couples chromosome segregation with mitotic exit. Low CDK activity in G1 is essential for assembly of prereplicative complexes at replication origins and, thus, for entry into another cell cycle.
Meiosis is a modified cell division in which a replicated diploid genome undergoes two subsequent rounds of chromosome segregation that produce four haploid nuclei. While the fundamental principles governing cell cycle progression are shared in meiosis and mitosis, meiosis-specific regulatory mechanisms have evolved to accomplish sequential segregation of homologous chromosomes in the first division and of sister chromatids in the second division. The most noticeable meiotic event is an extended prophase I, during which chromosomes pair and recombine to allow faithful segregation of homologous chromosome in anaphase I. Another important event is the suppression of DNA replication after the first meiotic division. This is achieved by precise fine-tuning of CDK activity that decreases to a level permissive for chromosome decondensation and spindle rearrangement but is still sufficient to prevent assembly of prereplicative complexes (Marston and Amon, 2004). Finally, in many organisms, including plants, meiosis is immediately followed by mitotic divisions, indicating that the meiotic regulatory network must be shut off to permit normal chromosome segregation in the following cell cycle.
Regulatory pathways that define meiotic chromosome segregation are understood to a much lesser extent than the regulation of mitosis. One layer of specificity may be conferred by utilization of meiotic CDK-cyclin complexes. Indeed, meiosis-specific cyclins have been identified in several organisms, such as Rem1 and Crs1 in fission yeast or the mouse cyclin A1 (Averbeck et al., 2005; Malapeira et al., 2005; Wolgemuth and Roberts, 2010). Nevertheless, it appears that most cyclins are shared in meiosis and mitosis. A recent study in budding yeast revealed that four out of six mitotic Clb cyclins are also expressed during meiosis. However, their meiotic expression is strictly regulated at the posttranscriptional and postranslational level; misregulation of these cyclins perturbs meiosis (Carlile and Amon, 2008). In Drosophila melanogaster, cyclin A is essential for mitosis, but it is also expressed in meiosis where it is restricted to prophase and prometaphase I (Vardy et al., 2009). Another important layer of meiotic regulation is conferred by inhibitors and activators of APC. Ama1, a meiosis-specific activator of APC, is important for meiotic exit and cytokinesis in budding yeast (Diamond et al., 2009). A germline-specific activator of APC is also required for meiotic exit in Drosophila (Page and Orr-Weaver, 1996). Progression into the second meiotic division in fission yeast depends on Mes1, which is a competitive inhibitor of APC that blocks cyclin destruction in interkinesis (Izawa et al., 2005). Vertebrate oocytes are arrested in metaphase II until fertilization to prevent parthenogenesis. Studies in Xenopus laevis indicate that this arrest is partially mediated by the APC inhibitors Emi1 and Erp1/Emi2 (Irniger, 2006). CDK activity is also controlled by inhibitors (CKI) that play a central role in cell cycle regulation. Their function in meiosis was implicated from studies of the Drosophila Roughex protein, which acts as an inhibitor of CDK and whose deficiency leads to an abortive third meiotic division (Gönczy et al., 1994; Foley et al., 1999).
There is a long tradition of meiotic research in plants, but the vast majority of studies are focused on meiotic recombination and chromosome segregation (Jones et al., 2003; Hamant et al., 2006; Liu and Makaroff, 2006; Mercier and Grelon, 2008). Nevertheless, recent efforts to understand meiotic mechanisms in Arabidopsis thaliana led to the identification of several genes implicated in cell cycle progression. Two cyclins, SDS and TARDY ASYNCHRONOUS MEIOSIS (TAM), with distinct functions in meiosis were discovered in forward genetic screens. SDS is a plant-specific cyclin required for homologous recombination and chromosome pairing (Azumi et al., 2002). TAM is an A-type cyclin (also known as CYCA1;2) that is important for the transition between the first and second meiotic divisions (Magnard et al., 2001; Wang et al., 2004b). A similar function has also been proposed for OSD1, a novel gene that was identified based on its meiosis-specific expression (d’Erfurth et al., 2009). Plants mutant for OSD1 skip the second meiotic division and form diploid gametes. Another interesting meiotic cell cycle regulator discovered in screens for mutants with impaired fertility is THREE DIVISION MUTANT1 (TDM1)/MS5/POLLENLESS3 (Ross et al., 1997; Glover et al., 1998; Sanders et al., 1999). Mutations in TDM1 lead to male sterility caused by a failed attempt to divide unreplicated haploid nuclei after meiosis II (Ross et al., 1997).
We recently described a unique meiotic cell cycle defect associated with inactivation of SUPPRESSOR WITH MORPHOGENETIC EFFECTS ON GENITALIA7 (SMG7), which is an evolutionary conserved protein essential for nonsense-mediated mRNA decay (Riehs et al., 2008). Arabidopsis smg7 mutants are infertile due to a meiotic arrest in anaphase II. In general, cell cycle arrest in anaphase is very unusual because entry into anaphase and exit from M-phase are coupled through the activation of APC. Anaphase arrest has so far been described only under conditions where cyclin destruction is impaired, either by expressing nondegradable forms of cyclin or by mutating an APC activation subunit (Holloway et al., 1993; Parry and O’Farrell, 2001; Swan et al., 2005; Wolf et al., 2006). The observation that Arabidopsis SMG7 is essential for progression through anaphase II and meiotic exit is particularly intriguing because it suggests a very specific link between the cell cycle machinery and RNA processing. In this study, we attempt to further understand the meiotic function of SMG7 by analyzing CDK activity in wild-type and smg7 meiocytes and by mapping genetic interactions with other genes implicated in meiotic progression. We show that the anaphase II arrest in smg7 mutants is likely caused by a failure to downregulate CDK activity after chromosome segregation in meiosis II and that its execution depends on the TDM1 protein. Our data further suggest that TAM is a meiosis I–specific cyclin that has both stimulatory as well as inhibitory effects on mechanisms that drive progression to meiosis II, but its activity is dispensable for entry into meiosis II in the absence of SMG7 or TDM1. We integrate our findings in a model suggesting that meiotic progression in pollen mother cells (PMCs) is primarily governed by a yet to be defined cyclin-CDK activity that is modulated by complex regulatory interactions between TDM1, SMG7, and TAM.
RESULTS
Analysis of CDKA;1 Phosphorylation in the Course of Meiosis
We have previously shown that meiosis in Arabidopsis smg7 mutants arrests in an anaphase II–like stage that is characterized by delayed chromosome decondensation, aberrant spindle rearrangement, and defective dephosphorylation of histone H3 at the Ser-10 residue. We hypothesized that SMG7 is important for downregulation of meiotic CDK activity or that it antagonizes CDK activity by promoting dephosphorylation of the cyclin-CDK substrates during anaphase II (Riehs et al., 2008). To directly test these predictions biochemically is difficult, mainly due to the limited amount of synchronized meiocytes that are available in Arabidopsis flowers. Therefore, we were looking for a marker that would allow detection of CDK activity indirectly using immunocytology. A hallmark of the active CDK-cyclin complexes is phosphorylation of the conserved Thr residues in the T-loop (Gould et al., 1991). Arabidopsis has several CDKs, among which CYCLIN DEPENDENT KINASE A;1 (CDKA;1) appears to be of key importance for the meiotic cell cycle since weak loss-of-function mutants exhibit aberrant meiosis (Dissmeyer et al., 2007). CDKA;1 contains a T-loop with the conserved Thr residue at position 161. Studies in synchronized tobacco BY2 cells demonstrated that Thr-161 phosphorylation is regulated in a cell cycle–dependent manner with the strongest appearance in M-phase (Harashima et al., 2007). Thus, we decided to examine Thr-161 phosphorylation of CDKA;1 in the course of meiosis.
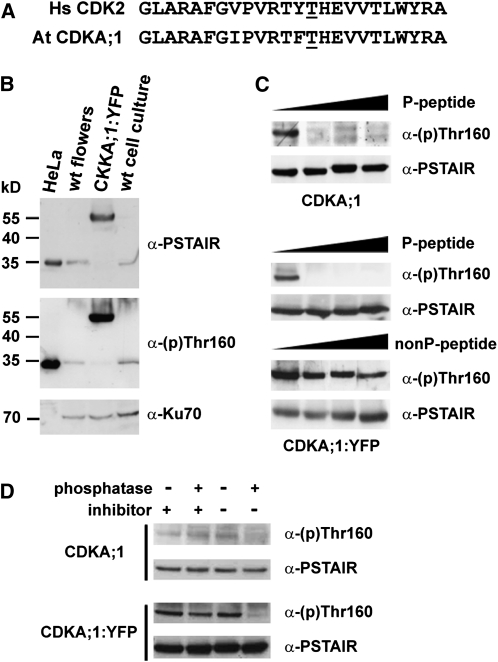
The T-loop of CDKA;1 is almost identical to the corresponding sequence in human CDK2 (Figure 1A). Indeed, an antibody raised against the phosphorylated T-loop of HsCDK2 [α-(p)Thr160] recognized an ~35-kD peptide in wild-type Arabidopsis protein extracts (Figure 1B). Importantly, only an ~55-kD protein was detected in plants in which the endogenous CDKA;1 gene was inactivated and complemented with a CDKA;1:yellow fluorescent protein (YFP) fusion construct (Figure 1B). Thus, the α-(p)Thr160 antibody detects only CDKA;1 and not any other Arabidopsis CDKs. Immunodetection was abolished by addition of a peptide corresponding to the phosphorylated T-loop but not in the presence of a nonphosphorylated peptide (Figure 1C). Furthermore, the detection was sensitive to the pretreatment of plant protein extracts to phosphatase (Figure 1D). Together, these data demonstrate that the α-(p)Thr160 antibody specifically recognizes CDKA;1 phosphorylated at Thr-161.
Figure 1.
Immunoblot Analysis of CDKA;1 Phosphorylation at Thr-161.
(A) Homology of Arabidopsis CDKA;1 and human CDK2 in the T-loop region with the indicated Thr residues at positions 161 and 160, respectively.
(B) Immunodetection of phosphorylated Hs CDK2, At CDKA;1, and At CDKA;1:YFP proteins in extracts from HeLa cells, Arabidopsis wild-type influorescences and suspension culture, and CDKA;1:YFP flowers.
(C) Immunodetection of Thr-161 phosphorylation of CDKA;1 and CDKA;1:YFP in the presence of increasing concentration (0, 1, 10, and 100 mg/mL) of phosphorylated (P) and nonphosphorylated (nonP) competitor peptides. Total CDKA;1 was detected with α-PSTAIR antibody as a loading control.
(D) The Thr-161 phosphorylation signal is sensitive to phosphatase treatment. Protein extracts from wild-type and CDKA;1:YFP plants were pretreated with the lambda protein phosphatase in the presence or absence of phosphatase inhibitors as indicated. Proteins were analyzed by immunoblot with α-phospho-CDK2(Thr160) and α-PSTAIR antibody.
To assay whether the antibody can be used for detection of phosphorylated CDKA;1 in situ, we performed immunocytology with mitotic cells prepared from floral buds of plants containing the CDKA;1:YFP fusion construct. To assure specificity of the detection, all experiments were performed in the presence of the competitive nonphosphorylated peptide. While the CDKA;1:YFP protein was readily visible in the majority of mitotic cells, a strong signal with the α-(p)Thr160 antibody that colocalized with the CDKA;1:YFP was detected only in metaphases (see Supplemental Figure 1 online). This result is in agreement with data obtained by immunoblot analysis of extracts from synchronized tobacco (Nicotiana tabacum) BY2 cells (Harashima et al., 2007) and demonstrates that T-loop phosphorylation in plant mitotic cells peaks at metaphase.
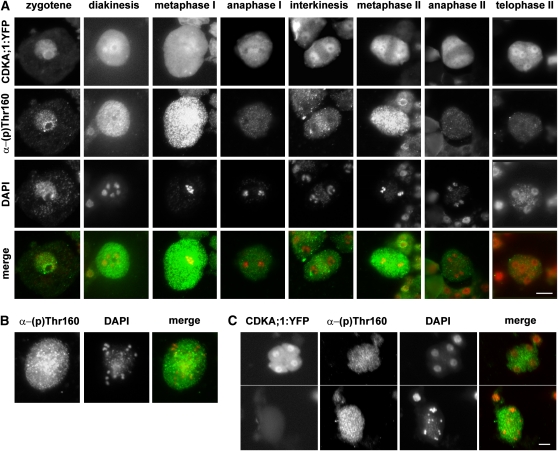
We used immunocytology to analyze CDKA;1 phosphorylation in the course of meiosis in PMCs (Figure 2A). Thr-161 phosphorylation was detectable already in early prophase I and colocalized with the CDKA;1:YFP signal primarily at chromatin (Figure 2A). While the overall level of CDKA;1 was approximately constant during meiosis, the phosphorylation signal peaked at metaphase I, decreased in anaphase I and interkinesis, peaked again in metaphase II, and disappeared in anaphase and telophase II (Figure 2A). In meiotic metaphases, CDKA;1:YFP and Thr-161 phosphorylation signals were distributed throughout whole meiocytes and did not localize to any particular structure. Interestingly, an opposite localization pattern was observed in interkinesis, where a weak Thr-161 signal was restricted to the organellar band that separates the two nuclei, whereas the bulk of CDKA;1 appeared primarily at the nuclei (Figure 2A). This suggests that a small amount of CDKA;1 complexes remain active in the midzone, presumably to inhibit cytokinesis between the two meiotic divisions. We also observed a similar phenomenon in binuclear cells of the tapetum, where phosphorylated CDKA;1 complexes formed a band separating the two nuclei, while most CDKA;1 was on chromatin (see Supplemental Figure 2 online).
Figure 2.
Immunolocalization of Thr-161 Phosphorylated CDKA;1 in the Course of Meiosis.
(A) Meiocytes from PMCs of CDKA;1:YFP plants stained with α-phospho-CDK2(Thr160) antibody. DNA was counterstained with DAPI. Bar = 10 μm.
(B) CDKA;1 Thr-161 phosphorylation in aberrant anaphase II of smg7 mutants.
(C) Comparison of CDKA;1 Thr-161 phosphorylation signal in telophase II of CDKA;1:YFP plants (top panel) with aberrant anaphase II in smg7 mutants (bottom panel). PMCs from these plants were prepared on the same slide, and pictures of the meiocytes represent two cropped sections of the same photograph. CDKA;1:YFP meiocytes are distinguished from smg7 meiocytes by the presence of the YFP signal. Bars = 5 μM.
We next examined CDKA;1 phosphorylation in smg7 meiocytes arrested at anaphase II. The aberrant anaphase II is characterized by the presence of 20 chromatids that do not decondense and are randomly distributed within the meiocyte due to defective spindle rearrangement (Riehs et al., 2008). While the phosphorylation signal disappeared in wild-type anaphase II (Figure 2A), strong staining remained in smg7 meiocytes arrested at anaphase II (Figures 2B and 2C). These data support our prediction that aberrant meiotic exit in SMG7-deficient plants is caused by a failure to downregulate CDK activity after chromosome segregation in anaphase II.
SMG7 Deficiency Suppresses Premature Meiotic Exit of tam Mutants
The smg7 phenotype could be mimicked by exposing meiocytes to the proteasome inhibitor MG115, suggesting that inefficient cyclin degradation may be a mechanism responsible for the anaphase arrest (Riehs et al., 2008). To test whether the smg7 phenotype can be alleviated by decreasing the level of a meiotic cyclin, we generated plants deficient in SMG7 carrying a temperature-sensitive tam-1 allele. tam-1 mutants grown at the restrictive temperature exhibit prolonged meiotic interkinesis caused by a delayed entry into meiosis II and premature formation of the cell wall as early as after the first meiotic division (Magnard et al., 2001; Wang et al., 2004b) (Figure 3A). Meiosis in smg7 tam-1 double mutants still arrested in anaphase II (Figure 3B), indicating that the meiotic arrest is not executed through cyclin A1;2. Surprisingly, however, SMG7 deficiency completely suppressed premature cell wall formation in tam-1 mutants. Thus, the meiotic role of SMG7 is not only restricted to anaphase II but already functions during interkinesis.
Figure 3.
Epistasis Analysis of smg7 and tam-1 Mutations.
(A) PMCs at interkinesis from wild-type and tam-1 mutants shown in phase contrast (top panel); nuclei are stained with DAPI (bottom panel).
(B) Irregular behavior of meiotic chromosomes in PMCs of smg7 tam-1 double mutants. DNA is counterstained by DAPI. Bars in (A) and (B) = 5 μm.
(C) Frequency (in %) of meiotic stages from metaphase I to metaphase II in wild-type, smg7-1, tam-1, and smg7 tam-1 plants. All plants were kept at 28°C during flowering. The total number of meiocytes is indicated below each pie chart.
We next asked whether SMG7 deficiency specifically affects cell wall formation or whether it has a broader effect on meiotic interkinesis. We examined progression through interkinesis by scoring the frequency of individual meiotic stages from metaphase I to metaphase II in PMCs (Figure 3C). Interkinesis was significantly prolonged in tam-1 mutants compared with the wild type as ~50% of the scored tam-1 meiocytes corresponded to this stage. Strikingly, cytologically defined interkinesis characterized by two fully decondensed nuclei was completely eliminated in smg7 tam-1 double mutants. Only 6 out of 478 meiocytes possessed two clearly defined nuclei, but even these were unusually condensed (Figure 3B). Furthermore, we detected a large proportion of meiocytes that contained fully condensed metaphase II–like chromosomes that were dispersed in the nuclei (Figure 3B, irregular metaphase II). Such a degree of condensation is only typical of chromosomes aligned on the metaphase plate. These data suggest that under low levels of cyclin A1;2, SMG7 is already required after the first meiotic division for full chromatin decondensation.
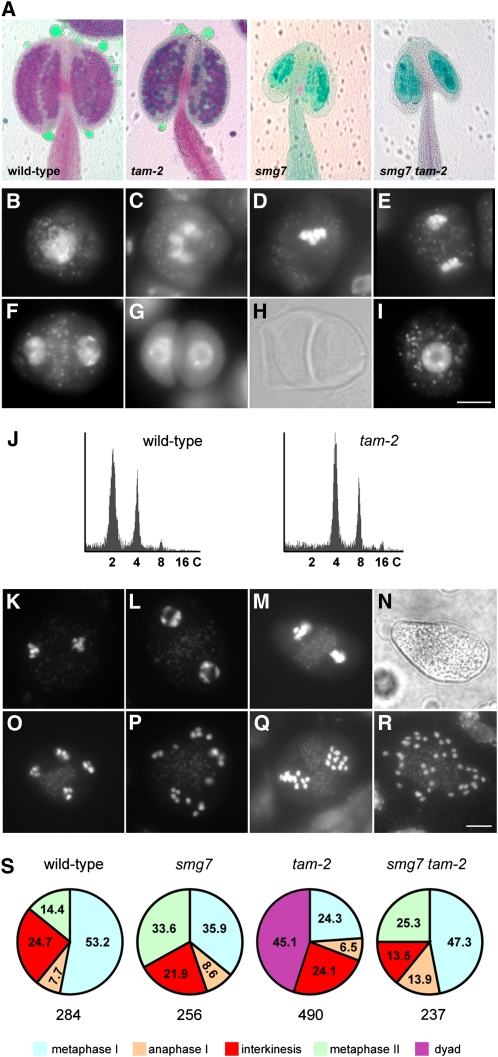
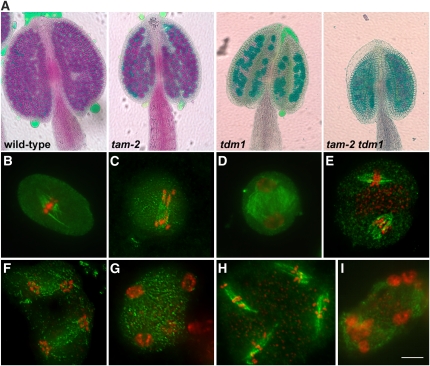
Because tam-1 is a temperature-sensitive allele likely retaining residual activity, we decided to confirm the genetic interaction with smg7 using a null tam allele. We characterized an Arabidopsis line carrying a T-DNA insertion that led to the deletion of the first half of the TAM gene, including the whole promoter (tam-2; see Supplemental Figure 3 online). Therefore, the tam-2 allele is likely null. Plants homozygous for the tam-2 allele are fertile, but their anthers contain fewer pollen grains, and these grains are bigger than in the wild type (Figure 4A). Cytogenetic examination of PMCs revealed that while meiosis progressed normally until telophase I, the second meiotic division was absent (Figures 4B to 4F). Instead, we observed numerous dyads with a fully developed cell wall, indicating exit from meiosis after the first division (Figures 4G and 4H). This should result in diploid spores and lead to polyploid progeny. Indeed, 15 out of 18 progeny plants obtained from selfed tam-2 mutants were tetraploid (Figure 4J). The occurrence of tetraploid progeny further suggested that TAM deficiency also affects female meiosis. An identical phenotype has recently been described for several additional tam alleles by other groups (d’Erfurth et al., 2010; Wang et al., 2010).
Figure 4.
SMG7 Deficiency Suppresses Premature Exit after Meiosis I Caused by the tam-2 Null Allele.
(A) Pollen viability determined by Alexander staining of anthers. Viable pollen in wild-type and tam-2 plants is stained in red. No pollen is detected in anthers of smg7 and smg7 tam-2 mutants.
(B) to (I) Meiotic chromosomes stained by DAPI in PMCs from tam-2 mutants. Zygotene (B), diakinesis (C), metaphase I (D), anaphase I (E), interkinesis (F), dyad visualized by DAPI staining (G) and by phase contrast (H), and microspore (I). Bar = 10 μm.
(J) Flow cytometry analysis of DNA content in nuclei prepared from inflorescences of progeny of wild-type and tam-2 plants.
(K) to (P) DAPI-stained PMCs from smg7 tam-2 double mutants. Anaphase I (K), interkinesis (L), fluorescence (M) and phase contrast (N) pictures of metaphase II, anaphase II (O), and irregular anaphase II (P).
(Q) and (R) Metaphase II (Q) and irregular anaphase II (R) from tetraploid smg7 tam-2 double mutants. Bar = 5 μm
(S) Frequency (in %) of meiotic stages from metaphase I to metaphase II in wild-type, smg7, tam-2, and smg7 tam-2 plants. The total number of meiocytes is indicated below each pie chart.
Strikingly, the premature meiotic exit and formation of dyads were completely suppressed in smg7 tam-2 double mutants. Meiosis appeared to progress normally until anaphase II (Figures 4K to 4O), and no cell wall formed after the first meiotic division (Figures 4M and 4N). Similarly to the situation in smg7 plants, meiosis in smg7 tam-2 mutants arrested at an irregular anaphase II that was characterized by 20 randomly distributed chromatids (Figure 4P). This phenotype was also observed in tetraploid smg7 tam-2 plants that contained 40 separated chromatids at an irregular anaphase II (Figures 4Q and 4R). This analysis confirmed data obtained with smg7 tam-1 mutants and demonstrated that the absence of SMG7 alleviates the requirement for TAM to enter into the second meiosis. Interestingly, in contrast with the situation in smg7 tam-1 plants, normal interkinesis appears to occur in smg7 tam-2 mutants (Figures 4L and 4S). This indicates that TAM plays a complex role in progression during interkinesis.
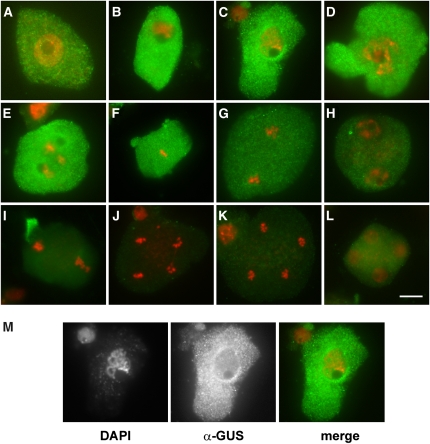
We further examined expression of TAM during meiosis. In a previous report, Wang et al. (2004b) used the fluorescence signal of a TAM:green fluorescent protein fusion to localize expression of the TAM gene to meiocytes. However, in their study, the identity of cells expressing TAM was determined only indirectly by staging anthers within the same floral bud. Such analysis does not permit accurate determination of TAM expression during meiosis. Therefore, we generated transgenic plants carrying the entire TAM gene fused to a β-glucuronidase (GUS) reporter at the C terminus and used immunodetection with GUS antibody to study the cellular localization of the fusion construct. GUS histochemical assays revealed that TAM is not restricted to meiosis, and it is expressed in tissues containing proliferating cells, such as root tips, shoot apical meristems, and young inflorescence buds (see Supplemental Figure 4 online). Immunolocalization experiments in mitotic cells revealed that TAM:GUS is most abundant in a fraction of interphase cells that are presumably in G2, and in prophase and prometaphase. Its level is lower in metaphase, and the signal disappears during anaphase (see Supplemental Figure 4 online). A similar localization pattern was also detected during the first meiosis where the TAM:GUS signal peaked in mid prophase I (Figures 5B to 5D), weakened during metaphase I, and disappeared in anaphase I (Figure 5G). The bulk of the TAM:GUS protein was localized to cytoplasm and was not detectable at the chromatin (Figure 5M). No specific signal was detected in later stages of meiosis, including interkinesis or metaphase II (Figures 5H to 5L). This analysis argues that TAM is a meiosis I–specific cyclin, although we cannot fully exclude the possibility that small undetectable levels of the cyclin are also present in interkinesis or in meiosis II.
Figure 5.
Immunolocalization of TAM:GUS in Meiosis.
Fluorescein isothiocyanate signal detecting TAM:GUS is indicated in green and DNA counterstained with DAPI in red. Leptotene (A), zygotene (B), pachytene (C), diplotene (D), diakinesis (E), metaphase I (F), anaphase I (G), interkinesis (H), metaphase II (I), anaphase II ([J] and [K]), and telophase II (L). The pachytene in (M) is the same as in (C), shown as unmerged DAPI and fluorescein isothiocyanate pictures. Bar = 10 μm.
Anaphase Arrest in smg7 Mutants Requires TDM1 Function
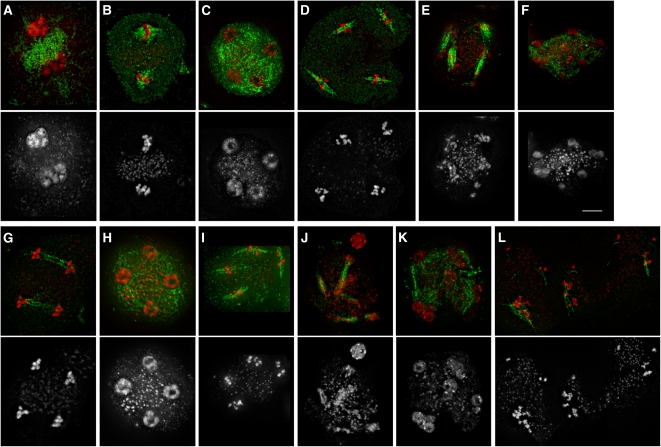
Another Arabidopsis gene implicated in meiotic progression is TDM1. TDM1 deficiency leads to formation of polyads and male sterility. Cytogenetic analysis of PMCs showed that chromatin in tdm1 mutants recondensed and stretched after meiosis II as if haploid nuclei entered a third meiotic division (Ross et al., 1997). To verify this suggestion, we examined whether chromatin recondensation in tdm1 mutants is accompanied by spindle formation. Indeed, we found that a dense microtubule network connecting haploid nuclei in the tetrad stage rearranged into four bipolar spindles as chromatids recondensed, resembling metaphase (Figures 6A to 6D). Chromatids then stretched along the spindle, likely as a consequence of the pulling forces of tubulin, and this process led to the formation of a variable number of micronuclei (Figures 6E and 6F). These observations suggest that TDM1 is essential for full completion of meiosis and transition to G1.
Figure 6.
Meiosis in PMCs of tdm1 and smg7 tdm1 Mutants.
The spindle was detected by immunostaining with anti-α-tubulin antibody (green), and DNA was counterstained with DAPI (red). Only DAPI staining is shown in the bottom panel.
(A) to (F) Meiosis in tdm1 mutants. Interkinesis (A), metaphase II (B), telophase II (C), metaphase III (D), telophase III (E), and polyads (F). Bar = 5 μm.
(G) to (L) Meiosis in smg7 tdm1 mutants. Anaphase II (G), telophase II (H), metaphase/anaphase III (I), anaphase III (J), polyads (K), and condensed chromatids aligned along spindles (L).
Because SMG7 is also required for meiotic exit (Riehs et al., 2008), we analyzed the epistatic interaction between SMG7 and TDM1. Phenotypic analysis indicated that SMG7 acts during anaphase II, while TDM1 function is important after telophase II, suggesting that TDM1 acts downstream of SMG7. Surprisingly, however, TDM1 was epistatic to SMG7 as the tdm1 mutation suppressed the smg7 phenotype. Meiosis in smg7 tdm1 double mutants normally progressed to telophase II and then reentered the third meiotic division forming polyads (Figures 6G to 6K). Occasionally, we observed meiocytes that contained condensed chromatids aligned along three to four irregular spindles as if there was a delay in chromatin decondensation after the third meiosis (Figure 6L). Two possible hypotheses can be proposed to explain the unexpected genetic interaction between SMG7 and TDM1. Either the anaphase II arrest in smg7 mutants depends on the function of TDM1 or anaphase II in tdm1 mutants resembles anaphase I that appears to be insensitive to the absence of SMG7. While SMG7 deficiency leads to male sterility and drastically reduced female fertility (Riehs et al., 2008), tdm1 mutants are female fertile, suggesting that the TDM1 protein acts only in male meiosis (Glover et al., 1998). However, we found that smg7 tdm1 plants are female fertile, suggesting that the absence of TDM1 recovers completion of female meiosis in smg7 mutants. This argues that TDM1 function is not only restricted to PMCs but that it also plays a role in female meiosis.
Since SMG7 is epistatic to TAM, we next examined the genetic interaction between TDM1 and TAM. Inspection of anthers revealed that similar to tdm1 mutants, tam-2 tdm1 double mutants produce no viable pollen (Figure 7A). Further cytogenetic analysis of tam-2 tdm1 PMCs showed no evidence for premature meiotic exit typical for tam-2 mutants. Meiosis normally progressed through the second division and continued to the third meiosis that is characteristic for tdm1 plants (Figures 7B to 7I). Thus, TDM1 is epistatic to TAM, and this genetic interaction argues that the early meiotic exit associated with the null tam-2 allele depends on TDM1 and that reentry into the third meiosis does not require TAM activity.
Figure 7.
TDM1 Deficiency Suppresses Premature Exit after Meiosis I Caused by tam-2 Null Allele.
(A) Pollen viability determined by Alexander staining of anthers. Viable pollen in wild-type and tam-2 plants is stained in red. No viable pollen is detected in tdm1 and tam-2 tdm1 mutants.
(B) to (I) Meiosis in tam-2 tdm1 mutants. Spindle is shown in green and DNA counterstained with DAPI in red. Metaphase I (B), anaphase I (C), interkinesis (D), metaphase II (E), anaphase II (F), telophase II (G), metaphase/anaphase III (H), and polyads (I). Bar = 5 μm.
DISCUSSION
Several major alterations of the chromosome segregation machinery must occur in meiosis to achieve formation of haploid nuclei in two consecutive rounds of nuclear division. These include homologous chromosome pairing and recombination in meiotic prophase I, monoorientation of sister kinetochores in metaphase I, protection of centromeric cohesion in anaphase I, and differential regulation of CDK activity after the first and second meiotic divisions (Petronczki et al., 2003; Marston and Amon, 2004). Mechanisms that regulate meiosis often vary between different phyla, reflecting diverse reproduction strategies. In angiosperm plants, entry into meiosis is controlled by developmental clues within the context of the flower, and it is preceded by mitotic divisions in developing reproductive organs. Meiosis is immediately followed by mitotic divisions that give rise to two different types of haploid gametophytes, the pollen and embryo sac (Ma, 2005; Sundaresan and Alandete-Saez, 2010). Thus, it is predictable that not only entry, but also exit, from meiosis is under strict regulation to permit rapid alteration from meiotic to mitotic chromosome segregation.
There is only rudimentary knowledge of the mechanisms that drive progression in, through, and out of meiosis in plants. Recent work in Arabidopsis led to identification of several genes important for meiotic progression, such as TDM1, OSD1, TAM, and SMG7, and their functional characterization indicated that these proteins may directly or indirectly modulate activity of meiotic CDKs (Ross et al., 1997; Glover et al., 1998; Sanders et al., 1999; Wang et al., 2004b; Riehs et al., 2008; d’Erfurth et al., 2009; d’Erfurth et al., 2010). However, the core components of the cell cycle machinery important for meiotic division in plants have not yet been defined. Already early experiments with Xenopus oocytes and temperature-sensitive Cdc28/Cdc2 mutants in yeast demonstrated that, as in mitosis, CDKs plays the central role in meiotic progression (reviewed in Perez-Hidalgo et al., 2007). In this study, we show that Arabidopsis CDKA;1, which is required for mitotic divisions (Iwakawa et al., 2006; Nowack et al., 2006), is also present in meiosis. Our data further indicate that while the level of CDKA;1 is not grossly altered, the activating phosphorylation in its T-loop oscillates during meiosis, with peaks in metaphases I and II. Experiments in synchronized tobacco cell culture demonstrated that T-loop phosphorylation peaks in M-phase and correlates with total CDK activity (Harashima et al., 2007). Together, these results are consistent with biochemical data from Xenopus oocytes and synchronized meiotic cultures in yeast (Iwabuchi et al., 2000; Carlile and Amon, 2008) and formally confirm that CDK activity oscillates during the course of plant meiosis. Immunocytogenetic approaches used in this study do not allow quantitative measurements of CDK activity; therefore, we cannot determine exactly what the residual level of CDK activity in interkinesis is. Nevertheless, we consistently observed phosphorylated CDKA;1 signal retained at organellar band between two separated nuclei. We suggest that a small amount of active CDKA;1 complexes localized to this region may inhibit cytokinesis. This notion is supported by the observation that Arabidopsis plants carrying the T161D substitution in the CDKA;1 T-loop exhibit a variety of meiotic defects, including cell wall formation in interkinesis (Dissmeyer et al., 2007). Our data suggest that such mechanisms may also be involved in inhibiting cytokinesis of binuclear tapetum cells.
The phenotypes associated with smg7 and tdm1 are consistent with the hypothesis that these genes act at the end of meiosis to downregulate CDK activity and promote transition to G1. Epistatic analysis indicates that SMG7 acts through TDM1, but it is currently unknown whether TDM1 is a direct target of SMG7 regulation. TDM1 belongs to a plant-specific protein family with a limited similarity to Xe-p9, a regulatory subunit of CDK from Xenopus (Glover et al., 1998). Xe-p9 is important for proteolytic destruction of cyclin B by enabling CDK-mediated phosphorylation of APC (Patra and Dunphy, 1998). The connection between SMG7 and meiotic progression is less obvious. SMG7 is a nonsense-mediated mRNA decay factor that is ubiquitously expressed in all plant tissues (Riehs et al., 2008). SMG7 carries an evolutionary conserved 14-3-3–like domain (Fukuhara et al., 2005); hence, it may act as an adaptor protein in cell cycle pathways. Alternatively, the meiotic function of SMG7 may indicate a so far not fully appreciated regulatory role of RNA processing in meiotic progression in plants. For example, in fission yeast, expression of cyclins and other cell cycle regulators is restricted to meiosis by elaborate posttranscriptional control mechanisms that involve splicing, polyadenylation, and mRNA stability (Malapeira et al., 2005; Moldón et al., 2008; McPheeters et al., 2009). Recent characterization of the Arabidopsis PS1 gene also supports the importance of RNA processing in plant meiosis (d’Erfurth et al., 2008). PS1 is a meiosis-specific protein containing an RNA degradation PIN domain and is required for proper spindle orientation in meiosis II.
Mutations in the Drosophila CDK inhibitor roughex lead to attempted third meiotic divisions, reminiscent of the phenotype observed in Arabidopsis tdm1 mutants (Gönczy et al., 1994). An anaphase arrest similar to that observed in Arabidopsis smg7 has been described in Drosophila deficient for the APC coactivator cortex (Page and Orr-Weaver, 1996). Both roughex and cortex appear to promote meiotic exit via downregulation of CDK-cyclin A activity, and the roughex mutation was suppressed by lowering the level of cyclin A (Gönczy et al., 1994; Swan et al., 2005). In an attempt to rescue the smg7 and tdm1 phenotypes, we sought to inactivate a meiotic cyclin and downregulate meiotic CDK. TAM (cyclin A1;2) appeared to be the obvious candidate, as its function is important for meiotic entry as well as for progression into the second meiosis (Wang et al., 2004b, 2010; d’Erfurth et al., 2010; this study). However, we found that neither the hypomorphic tam-1 nor the null tam-2 alleles alleviate the anaphase II arrest in smg7 mutants or the reentry into the third meiosis in tdm1 plants. By contrast, smg7 and tdm1 mutations completely abolished the premature meiotic exit in tam-2 plants, and meiosis in double mutants normally progressed through the second division. These data argue that TAM is not the core component of the oscillator that drives meiotic divisions, but rather that it plays a regulatory role. Further support for this notion was obtained from analysis of TAM localization. TAM is meiosis I–specific cyclin that does not appear to be expressed in meiosis II. Its level peaks in prophase of meiosis I, but the Thr-161 phosphorylation of CDKA;1 reaches a maximum in metaphase. This implies that other cyclins associate with CDKA;1 and drive progression through meiosis. Studies in animals established that mitotic M-phase depends mainly on B-type cyclins whose activity peaks in metaphase before a sharp decline caused by APCCdc20-mediated destruction during anaphase. The role of A-type cyclins in mitosis is less clear (Yam et al., 2002; Fung and Poon, 2005). Their destruction in animal cells is already initiated in mitotic prophase (van Zon and Wolthuis, 2010), and our immunodetection of TAM together with data from systematic localization of core cell cycle component in tobacco cells indicate that this also applies to plant A-type cyclins (Boruc et al., 2010). A-type cyclins appear to promote the onset of M-phase through activation of CDK-cyclin B complexes (Fung et al., 2007; Deibler and Kirschner, 2010).
To integrate our data with current knowledge on mitotic and meiotic progression, we propose a model that outlines a conceptual frame for regulation of meiosis in Arabidopsis pollen mother cells (Figure 8). As discussed above, our data suggest the existence of a so far unknown CDK-cyclin complex(es) that forms the core meiotic oscillator corresponding to the animal CDK-cyclin B. CDKA;1 is a likely component of this complex, although a role for other Arabidopsis CDKs cannot be excluded. Identity of the cyclins is still enigmatic. In silico analysis of the Arabidopsis genome predicted at least 21 A- and B-type cyclins (Wang et al., 2004a), but the majority of them have not been functionally characterized in meiosis. In fact, several cyclins may act in meiosis in a partially redundant manner, as is observed in other organisms. We further suggest that SMG7 and TDM1 are regulators that inhibit the major meiotic CDK activity, and their function is particularly important at meiotic exit to allow transition to mitotic G1 (Figure 8A). The role of TAM appears to be more complex. TAM is important for entry into the first and second meiotic divisions (Wang et al., 2004b, 2010; d’Erfurth et al., 2010), but its expression seems to be restricted to meiosis I. We suggest that TAM promotes the activities of the core CDK complexes that drive entry into the second meiosis by antagonizing the inhibitory effects of SMG7 and TDM1 (Figure 8A). Complete loss of TAM, together with the inhibitory functions of SMG7 and TDM1, brings CDK levels under a threshold that permits exit after the first meiotic division (Figure 8B). Alleviation of this inhibition in smg7 or tdm1 mutants allows normal interkinesis and entry into the second meiosis. An unexpected function of TAM was revealed by analysis of the tam-1 allele. While lower activity of the tam-1 allele causes delay in interkinesis expected from its weaker activation function (Figure 8C), the absence of interkinesis in smg7 tam-1 double mutants suggests a very rapid progression from the first to the second meiotic division (Figure 8D). Furthermore, lack of fully decondensed nuclei after the first meiosis indicates a higher residual level of CDK activity in comparison to wild-type interkinesis. Based on these observations, we postulate that besides carrying an activation function, TAM also has an inhibitory role in regulating meiotic CDK. Support for this hypothesis was obtained in Xenopus egg extracts where downregulation of cyclin A led to the earlier appearance of CDK-cyclin B activity and premature entry into mitosis (Walker and Maller, 1991). In the opposite experiment, cyclin A overexpression in HeLa cells was reported to cause a cell cycle delay in prometaphase, presumably by affecting the stability of cyclin B (den Elzen and Pines, 2001). The inhibitory function of TAM in CDK regulation may require higher TAM activity than the activating function. Such bimodal regulation can be implemented by phosphorylation of two substrates with different affinities to CDK-TAM complexes, one being an activator, while the other an inhibitor of the hypothetical meiotic CDK. We suggest that the hypomorphic tam-1 allele lacks the inhibitory activity, but in contrast with the null tam-2 allele, still retains some activating function, as inferred from the capability of tam-1 mutants to enter the second meiosis. Absence of SMG7-mediated inhibition in combination with the tam-1 allele could result in high CDK activity in interkinesis and rapid transition from meiosis I to meiosis II (Figure 8D). Thus, the key role of TAM may be fine-tuning of the total CDK activity to assure proper transition between meiotic divisions.
Figure 8.
Models of CDK Regulation in Male Meiosis in the Wild Type and in tam-2, tam-1, and smg7 tam-1 Mutants.
The wild type (A) and tam-2 (B), tam-1 (C), and smg7 tam-1 (D) mutants. The red line illustrates activity of the hypothetical CDK-cyclin kinase(s) in the course of meiosis. Green line depicts activity of TAM as inferred from TAM:GUS localization data. Stages of meioses and their approximate durations are indicated on the x axis (P1, prophase I; M1, metaphase I; A1, anaphase I; I/P2, interkinesis/prophase II; M2, metaphase II; A2, anaphase II). CDK activities and durations of meiotic stages reflect only hypothetical values and are not based on any exact measurements.
The key prediction of the model outlined in Figure 8 is the existence of a CDK oscillator that drives meiotic divisions in plants. We expect that identification of this oscillator will be essential for a full understanding of meiotic progression. This study also hints at the richness and complexity of regulatory mechanisms that govern meiotic regulation in plants. Further exploration of meiosis will undoubtedly bring novel mechanistic insights relevant not only to plant reproduction but also to a general understanding of M-phase, which is a rather poorly studied part of the plant cell cycle.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type (ecotype Columbia [Col-0]) and mutant plants were grown in soil at 21°C at 50 to 60% humidity under long-day (16 h light/8 h dark) conditions. Arabidopsis plants with the following mutations were used in this study: smg7-1 (Riehs et al., 2008), tdm1-1 also described as ms5-2 (Ross et al., 1997; Glover et al., 1998), tam-1 (Wang et al., 2004b), and tam-2 (SAIL_505_C06). Mutants carrying the smg7-1 allele were germinated and grown for the first 3 weeks at increased humidity (85 to 95%). The temperature-sensitive tam-1 allele was induced by keeping plants at 28°C during flowering.
PCR Genotyping
The tam-2 allele was genotyped using primers cycA1,2-1 (5′-ATGGAAAGAACTCAG-TCAAGCATCA-3′), cycA_r_nonsepec (5′-GAACAGAAGACTCCATCTCCAA-3′), and SAIL-LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). The single nucleotide mutation in the tam-1 allele was genotyped by two sets of PCR: the wild-type allele was amplified with primers cycA1,2-3 (5′-ATGGAGCGTATCGAAGCATAGCGT-3′) and cycA1,2-W (5′-GCGCGTGGAGGATTTCTGTTACATAAC-3′), while the tam-1 allele was identified with primers cycA1,2-3 and cycA1,2-M (5′-GCGCGTGGAGGATTTCTGTTACATAAT-3′). Each PCR reaction also included primers M1 and M3 (Puizina et al., 2004) as an internal amplification control. Plants carrying the smg7-1 and tdm1-1/ms5-2 alleles were genotyped as previously described (Glover et al., 1998; Riehs et al., 2008).
RT-PCR Analysis of TAM Expression
Total RNA was isolated from flowers with TriReagent solution (Sigma-Aldrich). One microgram of total RNA was treated with DNaseI (Fermentas) at 37°C for 30 min and after inactivation of DNase, reverse transcribed with RevertAid H Minus first-strand cDNA synthesis kit (Fermentas). The following primers were used for PCR: full-length TAM cDNA was amplified using primers 1 (5′-AACCCTAAATCTCACCGGAAAAC-3′) and 2 (5′-GAGGAAAAGCTCTTGCGGTA-3′), the region spanning the T-DNA border was amplified with primers 3 (5′-ATGGAAAGAACTCAGTCAAGCATCA-3′) and 4 (5′-GAACAGAAGACTCC-ATCTCCAA-3′), and the region downstream of the T-DNA insertion was amplified with primers 2 and 5 (5′-TGTTACCTGCATGATGATAGCA-3′). All PCR reactions were performed with GoTaq DNA Polymerase (Promega) using 32 cycles. Control PCR reactions were performed on ACTIN2 gene (At5g0980) with Actin 2-1 (5′-CTGCCGCTGTTGTTTCTCCT-3′) and Actin 2-2 (5′-CGTTGTAGAAAGTGTGATGCCA-3′) primers (24 cycles).
Flow Cytometry
Inflorescences of 4- to 5-week-old plants were chopped with a sharp razor blade in 250 μL CyStain UV Precise P nuclei extraction buffer and mixed with 750 μL of CyStain UV Precise P staining buffer (both Partec) containing propidium iodide. Released nuclei were purified by sieving through CellTrics filter columns. Fluorescence of the nuclei was measured by CyFlow space (Partec)
Generation of TAM:GUS Transgenic Lines
The 3974-bp TAM genomic region including 2 kb of a putative promoter was PCR amplified from Arabidopsis Col-0 genomic DNA using primers CycA1,2-F1 (5′-CACCATGACGAGAGGGGAGAGTGAT-3′) and CycA1,2-R1 (5′-GAGGAAAAGCTCTTGCGGTA-3′). The PCR product was cloned by directional TOPO cloning into the pENTR/D-TOPO vector (Invitrogen) and verified by sequencing. The insert was subcloned to the pMDC163 vector (Curtis and Grossniklaus, 2003) with Gateway LR Clonase enzyme mix (Invitrogen), which led to the TAM:GUS in-frame fusion construct. Arabidopsis Col-0 plants were transformed by the floral dip method. Transformed plants were selected on Grodan supplemented with 5 mg/L hygromycin (Calbiochem) according to Hadi et al. (2002). Seventeen independent T2 lines were analyzed by histochemical GUS assay (Jefferson et al., 1987); three representative lines were further used for immunocytology.
Protein Extraction and Phosphatase Treatment
Total proteins from wild-type and CDKA;1:YFP plants were extracted according to Peck (2006). Plant tissues were crushed in liquid nitrogen and mixed with extraction buffer (50 mM HEPES-KOH, pH 7.5, 5% glycerol, 50 mM sodium pyrophosphate, 1 mM sodium molybdate, 25 mM sodium fluoride, 10 mM EDTA, 0.5% polyvinylpyrrolidone, 1% Triton X-100, and 150 mM NaCl) supplemented with Complete Protease Inhibitor Cocktail Tablets (Roche) and with PhosSTOP Phosphatase Inhibitor Cocktail Tablets (Roche). Extracts were centrifuged for 10 min at 16,000 rcf at 4°C, and the protein-containing supernatant was transferred to a new tube. For phosphatase treatment, 200 to 400 μg of total protein was incubated with 1200 units of Lambda Protein Phosphatase (New England Biolabs) for 0 or 60 min at 30°C. The reaction was stopped by addition of 2× SDS loading buffer (100 mM Tris-Cl, pH 6.8, 20% glycerol, 0.01% bromphenol blue, 10% β-mercaptoethanol, and 5% SDS) and by boiling for 5 min at 95°C.
Immunoblot Analysis
Fifty micrograms of total proteins was separated in a 10% polyacrylamide gel and transferred to polyvinylidine fluoride transfer membrane (Thermo Scientific). Membranes were blocked in 1% milk in TBS-T for 1 h at room temperature. To detect total CDKA;1, membranes were incubated with the α-PSTAIR antibody (Sigma-Aldrich) diluted to 1:5000 in 1% milk in TBS-T buffer overnight at 4°C. KU70 was detected with α-AtKU70 antibody (B. Zellinger and K. Riha, unpublished data) diluted to 1:30,000 in 1% milk in TBS-T. For analysis of CDKA;1 phosphorylation at Thr-161, membranes were blocked in 5-10% BSA in 1× TBS-T for 1 h at room temperature. α-Phospho-CDK2(Thr160) antibody (Cell Signaling Technology) was diluted 1:1000 in blocking solution and incubated with membranes overnight at 4°C. In competition experiments, blocking peptides GIPVRTFTHEVVTLW or GIPVRTFT(PO3H2)HEVVTLW were added in concentrations 1, 10, or 100 μg/mL to the α-Phospho-CDK2(Thr160) antibody. Secondary α-mouse IgG + IgM, (H+L) antibody conjugated to HRP (Pierce) was used in a 1:50,000 dilution to detect α-PSTAIR antibody, while the anti-rabbit IgG, (H+L) antibody conjugated to HRP (Pierce) was used in a 1:100,000 dilution for detection of α-KU70 and α-phospho-CDK2(Thr160) antibodies. After immunolabeling, membranes were washed 3 × 10 min with TBS-T and incubated with the ECL Western Blotting Substrate (Pierce) for 5 min and then exposed to Hyperfilm ECL (Amersham).
Immunocytology
Preparation of meiotic spreads from PMCs and protein immunodetection was performed as previously described (Peirson et al., 1997; Riehs et al., 2008). The spindle was visualized using a rat anti-α-tubulin antibody (dilution 1:50; Serotec) and a Cy3-conjugated α-rat IgG antibody (1:500; Chemicon). TAM:GUS fusion protein was detected by primary rabbit α-GUS antibody (1:2000; Molecular Probes) and a fluorescein isothiocyanate–conjugated α-rabbit IgG antibody (1:200; Sigma-Aldrich). CDKA;1 phosphorylated at Thr-161 was visualized using α-phospho-CDK2(Thr160) antibody (1:50; Cell Signaling Technology) in the presence of GIPVRTFTHEVVTLW peptide (100 μg/mL) and a Cy3-conjugated α-rabbit IgG (H+L) antibody (1:2000; Jackson ImmunoResearch). DNA was counterstained with 4',6-diamidino-2-phenylindole (DAPI). Stained meiocytes were examined by epifluorescence microscopy using a Zeiss Axioscope fluorescence microscope equipped with a cooled CCD camera (Visitron). Pictures were acquired and analyzed using MetaVue software (Universal Imaging Corporation). For relative assessment of signal intensity of TAM:GUS and CDKA;1 Thr-161 phosphorylation, pictures from the same slide taken at equal exposure times (500 ms) were compared. Signal intensity in smg7 PMCs were compared with wild-type PMCs by squashing anthers from smg7-1 mutants with anthers from CDKA;1:YFP plants on the same slide. Meiocytes from smg7-1 and CDK:YFP plants were distinguished by scoring for the YFP signal. Alexander staining for pollen viability was performed as described (Alexander, 1969).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CDKA;1 (locus At3g48750), SMG7 (locus At5g19400), TAM (locus At1g77390), and TDM1 (locus At4g20900).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunolocalization of Thr-161 Phosphorylated CDKA;1 in Mitotic Cells from Flowers of CDKA;1:YFP Plants.
Supplemental Figure 2. Immunolocalization of Thr-161 Phosphorylated CDKA;1 in a Tapetum Cell from the CDKA;1:YFP Plant.
Supplemental Figure 3. Molecular Characterization of the tam-2 Allele.
Supplemental Figure 4. Expression of TAM:GUS Fusion Protein.
Acknowledgments
We thank Matt Watson for helpful comments on the manuscript. We also thank Susan Armstrong for providing tdm1-1 mutants and Ming Yang for tam-1 mutants. This work was supported by the Austrian Science Fund (Grant P19256-B03 to K.R.), by the Austrian Academy of Sciences, and by an Action Thématique et Incitative sur Programme Jeunes Chercheurs grant from the Centre National de la Recherche Scientifique and a European Research Council starting grant from the European Union to A.S.
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Averbeck N., Sunder S., Sample N., Wise J.A., Leatherwood J. (2005). Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol. Cell 18: 491–498 [DOI] [PubMed] [Google Scholar]
- Azumi Y., Liu D., Zhao D., Li W., Wang G., Hu Y., Ma H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21: 3081–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Mylle E., Duda M., De Clercq R., Rombauts S., Geelen D., Hilson P., Inzé D., Van Damme D., Russinova E. (2010). Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol. 152: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T.M., Amon A. (2008). Meiosis I is established through division-specific translational control of a cyclin. Cell 133: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gramont A., Cohen-Fix O. (2005). The many phases of anaphase. Trends Biochem. Sci. 30: 559–568 [DOI] [PubMed] [Google Scholar]
- Deibler R.W., Kirschner M.W. (2010). Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol. Cell 37: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen N., Pines J. (2001). Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 153: 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Cromer L., Jolivet S., Girard C., Horlow C., Sun Y., To J.P., Berchowitz L.E., Copenhaver G.P., Mercier R. (2010). The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 6: e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., Mercier R. (2009). Turning meiosis into mitosis. PLoS Biol. 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., Simon M., Jenczewski E., Mercier R. (2008). Mutations in AtPS1 (Arabidopsis thaliana parallel spindle 1) lead to the production of diploid pollen grains. PLoS Genet. 4: e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A.E., Park J.S., Inoue I., Tachikawa H., Neiman A.M. (2009). The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 20: 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N., Nowack M.K., Pusch S., Stals H., Inzé D., Grini P.E., Schnittger A. (2007). T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E., O’Farrell P.H., Sprenger F. (1999). Rux is a cyclin-dependent kinase inhibitor (CKI) specific for mitotic cyclin-Cdk complexes. Curr. Biol. 9: 1392–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N., Ebert J., Unterholzner L., Lindner D., Izaurralde E., Conti E. (2005). SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Fung T.K., Ma H.T., Poon R.Y. (2007). Specialized roles of the two mitotic cyclins in somatic cells: cyclin A as an activator of M phase-promoting factor. Mol. Biol. Cell 18: 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.K., Poon R.Y. (2005). A roller coaster ride with the mitotic cyclins. Semin. Cell Dev. Biol. 16: 335–342 [DOI] [PubMed] [Google Scholar]
- Glover J., Grelon M., Craig S., Chaudhury A., Dennis E. (1998). Cloning and characterization of MS5 from Arabidopsis: A gene critical in male meiosis. Plant J. 15: 345–356 [DOI] [PubMed] [Google Scholar]
- Gönczy P., Thomas B.J., DiNardo S. (1994). roughex is a dose-dependent regulator of the second meiotic division during Drosophila spermatogenesis. Cell 77: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Gould K.L., Moreno S., Owen D.J., Sazer S., Nurse P. (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10: 3297–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi M.Z., Kemper E., Wendeler E., Reiss B. (2002). Simple and versatile selection of Arabidopsis transformants. Plant Cell Rep. 21: 130–135 [Google Scholar]
- Hamant O., Ma H., Cande W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57: 267–302 [DOI] [PubMed] [Google Scholar]
- Harashima H., Shinmyo A., Sekine M. (2007). Phosphorylation of threonine 161 in plant cyclin-dependent kinase A is required for cell division by activation of its associated kinase. Plant J. 52: 435–448 [DOI] [PubMed] [Google Scholar]
- Holloway S.L., Glotzer M., King R.W., Murray A.W. (1993). Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 73: 1393–1402 [DOI] [PubMed] [Google Scholar]
- Irniger S. (2006). Preventing fatal destruction: inhibitors of the anaphase-promoting complex in meiosis. Cell Cycle 5: 405–415 [DOI] [PubMed] [Google Scholar]
- Iwabuchi M., Ohsumi K., Yamamoto T.M., Sawada W., Kishimoto T. (2000). Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M-M transition in Xenopus oocyte extracts. EMBO J. 19: 4513–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Shinmyo A., Sekine M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45: 819–831 [DOI] [PubMed] [Google Scholar]
- Izawa D., Goto M., Yamashita A., Yamano H., Yamamoto M. (2005). Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434: 529–533 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.H., Armstrong S.J., Caryl A.P., Franklin F.C. (2003). Meiotic chromosome synapsis and recombination in Arabidopsis thaliana; an integration of cytological and molecular approaches. Chromosome Res. 11: 205–215 [DOI] [PubMed] [Google Scholar]
- Liu Z., Makaroff C.A. (2006). Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Magnard J.L., Yang M., Chen Y.C., Leary M., McCormick S. (2001). The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis. Plant Physiol. 127: 1157–1166 [PMC free article] [PubMed] [Google Scholar]
- Malapeira J., Moldón A., Hidalgo E., Smith G.R., Nurse P., Ayté J. (2005). A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol. Cell. Biol. 25: 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A.L., Amon A. (2004). Meiosis: Cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5: 983–997 [DOI] [PubMed] [Google Scholar]
- McPheeters D.S., Cremona N., Sunder S., Chen H.M., Averbeck N., Leatherwood J., Wise J.A. (2009). A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat. Struct. Mol. Biol. 16: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Grelon M. (2008). Meiosis in plants: Ten years of gene discovery. Cytogenet. Genome Res. 120: 281–290 [DOI] [PubMed] [Google Scholar]
- Moldón A., Malapeira J., Gabrielli N., Gogol M., Gómez-Escoda B., Ivanova T., Seidel C., Ayté J. (2008). Promoter-driven splicing regulation in fission yeast. Nature 455: 997–1000 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Page A.W., Orr-Weaver T.L. (1996). The Drosophila genes grauzone and cortex are necessary for proper female meiosis. J. Cell Sci. 109: 1707–1715 [DOI] [PubMed] [Google Scholar]
- Parry D.H., O’Farrell P.H. (2001). The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr. Biol. 11: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D., Dunphy W.G. (1998). Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes Dev. 12: 2549–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck S.C. (2006). Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J. 45: 512–522 [DOI] [PubMed] [Google Scholar]
- Peirson B.N., Bowling S.E., Makaroff C.A. (1997). A defect in synapsis causes male sterility in a T-DNA-tagged Arabidopsis thaliana mutant. Plant J. 11: 659–669 [DOI] [PubMed] [Google Scholar]
- Perez-Hidalgo L., Moreno S., Martin-Castellanos C. (2007). Modified cell cycle regulation in meiosis. Genome Dyn. Stab. 2: 307–353 [Google Scholar]
- Petronczki M., Siomos M.F., Nasmyth K. (2003). Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell 112: 423–440 [DOI] [PubMed] [Google Scholar]
- Puizina J., Siroky J., Mokros P., Schweizer D., Riha K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehs N., Akimcheva S., Puizina J., Bulankova P., Idol R.A., Siroky J., Schleiffer A., Schweizer D., Shippen D.E., Riha K. (2008). Arabidopsis SMG7 protein is required for exit from meiosis. J. Cell Sci. 121: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz P., Armstrong S.J., Vizir I., Mulligan B., Franklin F.C., Jones G.H. (1997). Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res. 5: 551–559 [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Sundaresan V., Alandete-Saez M. (2010). Pattern formation in miniature: The female gametophyte of flowering plants. Development 137: 179–189 [DOI] [PubMed] [Google Scholar]
- Swan A., Barcelo G., Schupbach T. (2005). Drosophila Cks30A interacts with Cdk1 to target Cyclin A for destruction in the female germline. Development 132: 3669–3678 [DOI] [PubMed] [Google Scholar]
- van Zon W., Wolthuis R.M. (2010). Cyclin A and Nek2A: APC/C-Cdc20 substrates invisible to the mitotic spindle checkpoint. Biochem. Soc. Trans. 38: 72–77 [DOI] [PubMed] [Google Scholar]
- Vardy L., Pesin J.A., Orr-Weaver T.L. (2009). Regulation of Cyclin A protein in meiosis and early embryogenesis. Proc. Natl. Acad. Sci. USA 106: 1838–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.H., Maller J.L. (1991). Role for cyclin A in the dependence of mitosis on completion of DNA replication. Nature 354: 314–317 [DOI] [PubMed] [Google Scholar]
- Wang G., Kong H., Sun Y., Zhang X., Zhang W., Altman N., DePamphilis C.W., Ma H. (2004a). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135: 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jha A.K., Chen R., Doonan J.H., Yang M. (2010). Polyploidy-associated genomic instability in Arabidopsis thaliana. Genesis 48: 254–263 [DOI] [PubMed] [Google Scholar]
- Wang Y., Magnard J.L., McCormick S., Yang M. (2004b). Progression through meiosis I and meiosis II in Arabidopsis anthers is regulated by an A-type cyclin predominately expressed in prophase I. Plant Physiol. 136: 4127–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F., Wandke C., Isenberg N., Geley S. (2006). Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 25: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth D.J., Roberts S.S. (2010). Regulating mitosis and meiosis in the male germ line: Critical functions for cyclins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1653–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam C.H., Fung T.K., Poon R.Y. (2002). Cyclin A in cell cycle control and cancer. Cell. Mol. Life Sci. 59: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]