Fimbrins are major regulators of actin dynamics and organization. This article reports that Arabidopsis FIMBRIN5 (FIM5) mediates actin bundling and is required for proper spatial organization of actin filaments in pollen. Through these activities, FIM5 regulates pollen germination and pollen tube growth.
Abstract
Actin cables in pollen tubes serve as molecular tracks for cytoplasmic streaming and organelle movement and are formed by actin bundling factors like villins and fimbrins. However, the precise mechanisms by which actin cables are generated and maintained remain largely unknown. Fimbrins comprise a family of five members in Arabidopsis thaliana. Here, we characterized a fimbrin isoform, Arabidopsis FIMBRIN5 (FIM5). Our results show that FIM5 is required for the organization of actin cytoskeleton in pollen grains and pollen tubes, and FIM5 loss-of-function associates with a delay of pollen germination and inhibition of pollen tube growth. FIM5 decorates actin filaments throughout pollen grains and tubes. Actin filaments become redistributed in fim5 pollen grains and disorganized in fim5 pollen tubes. Specifically, actin cables protrude into the extreme tips, and their longitudinal arrangement is disrupted in the shank of fim5 pollen tubes. Consequently, the pattern and velocity of cytoplasmic streaming were altered in fim5 pollen tubes. Additionally, loss of FIM5 function rendered pollen germination and tube growth hypersensitive to the actin-depolymerizing drug latrunculin B. In vitro biochemical analyses indicated that FIM5 exhibits actin bundling activity and stabilizes actin filaments. Thus, we propose that FIM5 regulates actin dynamics and organization during pollen germination and tube growth via stabilizing actin filaments and organizing them into higher-order structures.
INTRODUCTION
In plants, the pollen tube extends rapidly via tip growth, thereby allowing the movement of two nonmotile sperm cells through the pistil for the purpose of double fertilization. Pharmacological disruption of actin and genetic manipulation of the actin regulatory system have revealed that the actin cytoskeleton is a major mediator of pollen tube growth (Gibbon et al., 1999; Chen et al., 2002; Cheung and Wu, 2004; Xiang et al., 2007). Furthermore, imaging of actin filaments has shown that distinct actin structures form in pollen tubes across different species. The consensus view is that actin filaments can be divided into at least three distinct structures in pollen tubes: longitudinal actin cables found in the shanks of pollen tubes; dense actin structures, such as collar-like structures or actin fringes, found at the subapexes; and highly dynamic, but less abundant, actin filaments found at the extreme tips (Kost et al., 1998; Fu et al., 2001; Lovy-Wheeler et al., 2005; Cheung et al., 2008; Chen et al., 2009; Vidali et al., 2009; Staiger et al., 2010). These distinct actin structures are believed to perform distinct functions. In particular, actin cables are believed to provide molecular tracks necessary for the intracellular trafficking events that support rapid tube extension. However, the mechanisms by which actin cables are generated and maintained remain largely unknown.
Actin cables are also present in other types of plant cells. It is generally accepted that actin cables are important for numerous cellular processes (Higaki et al., 2007; Thomas et al., 2009). In addition to functioning as tracks for cytoplasmic streaming and organelle movement (Tominaga et al., 2000a; Shimmen, 2007), actin cables are also important for positioning of organelles, such as nuclei (Ketelaar et al., 2002), and maintenance of cellular architecture (Tominaga et al., 2000a). In addition, actin cables have also been implicated in the response to biotic stress (Lipka and Panstruga, 2005). Several classes of actin binding proteins have been shown to play important roles in the formation of actin cables in plants, including fimbrins, LIM proteins, certain formins, and villins (Higaki et al., 2007; Thomas et al., 2009). However, more direct genetic and biochemical evidence is urgently needed to define the precise role of each bundling factor in the formation and maintenance of actin cables. Our recent work has shown that AFH3 is a major actin nucleator that is responsible for de novo actin nucleation events that regulate the formation of actin cables in the pollen tube (Ye et al., 2009). We proposed that actin bundling factors function in concert with AFH3 to organize AFH3-generated actin filaments into high-order structures (Ye et al., 2009).
Fimbrins, which were originally identified from the protein extracts of chicken intestinal microvilli (Bretscher and Weber, 1980), are well-characterized actin bundling proteins. Fimbrin/plastin family members all contain two highly conserved actin binding domains (ABDs). Each ABD contains two calponin-homology domains. The presence of two ABDs on the same polypeptide enables a single molecule to bundle or cross-link two adjacent actin filaments. The crystal structures of the core ABD domains of Arabidopsis thaliana and Schizosaccharomyces pombe fimbrin (fim1+) have been elucidated, and these structures reveal that the two ABDs are aligned in an approximately antiparallel fashion (Klein et al., 2004). The recently solved cryo-electron microscopy structure of the F-actin/L-plastin complex led the authors to propose that the two ABDs bind consecutively to two adjacent actin filaments. Specifically, binding of ABD2 to an actin filament subsequently activates ABD1 to bind an adjacent actin filament (Galkin et al., 2008).
Fimbrins, also known as plastins, play important roles in various actin-based processes across different species. For example, T-plastin has been shown to localize to the core actin bundles of microvilli and microspikes in epithelial cells (Bretscher and Weber, 1980; Daudet and Lebart, 2002), where it participates in assembly of these structures (Arpin et al., 1994). The fimbrin homolog in budding yeast, Sac6p, localizes to actin bundles and cortical actin patches (Drubin et al., 1988). Mutations in SAC6 affect the distribution and stability of actin cables, as well as actin patches, consequently affecting morphogenesis and endocytosis (Adams et al., 1991; Kübler and Riezman, 1993; Brower et al., 1995; Karpova et al., 1995; Young et al., 2004). In vitro experiments have revealed that recombinant Sac6p stabilizes actin filaments, preventing depolymerization (Goode et al., 1999). These results support an in vivo role for Sac6p in maintaining actin cables and actin patches. Disruption of fimbrin in Aspergillus nidulans results in delay of the establishment of cell polarity during condium germination and induces abnormal hyphal growth (Upadhyay and Shaw, 2008), suggesting that this protein plays a role in regulation of polarized cell growth. In addition, fimbrin has been shown to localize to the cortical region of the cleavage furrow in Tetrahymena pyriformis and to the cortical actin patches and the medial ring in the fission yeast S. pombe, suggesting a possible role for fimbrin in cytokinesis (Wu et al., 2001; Shirayama and Numata, 2003; Wu and Pollard, 2005). Fission yeast fim1+ organizes actin filaments into bundles in vitro, and loss-of-function of S. pombe fim1+ increases the sensitivity of actin filaments to latrunculin A treatment (Nakano et al., 2001), suggesting that fim1+ is involved in stabilization of actin filaments in cytokinesis. Interestingly, mutation of fimbrin in Dictyostelium does not affect growth, endocytosis, exocytosis, or chemotaxis but causes a small cell phenotype (Pikzack et al., 2005).
Five fimbrin-like genes are present in the Arabidopsis genome, termed FIM1-5 (Staiger and Hussey, 2004). FIM1 has been shown to bind and bundle actin filaments and stabilize them from profilin-mediated actin depolymerization in vitro and in vivo (Kovar et al., 2000). Compared with nonplant fimbrins, which have two calmodulin-like calcium binding domains in their N-terminal regions (de Arruda et al., 1990), Arabidopsis fimbrins have poorly conserved calmodulin-like calcium binding domains at their N terminus (McCurdy and Kim, 1998; Staiger and Hussey, 2004), implying that the activity of Arabidopsis fimbrins may not be regulated by calcium. Indeed, Arabidopsis FIM1 has been demonstrated to be a calcium-insensitive cross-linking factor (Kovar et al., 2000). Furthermore, in contrast with nonplant fimbrins, Arabidopsis fimbrins exhibit increased diversification of the C termini (McCurdy and Kim, 1998; Staiger and Hussey, 2004), suggesting that Arabidopsis fimbrins may perform specific functions or be differentially regulated. However, the cellular and developmental functions of fimbrins in plants remain uncharacterized.
In this study, we functionally characterized the Arabidopsis FIM5, which is expressed preferentially in pollen. FIM5 was found to decorate actin filaments in both pollen grains and pollen tubes. fim5 mutants exhibit disorganization of actin filaments in pollen grains and pollen tubes, resulting in a consequent delay in pollen germination and inhibition of pollen tube growth. Loss of FIM5 function also renders pollen germination and pollen tube growth hypersensitive to latrunculin B (LatB). Importantly, in vitro biochemical analyses indicated that FIM5 is a bona fide actin cross-linking factor that stabilizes actin filaments in vitro. Based on these observations, we propose that FIM5 is a major regulator of pollen germination and pollen tube growth through stabilization of actin filaments and organization of these filaments into higher-order actin structures.
RESULTS
FIM5 Contains Conserved ABDs
FIM5 initially caught our attention because it is expressed preferentially in mature pollen (Honys and Twell, 2003; Pina et al., 2005; see Supplemental Figure 1 online). We began by obtaining the FIM5 protein sequence (accession number At5g35700) from The Arabidopsis Information Resource database. Analysis of the protein sequence revealed that the protein consisted of 687 amino acids. Alignment of FIM5 with FIM1 and fimbrins from other species showed that FIM5 is relatively conserved and contains the core ABD. FIM5 was found to share 64.7, 33.3, and 30.5% protein sequence identity and 79.1, 49.2, and 49.4% protein sequence similarity with FIM1, Sac6p, and L-plastin, respectively (see Supplemental Figure 2 online). The presence of conserved actin binding sites in FIM5 (see Supplemental Figure 2 online) implies that FIM5 may play a role in conserved actin regulatory functions. Additionally, similar to other Arabidopsis fimbrins, FIM5 possesses an extended C-terminal region, which could be required for a specialized activity or a distinct means of regulation.
FIM5 Loss-of-Function Mutants Exhibit Defects in the Male Gametophyte
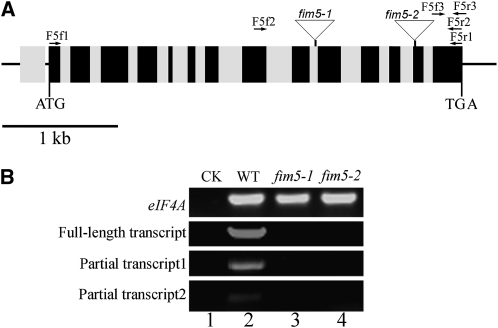
The biological function of FIM5 was investigated by characterizing two T-DNA insertion lines, termed fim5-1 and fim5-2, respectively (Figure 1A). Three independent pairs of primers were designed to assess the levels of FIM5 transcripts in fim5 mutants. No full-length FIM5 cDNA or partial cDNAs were detected in the fim5-1 and fim5-2 mutants (Figure 1B), indicating that both insertion lines are null alleles.
Figure 1.
T-DNA Insertions in the fim5-1 and fim5-2 Lines Generated Null fim5 Alleles.
(A) Structure of the FIM5 gene (At5g35700). A total of 14 exons and 14 introns are present in the gene. Introns, exons, and untranslated regions are defined by gray boxes, black boxes, and black lines, respectively. The two T-DNA insertion lines, CS856909 (designated as fim5-1) and CS853476 (designated as fim5-2), contained insertions in the 10th intron and 13th exon, respectively.
(B) RT-PCR results demonstrated that the fim5-1 and fim5-2 lines contain fim5 null alleles. Transcript expression levels in the insertion lines were determined by RT-PCR using three independent pairs of primers. eIF4A was used as an internal control. The first pair of primers (F5f1 and F5r1) was designed to amplify the FIM5 full-length cDNA, which was not present in fim5-1 and fim5-2 mutant plants. The second and third primer pairs, F5f2/F5r2 and F5f3/F5r3, were used to amplify partial transcript 1 and partial transcript 2, respectively. RT-PCR results showed that no partial FIM5 cDNAs were detected in fim5-1 and fim5-2 mutant plants. Lane 1, water (negative control; CK); lane 2, wild-type (WT) Col-0; lane 3, fim5-1; lane 4, fim5-2.
Given that FIM5 is expressed preferentially in pollen, it is reasonable to speculate that FIM5 deficiency may alter the function of pollen. To test this hypothesis, genetic segregation experiments were performed. Self-crossing of plants heterozygous for fim5 led to a distorted segregation ratio that differed significantly from the expected Mendelian ratio of 1:3 (Table 1; P < 0.001), suggesting a potential defect in male or/and female gametophyte transmission. To distinguish between these possibilities, reciprocal crosses were performed using plants heterozygous for fim5 as either the male or female parent. Use of heterozygous females in crosses with wild-type plants resulted in no significant deviation in the transmission efficiency of fim5 (Table 2; P = 0.896 for fim5-1 and P > 0.9 for fim5-2). However, when heterozygotes were used as male parents, strongly decreased transmission of fim5 was observed in the progeny (Table 2; P < 0.001), indicating that distorted segregation ratios were due to defects in male gametophytes. In summary, loss of FIM5 function results in defects in male gametophytic function but not female gametophytic function.
Table 1.
Segregation Analysis of the fim5 Mutant Allele in Self-Crosses of fim5 Heterozygous Plants
| Progeny Genotype |
||||
| Parental Genotype | FIM5/FIM5 | FIM5/fim5 and fim5/fim5 | Ratio | P Value |
| FIM5/fim5-1 | 248 | 332 | 1:1.34 | <0.001a |
| FIM5/fim5-2 | 168 | 248 | 1:1.48 | <0.001a |
fim5 heterozygous plants were allowed to self-cross, and the observed segregation ratios of the progeny were compared with the expected Medelian ratio 1:3. P values were calculated using the χ2 test.
Significant difference between the observed ratio and the expected ratio.
Table 2.
Transmission Efficiency Analysis Using Reciprocal Crosses
| Parental Genotype |
Progeny Genotype |
||||
| Female × Male | FIM5/fim5 | FIM5/FIM5 | TEFa | TEMa | P Value |
| FIM5/fim5-1 × wild type | 117 | 115 | 101.7% | NAb | 0.896 |
| Wild type × FIM5/fim5-1 | 49 | 373 | NAb | 13.1% | <0.001c |
| FIM5/fim5-2 × wild ype | 32 | 32 | 100% | NAb | >0.900 |
| Wild-type × FIM5/fim5-2 | 13 | 185 | NAb | 7% | <0.001c |
Reciprocal crosses were performed between fim5 heterozygous plants and wild-type plants. The transmission efficiency (TE) of gametes was calculated as folows: TE = number of progeny with the fim5 allele/number of wild-type progeny × 100%. The observed TE was compared with the expected TE for normal gametes (100%). P values were calculated using the χ2 test.
TEF, female transmission efficiency; TEM, male transmission efficiency.
NA, not applicable.
Significant difference between the observed TE and the expected TE.
Loss of FIM5 Function Leads to Delayed Pollen Germination, Inhibition of Pollen Tube Growth, and Pollen Tube Morphological Abnormalities
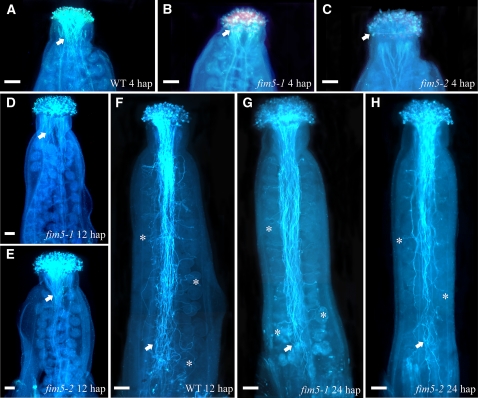
The aforementioned results suggested that male gametophytes in fim5 plants are abnormal. Therefore, to directly observe the behavior of male gametophytes of fim5 mutants, wild-type stigmas were pollinated with pollen derived from wild-type Columbia-0 (Col-0) and fim5 mutants. As shown in Figure 2, pollen tubes of wild-type Col-0 had penetrated through style and reached the top of transmitting tract 4 h after pollination (Figure 2A). By contrast, pollen tubes from fim5 mutants did not begin to penetrate into the style at 4 h after pollination (Figures 2B and 2C). By 12 h after pollination, wild-type Col-0 pollen tubes reached the bottom of the transmitting tract and entered the ovules (Figure 2F). By contrast, at the same time point, fim5 pollen tubes had just passed by the style and intruded the transmitting tract (Figures 2D and 2E). Further observation revealed that fim5 pollen tubes were capable of reaching the bottom of the transmitting tract and entering the ovules, as they did so by 24 h after pollination (Figures 2G and 2H). Together, data from these pollination experiments suggest that pollen germination on the stigma is delayed and growth of the pollen tube in the style and transmitting tract is inhibited in fim5 mutants.
Figure 2.
Loss of FIM5 Function Results in Delayed Pollen Germination and Inhibited Elongation of Pollen Tubes.
Pollen grains from wild-type Col-0 and fim5 mutants were used to pollinate wild-type stigma. Pollen tubes were visualized by aniline blue staining. Arrows indicate pollen tubes in transmitting tracts. Asterisks mark representative pollen tubes that invade the ovule. hap, hours after pollination. Bars = 100 μm.
(A) Pollen from wild-type (WT) Col-0 germinated for 4 h.
(B) Pollen from fim5-1 germinated for 4 h.
(C) Pollen from fim5-2 germinated for 4 h.
(D) Pollen from fim5-1 germinated for 12 h.
(E) Pollen from fim5-2 germinated for 12 h.
(F) Pollen from wild-type Col-0 germinated for 12 h.
(G) Pollen from fim5-1 germinated for 24 h.
(H) Pollen from fim5-2 germinated for 24 h.
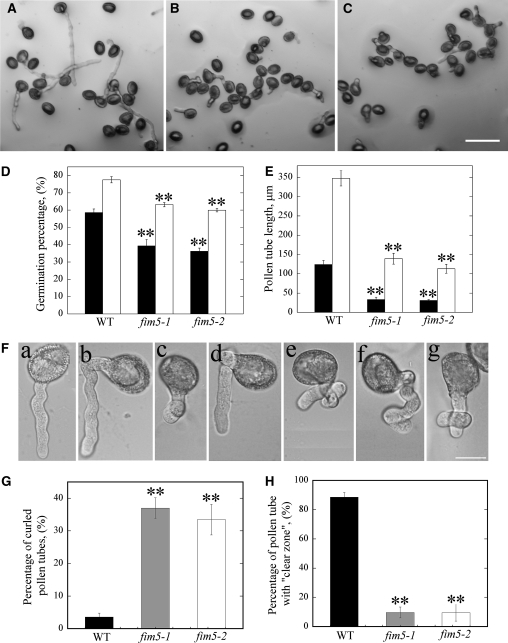
Behavior of pollen from wild-type and mutant plants was also assessed in vitro. Pollen derived from mutant plants exhibited a reduction in the pollen germination percentage (Figures 3B and 3C) in comparison to wild-type Col-0 pollen (Figure 3A). Quantification of these results revealed that the average germination percentage was significantly reduced in fim5 mutants (P < 0.01; Figure 3D). In addition to defects in pollen germination, the average pollen tube length was also reduced in fim5 mutants (P < 0.01; Figure 3E). Additionally, the frequency of abnormal pollen tubes was increased in fim5 mutants. A majority of the abnormal pollen tubes comprised curled pollen tubes (Figure 3F). The frequency of curled pollen tubes in fim5 mutants was significantly higher than that of wild-type Col-0 plants (P < 0.01; Figure 3G). Furthermore, the percentage of pollen tubes with clear zone, which is one of the characteristic features of an elongating pollen tube, is also reduced in the mutant (P < 0.01; Figure 3H). Loss of the clear zone may at least partially account for growth defects present in fim5 pollen tubes.
Figure 3.
Loss of FIM5 Function Decreases Pollen Germination, Inhibits Pollen Tube Growth, and Increases the Frequency of Morphologically Abnormal Pollen Tubes in Vitro.
Pollen grains from wild-type Col-0 and fim5 mutants were germinated on Arabidopsis pollen germination medium; the germination percentage, pollen tube length, and morphology were analyzed.
(A) Representative image of wild-type Col-0 pollen germinated for 3 h.
(B) Representative image of fim5-1 pollen germinated for 3 h.
(C) Representative image of fim5-2 pollen germinated for 3 h. Bar = 100 μm.
(D) Graph of the pollen germination percentage at 3 and 6 h. Black columns represent germination percentage of pollen at 3 h; white columns represent germination percentage of pollen at 6 h. Error bars represent ± se, **P < 0.01, by χ2 test. WT, wild type.
(E) Graph of pollen tube length at 3 and 6 h. Black columns represent lengths of pollen tubes at 3 h; white columns represent lengths of pollen tubes at 6 h. Error bars represent ± se, **P < 0.01, by Kruskal-Wallis test.
(F) Enlarged images of pollen tubes. (a) Normal pollen tube from wild-type Col-0; (b) to (g) different types of curled pollen tubes from fim5 mutants. Bar = 20 μm.
(G) Graph showing the frequency of curled pollen tubes. Black bar represents wild-type Col-0, gray bar represents fim5-1, and white bar represents fim5-2. Error bars represent ± sd, **P < 0.01, by χ2 test.
(H) Graph showing the percentage of pollen tubes with normal clear zones. Black bar represents wild-type Col-0, gray bar represents fim5-1, and white bar represents fim5-2. Error bars represent ± sd, **P < 0.01, by χ2 test.
Defects in Pollen Tube Growth Are Rescued by Complementation with FIM5-GFP
To determine whether defects in male gametophytes are indeed due to loss of function of FIM5, complementation experiments were performed by driving expression of FIM5-GFP (for green fluorescent protein) under control of the native FIM5 promoter in fim5 plants. As shown in Supplemental Figure 3A online, expression of FIM5 transcripts was restored in complemented lines, indicating that the fusion protein was expressed. To determine whether expression of FIM5-GFP rescues the phenotype of fim5 mutants, the in vitro pollen germination assay was performed, and pollen tube lengths were measured. Results from these analyses showed that the average pollen tube length of fim5 plants carrying the complementation construct was not significantly different from that of wild-type Col-0 plants (see Supplemental Figure 3B online), indicating that defects in male gametophytes are indeed caused by loss of FIM5 expression.
FIM5-GFP Decorates Actin Filaments in Pollen Grains and Pollen Tubes
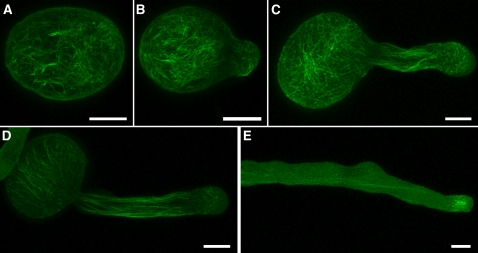
Results from complementation experiments indicated that the FIM5-GFP protein is functional, providing suitable means to characterize the subcellular localization of FIM5. As shown in Figure 4, FIM5-GFP decorated filamentous structures in both pollen grains and pollen tubes. The filamentous structures were distributed throughout the pollen grain (Figure 4A; see Supplemental Figures 4A and 4B online). In addition, the filamentous structures appeared in pollen tubes of different lengths (Figures 4B to 4E; see Supplemental Figures 4C to 4I online). FIM5-GFP decorated longitudinal filamentous structures in the shanks of pollen tubes (Figures 4C to 4E) that were reminiscent of actin cables previously reported to be present in Arabidopsis and lily (Lilium longiflorum) pollen tubes (Lovy-Wheeler et al., 2005; Ye et al., 2009). In addition, filamentous structures were also evident at the subapexes of pollen tubes (Figures 4C to 4E). However, fluorescence was less abundant at the extreme tips of pollen tubes (Figures 4C to 4E). The FIM5-GFP filamentous structures dispersed after treatment with LatB (see Supplemental Figure 5 online), suggesting that the filamentous structures are indeed actin filaments. This further confirmed, by visualizing actin filaments and FIM5-GFP simultaneously, that FIM5-GFP colocalizes with actin filaments in pollen grains and pollen tubes (see Supplemental Figure 6 online). Together, these results suggest that FIM5-GFP decorates actin filaments in both pollen grains and pollen tubes. This localization pattern implies that FIM5 may be an important regulator of actin organization and/or actin dynamics in pollen grains and pollen tubes.
Figure 4.
FIM5-GFP, Expressed under Control of the FIM5 Promoter, Decorates Actin Filaments in Pollen Grains and Pollen Tubes.
Representative images of complemented fim5 plants showing that FIM5-GFP decorates actin filaments in pollen grains (A) and in pollen tubes of differing lengths ([B] to [E]). About 50% of pollen tubes show accumulation of GFP signal at the subapical region, as shown in (E). The step size of optical section is 0.5 μm for pollen grains and 0.4 μm for pollen tubes. Maximal projection of confocal optical sections throughout the entire pollen grain or pollen tube was performed. See more examples in Supplemental Figure 4 online. Bars = 10 μm.
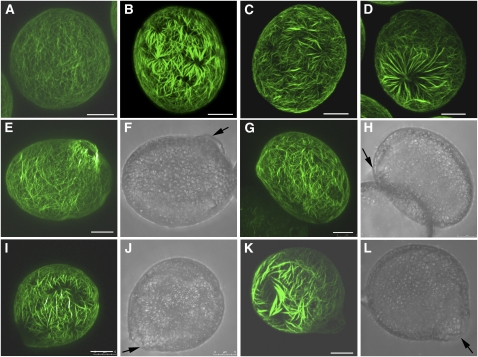
Thicker Actin Bundles Are Distributed Randomly and Irregularly in fim5 Pollen Grains
Given that FIM5-GFP decorates actin structures in pollen grains and pollen tubes, it is imperative to know whether loss of FIM5 function affects the organization of actin filaments in vivo. As shown in Figure 5, actin filaments were distributed uniformly throughout wild-type Col-0 pollen grains (Figure 5A). By contrast, redistribution of actin filaments occurred in fim5 pollen grains (Figures 5B to 5D). Specifically, thicker actin bundles were present, and these bundles were distributed randomly and irregularly in fim5 pollen grains (Figures 5B to 5D; see Supplemental Figure 7 online). The presence of thicker actin bundles in fim5 pollen was confirmed by analyzing the fluorescence pixel intensity of actin staining along the axis of pollen grains. Indeed, the widths of the fluorescence peaks were increased in fim5 pollen grains compared with those of wild-type Col-0 pollen grains (see Supplemental Figure 8D online). More thick actin bundles appeared in fim5 pollen when they were subjected to the fixation with paraformaldehyde followed by Alexa-488 phalloidin staining (see Supplemental Figure 9 online), suggesting that the occurrence of thick actin bundles in fim5 pollen is not because actin filaments in fim5 pollen are more sensitive to the cross-linking effect of 3-maleimidobenzoic acid N-hydroxysuccinimide ester.
Figure 5.
Actin Filaments become Redistributed and Form Thick Bundles in fim5-1 Pollen Grains and Nascent Germinating Pollen.
As the pattern of actin organization was similar in fim5-1 and fim5-2 pollen grains, only fim5-1 pollen grains are shown in this figure. Arrows in (F), (H), (J), and (L) indicate the germination aperture. Bars = 5 μm.
(A) Wild-type Col-0 pollen grains.
(B) to (D) fim5-1 pollen grains. The frequency of actin staining pattern shown in (D) is ~27%. See Supplemental Figure 7 online for more images of actin cytoskeleton redistribution in ungerminated fim5 mutant pollen grains.
(E) Germinating wild-type Col-0 pollen.
(F) Corresponding bright-field image of (E).
(G) Germinating fim5-1 pollen.
(H) Corresponding bright-field image of (G).
(I) Germinating fim5-1 pollen.
(J) Corresponding bright-field image of (I).
(K) Germinating fim5-1 pollen.
(L) Corresponding bright field image of (K).
To determine whether redistribution of actin filaments and presence of thick actin bundles were sustained throughout pollen germination in fim5 mutants, we visualized actin filaments in germinating pollen. As shown in Figure 5E, actin filaments were distributed uniformly throughout the pollen grains and converged at the germination aperture in germinating wild-type Col-0 pollen grains. However, more randomly distributed and irregular actin bundles were present in germinating fim5-1 pollen (Figures 5G, 5I, and 5K). Importantly, actin filaments were distributed uniformly in pollen grains from fim5 plants complemented with FIM5-GFP (see Supplemental Figures 10C and 10D online), indicating that changes in actin filament organization and morphology are indeed due to loss of FIM5 function. Together, these results indicate that loss of FIM5 function induces formation of thick, irregular actin bundles that are sustained throughout pollen germination.
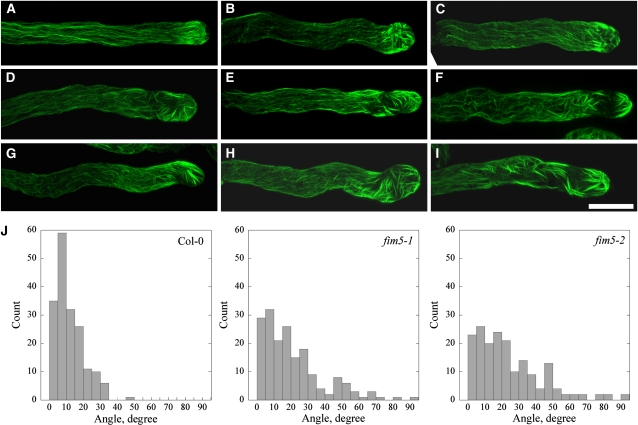
Actin Cables Are Disorganized and Protrude into the Tips of Pollen Tubes in fim5 Mutants
The distribution of the actin cytoskeleton in fim5 pollen tubes was also investigated. In wild-type Col-0 pollen tubes, actin filaments were present as distinct structures, dependent on their location within the tubes. Specifically, actin filaments were less abundant in the apical region of the pollen tube but formed a dense structure at the subapical region with numerous longitudinal actin cables extending throughout the shank (Figure 6A). By contrast, the overall distribution of the actin cytoskeleton was disrupted in fim5 pollen tubes, although the extent of disruption varied greatly. In some fim5-1 pollen tubes, longitudinal actin cables appeared relatively normal, but they invaded into the extreme tip, rather than terminating at the base of the tip of the pollen tube. The actin filaments at the extreme tips of the pollen tubes also assumed different organizations (Figures 6B and 6C). In other fim5-1 pollen tubes, besides actin cables protruding into the apical region, the dense structures normally present at the subapex were absent (Figures 6D and 6E). In a third group of fim5-1 pollen tubes, actin filaments were disorganized throughout the entire pollen tube (Figures 6F to 6I). For example, actin cables in these pollen tubes did not assume a strict longitudinal arrangement parallel to the growth axis, and actin filaments in the subapical and apical regions were also disorganized.
Figure 6.
Actin Cables Are Disorganized and Protrude into the Extreme Tips of fim5-1 Pollen Tubes.
Actin filaments were stained with Alexa-488 phalloidin. As the pattern of actin organization was similar in fim5-1 and fim5-2 pollen tubes, only fim5-1 pollen tubes are shown.
(A) Wild-type Col-0 pollen tube.
(B) to (I) Representative images of fim5-1 mutant pollen tubes showing different extents of actin filament disorganization.
(B) and (C) Actin cables in the shanks of the pollen tubes are relatively normal, but actin filaments protrude into the tips of pollen tubes and exhibit aberrant structures.
(D) and (E) Actin cables are relatively normal, but actin filaments in the subapical and apical regions of the pollen tubes are disorganized.
(F) to (I) Actin filaments are disorganized throughout the entire pollen tube. Bar = 10 μm.
(J) Loss of FIM5 function leads to an increase in the average angle between actin cables and the pollen tube elongation axis. Actin cables visible in the shanks of pollen tubes were selected, and the angles formed by each actin cable and the elongation axis were determined. A total of 180 actin cables were analyzed. As shown in the histogram, the majority of actin cables in wild-type Col-0 pollen tubes formed angles <20° with the elongation axis, and the angles were seldom larger than 35°. In fim5 pollen tubes, however, approximately half of the actin cables formed angles >20° with the elongation axis. Additionally, angles >45° were also frequently observed in the shanks of fim5 pollen tubes.
To quantify irregularities in the longitudinal arrangement of actin cables in the shanks of pollen tubes, we determined the angles formed between actin cables and the elongation axis of the pollen tube. These angles in wild-type Col pollen tubes were generally smaller than 20°, and no angles were determined to be larger than 50°. However, in the shanks of both fim5-1 and fim5-2 pollen tubes, the angles formed between actin cables and the elongation axes were substantially increased, with some angles greater than 60° (Figure 6J). In summary, these data suggest that FIM5 is required for proper organization of actin cables in pollen tubes
The Pattern and Velocity of Cytoplasmic Streaming Are Altered in fim5 Pollen Tubes
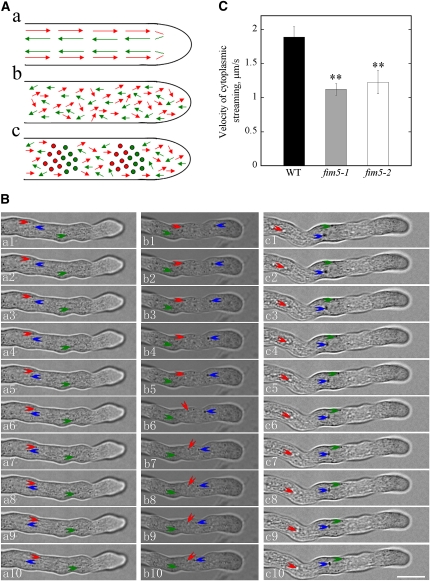
Actin filaments are believed to provide tracks for intracellular trafficking events, including cytoplasmic streaming, which was also defined as the coordinated flow of cytosol that carries small organelles (Avisar et al., 2008; Ueda et al., 2010). Therefore, the disorganization of actin filaments in fim5 pollen tubes suggests that cytoplasmic streaming could be affected in these mutants. As shown in the supplemental movies (see Supplemental Movies 1 and 2 online), cytoplasmic streaming occurred in a reverse fountain pattern in a large number of wild-type Col-0 pollen tubes. Specifically, cytosolic organelles moved to the tip of the pollen tube by traveling along the cortex (red arrows) and then reversed direction and moved back into the inner part of the pollen tube (green arrows) after reaching the subapex (Figure 7A, a).
Figure 7.
The Pattern and Velocity of Cytoplasmic Streaming Were Altered in fim5 Pollen Tubes.
(A) Schematic representations of cytoplasmic streaming patterns in wild-type and mutant pollen tubes. (a) Patterns in wild-type Col-0 pollen tubes. (b) and (c) Patterns in fim5 pollen tubes. Patterns shown in (b) and (c) occurred in ~80 and 20% of fim5 pollen tubes, respectively. Red arrows and green arrows represent the directions of cytosolic organelles moving acropetally and basipetally, respectively. Filled circles indicate cytosolic organelles that crowd together and cannot move forward. Large cytosolic organelles in wild-type Col-0 pollen tubes seldom enter the extreme apex, whereas those in fim5 pollen tubes enter the tip frequently.
(B) Single frames at 1-s intervals are shown. Three independent cytosolic organelles that show continuous movement were labeled with three different colored arrows. a1 to a10, wild-type Col-0; b1 to b10, fim5-1; c1 to c10, fim5-2. Bar = 10 μm in c10.
(C) Velocity of cytoplasmic streaming was significantly reduced in fim5 pollen tubes. Error bars represent mean ± sd. **P < 0.01, ANOVA test followed by Dunnett post-hoc multiple comparisons. Only the cytosolic organelles with continuous movement were selected for velocity analysis. Black column, wild-type (WT) Col-0; gray column, fim5-1; white column, fim5-2.
In contrast with wild-type plants, the reverse fountain pattern of cytoplasmic streaming was disrupted in fim5 pollen tubes. One of the most obvious and common phenotypes was that large cytosolic organelles invaded the extreme tip of the fim5 pollen tubes, leading to a loss of the clear zone normally found at the tip (Figure 3H). Furthermore, the overall movement of cytosolic organelles appeared to be random and irregular in fim5 pollen tubes. Two primary types of cytoplasmic streaming patterns were often observed in fim5 pollen tubes (shown in Figure 7A, b and c). The first pattern resulted in complete disruption of the reverse fountain pattern, as the movement of cytosolic organelles was random and cytosolic organelles moving toward different directions can be seen at a specific location; some cytosolic organelles show oscillatory movement (Figure 7A, b; see Supplemental Movies 3 and 4 online). This pattern was present in >80% of fim5 pollen tubes. The second pattern appeared similar to the first pattern, but in some regions of the pollen tubes, cytosolic organelles were crowded together (filled circles) (Figure 7A, c; see Supplemental Movies 5 and 6 online). The second pattern was present in ~20% of fim5 pollen tubes.
In addition to characterizing the pattern of cytoplasmic streaming, we also selected cytosolic organelles from several independent pollen tubes that exhibited continuous movement (Figure 7B), at least over a detectable distance, to determine the velocity of cytoplasmic streaming. Results from this analysis showed that the velocity of cytoplasmic streaming was significantly reduced in fim5 pollen tubes compared with wild-type pollen tubes (P < 0.001; Figure 7C). Based on these observations, loss of FIM5 function appears to alter the pattern and reduce the rate of cytoplasmic streaming in pollen tubes.
Pollen Germination and Pollen Tube Growth Are Hypersensitive to LatB in fim5 Mutants
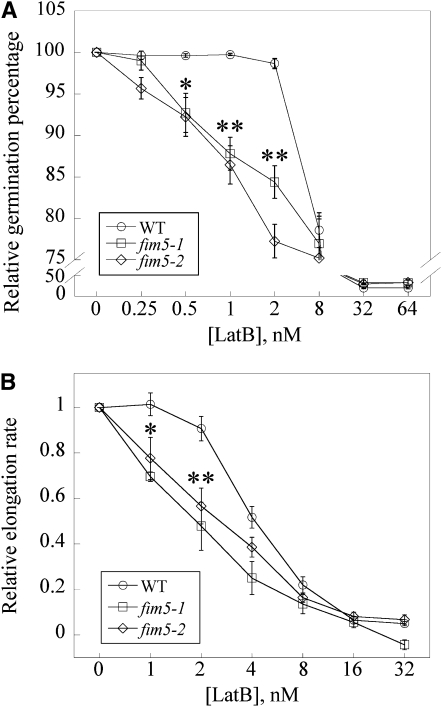
The aforementioned results suggest that FIM5 is a major regulator of actin organization in pollen grains and pollen tubes. We next sought to assess the effects of LatB treatment on fim5 pollen. To determine the effects of LatB on pollen germination, pollen derived from wild-type Col-0 plants and fim5 mutants was germinated on medium containing varying concentrations of LatB. For wild-type Col-0 pollen, the germination percentage was minimally affected at LatB concentrations below 2 nM (Figure 8A). However, the pollen germination percentage of fim5 mutants decreased in a dose-dependent manner across a range of LatB concentrations below 2 nM (Figure 8A). These results suggest that germination of fim5 pollen was more sensitive to LatB treatment than that of the wild type.
Figure 8.
Loss of FIM5 Function Increases the Sensitivity of Pollen Germination and Pollen Tube Elongation to LatB.
(A) Pollen germination is more sensitive to LatB in the absence of FIM5. The germination percentage after 12 h for wild-type (WT) Col-0, fim5-1, and fim5-2 treated with the vehicle (DMSO) were 98, 90, and 92%, respectively. All results were normalized to these percentages to generate a relative germination percentage of 100% for all controls. The relative germination percentage was plotted versus the concentration of LatB. A minimum of four independent experiments was conducted for each concentration. Error bars represent mean ± se. *P < 0.05 and **P < 0.01, by χ2 test.
(B) Pollen tube elongation was more sensitive to LatB treatment in the absence of FIM5. The average elongation rates for vehicle (DMSO) controls were 126, 46, and 41 μm/h for wild-type Col-0, fim5-1, and fim5-2, respectively. All results were normalized to these rates to generate a relative elongation rate of 1.0 for all controls. Relative pollen tube elongation rate was plotted versus concentration of LatB. A minimum of three independent experiments was conducted for each concentration. Half-maximal inhibition of elongation occurred at ~4 nM for wild-type Col-0 and ~2 nM for fim5. Error bars represent mean ± se. *P < 0.05 and **P < 0.01 (ANOVA test followed by Dunnett post-hoc multiple comparisons).
In addition to assessing pollen germination, we also determined whether growth rates of wild-type Col-0 and fim5 pollen tubes are differentially affected by LatB treatment. As shown in Figure 8B, the growth rate of both wild-type Col-0 and fim5 pollen tubes was inhibited by treatment with LatB in a dose-dependent manner. Half-maximal inhibition of growth occurred at ~4 nM LatB for wild-type Col-0 pollen tubes. In comparison, the growth rate of fim5 pollen tubes was more strongly affected at lower concentrations of LatB compared with wild-type Col-0 pollen tubes, and half-maximal inhibition of growth occurred at ~2 nM for fim5 pollen tubes (Figure 8B), which represented an estimated twofold decrease. Together, these results suggest that loss of FIM5 function increases the sensitivity of both pollen germination and pollen tube growth to LatB, implying that FIM5 regulates pollen germination and pollen tube growth through stabilization of actin filaments.
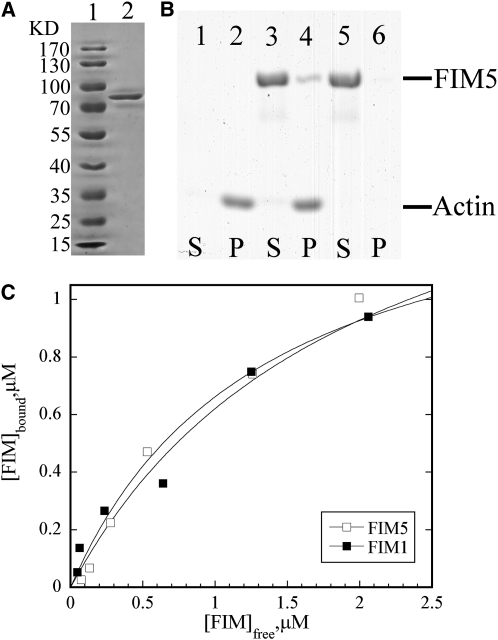
FIM5 Binds Actin Filaments with High Affinity
As the preponderance of evidence indicated that FIM5 is a major regulator of actin organization in pollen, we next sought to investigate the biochemical basis for the function of FIM5. To determine the actin regulatory activities of FIM5 in vitro, recombinant FIM5 was produced in Escherichia coli and purified to homogeneity (Figure 9A). The ability of the recombinant protein to bind to actin filaments was initially determined using a high speed cosedimentation assay. As shown in Figure 9B, FIM5 binds and cosediments with actin filaments. To determine the equilibrium dissociation constant (Kd) for this interaction, the amount of unbound FIM5 in the supernatant was plotted versus the amount of bound FIM5 in the pellet, and the data were fitted with a hyperbolic function. This analysis yielded a representative Kd value of 0.75 μM (Figure 9C). Additional experiments yielded a mean Kd value (±sd) of 0.71 ± 0.04 μM for binding of FIM5 to actin filaments. For comparative purposes, the representative Kd value was determined to be 0.51 μM for FIM1 (Figure 9C), which is similar to the published value for this interaction (Kovar et al., 2000). These results indicate that FIM5 binds to actin filaments with high affinity.
Figure 9.
FIM5 Binds Actin Filaments with High Affinity.
(A) Production of recombinant FIM5 protein. Lane 1, molecular mass standard; lane 2, purified FIM5 protein.
(B) A high-speed cosedimentation assay was used to assess FIM5 binding to actin filaments. Lanes 1 and 2 contain actin alone, lanes 3 and 4 contain actin and 2 μM FIM5, and lanes 5 and 6 contain 2 μM FIM5 alone. S, supernatant; P, pellet.
(C) The concentration of bound FIM5 or FIM1 was plotted against the concentration of free FIM5 or FIM1 and fitted with a hyperbolic function. The representative Kd values for this particular experiment were 0.75 and 0.51 μM for FIM5 and FIM1, respectively.
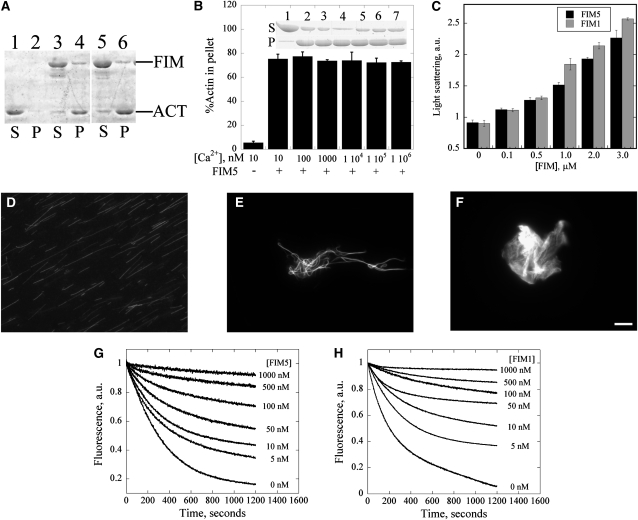
FIM5 Bundles and Stabilizes Actin Filaments in Vitro
It is a general feature of fimbrin family members to bundle actin filaments. Therefore, to determine whether FIM5 bundles actin filaments, a low-speed cosedimentation assay was employed. As shown in Figure 10, a majority of the actin remained in the supernatant in samples containing actin alone (Figure 10A, lane 1). By contrast, most of the actin sedimented in the presence of FIM5 (Figure 10A, lane 4), indicating that FIM5 bundles actin filaments. To determine whether FIM5 bundling activity was regulated by calcium, actin filaments were incubated with 1 μM FIM5 in the presence of varying concentrations of calcium prior to sedimentation. As shown in Figure 10B, the amount of pelleted actin was not substantially different in the presence of varying concentrations of calcium, suggesting that the bundling activity of FIM5 is calcium insensitive.
Figure 10.
FIM5 Bundles and Stabilizes Actin Filaments.
(A) A low-speed cosedimentation assay was employed to assess the actin bundling activity of FIM5. Lanes 1 and 2 contain actin alone, lanes 3 and 4 contain actin +1 μM FIM1, and lanes 5 and 6 contain actin + 1 μM FIM5. S, supernatant; P, pellet.
(B) FIM5 bundles actin filaments in a calcium-insensitive manner. F-actin (3 μM) was incubated with 1 μM FIM5 in the presence of the indicated concentrations of free calcium. Graph shows the percentage of sedimented actin for each [Ca2+]free value. Error bars represent ± se. Inset: SDS-PAGE gel showing the amount of actin in the supernatant and pellet in the presence of the indicated concentrations of Ca2+ after sedimentation at 13,600g for 30 min at 4°C.
(C) A light scattering assay was used to characterize the formation of higher-order actin structures induced by FIM5. The absorbance of the mixture was monitored at 400 nm after 3 μM preassembled actin filaments were incubated with the indicated concentrations of FIM5 (black) or FIM1 (gray) for 30 min at room temperature. Error bars represent ± se.
(D) to (F) Cross-linking of actin filaments was visualized directly by fluorescence microscopy for samples containing actin alone (D), actin + 1 μM FIM5 (E), and actin + 1 μM FIM1 (F). Bar = 5 μm.
(G) and (H) FIM5 and FIM1 inhibit dilution-mediated actin depolymerization. Graphs show representative results for the indicated concentrations of FIM5 (G) and FIM1 (H). a.u., arbitrary units.
Formation of actin higher-order structures induced by FIM5 was also characterized by light scattering, which has previously been used to assess the bundling activity of other actin binding proteins (Huang et al., 2005; Michelot et al., 2005). In agreement with results from cosedimentation assays, we found that addition of FIM5 increased the absorbance of actin filaments in a dose-dependent manner (Figure 10C), indicating that FIM5 induces formation of higher-order actin structures, similar to FIM1 (Figure 10C), VLN1 (Huang et al., 2005), and Formin1 (Michelot et al., 2005).
The formation of higher-order actin structures was also characterized by direct visualization of actin samples stained with rhodamine-phalloidin using fluorescence microscopy. Actin filaments behaved as single filaments in the absence of FIM5 (Figure 10D). However, addition of 1 μM FIM5 led to the organization of actin filaments into bundles (Figure 10E). Like FIM5, FIM1 also organized actin filaments into higher-order structures, but these structures appeared to be more consistent with aggregates than with bundles (Figure 10F), in agreement with a previous report (Kovar et al., 2000). These observations suggest that two phylogenetically distinct fimbrins form different types of actin higher-order structures.
To determine whether FIM5 stabilizes actin filaments, a dilution-mediated actin depolymerization assay was employed. As shown in Figure 10G, FIM5 inhibited dilution-mediated actin depolymerization in a dose-dependent manner, similar to FIM1 (Figure 10H). Taken together, these results suggest that FIM5 bundles and stabilizes actin filaments.
DISCUSSION
Here, we functionally characterized Arabidopsis FIM5, a fimbrin family gene that is expressed preferentially in pollen. Loss of FIM5 function was shown to induce disorganization of actin filaments in pollen, causing delayed pollen germination and retarded pollen tube growth. In vitro biochemical analyses revealed that FIM5 exhibits actin bundling activity and stabilizes actin filaments. Consistent with its biochemical activities, FIM5 loss-of-function increases the sensitivity of pollen germination and pollen tube growth to LatB treatment. In summary, our data suggest that FIM5 regulates pollen germination and pollen tube growth through stabilization of actin filaments and organization of these filaments into higher-order structures. Thus, this study provides key genetic evidence supporting a function for fimbrins in regulation of actin organization in plants.
FIM5 Is a Bona Fide Actin Bundling Factor
The conservation of actin binding residues in FIM5 (see Supplemental Figure 2 online) led us to predict that this protein may perform conserved actin-related functions. Indeed, several lines of evidence support this hypothesis. First, high-speed cosedimentation experiments showed that FIM5 binds actin filaments with high affinity (Figure 9), with a Kd value very close to that of FIM1 (Kovar et al., 2000). Second, a combination of low-speed cosedimentation, light scattering, and fluorescence microscopy assays unambiguously demonstrated that FIM5 bundles actin filaments in vitro (Figures 10A and 10E).
Although both FIM1 and FIM5 clearly bind actin and promote the formation of higher-order actin structures, notable differences were present in the morphology of low-speed sedimented actin pellets in samples containing FIM1 and FIM5. In FIM1-containing samples, the pellet was large and loose, as opposed to the small, tight pellets found in FIM5-containing samples. These differences suggest that FIM5 packs actin filaments into more compact higher-order structures than FIM1. This finding is consistent with the results of direct fluorescence light microscopy visualization of these actin higher-order structures. FIM5 induced the formation of actin bundles (Figure 10E), whereas FIM1 induced the formation of actin aggregates (Figure 10F; Kovar et al., 2000) in these experiments. The molecular mechanisms underlying the formation of these distinct actin higher-order structures by two different fimbrin isoforms should be further characterized in future studies.
Interestingly, FIM5 was also shown to bundle actin filaments in a calcium-insensitive manner (Figure 10B), similar to FIM1 (Kovar et al., 2000). Calcium insensitivity could be due to poor conservation of the N-terminal calmodulin-like calcium binding domain (McCurdy and Kim, 1998; Staiger and Hussey, 2004). Calcium insensitivity was also reported for chicken fimbrin (Bretscher, 1981) and S. pombe Fim1p (Nakano et al., 2001). By contrast, plastins from mammalian cells (Namba et al., 1992; Lin et al., 1994) and Dictyostelium (Prassler et al., 1997) have been shown to be calcium sensitive. Another study reported that a fimbrin-like protein from T. pyriformis lacks EF-hand domains (Watanabe et al., 2000). These observations suggest that the effect of calcium on fimbrin activity varies across species, implying that an alternative mechanism likely regulates the activity of Arabidopsis fimbrins. The extended C terminus present in both FIM5 and FIM1 (see Supplemental Figure 2 online; McCurdy and Kim, 1998), as well as other Arabidopsis fimbrins (Staiger and Hussey, 2004), could be important for fimbrin regulation. However, currently, the functional significance of the extended C terminus in plant fimbrins remains unclear.
FIM5 Is Required for the Organization of Actin Filaments in Pollen Grains and Pollen Tubes
Actin filaments exhibit distinct distributions in pollen grains and pollen tubes. The mechanisms by which these dynamic actin structures are generated and maintained remain a mystery. In this study, FIM5 was shown to decorate actin filaments throughout pollen grains and pollen tubes (Figure 4), implying that FIM5 may be an important regulator of actin organization in pollen. Consistent with this hypothesis, fim5 loss-of-function mutants exhibited redistribution of actin filaments in pollen grains. Thick, irregular actin bundles were present in fim5 pollen (Figures 5B to 5D) in comparison to wild-type Col-0 pollen (Figure 5A). This phenotype resembles that of multiple myosin knockouts in which more thick actin bundles appeared in the cells of midvein epidermis (Peremyslov et al., 2010). Actin redistribution was sustained throughout pollen germination and was also present in germinating pollen (Figures 5G, 5I, and 5K). Importantly, expression of FIM5-GFP under control of the native FIM5 promoter rescued actin organization defects in fim5 pollen grains (see Supplemental Figures 10C and 10D), indicating that actin redistribution is indeed due to loss of FIM5 function.
Convergence of actin filaments at the germination aperture and establishment of distinct polarity in wild-type Col-0 pollen grains (Figure 5E) facilitates the delivery of material necessary for cell wall synthesis and membrane expansion, which are required for emergence and elongation of the pollen tube. Redistribution of actin filaments in fim5 pollen grains could affect transport of these materials. Defect in transport may, therefore, underlie the delay in pollen germination observed in fim5 mutants (Figure 3D).
In addition to pollen grains, actin filaments also become disorganized in fim5 pollen tubes. The most obvious phenotype was protrusion of actin bundles into the extreme tips of fim5 pollen tubes (Figure 6), which was associated with loss of the clear zone (Figure 3H). This phenomenon resembles that of loss of function of multiple myosin genes during root hair tip growth that actin cables invaded into the tip of root hairs (Peremyslov et al., 2010), implying that myosin and fimbrin may play similar roles in maintaining actin cables away from the tip. In addition to protrusion of the actin cables, actin filaments at the tip of the pollen tube also assume different distributions in fim5 mutants (Figure 6), which may affect tip-targeted vesicle fusion and docking events. Changes in vesicle docking may in turn alter the direction of pollen tube growth and induce morphological changes, such as the appearance of curled pollen tubes (Figures 3F, c to g, and 3G). The dense actin structures at the subapex also become disorganized in fim5 pollen tubes (Figures 6D to 6I), suggesting that FIM5 is also important for generating and/or maintaining actin structures in this region.
Logically, it is difficult to understand why loss-of-function of an actin bundling factor leads to an increase in filamentous actin at the tips of pollen tubes. It could be that loss of FIM5 function increases the action of actin depolymerizing factors, such as profilin and ADF, which induces instability of actin filament in the shank of pollen tubes. Consequently, it may lead to elevation of cytosolic concentrations of actin monomers, which could be subsequently used for polymerization of actin at the tips of pollen tubes. Indeed, FIM1 was previously reported to prevent profilin-mediated actin depolymerization in vitro and in vivo (Kovar et al., 2000); another report demonstrated that T-plastin prohibited cofilin-mediated depolymerization of F-actin in vitro (Giganti et al., 2005). In this study, FIM5 was shown to protect actin filaments from dilution-mediated actin depolymerization (Figure 10G), suggesting that this protein stabilizes actin filaments. Similar results were observed for FIM1 (Figure 10H). In addition, loss of FIM5 function renders pollen germination and pollen tube growth hypersensitive to LatB (Figures 8A and 8B), further supporting the hypothesis that FIM5 is involved in these processes through stabilization of actin filaments.
We also found that actin cables in the shanks of fim5 pollen tubes were disorganized. The majority of the actin cables were not parallel to the growth axis of the pollen tubes, and the angles formed between the actin cables and the elongation axis increased substantially in fim5 pollen tubes (Figure 6J). These results suggest that FIM5 plays an important role in maintaining the strict parallel arrangement of actin cables in the pollen tube. Actin cables were also distributed irregularly and randomly in fim5 pollen grains (Figures 5B to 5D, 5G, 5I, and 5K). Collectively, these observations suggest that FIM5 mediates targeting of actin cables to specific locations in the cytoplasm of pollen through an unknown mechanism.
The thick actin cables present in fim5 pollen (see Supplemental Figure 8B online) could be due to an increase in the distance between adjacent actin filaments within individual cables and/or an increase in the number of actin filaments per cable. Loss of FIM5 function may lead to functional substitution of other bundling factors, such as other pollen-expressed fimbrin isoforms (FIM4 and FIM3; Pina et al., 2005), villins (VLN5 [Zhang et al., 2010], VLN2, and VLN1 [Pina et al., 2005]), or LIM domain-containing proteins (Wang et al., 2008; Thomas et al., 2009). Actin cables formed in the presence of these other factors could have longer distances between adjacent actin filaments. To support this, we found that actin cables formed in vitro by FIM5 (Figure 10E) are relative compact compared with those formed by FIM1 (Figure 10F; Kovar et al., 2000). Additionally, Cooley and colleagues have proposed that assembly of actin cables may be stepwise (Matova et al., 1999) and that villin initially brings actin filaments together, followed by subsequent cable contraction by fimbrin. It is not clear whether a stepwise assembly mechanism also applies to assembly of actin cables in the pollen tube. If this model holds true, FIM5 may function to tighten actin cables initiated by pollen-expressed villins, such as VLN5 (Zhang et al., 2010), VLN2, or VLN1 (Honys and Twell, 2003; Pina et al., 2005). Based on these observations, it is reasonable to propose that loss of function of FIM5 may induce a defect at the step of tightening actin cables initiated by villins, leading to the appearance of thicker actin cables. However, further genetic and biochemical evidence is needed to validate this hypothesis. Nonetheless, the possibility of increase in the number of actin filaments per cable cannot be ruled out completely here.
Loss of FIM5 Function Affects Cytoplasmic Streaming in the Pollen Tube
It is generally believed that the acto-myosin system powers cytoplasmic streaming. Within this model, actin cables provide molecular tracks; therefore, the pattern of cytoplasmic streaming mirrors the arrangement of actin filaments, and actin cables have been shown to be organized such that the plus ends face the tip at the cortex, with opposite polarity in the middle of root hairs and pollen tubes (Tominaga et al., 2000b; Lenartowska and Michalska, 2008).
One of the most obvious phenotypes observed in fim5 mutants was the entry of cytoplasmic streaming into the tips of the pollen tubes (Figure 7A, b and c; see Supplemental Movies 3 to 6 online), leading to the loss of the clear zone (Figure 3H). This observation is consistent with results showing that actin cables protrude into the apical region of fim5 pollen tubes (Figure 6). Entrance of cytoplasmic streaming into the tips of fim5 pollen tubes could also be due to disorganization of actin filaments at the subapex (Figures 6D to 6I). Given that the subapex contains the location at which cytoplasmic streaming normally reverses direction, disorganization of actin filaments in this region in fim5 pollen tubes may cause cytosolic organelles to proceed into the tip. Movement of cytosolic organelles in the shanks of fim5 pollen tubes was also altered. Compared with those in wild-type Col-0 pollen tubes (see Supplemental Movies 1 and 2 online), cytosolic organelles did not move in a straight line toward the tip or the base, suggesting that the tracks necessary for proper movement were altered. These changes in movement were consistent with results showing that actin cables are distributed randomly and irregularly in fim5 pollen tubes (Figure 6).
In some regions of the fim5 pollen tubes, cytosolic organelles appear to exhibit oscillatory movement and crowding, suggesting that loss of FIM5 may alter the polarity of actin bundles. Oscillatory movement of cytosolic organelles in fim5 pollen tubes could result from alterations in the polarity of actin bundles in these regions. Neighboring actin cables could have opposite polarity, or actin filaments could have mixed polarity within individual cables.
In addition to changes in the pattern of streaming, the velocity of cytoplasmic streaming was significantly decreased in fim5 pollen tubes (Figure 7B). These results suggest that alterations in the arrangement of actin cables impair myosin-based particle movement. Certainly, contributions from alterations in actin dynamics cannot be excluded as a possibility as well. The decrease in the velocity of cytoplasmic streaming may contribute to inhibition of pollen tube growth (Figure 3E). Collectively, these results demonstrate that loss of FIM5 function reduces the velocity and alters the direction of cytoplasmic streaming, which may result from the alteration of the arrangement of actin cables in fim5 pollen tubes.
METHODS
Identification of FIM5 T-DNA Insertion Lines
Two T-DNA insertion lines, fim5-1 (CS856909) and fim5-2 (CS853476), with insertions in the FIM5 gene, were obtained from the Nottingham Arabidopsis Stock Centre. Arabidopsis thaliana ecotype Col-0 was used as the wild-type plant (wild-type Col-0). PCR from genomic DNA was used to identify homozygous lines. Two sets of gene-specific primers (fim5-1 LP, fim5-1 RP, fim5-2 LP, and fim5-2 RP), together with left border primer (see Supplemental Table 1 online), were used to genotype T-DNA insertion lines. Homozygous plants were identified and used for the following phenotypic analyses.
To assess FIM5 transcript levels in fim5 mutants, RT-PCR was performed. Total RNA was extracted from seedlings (7 d old) using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. To synthesize cDNA, total RNA was reverse transcribed using MMLV reverse transcriptase (Promega) according to the manufacturer’s instructions. Three independent pairs of primers (F5f1/F5r1, F5f2/F5r2, and F5f13F5r3; see Supplemental Table 1 online) were used to determine the levels of full-length and partial FIM5 transcripts. eIF4A was amplified as a control for RT-PCR analysis.
Complementation Analysis
To complement fim5 mutants, we generated a FIM5-GFP expression construct using the following procedures. Briefly, the FIM5 promoter (including 1090 bp upstream of the start codon) was amplified using the FIM5proFor and FIM5proRev primers (see Supplemental Table 1 online). The resulting PCR product was cloned into the pMD19-T vector (TaKaRa Biotechnology) and was subsequently inserted into the binary vector pCambia1301-NOS between the SalI and BamHI sites. The EGFP and FIM5 coding sequences were then sequentially transferred to the pCambia1301-NOS vector containing the FIM5 promoter via digestion with SmaI/EcoRI and BamHI/SmaI, respectively. The resulting expression vector was termed pCambia1301 FIM5 pro:FIM5-EGFP-NOS. Primers used for amplification of FIM5 and EGFP were FIM5For/FIM5Rev and EGFPFor/EGFPRev, respectively (see Supplemental Table 1 online). The EGFP-FIM5 fusion construct was then introduced into fim5 plants using the Agrobacterium tumefaciens strain GV3101 by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog culture medium containing 25 μg/mL hygromycin.
Segregation Analysis
Self-crosses between FIM5 heterozygous mutants and reciprocal crosses between FIM5 heterozygous mutants and wild-type plants were performed as described previously (Howden et al., 1998). The T-DNA was linked to the BAR gene, which conferred resistance to the herbicide BASTA. Resistant seedlings were selected by spraying Finale (AgrEvo) at a 1:1000 dilution after plants had germinated in soil for 5 d. T-DNA–tagged male and female gametophyte segregation ratios were calculated by counting BASTA-resistant and sensitive plants.
Aniline Blue Staining Assay
Aniline blue staining of pollen tubes in pistils was performed as previously described (Ishiguro et al., 2001). Briefly, pre-emasculated mature wild-type flowers were pollinated either with wild-type, fim5-1, or fim5-2 pollen. After 4, 12, and 24 h, the pollinated pistils were incubated in fixing solution containing ethanol:acetic acid (3:1) for 2 h at room temperature. The fixed pistils were then washed with distilled water three times for 5 min each, followed by incubation in 8 M NaOH softening solution overnight. The following day, the pistils were washed in distilled water three times for 1 h each. The pistils were then stained in aniline blue solution (0.1% aniline blue in 0.1 M K2HPO4-KOH buffer, pH 11) for 3 to 5 h in the absence of light and were observed using a Leica DM4500B fluorescence light microscope with a ×10 objective (Leica Microsystems).
In Vitro Pollen Germination and Characterization of Pollen Grains and Tubes
In vitro pollen germination was performed essentially according to previously published methods (Ye et al., 2009). Briefly, pollen was isolated from newly opened flowers and was placed on pollen germination medium [1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 0.01% (w/v) H3BO3, and 18% (w/v) sucrose solidified with 0.5% (w/v) agar, pH 7.0]. The plates were cultured at 25°C under moist conditions. To determine the pollen germination percentage and the length of the pollen tubes, pollen grains and pollen tubes were observed using an IX71 microscope (Olympus) equipped with a ×10 objective. Digital images were collected with a Retiga EXi Fast 1394 CCD camera (Qimaging) using Image-Pro Express 6.3 software (Media Cybernetics). To calculate germination percentage, a minimum of 500 pollen grains was counted in each experiment. At least three experiments were conducted. The criteria for counting germination percentage were according to the previous published method that protrusion from the germination aperture was regarded as positive germination (Gibbon et al., 1999). To calculate average pollen tube length, 100 pollen tubes were measured in each experiment, and at least three experiments were performed.
To assess morphological changes in pollen tubes caused by loss of FIM5 function, wild-type and fim5 pollens were germinated for 2 h and 4 h in vitro, respectively, at which time the average length of the pollen tubes was ~100 μm. Pollen tubes were then observed under a microscope (IX71; Olympus) equipped with a ×20 objective. Digital images were acquired with a Retiga EXi Fast 1394 CCD camera using Image-Pro Express 6.3 software. One hundred pollen tubes were included in each replicate, and four independent experiments were conducted. To calculate the percentage of pollen tubes with clear zone, wild-type and fim5 pollen tubes were germinated as described above and observed with a ×60 1.42 numerical aperture (NA) objective. Pollen tubes with apical regions free of large organelles were counted as having clear zone, and invasion of large organelles was scored as loss of clear zone. A minimum of 70 pollen tubes was included in each experiment, and at least four experiments were conducted.
To characterize the effects of LatB on pollen germination, the indicated concentrations of LatB were added to pollen germination medium. Pollen grains obtained from wild-type Col-0 and fim5 mutant plants were germinated for 12 h in vitro. A minimum of 500 pollen grains was counted to determine the germination percentage. To assess the effects of LatB on pollen tube growth, pollen grains derived from wild-type Col-0 and fim5 mutants were germinated for 2 and 4 h in normal germination medium, respectively. Liquid germination medium containing the indicated concentrations of LatB was then added to the surface of the solid germination medium. Images of pollen tubes were taken as described above at different time points after the addition of LatB. A minimum of 20 pollen tubes was examined to determine the average growth rate in each experiment, and at least three independent experiments were performed.
F-Actin Staining
The actin cytoskeleton was visualized according to previously described methods (Gibbon et al., 1999; Snowman et al., 2002) with slight modifications. Briefly, pollen grains were spread on the surface of pollen germination medium and were fixed for 1 h in 300 μM 3-maleimidobenzoic acid N-hydroxysuccinimide ester in liquid pollen germination medium. The pollen grains were subsequently extracted with 0.05% Nonidet P-40 in liquid germination medium for 10 min. Fixed pollen grains were then rinsed three times in TBSS (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 400 mM sucrose) containing 0.05% Nonidet P-40 for 10 min each. The pollen was then stained with 200 nM Alexa-488 phalloidin in TBSS containing 0.05% Nonidet P-40 overnight at 4°C. To visualize the actin cytoskeleton in pollen tubes, pollen grains from wild-type and mutant plants were germinated for 2 and 4 h in vitro, respectively. The germinated pollen tubes were then fixed and stained with Alexa-488 phalloidin following the procedure described above for pollen grains. Images were collected with a Leica TCS SP5 confocal laser scanning microscope equipped with a ×100 1.46 NA HC PLAN objective. The fluorescent phalloidin was excited using the 488-nm line of an argon laser, optical sections were scanned and captured, and three Kalman-filtered scans were averaged for each optical section. Images were prepared by generating projections of the optical sections through an individual pollen grain or pollen tube.
Quantification of the Angles Formed between Actin Cables and the Pollen Tube Growth Axis
The angles formed between each actin cable and the growth axis of individual pollen tubes were analyzed using ImageJ (http://rsbweb.nih.gov/ij/, version 1.38). Curled pollen tubes were excluded from this analysis because their growth axes were difficult to define. As actin cables were nearly parallel to the growth axis in the shanks of wild-type pollen tubes, only this region was used for quantification. For each pollen tube, three to four optical sections were excluded between sections used for analysis to ensure that each actin cable was analyzed only once. For wave-like actin cables that formed more than one angle with the growth axis, only the largest angle was used for quantification. A minimum of five pollen tubes was selected randomly to obtain 180 cables in each experiment and three replicates were conducted.
Analysis of Cytoplasmic Streaming
Wild-type and mutant pollens were germinated for 2 and 4 h, respectively, as described above. Time-lapse images of cytoplasmic streaming were collected at 1-s intervals using an Olympus IX71 microscope with differential interference contrast optics equipped with a ×60 1.42 NA objective. Image capture was realized with a Retiga EXi Fast 1394 CCD camera using Image-Pro Express 6.3 software. A minimum of 15 cytosolic organelles from at least five pollen tubes was selected randomly for cytoplasmic streaming velocity determination in each replicate, and three replicates were conducted. Only cytosolic organelles undergoing continuous movements were selected for velocity analysis. The distance traveled by a selected cytosolic organelle during a given time was determined with Image J (http://rsbweb.nih.gov/ij/; version 1.38).
Statistical Analysis
Several statistical analysis methods were applied for characterizing the phenotypic difference between the wild type and fim5 mutant. Proportion data, such as segregation ratio and pollen germination percentage, are analyzed by χ2 test. Other data that followed normal distribution were analyzed by Student’s t test for comparison between two groups or analysis of variance (ANOVA) test among three groups, followed by Dunnett post-hoc multiple comparisons. Data that did not follow a normal distribution were normalized (e.g., by square root transformation). If the data still did not follow a normal distribution, they were analyzed with a Kruskal-Wallis test followed by multiple comparison test, taking into account Bonferroni’s correction to determine significant levels. Kruskal-Wallis tests were conducted in R programming language (http://www.R-project.org; version 2.10.1), and the following multiple comparison tests were performed with kruskalmc function in the R package pgirmess (http://CRAN.R-project.org/package=pgirmess; version 1.4.5).
Protein Purification
The full-length FIM5 cDNA was amplified by RT-PCR. Briefly, Arabidopsis total RNA was purified from flowers using the Total RNA Isolation kit (Promega). The FIM5 cDNA sequence was used as a reference to design primers for RT-PCR (FIM5KGFor and FIM5KGRev; see Supplemental Table 1 online). Amplified sequences were cloned into the pMD19-T vector (TaKaRa Biotechnology). The resulting cDNA sequences were verified by sequencing. For expression of FIM5 in Escherichia coli, the pGEX-KG-FIM5 vector was produced by cloning the FIM5 coding sequence into the pGEX-KG vector after digestion of both DNA sequences using EcoRI. Fusion proteins were expressed in the E. coli BL21 DE3 strain by induction with 0.2 mM isopropyl β-d-thiogalactopyranoside overnight at 16°C. GST-FIM5 was affinity purified using glutathione-sepharose resin according to the manufacturer's recommended protocol (Amersham Biosciences). After extensive washing of the column, thrombin (Sigma-Aldrich) was added to cleave GST, releasing FIM5. Fimbrin-containing fractions were further purified by anion exchange chromatography (Q-Sepharose; Amersham Biosciences). The resulting purified protein was concentrated by ultrafiltration, aliquoted, flash-frozen, and stored at −80°C. Prior to use, the protein was further clarified by centrifugation at 200,000g for 1 h.
Actin was purified from acetone powder of rabbit skeletal muscle according to previous methods (Spudich and Watt, 1971). Monomeric Ca-ATP-actin was further purified using Sephacryl S-300 chromatography at 4°C in G buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, and 0.1 mM imidazole) (Pollard, 1984). Pyrene-actin was prepared by labeling actin at Cys-374 with pyrene iodoacetamide as previously described for kinetic analysis (Pollard, 1984). Recombinant Arabidopsis FIM1 was purified as previously described (Kovar et al., 2000).
Low- and High-Speed Cosedimentation Assays
Low- and high-speed cosedimentation experiments were performed as described previously (Kovar et al., 2000). Briefly, for the low-speed cosedimentation assay, varying concentrations of FIM5 were incubated with 3 μM preassembled rabbit muscle actin in F buffer (0.1 M KCl, 5 mM MgCl2, 0.5 mM DTT, 0.5 mM ATP, and 5 mM Tris-HCl, pH 7.5) in the presence or absence of calcium for 60 min at room temperature. The resulting mixtures were then centrifuged at 4°C for 30 min at 13,600g. After centrifugation, the supernatant (S) and pellet (P) fractions were resolved on SDS-PAGE gels and stained with Coomassie Brilliant Blue.
To assess F-actin binding activity, varying concentrations of FIM5 (0.1 to 3.0 μM) were incubated with 3 μM F-actin for 60 min, followed by centrifugation at 4°C for 30 min at 200,000g. After supernatants (S) and pellets (P) were resolved on SDS-PAGE gels and stained as described above, the intensities of the resulting bands were quantified by densitometry using Image J software (http://rsbweb.nih.gov/ij/; version 1.38). The apparent dissociation constant (Kd) value was determined by plotting the amount of bound FIM5 versus free FIM5, followed by fitting the data with a rectangular hyperbolic curve using KaleidaGraph software (Synergy Software).
Visualization of Actin Filament Cross-Linking by Fluorescence Microscopy
Fluorescence microscopy was used to visualize the effects of FIM5 on the organization of actin filaments into higher-order structures. The indicated concentrations of FIM5 or FIM1 were incubated with 4 μM preformed F-actin at room temperature for 30 min. Subsequently, actin filaments were labeled with an equimolar concentration of rhodamine-phalloidin. Actin filaments were then diluted to a final concentration of 10 nM in fluorescence buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 100 mM DTT, 100 μg/mL glucose oxidase, 15 mg/mL glucose, 20 μg/mL catalase, and 0.5% methylcellulose) as described previously (Huang et al., 2003). The diluted samples were visualized using a 1.42 NA ×60 oil immersion lens mounted on an Olympus IX71 microscope. Images were collected with a Retiga EXi Fast 1394 CCD camera using Image-Pro Express 6.3 software.
Dilution-Mediated Actin Depolymerization Assay
To test the ability of FIM5 to stabilize actin filaments, a dilution-mediated actin depolymerization assay was performed in a manner similar to a previous report (Huang et al., 2003). Briefly, varying concentrations of FIM5 or FIM1 were incubated with 5 μM preassembled actin filaments (50% pyrene-labeled) at room temperature for 5 min. The mixtures were then diluted 25-fold in G buffer. Actin depolymerization was tracked by monitoring the decrease in pyrene fluorescence for 1200 s at room temperature using a spectrofluorometer (Photon Technology International).
Light Scattering Assay to Characterize Filament Bundling Activity
Actin bundling activity of FIM5 was also analyzed using a light scattering assay, which was essentially performed as previously described (Michelot et al., 2005). Briefly, the indicated concentrations of FIM5 or FIM1 were added to a solution containing 3 μM preassembled actin filaments, and the resulting mixtures were incubated for 30 min at room temperature. The samples were then analyzed for 90° light scattering of unlabeled actin at 400 nm with a spectrofluorometer (Photon Technology International).
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL or Arabidopsis Genome Initiative database under the following accession numbers: FIM1 (Arabidopsis fimbrin1, NP_194400; At4g26700), FIM5 (NP_198420; At5g35700), Sac6p (Saccharomyces cerevisiae, NP_010414), and L-plastin (NP_002289).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FIM5 Is Preferentially Expressed in Stamen and Mature Pollen.
Supplemental Figure 2. Alignment of the FIM5 Protein Sequence with Sequences of Other Fimbrin Family Members.
Supplemental Figure 3. Pollen Tube Growth Defects in fim5 Mutants Were Rescued by Expression of FIM5-GFP Driven by the FIM5 Native Promoter.
Supplemental Figure 4. FIM5-GFP, Driven by the FIM5 Promoter, Decorates Actin Filaments in Both Pollen Grains and Pollen Tubes.
Supplemental Figure 5. Filamentous Structures Decorated by FIM5-GFP Are Dispersed by LatB Treatment.
Supplemental Figure 6. FIM5-GFP Colocalizes with Actin Filaments in Pollen Grains and Pollen Tubes.
Supplemental Figure 7. Actin Filaments Become Redistributed, Form Thick Bundles, and Display Diverse Patterns in fim5 Pollen Grains.
Supplemental Figure 8. Thicker Actin Bundles Are Present in fim5-1 Pollen Grains.
Supplemental Figure 9. Actin Filaments Are Reorganized and Form Thicker Bundles in fim5-1 Pollen Grains Revealed by Fluorescent Phalloidin Staining Fixed with Paraformaldehyde.
Supplemental Figure 10. Uniform Normal Distribution of Actin Filaments Was Restored in Complemented fim5-1 Pollen Grains Carrying the FIM5-GFP Construct.
Supplemental Table 1. Primers Used in This Study.
Supplemental Movie 1. Cytoplasmic Streaming in a Wild-Type Pollen Tube.
Supplemental Movie 2. Cytoplasmic Streaming in a Wild-Type Pollen Tube.
Supplemental Movie 3. Cytoplasmic Streaming in a fim5 Pollen Tube.
Supplemental Movie 4. Cytoplasmic Streaming in a fim5 Pollen Tube.
Supplemental Movie 5. Cytoplasmic Streaming in a fim5 Pollen Tube.
Supplemental Movie 6. Cytoplasmic Streaming in a fim5 Pollen Tube.
Supplementary Material
Acknowledgments
We thank Ming Yuan (China Agricultural University) for critical reading and constructive comments on this manuscript. We also thank Christopher J. Staiger (Purdue University) for providing Arabidopsis FIM1 expression plasmid and the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion lines. This work was supported by grants from the Ministry of Science and Technology (2007CB947600 and 2011CB944600), the National Natural Science Foundation of China (30900721, 30821007, and 30771088), and the Chinese Academy of Sciences (Hundred Talents Program and KJCX2-YW-L08). This work was also partially supported by the Knowledge Innovation Program of Chinese Academy of Sciences (110500Q002).
References
- Adams A.E., Botstein D., Drubin D.G. (1991). Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature 354: 404–408 [DOI] [PubMed] [Google Scholar]
- Arpin M., Algrain M., Louvard D. (1994). Membrane-actin microfilament connections: An increasing diversity of players related to band 4.1. Curr. Opin. Cell Biol. 6: 136–141 [DOI] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A.I., Makarova K.S., Koonin E.V., Dolja V.V. (2008). Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. (1981). Fimbrin is a cytoskeletal protein that crosslinks F-actin in vitro. Proc. Natl. Acad. Sci. USA 78: 6849–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A., Weber K. (1980). Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J. Cell Biol. 86: 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower S.M., Honts J.E., Adams A.E. (1995). Genetic analysis of the fimbrin-actin binding interaction in Saccharomyces cerevisiae. Genetics 140: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Wong E.I., Vidali L., Estavillo A., Hepler P.K., Wu H.M., Cheung A.Y. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Qu X., Wu Y., Huang S. (2009). Regulation of actin dynamics in pollen tubes: control of actin polymer level. J. Integr. Plant Biol. 51: 740–750 [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Duan Q.H., Costa S.S., de Graaf B.H., Di Stilio V.S., Feijo J., Wu H.M. (2008). The dynamic pollen tube cytoskeleton: Live cell studies using actin-binding and microtubule-binding reporter proteins. Mol. Plant 1: 686–702 [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Daudet N., Lebart M.C. (2002). Transient expression of the t-isoform of plastins/fimbrin in the stereocilia of developing auditory hair cells. Cell Motil. Cytoskeleton 53: 326–336 [DOI] [PubMed] [Google Scholar]
- de Arruda M.V., Watson S., Lin C.S., Leavitt J., Matsudaira P. (1990). Fimbrin is a homologue of the cytoplasmic phosphoprotein plastin and has domains homologous with calmodulin and actin gelation proteins. J. Cell Biol. 111: 1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D.G., Miller K.G., Botstein D. (1988). Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107: 2551–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V.E., Orlova A., Cherepanova O., Lebart M.C., Egelman E.H. (2008). High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. USA 105: 1494–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giganti A., Plastino J., Janji B., Van Troys M., Lentz D., Ampe C., Sykes C., Friederich E. (2005). Actin-filament cross-linking protein T-plastin increases Arp2/3-mediated actin-based movement. J. Cell Sci. 118: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Goode B.L., Wong J.J., Butty A.C., Peter M., McCormack A.L., Yates J.R., Drubin D.G., Barnes G. (1999). Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144: 83–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Sano T., Hasezawa S. (2007). Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 10: 549–556 [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova T.S., Tatchell K., Cooper J.A. (1995). Actin filaments in yeast are unstable in the absence of capping protein or fimbrin. J. Cell Biol. 131: 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Faivre-Moskalenko C., Esseling J.J., de Ruijter N.C., Grierson C.S., Dogterom M., Emons A.M. (2002). Positioning of nuclei in Arabidopsis root hairs: An actin-regulated process of tip growth. Plant Cell 14: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M.G., Shi W., Ramagopal U., Tseng Y., Wirtz D., Kovar D.R., Staiger C.J., Almo S.C. (2004). Structure of the actin crosslinking core of fimbrin. Structure 12: 999–1013 [DOI] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Staiger C.J., Weaver E.A., McCurdy D.W. (2000). AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Kübler E., Riezman H. (1993). Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 12: 2855–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowska M., Michalska A. (2008). Actin filament organization and polarity in pollen tubes revealed by myosin II subfragment 1 decoration. Planta 228: 891–896 [DOI] [PubMed] [Google Scholar]
- Lin C.S., Shen W., Chen Z.P., Tu Y.H., Matsudaira P. (1994). Identification of I-plastin, a human fimbrin isoform expressed in intestine and kidney. Mol. Cell. Biol. 14: 2457–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V., Panstruga R. (2005). Dynamic cellular responses in plant-microbe interactions. Curr. Opin. Plant Biol. 8: 625–631 [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Wilsen K.L., Baskin T.I., Hepler P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221: 95–104 [DOI] [PubMed] [Google Scholar]
- Matova N., Mahajan-Miklos S., Mooseker M.S., Cooley L. (1999). Drosophila quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development 126: 5645–5657 [DOI] [PubMed] [Google Scholar]
- McCurdy D.W., Kim M. (1998). Molecular cloning of a novel fimbrin-like cDNA from Arabidopsis thaliana. Plant Mol. Biol. 36: 23–31 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Satoh K., Morimatsu A., Ohnuma M., Mabuchi I. (2001). Interactions among a fimbrin, a capping protein, and an actin-depolymerizing factor in organization of the fission yeast actin cytoskeleton. Mol. Biol. Cell 12: 3515–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba Y., Ito M., Zu Y., Shigesada K., Maruyama K. (1992). Human T cell L-plastin bundles actin filaments in a calcium-dependent manner. J. Biochem. 112: 503–507 [DOI] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Dolja V.V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikzack C., Prassler J., Furukawa R., Fechheimer M., Rivero F. (2005). Role of calcium-dependent actin-bundling proteins: Characterization of Dictyostelium mutants lacking fimbrin and the 34-kilodalton protein. Cell Motil. Cytoskeleton 62: 210–231 [DOI] [PubMed] [Google Scholar]
- Pina C., Pinto F., Feijó J.A., Becker J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassler J., Stocker S., Marriott G., Heidecker M., Kellermann J., Gerisch G. (1997). Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin-myosin complexes. Mol. Biol. Cell 8: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T. (2007). The sliding theory of cytoplasmic streaming: fifty years of progress. J. Plant Res. 120: 31–43 [DOI] [PubMed] [Google Scholar]
- Shirayama S., Numata O. (2003). Tetrahymena fimbrin localized in the division furrow bundles actin filaments in a calcium-independent manner. J. Biochem. 134: 591–598 [DOI] [PubMed] [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246: 4866–4871 [PubMed] [Google Scholar]
- Staiger C.J., Hussey P.J. (2004). Actin and actin-modulating proteins. In The Plant Cytoskeleton in Cell Differentiation and Development, Hussey P.J., (Oxford, UK: Blackwell Publishing; ), pp. 32–80 [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton 66: 940–957 [DOI] [PubMed] [Google Scholar]
- Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S.A., Alberts A.S. (2000b). Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell 5: 13–25 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P.K., Shimmen T. (2000a). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta 210: 836–843 [DOI] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., Hasezawa S., Dolja V.V., Hara-Nishimura I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S., Shaw B.D. (2008). The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans. Mol. Microbiol. 68: 690–705 [DOI] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.J., Wan A.R., Jauh G.Y. (2008). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147: 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Yonemura I., Gonda K., Numata O. (2000). Cloning and sequencing of the gene for a Tetrahymena fimbrin-like protein. J. Biochem. 127: 85–94 [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Bähler J., Pringle J.R. (2001). Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell 12: 1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Pollard T.D. (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science 310: 310–314 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.E., Cooper J.A., Bridgman P.C. (2004). Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.