Chloroplast development requires protein quality control in which the ATP-dependent protease FtsH plays a versatile role. This study finds that FtsH, composed of a heterocomplex by Type A and Type B isomers in the thylakoid membrane, can function without the protease activity provided by Type B isomers, implying that it has more protease sites than are needed for proper chloroplast development.
Abstract
FtsH is an ATP-dependent metalloprotease present as a hexameric heterocomplex in thylakoid membranes. Encoded in the Arabidopsis thaliana YELLOW VARIEGATED2 (VAR2) locus, FtsH2 is one isoform among major Type A (FtsH1/5) and Type B (FtsH2/8) isomers. Mutants lacking FtsH2 (var2) and FtsH5 (var1) are characterized by a typical leaf-variegated phenotype. The functional importance of the catalytic center (comprised by the zinc binding domain) in FtsH2 was assessed in this study by generating transgenic plants that ectopically expressed FtsH2(488), a proteolytically inactive version of FtsH2. The resulting amino acid substitution inhibited FtsH protease activity in vivo when introduced into Escherichia coli FtsH. By contrast, expression of FtsH2(488) rescued not only leaf variegation in var2 but also seedling lethality in var2 ftsh8, suggesting that the protease activity of Type B isomers is completely dispensable, which implies that the chloroplastic FtsH complex has protease sites in excess and that they act redundantly rather than coordinately. However, expression of FtsH2(488) did not fully rescue leaf variegation in var1 var2 because the overall FtsH levels were reduced under this background. Applying an inducible promoter to our complementation analysis revealed that rescue of leaf variegation indeed depends on the overall amount of FtsH. Our results elucidate protein activity and its amount as important factors for the function of FtsH heterocomplexes that are composed of multiple isoforms in the thylakoid membrane.
INTRODUCTION
Differentiation of proplastids into chloroplasts is a light-dependent and organ-specific process. This process accompanies rapid development of the thylakoid membranes, where protein complexes for pigment-containing antenna, photosynthetic electron transfer, and ATP synthesis are assembled (López-Juez, 2007; Sakamoto et al., 2008). These complexes are made from subunits encoded by both nuclear and chloroplastic genomes. Therefore, coordinated synthesis/import and assembly of the subunit proteins is necessary (Wollman et al., 1999). Some of these regulatory mechanisms have a prokaryotic nature because of endosymbiotic origin of chloroplasts from ancestral cyanobacterium (Møller, 2005; Wise and Hoober, 2006). Once assembled, the complexes must be maintained through protein quality control (Bukau et al., 2006; van der Hoorn, 2008). Although the underlying mechanism in protein quality control is not fully understood, recent findings imply that the proteases of prokaryotic origin play critical roles in chloroplast development and maintenance (Sokolenko et al., 2002; Adam et al., 2006; Sakamoto, 2006; Kato and Sakamoto, 2010). Among the chloroplastic proteases, we specifically examine FtsH, which is an ATP-dependent metalloprotease localized in thylakoid membranes.
FtsH belongs to the AAA (ATPases associated with diverse cellular activities) protein family, whose members include the conserved Walker A and B and the second region of homology (SRH) motifs (Ogura and Wilkinson, 2001; Ito and Akiyama, 2005; Baker and Sauer, 2006; Narberhaus et al., 2009). In addition to these motifs, FtsH contains N-terminal transmembrane domains and the C-terminal protease domain (also known as the zinc binding domain [ZnBD]), which acts as a catalytic center of peptide hydrolysis. A crystal structure of the FtsH protease domain was recently solved in Aquifex aeolicus (Suno et al., 2006) and Thermotoga maritima (Bieniossek et al., 2006, 2009). Based on these structures, a threefold-symmetric open-to-closed transition model of the protease domain was proposed. However, in chloroplasts, the detailed proteolysis mechanism driven by FtsH remains unclear. Particularly, chloroplast FtsHs are unique in that they are produced as heteromeric isomers (Sakamoto, 2006).
Notable roles of FtsH in chloroplast homeostasis have emerged from several earlier studies (Lindahl et al., 1996, 2000; Ostersetzer and Adam, 1997; Bailey et al., 2002). A series of biochemical studies demonstrated that thylakoid-embedded FtsH degrades photodamaged D1 reaction center proteins in the photosystem II (PSII) repair cycle (Kato and Sakamoto, 2009; Nixon et al., 2010). In addition, many genetic studies have focused on determining the function of chloroplastic FtsH (Sakamoto, 2003; Aluru et al., 2006; Liu et al., 2010a). Loss of several FtsHs results in leaf-variegated phenotypes in Arabidopsis thaliana: yellow variegated1 (var1) and var2, caused by the lack of FtsH5 and FtsH2, respectively (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002).
In Escherichia coli, FtsH forms a hexameric homocomplex and functions to degrade misassembled membrane proteins (Ito and Akiyama, 2005; Narberhaus et al., 2009). However, in plants, FtsH exists as a heterocomplex comprising isomers of two types: FtsH5/FtsH1 (Type A) and FtsH2/FtsH8 (Type B) (Yu et al., 2004, 2005; Zaltsman et al., 2005a). The major isoforms representing each type are FtsH2 and FtsH5, as evidenced by the variegation phenotype in var2 and var1; FtsH1 and FtsH8 appear to function as minor isomers in the heterocomplex because a knockout of the genes corresponding to these isomers results in no obvious phenotype (Sakamoto et al., 2003; Zaltsman et al., 2005a). The variegation phenotype, generating green sectors containing normal-appearing chloroplasts and white sectors containing abnormal plastids, also implies that a threshold level of FtsHs is necessary for proper thylakoid formation (Yu et al., 2005; Zaltsman et al., 2005b; Kato et al., 2007).
We previously characterized many var2 mutant alleles in which amino acid substitutions were identified: three of five amino acid substitutions occurred in the Walker A and Walker B motifs (Sakamoto et al., 2004). Specifically, one occurred in the central pore region of ATPase (formed by conserved residues MFVG in the case of FtsH) and one occurred in the GAD motif (between the SRH and protease domain). By contrast, no mutation has been detected in ZnBD, raising the possibility that the leaf-variegated phenotype requires no protease activity. Along with these observations, it is suggested that the overall levels of FtsH2 are correlated with the degree of leaf variegation in most of the var2 alleles (Chen et al., 2000). These results prompted us to study the relevance of the variegated phenotype to protease activity and accumulation level of FtsH2 systematically. In this study, we performed multiple complementation tests of the variegated phenotype in var1, var2, var1 var2, and var2 ftsh8 mutants with a mutant form of FtsH2 that is proteolytically inactive. To our surprise, the mutant version of FtsH2 fully rescued not only leaf variegation in var2 but also seedling lethality in var2 ftsh8 (lacking both B-type isomers). Therefore, the FtsH heterocomplex functions without the protease activity provided by Type B isomers, implying that it has more protease sites than are needed for proper chloroplast development. Furthermore, we show that the variegation of var2 was rescued by expressing FtsH2 under a promoter that is inducible with estrogen hormone. The degree of leaf variegation depends on the expression level of FtsH2, allowing us to manipulate chloroplast development in our system. These results collectively provide insight into the function of the FtsH heterocomplex that is characteristic of photosynthetic organisms.
RESULTS
Site-directed Mutagenesis of ZnBD in FtsH
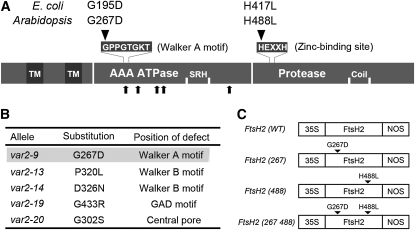
To determine whether protease activity in FtsH2 influences leaf variegation, we introduced an amino acid change within the conserved ZnBD motif (composed of HEXXH, where X represents any residue) and expressed the mutant FtsH2 in var2. As shown in Figure 1A, we replaced the first His (His-488) of ZnBD with Leu (designated as H488L). As a negative control, we generated an amino acid change (from Gly to Asp) at the first Gly (Gly-267) residue within the Walker A motif (GPPGTGKT). This mutation (termed as G267D) is identical to var2-9 (Figure 1B) and was shown not to rescue leaf variegation when expressed in var2 (Sakamoto et al., 2004). As a positive control, we used wild-type FtsH2 (termed as WT) with no mutations. These three versions of FtsH2, termed FtsH2(488), FtsH2(267), and FtsH2(WT), together with the double mutant FtsH2(267 488), were expressed under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase terminator (Figure 1C). The corresponding genes were first transformed into wild-type Columbia (Col) and subsequently introduced into var1, var2, and ftsh8 mutants. More than three independent transgenic lines for expressing each FtsH2 were obtained and subjected to further analysis.
Figure 1.
Summary of Amino Acid Substitutions Introduced into FtsH2.
(A) Schematic view of FtsH/FtsH2 protein in E. coli and Arabidopsis. The protein sequence is shown as a horizontal bar with illustrations of the transmembrane domain (TM), ATPase domain (AAA ATPase), second region of homology (SRH), Zn2+ metalloprotease domain (Protease), and coiled-coil region (Coil). The N-terminal variation between E. coli and Arabidopsis is not included. The conserved amino acid sequences for the Walker A motif and zinc binding site are shown above the protein sequence. The amino acid substitutions characterized in this study are also shown. Five other amino acid substitutions identified from different alleles (B) are indicated below the protein sequence by arrows.
(B) Amino acid substitutions in var2 alleles. The amino acid substitution in var2-9 (highlighted) is identical to that in (A) (G267D).
(C) Schematic representation of gene constructs used for this study. 35S, CaMV 35S promoter; NOS, nopaline synthase terminator. Amino acid substitutions are indicated by arrowheads.
Mutation in ZnBD Inhibits FtsH Protease Activity in E. coli
Although it is likely that the mutation in FtsH2(488) inhibits zinc binding and inactivates proteolysis, it was necessary to verify this hypothesis. Using in vitro studies, FtsH2 has not been shown to exhibit protease activity, nor has it been shown to complement E. coli FtsH mutants. Furthermore, our attempt to express a chimeric FtsH protein derived from an N-terminal E. coli ATPase domain and C-terminal FtsH2 protease domain has not been successful. Therefore, we introduced the amino acid changes corresponding to H488L and G267D into E. coli FtsH (termed as H417L and G195D; Figure 2A) and performed complementation analysis of E. coli ftsH mutants using these mutant FtsHs. We used this strategy in an earlier study to reveal an important motif involved in ATP binding (Sakamoto et al., 2004).
Figure 2.
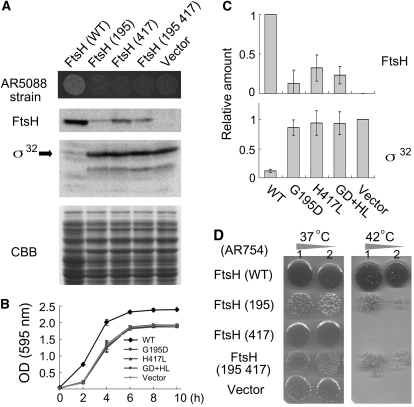
Inhibition of Protease Activity in E. coli FtsH with Amino Acid Substitutions.
(A) In vivo protease activity of wild-type and mutant FtsH proteins in E. coli. AR5088 (sfhC ΔftsH) was transformed with pIFH108 containing wild-type FtsH (WT) or containing mutated FtsHs (G195D, H417L, or G195D+H417L). Vector indicates an empty vector without FtsH. Cells harboring plasmids of mutated FtsHs (G195D, H417L, and G195D+H417L) or without FtsH displayed smaller colonies, reflecting deficient growth (top panel). Immunoblots of E. coli extracts with anti-FtsH and anti-σ32 (a substrate of FtsH) are shown together with the CBB-stained gel image of the same samples.
(B) Growth curves of E. coli AR5088. Cells transformed with empty vector pER15b (Vector) or with pIFH108 expressing wild-type FtsH or mutated FtsHs (G195D, H417L, and G195D+H417L [abbreviated as GD+HL]) were grown in LB medium. Cell densities of bacteria were measured according to photometric detection with OD (595 nm) readings (bars represent sd; five biological repeats).
(C) Relative amounts of FtsH and σ32 estimated by immunoblotting (bars represents sd; six biological repeats). To normalize the expression values, the pIFH108 (WT) and empty vector control values were set to 1 for FtsH and σ32, respectively
(D) Complementation of ftsH1 (Ts) by wild-type and mutated FtsH proteins. ftsH1 (AR754) is a temperature-sensitive mutant that was transformed with pIFH108 containing FtsH (WT), FtsH (195), FtsH (417), or FtsH (195 417) or without an insert (Vector). Growth of each strain on LB agar at 37°C (left) and 42°C (right) is shown. Rows 1 and 2 present data of 1-fold and 10-fold dilutions of cultures, respectively.
For our complementation studies, we used E. coli strain AR5088 as a recipient host carrying ΔftsH and sfhC (a suppressor mutation of ftsH that allows survival without FtsH) and generating small colonies (Qu et al., 1996). Expression of the wild-type FtsH complemented this strain, but expression of H417L, G195D, or H417L G195D failed to rescue the slow growth phenotype (Figure 2B). When cell densities were measured over a 10-h culture period, clear differences were observed between wild-type and mutated versions. All of these mutations appeared to affect accumulation of FtsH in vivo: H417L accumulated ~20% relative to the wild type. Additional mutations occurring within ATP binding and protease domains were shown to destabilize FtsHs in E. coli dramatically. To assess protease activity, accumulation of σ32, a soluble substrate of FtsH, was measured using immunoblot analysis with anti-σ32 antibodies. Both G195D and H417L retained σ32 levels equivalent to that in the negative control (vector only) (Figure 2C). These results suggested that the substitution of the first His residue in ZnBD to Leu leads to drastic or complete loss of FtsH activity, most likely attributable to prevention of zinc binding.
We also performed complementation using another ftshH strain (AR754) that exhibits temperature sensitivity to growth at 42°C (Begg et al., 1992). As shown in Figure 2D, none of the tested FtsH mutants completely rescued the temperature-sensitive growth when transformed into AR754. We found that G195D has a profound effect in AR754 and causes slower growth even at the permissive temperature, although it allowed very slow growth at 42°C. The precise reason for this effect of G195D is not known. Overall, though, these complementation data support our observation that H417L inactivates the protease activity of FtsH.
Expression of FtsH2(488) Rescues Leaf Variegation in var2
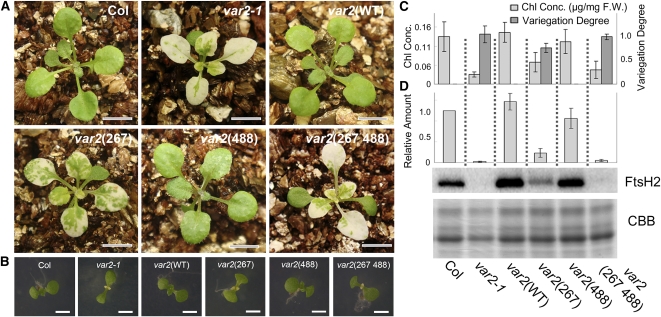
To test if the protease activity of FtsH2 is necessary for rescuing the leaf variegation phenotype, transgenes coding for FtsH2(WT), FtsH2(488), and FtsH2(267) were introduced into var2-1 (a null mutant of FtsH2) and evaluated for variegated phenotypes. The resulting transgenic lines were designated as var2(WT), var2(488), and var2(267), respectively. Figure 3A shows that var2(WT) rescued leaf variegation, demonstrating that our complementation assay was successful. However, we observed that var2(488) also rescued leaf variegation. This recovery was in strong contrast with that observed with E. coli H417L (Figure 2). Such recovery of leaf variegation occurred not only in mature leaves but also at the early stage of development for the first true leaves (Figure 3B). Moreover, var2(488) plants grew normally and displayed rosette leaves that were comparable between Col and var2(WT) plants after 6 weeks (see Supplemental Figure 1 online). No recovery of variegation was observed in negative controls var2(267) and var2(267 488). Therefore, we excluded the possibility that the recovered variegation in var2(488) was attributable to a secondary effect of overexpressing a nonfunctional FtsH2. It is noteworthy that var2(267) displayed variegation very similarly to the weak allele var2-9, although var2(267 488) resembled the strong allele var2-1 (Figure 3B). These data suggest that although FtsH2(488) does not cause variegation by itself, it can enhance the variegation phenotype caused by a mutation in the ATPase domain.
Figure 3.
Complementation Analysis of var2 with Wild-type and Mutated FtsH2.
(A) Photographs of 3-week-old Col, var2-1, var2(WT), var2(267), var2(488), and var2(267 488). Bars = 6 mm.
(B) Photographs of 8-d-old Col, var2-1, var2(WT), var2(267), var2(488), and var2(267 488). Cotyledons are normal in all lines. The first pair of true leaves displayed variegation in var2-1, var2(267), and var2(267 488) but not in Col, var2(WT), and var2(488). Bars = 3 mm.
(C) Evaluation of leaf variegation by chlorophyll content and digital imaging. Chlorophyll concentrations (Chl Conc.) of 3-week-old seedlings are shown based on fresh weight (F.W.). The degree of leaf variegation was estimated from images of the leaves as described in Methods. Data were generated from three biological replicates (bars represent sd).
(D) Immunoblot analysis of FtsH2. Total proteins from 3-week-old seedlings were probed with anti-FtsH2 (normalized by fresh weight). A CBB-stained gel of the samples is shown on the bottom. Relative levels of FtsH2 (normalized by LHCII, and Col was adjusted as 1) are shown on the top (bars represent sd; data were obtained from five biological replicates).
To elucidate the degree of variegation observed in the transgenic lines described above, we measured chlorophyll contents in mature leaves (Figure 3C). We also measured the degree of variegation based on the image analysis of the first three true leaves (Figure 3C), as described in Methods. Collectively, the results from these experiments confirmed our conclusions that var2(488) is indistinguishable from Col and var2(WT) and that the protease activity from FtsH2 is dispensable for the FtsH function.
Accumulation of FtsH2 in var2(WT) and var2(488)
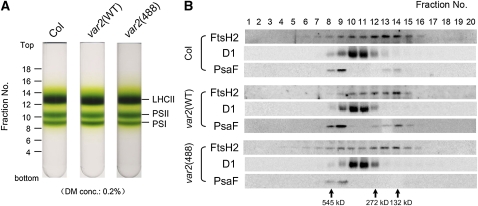
Immunoblot analysis was performed to examine the accumulation of FtsH2 in vivo. Total proteins from 3-week-old leaves in Col and transgenic lines were probed with anti-FtsH2 antisera. The FtsH2 levels were similar between Col, var2(WT), and var2(488) (Figure 3D). When 6-week-old plants were subjected to immunoblot analysis, FtsH2 levels were again similar between FtsH2(WT) and FtsH2(488) (see Supplemental Figure 2 online). By contrast, accumulation of FtsH2(267) was apparently reduced and had an epistatic effect over FtsH2(488) (Figure 3D). Consequently, the effect of G267D is consistent with G195D in E. coli and suggests a deficiency in protein stability/complex formation. Next, we tested the accumulation and formation of the FtsH complex in var2(488). Thylakoid membranes purified from total cell extracts were solubilized by 0.2% dodecylmaltoside (DM) and separated by sucrose density gradient centrifugation (Figure 4A). Fractions were collected and subsequently subjected to immunoblot analysis (Figure 4B). Anti-FtsH2, Anti-D1, and Anti-PsaF antibodies were used to locate the relative position of the FtsH complex, PSII, and photosystem I, respectively. FtsH2 was detected broadly in the sucrose gradients from Col, var2(WT), and var2(488), with the peak at ~200 kb, which was apparently smaller than the size of a hexameric form (450 kD) and likely represented oligomeric forms. Nevertheless, the distribution profile of the FtsH2 oligomers in the sucrose density gradient was similar between var2(WT) and var2(488). We therefore considered that the lack of zinc binding does not prevent FtsH2(488) from forming the complex. Together, these results indicate that FtsH(488) accumulates and forms a complex comparably to FtsH2(WT) in thylakoid membranes.
Figure 4.
Sucrose Density Gradient Analysis of Thylakoid Membrane Proteins.
(A) Photographs of 0.1 to 1.3 M sucrose density gradients. Thylakoid membranes from Col, var2(WT), and var2(488) were solubilized using 0.2% DM. Relative positions of the fractions and the locations of LHCII monomer/trimer (LHCII), PSII monomer (PSII), and photosystem I complex (PSI) are indicated.
(B) Immunoblot analysis of the sucrose density gradient fractions. The fractions were collected from the tubes of the sucrose density gradients shown in (A). Equal amounts of the fractions were subjected to SDS-PAGE and probed with anti-FtsH2, anti-D1, and anti-PsaF. Immunoblot analysis data from Col, var2(WT), and var2(488) are shown.
Protease Activity and Photoinhibitory Status in var2(488)
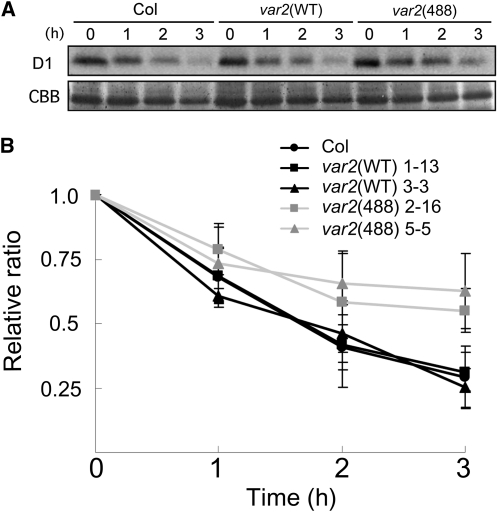
The normal phenotype of var2(488) strongly implied that the FtsH complex can retain its protease activity despite the existence of proteolytically inactive FtsH2(488). To test whether the presence of FtsH2(488) affects the protease activity of the FtsH complex, var2(488) and other control plants were subjected to an in vivo protease assay. We set up a system of testing D1 degradation in vivo using intact leaf discs that had been pretreated with lincomycin, an inhibitor of chloroplast protein synthesis (Kato et al., 2009). Photodamaged D1 is one substrate for FtsH in thylakoid membranes; its rapid turnover enabled assessment of the proteolytic activity of FtsH. After high-light irradiation (1200 μmol photons m−2 s−1), the D1 levels were monitored carefully using immunoblot analysis and normalized based on the level of LHCII (light-harvesting complex in PSII). The results from Col, var2(WT), and var2(488) indicated that the level of remaining D1 protein in var2(488) was slightly higher (~20%) than that in Col and var2(WT) after 3 h of high-light irradiation (Figure 5). Leaves from independent transgenic lines were also used, yielding similar data results (see Supplemental Figure 3 online). Based on these results, we inferred that the FtsH complex with FtsH2(488) can degrade D1 protein, but not at levels similar to the wild type. Thus, it was possible that FtsH2(488) indeed affected protease activity of the FtsH complex.
Figure 5.
In Vivo D1 Degradation Assay under High-Light Irradiation.
(A) Immunoblot analysis. Total proteins from Col, var2(WT), and var2(488) leaf discs (8 weeks) were incubated with anti-D1 (top panel). The CBB-stained gel of the same samples (in the area corresponding to LHC) is shown on the bottom. Prior to extraction, discs were infiltrated with 5 mM lincomycin and placed under high light (1200 μmol photons m−2 s−1) for 3 h.
(B) Relative ratio of D1 levels during high-light irradiation shown in (A). D1 levels were estimated from immunoblots and normalized based on the CBB-stained LHC (bars indicate sd; n = 5; five biological replicates). The ratio at 0 h was adjusted to 1. Calculations from two independent transgenic lines are presented for var2(WT) (lines 1-13 and 3-3) and var2(488) (lines 2-16 and 5-5).
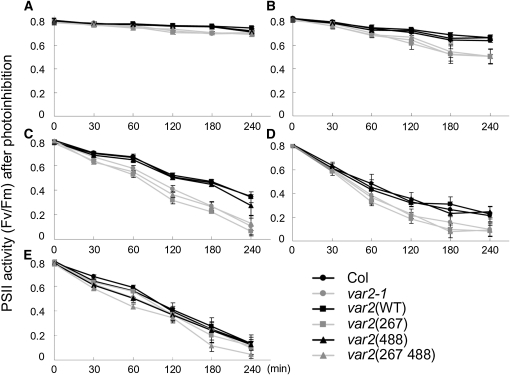
To test whether altered FtsH protease activity for degrading D1 influences the PSII repair cycle, susceptibility of var2(488) and other control plants to photoinhibition of PSII was measured using chlorophyll fluorescence analysis. The maximum quantum yields of PSII (Fv/Fm) were scored in each line, as previously reported (Figure 6). An increased PSII photosensitivity was detected in var2-1, var2(267), and var2(267 488). By contrast, var2(WT) and var2(488) showed Fv/Fm profiles similar to Col. This trend was true under several different light conditions (Figures 6A to 6D). In the presence of lincomycin, PSII sensitivity to high light was similar among all plants tested (Figure 6E). These data suggest that the reduced protease activity in var2(488) is too slight to influence the photoinhibition status of PSII. It is noteworthy that our D1 degradation assay was performed in the presence of lincomycin, which inhibits PSII repair equally in all the tested lines (Figure 6E). This condition means that we detected the protease activity of FtsH, irrespective of the impairment in PSII repair. Based on the measurement of Fv/Fm, we considered that var2(488) retains FtsH complexes capable of D1 degradation within the capacity of PSII repair, most likely because protease sites are provided by other isomers. Alternatively, our result with var2(488) raises the possibility that its chaperone activity rather than its protease activity plays a role in the PSII repair because FtsH2(488) retains its functional ATPase domain (see Discussion).
Figure 6.
Measurement of PSII Activity Estimated Using Chlorophyll Fluorescence during High-Light Irradiation.
(A) to (D) Relative ratio of PSII activity without lincomycin. Mature leaf discs from Col, var2-1, var2(WT), var2(267), var2(488), and var2(267 488) were exposed to low light (250 μmol photons m−2 s−1) (A), moderate light (500 μmol photons m−2 s−1) (B), high light (1000 μmol photons m−2 s−1) (C), or strong light (1500 μmol photons m−2 s−1) (D); the maximum photochemical efficiency (Fv/Fm) was measured at 0, 30, 60, 120, 180, and 240 min after irradiation (mean ± sd; data were obtained from five biological replicates).
(E) An identical experiment to that described in (C) under strong light (1500 μmol photons m−2 s−1) except plants were infiltrated with lincomycin prior to irradiation, as shown in Figure 5.
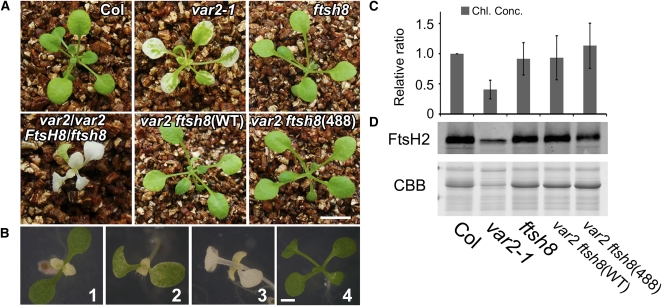
Expression of FtsH2(488) Fully Rescues Seedling Lethality in var2 ftsh8
It has been shown that normal chloroplast development requires at least one isomer from each FtsH type. Loss of the two isomers from either type (i.e., var1 ftsh1 or var2 ftsh8) completely impairs chloroplast development and engenders seedling lethality (Zaltsman et al., 2005a). We examined whether FtsH2(488) can rescue this seedling lethality in the var2 ftsh8 background. To perform this experiment, var2 ftsh8(488) was generated by crossing var2(488) with an ftsh8 mutant in which the FtsH8 gene is disrupted by a T-DNA insertion (Sakamoto et al., 2003). We also used var2(WT) as a control. The resulting F2 plants were subjected to PCR to verify genotypes. As reported previously, var2 ftsh8 double mutants showed a severely impaired growth defect, although pale plants whose genotypes were var2/var2 FtsH8/ftsh8 were able to grow after they were transferred to soil. The result demonstrated that not only FtsH2(WT) but also FtsH2(488) rescued seedling lethality in var2 ftsh8 (Figure 7A). Similarly to the case in var2(488), such a rescue of leaf variegation occurred not only in mature leaves but also at the early stage of development for the first true leaves (Figure 7B). Measurement of chlorophyll contents supported that chloroplast development was comparable between Col, ftsh8, var2 ftsh8(WT), and var2 ftsh8(488) (Figure 7C). Immunoblot analysis also supported that FtsH2 accumulated substantially in var2 ftsh8(488) (Figure 7D). Based on these results, we concluded that the FtsH heterocomplex does not require protease activity from Type B isomers (FtsH2 and FtsH8).
Figure 7.
Complementation Analysis of var2 ftsh8 with Wild-type and Mutated FtsH2.
(A) Photographs of 3-week-old Col, var2-1, ftsh8, var2/var2 FtsH8/ftsh8, var2 ftsh8(WT), and var2 ftsh8 (488). Not only FtsH(WT) but also FtsH2(488) rescues seedling lethality of var2 ftsh8. Bars = 6 mm.
(B) Photographs of 8-d-old var2 (1), var2/var2 FtsH8/ftsh8 (2), var2 ftsh8 (3), and var2 ftsh8(488) (4) plants. Bars = 3 mm.
(C) Chlorophyll content of 3-week-old seedlings (estimated based on fresh weight). The relative ratios of chlorophyll concentrations (Chl. Conc.) were adjusted according to Col (Col was set to 1). Data were generated from five biological replicates (bars represent sd).
(D) Immunoblot analysis of FtsH2. Total proteins from 4-week-old seedlings were probed with anti-FtsH2 (normalized by fresh weight). A CBB-stained gel of the samples is shown on the bottom. The experiment was repeated five times, with results obtained from five biological replicates.
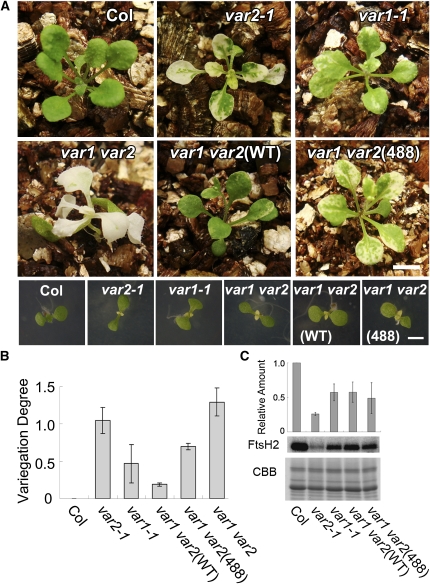
Expression of FtsH2(488) Partially Rescues Leaf Variegation in var1 var2
The aforementioned dispensability of the Type B protease activity is probably explainable in light of the existence of Type A isomers, such as FtsH5, that retain the protease domain. Next, we introduced FtsH2(WT) and FtsH2(488) into the var1 var2 background (lacking both FtsH2 [Type B] and FtsH5 [Type A]) and assessed leaf variegation phenotypes. We hypothesized that the protease activity in FtsH2 would become more important in var1 var2 than in var2 ftsh8 because the coordination of the accumulation of both isoforms in the FtsH heterocomplex would lead to decreased overall FtsH levels in var1 var2.
The variegated phenotype observed in var1 var2 is more severe than that in var2. If the expression of FtsH2(WT) or FtsH2(488) rescues variegation, then the plants should resemble var1 and accumulate FtsH to a level similar to that of var1. This prediction is based on the previous observation that Type A and Type B isomers do not mutually complement each other’s function. In fact, we observed that var1 var2(WT) plants showed weak variegation similar to that of var1. By contrast, var1 var2(488) plants exhibited variegation that is stronger than var1-1 (Figure 8A). When the degree of variegation was measured using image analysis, the difference was clear (Figure 8B). Furthermore, the differential degree of variegation was remarkable when these plants were grown at low temperature (17°C) (see Supplemental Figure 4 online). Leaf variegation has been shown to be enhanced at low temperature in var2 but not in var1 (Sakamoto et al., 2002). Growth at 17°C indicated that var1 var2(WT) was similar to var1, although var1 var2(488) exhibited enhanced variegation similar to that of var2 (see Supplemental Figure 4 online). Together, these data indicate that the loss of protease activity in FtsH2 affects the variegation phenotype in var1 var2(488), in contrast with the complete rescue observed in var2(488) and in var2 ftsh8(448). Immunoblot analysis showed that the observed difference in the variegation degree between var1 var2(488) and other control plants was not attributable to the substantial difference in the accumulation of FtsH2 (Figure 8C). As expected, var1 var2(488) accumulated FtsH2 to a similar level as var1-1, var1 var2(WT): these lines accumulated ~60% of FtsH2 compared with Col, whereas var2 accumulated ~20%. Together, we concluded from this data that in the absence of FtsH5, the loss of FtsH2 protease activity is sufficient to cause leaf variegation.
Figure 8.
Complementation Analysis of var1 var2 with Wild-Type and Mutated FtsH2.
(A) Photographs of 3-week-old (top panels) and 8-d-old (bottom panels) Col, var2-1, var1-1, var1 var2, var1 var2(WT), and var1 var2(488). Bars = 6 mm in the top panels and 3 mm in the bottom panels.
(B) Evaluation of leaf variegation using digital image analysis. The degree of leaf variegation of the first three true leaves (at 3 weeks) was estimated as described in Methods. The value was estimated from three independent experiments (bars indicate sd; n = 3) and normalized according to Col (Col was set to 0).
(C) Immunoblot analysis of FtsH2. Total proteins from 3-week-old seedlings were probed with anti-FtsH2 antisera (normalized by fresh weight). A CBB-stained gel of the samples is shown on the bottom. Relative levels of FtsH2 (normalized by LHCII; Col was set to 1) are shown at the top (bars represent sd; data from three biological repeats).
An Alternative Approach to Rescue Leaf Variegation in var2 Using an Inducible Promoter
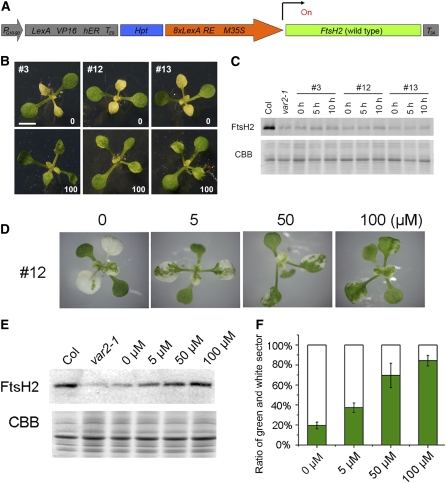
Given the sufficient protease activity in the wild-type FtsH heterocomplex, we next attempted to ascertain whether the overall amount of FtsH, rather than the protease activity, influences leaf variegation in var2. Previous studies and this study used only the constitutive CaMV 35S promoter to drive FtsH expression. In no transgenic lines examined do FtsH2 protein levels appear to exceed those in Col, suggesting that the accumulation of FtsH is coordinately regulated posttranslationally between Type A and Type B isomers. In this study, we therefore tested whether the different levels of FtsH2(WT) expression affect leaf variegation in var2. To perform this experiment, we introduced a chemical-inducible transactivation system: XVE (Zuo et al., 2000). The vector in this system (pER8, summarized in Figure 9A) contains the chimeric gene that drives constitutive expression of XVE (a transactivator consisting of the LexA DNA binding domain, VP16 activation domain, and human estrogen receptor C-terminal region). The XVE transactivation of the target gene depends on external application of estrogen hormone. We cloned FtsH2(WT) into pER8 to place it under the control of the target 8xLexA-35S minimal promoter and to allow expression of FtsH2(WT) in a 17-β-estradiol dose-dependent manner (Figure 9A). The resulting construct pER8-FtsH2(WT) was transformed into var2; then, hygromycin-resistant transgenic Arabidopsis lines responding to hormone were selected for additional analysis in the T4 generation. We obtained at least three lines [designated as pER8-var2(WT), and independent lines were #3, #12, and #13] that showed rescue of leaf variegation when plants were grown on plates supplemented with 100 μM 17-β-estradiol (Figure 9B). Under this condition, upregulation of FtsH2 at the protein level was confirmed using immunoblots (see Supplemental Figure 5 online).
Figure 9.
Rescue of Leaf Variegation in var2 by Inducible Expression of FtsH2(WT).
(A) Schematic diagram of the pER8-FtsH2(WT) construct. The region corresponding to the T-DNA region is shown. FtsH2 cDNA (shown in green) was cloned between the target promoter (8xLexA RE M35S, shown as an orange arrow) and rbcS3A poly(A) addition sequence (T3A, shown in gray). The chimeric gene (LexA VP16 hER TE9 shown in gray) is constitutively expressed by the PG10-90 promoter (gray arrow) and transactivates FtsH2(WT) in an estradiol dose-dependent manner. Hpt (shown in blue) indicates hygromycin-resistant gene as a selectable marker.
(B) Photographs of pER8-var2(WT) transgenic lines grown on Murashige and Skoog plates. Three independent lines (#3, #12, and #13) were sown on hygromycin plates with or without supplementation of 100 μM 17-β-estradiol (shown as 0 or 100, respectively). Representative plants are shown 14 d after sowing. Bar = 6 mm.
(C) Induction of FtsH2 in excised mature leaves from pER8-var2(WT). Leaflets from the three pER8-var2(WT) lines were incubated on 100 μM 17-β-estradiol solution for 0, 5, and 10 h (as shown on the immunoblot). Three leaflets were collected and subjected to immunoblot analysis with FtsH2 antisera (top panel). A CBB-stained gel of the samples (in the area corresponding to LHC) is shown on the bottom panel (samples were normalized based on fresh weight). Experiments were performed with four biological repeats.
(D) Rescue of leaf variegation in pER8-var2(WT) with different concentrations of 17-β-estradiol. Photographs of plants from line #12 with 0, 5, 50, or 100 μM 17-β-estradiol are shown.
(E) Induction of FtsH2 correlates with 17-β-estradiol concentration. Immunoblots of total proteins from line #12 grown in different concentrations of 17-β-estradiol (shown on top of the blot) were probed with anti-FtsH2 antisera (samples were normalized by chlorophyll content). A CBB-stained gel of the samples (in the area corresponding to LHC) is shown at the bottom. Experiments were performed with three biological repeats.
(F) Degree of leaf variegation in pER8-var2(WT) #12 lines grown in different concentrations of 17-β-estradiol (bars represent sd; data from 10 leaves with five different individuals).
Recovery of Leaf Variegation in var2 Depends on FtsH Amounts
We first tested whether our system rescues leaf variegation in mature leaves. However, incubation of 6-week-old excised leaves with 100 μM 17-β-estradiol solution neither allowed FtsH2 accumulation on immunoblots nor rescued leaf variegation (Figure 9C). We reasoned in part that thylakoid formation in mature leaves is irreversibly impaired in plastids of white sectors, irrespective of FtsH levels (Sakamoto et al., 2009). We then attempted periodical application of 17-β-estradiol into specific tissues like shoot meristem and juvenile leaves, but technical difficulties prevented us from detecting increased FtsH2 levels in these attempts. To assess the relationship between FtsH levels and leaf variegation, we therefore sowed and grew pER8-var2(WT) transgenic lines on plates supplemented with different concentrations of 17-β-estradiol (0, 5, 50, and 100 μM; Figure 9D). The results showed that FtsH2 indeed accumulates according to 17-β-estradiol concentration, confirming dose-dependent XVE transactivation (Figure 9E). More importantly, the degree of leaf variegation was shown to correlate well with the inducer concentration (Figure 9F). These observations in pER8-var2(WT) are consistent with our assumption that the overall FtsH level is a primary factor in var2 leaf variegation. Leaf variegation in pER8-var2(WT) was not fully rescued in 100 μM 17-β-estradiol despite substantial FtsH levels. We considered that prolonged exposure to 17-β-estradiol had a secondary effect or that FtsH levels in tissues other than mature leaves (e.g., shoot meristem) are important for proper chloroplast development.
DISCUSSION
Protease Activities in the FtsH Heterocomplex
The importance of the FtsH protease during chloroplast biogenesis has been well documented. However, very little information is available with regard to how the protease activity is controlled, or mutually affected, within the FtsH complex. In this study, we performed multiple complementation analyses with a combination of various ftsh mutants and constructs, attempting to gain an insight into the relationship between the protease activity and chloroplast development. Our finding demonstrates that the number of protease sites is not a limiting factor for manifesting FtsH function, as represented by the dispensability of Type B protease activity. However, several important questions remain to be answered in this study, such as, how many protease sites are required, whether the complete loss of protease activity in the FtsH complex can be tolerated, and why FtsH2(488) did not complement the variegation in var1 var2. Further studies are apparently required to answer these questions.
Distinctive ATPase and protease domains are found in a major type of the proteases involved in the processive degradation of chloroplast proteins (breaking polypeptides down to oligopeptides or completely to amino acids), such as Clp and FtsH (Adam et al., 2006; Sakamoto, 2006). A common feature in these proteases is that they form a complex with different subunits/isoforms. Clp has separate subunits for its ATPase and its core protease, whereas FtsH has isoforms that contain both ATPase and protease domains in a single peptide. In this study, we assessed the importance of the ZnBD acting as a catalytic center for zinc-metalloprotease in FtsH2. The ZnBD includes consensus HEXXH residues within its C-terminal region (van der Hoorn, 2008), and both His residues are required as a ligand of zinc. We generated a proteolytically inactive version of FtsH2 [FtsH2(488) lacking its first His residue] and showed that the equivalent mutation in E. coli FtsH abolished protease activity. Importance of the zinc binding site is also suggested in E. coli FtsH (Karata et al., 1999). A study on the Site-2 protease family (a member of intramembrane metalloproteases) suggests that a mutation in this His residue can engender a severe zinc binding defect and can completely abolish its catalytic activity (Feng et al., 2007). In contrast with these observations, FtsH2(488) had little impact on var2 variegation or even on var2 ftsh8 lethality because chloroplast FtsHs are formed by four isomers. Consequently, unlike the authentic bacterial homocomplex, the FtsH heterocomplex in chloroplasts can accommodate inactive protease sites, suggesting that protease domains in the complex function redundantly rather than coordinately. Such independence of the proteolytic sites in the FtsH protease chamber has been predicted based on the proposed crystal structure of homomeric bacterial FtsH complexes (Bieniossek et al., 2009). Our current study on the heteromeric FtsH in chloroplasts verified this prediction. By contrast, a mutation in the ATPase domain, which coordinately forms a single ATPase unit within the hexamer, might directly affect ATP binding.
FtsH is suggested to degrade processively through both N- and C-terminal regions (Chiba et al., 2002). Substrates are first unfolded/translocated by passing through the central pore of the ATPase domain (Suno et al., 2006). Substrates are subsequently delivered to the proteolytic chamber where hydrolysis of peptide bonds takes place at any of the catalytic centers and generates free amino acids. Coupled unfolding/translocation and proteolysis are similar, and evolutionarily related, processes in FtsH, Clp, and the eukaryotic 26S proteasomes (Wickner et al., 1999; Moon et al., 2004; Baker and Sauer, 2006; Bukau et al., 2006). In E. coli, FtsH was shown to lack robust unfoldase activity, which might partially account for its substrate specificity compared with other proteases (Herman et al., 2003). This observation implies that unfoldase, rather than protease, activity is a rate-limiting step for FtsH protein degradation. Consistent with this, our results from var2(488) and var2 ftsh8(488) reveal that the protease sites exist in excess and that some of them are dispensable.
FtsH plays a predominant role in degrading photodamaged D1 protein in the PSII repair cycle. This degradation by FtsH reportedly proceeds in concert with another endopeptidase (Deg) that is attached peripherally to both sides of thylakoid membranes (Kapri-Pardes et al., 2007; Sun et al., 2007, 2010). In the transgenic line var2(488), in which the number of protease sites was limited, we observed increased accumulation of D1 protein after 3 h of high-light irradiation (Figure 5). Our results therefore provide supportive evidence that FtsH is involved in processive degradation of D1 protein. Unlike var1 and var2 mutants, however, var2(488) showed no impaired PSII repair, as measured by Fv/Fm. One possibility to explain this inconsistency is that a partial loss of protease sites had very little influence on the PSII repair despite a detectable reduction in D1 degradation. Because D1 degradation proceeds even under nonphotoinhibitory light conditions (e.g., Kato et al., 2009), a fraction of D1 levels detected in our assay represents D1 degradation not directly related to PSII repair cycle. It is reasonable to assume that var2(488) retains sufficient activity to degrade D1 during PSII repair, although var2 does not. We therefore considered that the reduced D1 degradation in var2(488) was within the capacity of the repair cycle under our conditions (250 to 1500 μmol photons m−2 s−1). We raise the alternative possibility that FtsH is involved in not only degradation but also refolding of D1 protein. FtsH2(488), despite its defective protease activity, forms the FtsH complex with Type A isoforms and retains its functional ATPase domain that confers unfoldase activity. Misfolded D1 could be recruited into this proteolytically deficient complex and unfolded, but subsequently refolded into PSII without processive degradation. To address this possibility, further analysis is required.
Composition of the FtsH Hetero-Complex with Type A and Type B Isomers
FtsH seems unstable when purified from thylakoid membranes. Nevertheless, chloroplastic FtsH is very similar to bacterial homologs and is predicted to form a hexamer. Although the chloroplastic FtsH complex has not been characterized biochemically, this genetic study along with previous data provided several important features on its composition. Because of the lethal phenotype in ftsh1 var1 (lacking Type A) or in var2 ftsh8 (lacking Type B; Zaltsman et al., 2005a), the major form of chloroplastic FtsH is presumed to be solely heteromeric with both types. The two isomers within each type (i.e., FtsH1/5 in Type A or FtsH2/8 in Type B) are functionally interchangeable, as evidenced by transcomplementation analysis (Yu et al., 2004, 2005). Based on transcript abundance, Zaltsman et al. (2005a) predict that Type B isomers outnumber Type A isomers in the heterocomplex. However, this abundance might not reflect protein stoichiometry because FtsH levels are regulated posttranscriptionally. Investigation of the protein levels of FtsH isomers in vivo suggests that the ratio of Type A and Type B isomers is ~1:1 (Yu et al., 2004). We therefore predict that a hexameric FtsH complex has three protease sites derived from each type. A complete rescue of var2 ftsh8 seedling lethality with FtsH2(488) indicates that three protease sites from Type A isomers in the proteolytic chamber are sufficient to allow minimal proteolysis by FtsH. The reason that var1 var2(488) shows more severe variegation than var1 is explainable by decreased FtsH levels (see below), raising the possibility that the overall FtsH amount becomes an important factor when the protease sites are limited.
The fact that all FtsH isomers have consensus ZnBDs contrasts with the protease domain of the Clp complex, in which two isoforms termed ClpP and ClpR are present (Clarke et al., 2005). In Arabidopsis, five ClpP isomers (ClpP1, P3, P4, P5, and P6) contain all the necessary catalytic residues for Ser proteases, whereas four ClpR isomers (ClpR1, R2, R3, and R4) do not and, therefore, likely lack protease activity. Furthermore, the protease core contains another subunit ClpT (ClpT1 and T2) that is plant specific and attached peripherally to the protease core complex (Peltier et al., 2001, 2004). Similar to the case in FtsH, multimeric isoforms present in Clp are characteristic to photosynthetic organisms. Analysis of clpp and clpr mutants revealed involvement of Clp in various steps of plant growth. Functional redundancy among the isomers was also investigated. For example, Kim et al. (2009) recently reported transcomplementation analysis of mutants lacking ClpR isomers (clpr1, clpr2, and clpr4). Based on the rescue of defective growth in clpr mutants, only ClpR1 and ClpR3 appear to function redundantly in the Clp core complex. Consequently, FtsH and Clp share the complexity of having multiple isomers, but they differ considerably in their numbers of isomers, their numbers of nonproteolytic isomers, and functional redundancy between the isomers. Despite such diversity, the presence of FtsH isomers of two types seems well conserved in green and red algae, moss, and flowering plants (Sakamoto et al., 2003; Yu et al., 2004). Chloroplasts constantly receive oxidative damage; such heterocomplexes are perhaps more advantageous than homocomplexes for adapting to various environmental conditions. Regarding FtsH isoforms, it is noteworthy that the Arabidopsis genome contains four genes potentially coding for FtsH homologs that apparently lack an active zinc binding domain (termed as FtsHi; Sokolenko et al., 2002). Relevance of these FtsHi homologs to the chloroplastic FtsH complex is currently unclear.
Factors Affecting Leaf Variegation in var2
Our study reveals that two factors of FtsH, protease activity and protein accumulation level, apparently together affect formation of leaf variegation in var2. Leaf variegation predicts a threshold of unknown factors that determine chloroplast development in a cell-autonomous fashion (e.g., Rosso et al., 2009). We demonstrate that the threshold is determined by the total amount of the FtsH heterocomplex rather than the total number of protease sites. The overall FtsH level as a predominant factor for var2 leaf variegation is supported by two complementary experiments. First, it has been shown that the degree of leaf variegation in most var2 alleles correlates with the FtsH level (Chen et al., 2000; Sakamoto et al., 2004). Second, the results from pER8-FtsH2(WT) study demonstrated that rescue of leaf variegation depends on the FtsH expression level. Manipulation of leaf variegation with the inducible promoter enables us to dissect factors involved in chloroplast development in future research.
Earlier studies suggest that the overall FtsH levels are coordinately regulated posttranslationally. Lack of FtsH2 results in a reduced FtsH5 level and vice versa (Sakamoto et al., 2003). Ectopic expression of FtsH2 and other isomers does not allow upregulation of the overall FtsH levels beyond the level of the wild type (Yu et al., 2004, 2005). Based on these observations, we hypothesize that the overall FtsH level is determined by whichever of the types is reduced relative to the proper stoichiometry (likely to be 3:3). For example, the FtsH8 level determines the total FtsH level in var2, and FtsH1 level determines the total FtsH level in var1. Similarly, complete rescue of seedling lethality in var2 ftsh8(488) is explained by this assumption (the total FtsH level is determined by FtsH1/5 levels, which is equivalent to that in the wild type). Enhanced variegation in var1 var2(488) is explainable by the prediction that the overall FtsH level is determined by the amount of FtsH1. The FtsH2 level is lowered in this line (~60%; Figure 8). We predict that under the lowered FtsH level, the number of protease sites becomes critical and enhances leaf variegation. Complementation of var1 var2 ftsh8 with FtsH2(488) was not investigated in this study, but our assumption leads to the prediction that the resulting transgenic line has an FtsH level equivalent to that of var1 var2(488) but with more variegated sectors than var1 var2(488).
Characterization of the transacting mutations that rescue leaf variegation was reported previously (Park and Rodermel, 2004; Miura et al., 2007; Yu et al., 2008; Liu et al., 2010b). Most suppressor mutations resulted from or were associated with a defect in chloroplast translation. Based on these investigations, leaf variegation was accounted for by the balance between protein synthesis and degradation in chloroplasts (Miura et al., 2007). Our study demonstrates that leaf variegation in var2 is predominantly determined by the overall FtsH level but that the protease activity becomes important when FtsH levels are limited. We therefore consider that although the FtsH complex has excess protease sites, minimal protease activity is essential for proper chloroplast development. Based on our recent observation, we also consider that the rescue of leaf variegation in var2 suppressors can be explained by retarded chloroplast development during leaf morphogenesis (Sakamoto et al., 2009). Retarded chloroplast development might lower the threshold FtsH level: the required level for proper chloroplast development. Most mutations associated with chloroplast translation indeed appear to retard chloroplast development (e.g., fu-gaeri1 has a nonlethal defect in chloroplast translation initiation factor cpIF2 and shows a slightly virescent phenotype; Miura et al., 2007). We consider that defects in chloroplast translation are related indirectly to leaf variegation.
In conclusion, we experimentally identified ZnBD in FtsH2 as a catalytic site for proteolysis that acts redundantly in the proteolytic chamber. Unlike the bacterial homohexamers, the chloroplastic FtsH has the capacity to accommodate proteolytically inactive isoforms. Dispensability of Type B isomers in the heterocomplex suggests that protease activity does not limit the function of FtsH for chloroplast development. However, our complementation with an inducible promoter suggests that the overall FtsH level predominantly determines the threshold of chloroplast development when the protease activity is in excess. Our study therefore revealed unique features of the heteromeric FtsH complex that are characteristic of photosynthetic organisms.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col was used as the source for a wild-type plant. We also used the FtsH-related leaf variegation mutants (var2-1, var2-9, var1-1, and var1-1 var2-1) in this study, as described previously (Takechi et al., 2000; Sakamoto et al., 2002, 2004). The plants were germinated on 0.7% (w/v) agar plates containing Murashige and Skoog medium supplemented with Gamborg’s vitamins (Sigma-Aldrich) and 1.5% (w/v) sucrose. For induction with estrogen hormone, 17-β-estradiol (Wako Pure Chemical Industries) was added to media at an appropriate concentration. The plants were grown under 12-h light (60 μmol photons m−2 s−1) at 22°C, transferred to soil after 2 weeks, and maintained under the same conditions. FtsH2(WT) and other mutated FtsH2 genes were cloned into the binary vector pBI121 (Clontech) as described below and transformed into Col. Agrobacterium tumefaciens–mediated transformation was performed as described previously (Miura et al., 2007). More than three independent transgenic lines were generated for each construct. For selecting the transformants homozygous for var2, DNAs were isolated from F2 kanamycin-resistant plants. To verify the genotype for var2, the genomic region corresponding to the var2-1 mutation was amplified by PCR using primers KT204 and KT402. Sequences of all primers used for this study are listed in Supplemental Table 1 online. The amplified DNA was subsequently sequenced using the primer KT304. For selecting homozygous transformants for both var1 and var2-1, two independent lines of var2(WT) and var2(488) were crossed with var1-1. To verify the var1 genotype, the genomic region surrounding the var1 mutation was amplified by PCR using SQ102F and SQ110R. It was sequenced using the primer SQ103F. For selecting homozygous transformants for var2 ftsh8, var2(WT) and var2(488) were crossed with ftsh8 (a T-DNA insert mutant, which was inserted in intron 1; Sakamoto et al., 2003). To verify the ftsh8 genotype, the T-DNA insertion was amplified using PCR with PCR primers F2-102 and F2-101 and T-DNA left border primer TMRI-LB2.
Site-Directed Mutagenesis and Plasmid Construction
The original VAR2 cDNA (Takechi et al., 2000) was cloned into BamHI and SacI sites of the pBluescript KS+ vector (Stratagene). This clone was subjected to site-directed mutagenesis using the QuickChange Multi site-directed mutagenesis kit (Stratagene). FtsH2(488) was created using the primer ZnBD-A1, resulting in the substitution of the CAT to GCT codon to generate H488L. Primer sequences are given in Supplemental Table 1 online. Similarly, FtsH(267) was created using the primer G267D-A1, resulting in the substitution of GGT to GAT codon to generate G267D. FtsH2(488 267) was created using both ZnBD-A1 and G267D-A1 primers. After sequences were verified, the clones were finally cloned into BamHI and SacI sites of the binary vector pBI121 so that the β-glucuronidase gene in PBI121 was replaced with the mutated FtsH2 genes. To mutagenize Escherichia coli ftsH genes, we used the primer H417L to create a codon change from CAC to CTC and the G195D primer to create a codon change from GGT to GAT. The original E. coli ftsH gene was cloned into a pET backbone vector pIFH108 (Karata et al., 1999); then this plasmid was used for site-directed mutagenesis. The resulting plasmids (pIFH108 containing wild-type ftsH and mutated ftsH, as indicated in Figure 2) were subjected to complementation analysis. To construct pER8-FtsH2(WT), the XVE-inducible binary vector pER8 (Zuo et al., 2000) was digested with XhoI and SpeI, and a full-length FtsH2 cDNA was cloned into these sites using an In-fusion advantage PCR cloning kit (Clontech).
Protease Assay and Complementation of ftsH Mutants in E. coli
A derivative of the BL21 (DE3) strain [AR5088 (sfhC ΔftsH)] was used as a host for our bacterial complementation experiment (Qu et al., 1996). AR5088 cells carrying pIFH108 and other derivatives were cultured in 3 mL Luria-Bertani (LB) liquid medium containing ampicillin (100 μg/mL) and kanamycin (50 μg/mL) without isopropyl β-d-1-thiogalactopyranoside (IPTG) for 10 h. Harvested samples were subsequently subjected to SDS-PAGE and immunoblot analysis. For immunoblots using σ32 antibodies (kindly provided by Teru Ogura, Kumamoto University), cells were resuspended in SDS loading buffer (100 mM Tris-Cl, 4% [w/v] SDS, 200 mM DTT, 20% [v/v] glycerol, and 0.05% [w/v] bromophenol blue) and boiled at 100°C for 5 min prior to SDS-PAGE and subsequent Coomassie Brilliant Blue (CBB) staining. Signals were measured using the NIH Image program (http://rsbweb.nih.gov/). The experiments were repeated with six biological replicates and subjected to statistical analysis according to standard deviations. Also, ftsH1 (AR754), a temperature-sensitive mutant of ftsH, was used as a host for complementation experiments. AR754 carrying a mutation in ftsH (Ts) showed a colony-forming defect at 42°C, but it grew normally at 37°C (Begg et al., 1992). The AR754 cells were first cultured in 3 mL LB liquid medium containing ampicillin without IPTG at 37°C for 10 h. For testing colony growth, cell cultures at the same density were plated on LB agar plates containing ampicillin without IPTG at 37 and 42°C overnight in five replicate experiments.
Measurement of Leaf Variegation Degree and Chlorophyll Content
The degree of leaf variegation was quantified based on the method previously described by Zaltsman et al. (2005b) with slight modifications. Intensity of the color image in the first three true leaves was measured using the NIH Image program at 3 weeks after sowing. Photographs of the leaves were taken under the same light conditions and with the same digital camera settings. Relative variegation degree was calculated using a mathematical formula: (Vn-Vc)/Vc. Therein, Vc represents the color image intensity of Col (100% green plant), Vn denotes the color image intensity of variegated mutants and transgenic lines, and Vn-Vc signifies the variegation degree. Total chlorophyll was extracted from shoot tissues harvested from 3-week-old seedlings with 80% v/v acetone. The chlorophyll concentration (based on fresh weight) was subsequently quantified according to Porra et al. (1989). Measurements of the degree of variegation and chlorophyll content were performed using three biological replicates; statistical analyses were done according to the standard deviation.
Protein Extraction, CBB Staining, and Immunoblot Analysis
For immunoblots, leaves were harvested from 3-week-old and 6-week-old plants. Protein extraction and SDS-PAGE were conducted as described previously (Kato et al., 2007). Loading was normalized by fresh weight or chlorophyll, as described in the text. For visualizing protein bands, SDS-PAGE gels were stained with CBB solution (1% [w/v] CBB [G 250], 50% [v/v] methanol, and 7% [v/v] acetic acid) for 15 min at room temperature and destained with destain solution (15% methanol and 7% acetic acid). For immunoblot analysis, SDS-PAGE gels were electroblotted to the polyvinylidene difluoride membrane (ATTO) and blocked by 5% (w/v) skin milk in PBST buffer (50 mM sodium phosphate buffer, pH 7.5, containing 155 mM NaCl and 0.05% [v/v] Tween 20). Then the membranes were incubated with anti-FtsH2 (Sakamoto et al., 2003), anti-D1 (AS 05084; Agrisera), or anti-PsaF (Farah et al., 1995) (dilution 1:5000). Signals from immunoblotting were detected using the ECL system (GE Healthcare) and recorded with a LAS100-mini system (Fuji Photo Film Co.). Experiments employed at least three biological repeats (indicated in each figure); data were subjected to statistical analysis according to standard deviation.
Preparation of the Thylakoid Membrane and Sucrose Density Gradient
For isolation of chloroplasts, fresh rosette leaves were harvested from 8-week-old plants and homogenized using a blender with homogenization buffer (330 mM mannitol, 50 mM HEPES, and 2 mM Na2EDTA, pH 7.0). The homogenate was filtered with gauze. Then, the filtrate was centrifuged at 80g for 3 min. Pellets were resuspended in 1 mL homogenization buffer and loaded on three layers of Percoll gradient (10, 40, and 80% [v/v]). The gradients were centrifuged at 2500g for 20 min. The sediment at the interface between the 40 and 80% gradient (intact chloroplasts) and the interface between the 10 and 40% gradient (broken thylakoids) was used for additional preparation of thylakoid membranes and diluted 10 times in extraction buffer (50 mM HEPES and 5 mM mannitol, pH 7.5). After centrifugation at 80g for 2 min, pellets were resuspended in PBS buffer (137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, and 1.5 mM KH2PO4) and adjusted to a concentration of 0.4 mg/mL chlorophyll. DM was added to the suspension to a final concentration of 0.2% (w/v). Then, thylakoid membranes were solubilized for 30 min on ice. After 15-min centrifugation at 15,000g, supernatants were loaded onto a linear sucrose density gradient (0.1 to 1.3 M, in 1× PBS and 0.05% [w/v] DM). Gradients were centrifuged (SW41-Ti rotor; Beckman) at 40,000g for 15 h at 4°C. Then, 20 fractions were collected and subjected to immunoblot analyses as described (Sakamoto et al., 2003). Experiments were performed using at least three biological replicates.
Fluorescence Measurement and D1 Degradation Assay
Maximum quantum efficiency of PSII photochemistry (Fv/Fm) was regarded as the PSII capacity. Leaf discs from mature rosette leaves (treated and not treated with lincomycin) were exposed to 250, 500, 1000, and 1500 μmol photons m−2 s−1 for appropriate times; their Fv/Fm was measured using a chlorophyll fluorometer junior-PAM (Heinz Walz). Before measurement, leaf discs were maintained in the dark for 10 min to oxidize the plastoquinone pool. The D1 degradation assay was conducted as described previously (Kato et al., 2009). Leaf discs (5-mm diameter) were harvested from mature rosette leaves of 8-week-old plants and transferred to a glass vial containing 5 mM lincomycin dissolved in 0.2% (v/v) Tween 20. Lincomycin was infiltrated for 1 min into the leaf discs using a syringe needle with a rubber cap. The lincomycin-treated leaf discs were subsequently placed on wet filter paper and irradiated with the light intensities and durations indicated in the figure legends. For preparation of thylakoid membranes, three leaf discs were collected at each time point and subjected to immunoblot analysis. Samples were loaded based on fresh weight. The quantification of signals was normalized by CBB staining of LHCII. Experiments were performed with five biological replicates and subjected to statistical analysis according to the standard deviation.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: FtsH1, At1g50250; FtsH2 (also annotated as VAR2), At2g30950; FtsH5 (also annotated as VAR1), At5g42270; and FtsH8, At1g06430.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Photographs of 6-Week-Old Rosette Leaves from Col, var2-1, var2(WT), var2(267), var2(488), and var2(267 488).
Supplemental Figure 2. Immunoblot Analysis of FtsH Proteins in Mature Leaves from var2(WT) and var2(488).
Supplemental Figure 3. In Vivo D1 Degradation Assay from Independent var2(WT) and var2(488) Transgenic Lines.
Supplemental Figure 4. Effect of Low Temperature on the Variegated Phenotype of var1 var2(WT) and var1 var2(488).
Supplemental Figure 5. Estrogen-Dependent Accumulation of FtsH2 in Independent pER8-var2(WT) Lines.
Supplemental Table 1. List of Primers Used for This Study.
Supplementary Material
Acknowledgments
We thank Teru Ogura for providing E. coli ftsH mutants and the plasmid clone pIFH108, Yuichiro Takahashi for PsaF antibodies and input on sucrose density gradient centrifugation, and Nam-Hai Chua for providing pER8. We also thank Rie Hijiya for her technical help. This work was supported by a Grant-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science and Technology (16085207 to W.S.), by the Asahi Glass Foundation, and by the Oohara Foundation.
References
- Adam Z., Rudella A., van Wijk K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Aluru M.R., Yu F., Fu A., Rodermel S. (2006). Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 57: 1871–1881 [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P.J., Horton P., Mullineaux C.W., Robinson C., Mann N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Baker T.A., Sauer R.T. (2006). ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K.J., Tomoyasu T., Donachie W.D., Khattar M., Niki H., Yamanaka K., Hiraga S., Ogura T. (1992). Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsI mutations. J. Bacteriol. 174: 2416–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C., Niederhauser B., Baumann U.M. (2009). The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation. Proc. Natl. Acad. Sci. USA 106: 21579–21584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C., Schalch T., Bumann M., Meister M., Meier R., Baumann U. (2006). The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103: 3066–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. (2006). Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Chen M., Choi Y., Voytas D.F., Rodermel S. (2000). Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Chiba S., Akiyama Y., Ito K. (2002). Membrane protein degradation by FtsH can be initiated from either end. J. Bacteriol. 184: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.K., MacDonald T.M., Sjögren L.L.E. (2005). The ATP-dependent Clp protease in chloroplasts of higher plants. Physiol. Plant. 123: 406–412 [Google Scholar]
- Farah J., Rappaport F., Choquet Y., Joliot P., Rochaix J.D. (1995). Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: Efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 14: 4976–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Yan H., Wu Z., Yan N., Wang Z., Jeffrey P.D., Shi Y. (2007). Structure of a site-2 protease family intramembrane metalloprotease. Science 318: 1608–1612 [DOI] [PubMed] [Google Scholar]
- Herman C., Prakash S., Lu C.Z., Matouschek A., Gross C.A. (2003). Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell 11: 659–669 [DOI] [PubMed] [Google Scholar]
- Ito K., Akiyama Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E., Naveh L., Adam Z. (2007). The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K., Inagawa T., Wilkinson A.J., Tatsuta T., Ogura T. (1999). Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem. 274: 26225–26232 [DOI] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. (2009). The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Miura E., Matsushima R., Sakamoto W. (2007). White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 144: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2009). Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2010). New insights into the types and function of proteases in plastids. Int. Rev. Cell Mol. Biol. 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D., van Wijk K.J. (2009). Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Spetea C., Hundal T., Oppenheim A.B., Adam Z., Andersson B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Tabak S., Cseke L., Pichersky E., Andersson B., Adam Z. (1996). Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 271: 29329–29334 [DOI] [PubMed] [Google Scholar]
- Liu X., Yu F., Rodermel S. (2010a). Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: A review. J. Integr. Plant Biol. 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Liu X., Yu F., Rodermel S. (2010b). An Arabidopsis pentatricopeptide repeat protein, SVR7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. http://dx.doi.org/10.1104/pp.110.164111 [DOI] [PMC free article] [PubMed]
- López-Juez E. (2007). Plastid biogenesis, between light and shadows. J. Exp. Bot. 58: 11–26 [DOI] [PubMed] [Google Scholar]
- Miura E., Kato Y., Matsushima R., Albrecht V., Laalami S., Sakamoto W. (2007). The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller S.G. (2005). Plastids. (Oxford: Blackwell; ). [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F., Obrist M., Führer F., Langklotz S. (2009). Degradation of cytoplasmic substrates by FtsH, a membrane-anchored protease with many talents. Res. Microbiol. 160: 652–659 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Wilkinson A.J. (2001). AAA+ superfamily ATPases: Common structure—diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O., Adam Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell 9: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Rodermel S.R. (2004). Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. USA 101: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.B., Ripoll D.R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg J., Liberles D.A., Roepstorff P., van Wijk K.J. (2001). Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276: 16318–16327 [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b edxtracted with four different solvents; verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Qu J.N., Makino S.I., Adachi H., Koyama Y., Akiyama Y., Ito K., Tomoyasu T., Ogura T., Matsuzawa H. (1996). The tolZ gene of Escherichia coli is identified as the ftsH gene. J. Bacteriol. 178: 3457–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D., Bode R., Li W., Krol M., Saccon D., Wang S., Schillaci L.A., Rodermel S.R., Maxwell D.P., Hüner N.P. (2009). Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21: 3473–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. (2003). Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 78: 1–9 [DOI] [PubMed] [Google Scholar]
- Sakamoto W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Miura E., Kaji Y., Okuno T., Nishizono M., Ogura T. (2004). Allelic characterization of the leaf-variegated mutation var2 identifies the conserved amino acid residues of FtsH that are important for ATP hydrolysis and proteolysis. Plant Mol. Biol. 56: 705–716 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Miyagishima S.-y., Jarvis P. (2008). Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Physiologists; ), doi/10.1199/tab.0110, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Tamura T., Hanba-Tomita Y., Murata M., Sodmergen (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7: 769–780 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Uno Y., Zhang Q., Miura E., Kato Y., Sodmergen (2009). Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol. 50: 2069–2083 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko A., Pojidaeva E., Zinchenko V., Panichkin V., Glaser V.M., Herrmann R.G., Shestakov S.V. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41: 291–310 [DOI] [PubMed] [Google Scholar]
- Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. (2010). The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. (2007). Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suno R., Niwa H., Tsuchiya D., Zhang X., Yoshida M., Morikawa K. (2006). Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol. Cell 22: 575–585 [DOI] [PubMed] [Google Scholar]
- Takechi K., Sodmergen, Murata M., Motoyoshi F., Sakamoto W. (2000). The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 41: 1334–1346 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Wickner S., Maurizi M.R., Gottesman S. (1999). Posttranslational quality control: Folding, refolding, and degrading proteins. Science 286: 1888–1893 [DOI] [PubMed] [Google Scholar]
- Wise R.R., Hoober J.K. (2006). The Structure and Function of Plastids. (Dortrecht, The Netherlands: Springer; ). [Google Scholar]
- Wollman F.A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Yu F., Liu X., Alsheikh M., Park S., Rodermel S. (2008). Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Park S., Rodermel S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Yu F., Park S., Rodermel S.R. (2005). Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Feder A., Adam Z. (2005b). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—Implications for thylakoid formation and photosystem II maintenance. Plant J. 42: 609–617 [DOI] [PubMed] [Google Scholar]
- Zaltsman A., Ori N., Adam Z. (2005a). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.