Abstract
Background
Due to advances in viral design, oncolytic adenoviruses have emerged as a promising approach for treatment of breast cancer. Tumor tissue slices offer a stringent model system for preclinical evaluation of adenovirus therapies, since the slices retain a morphology and phenotype that more closely resembles the in vivo setting than cell line cultures, and it has been shown to have utility in the evaluation of viral infectivity and replication. In this study, we evaluated the efficacy of viral infection and replication using a tropism-modified oncolytic adenovirus.
Methods
Breast tumor tissue slices were infected with a tropism-modified oncolytic adenovirus, and a wild-type adenovirus for comparison. Efficiency of infection was evaluated using fluorescent microscopy, as the viruses used have been modified to express red fluorescent protein. Replication of the viruses was evaluated with quantitative real-time PCR to assay viral E4 genome copy number, a surrogate indicator for the number of virions. The breast tumor tissue slices were evaluated for the expression of CD46 expression by immunohistochemistry.
Results
Infection and replication of our tropism modified oncolytic virus has been observed in breast cancer tissue slice model system and is comparative to wild-type virus. A qualitative increase in the number of cells showing RFP expression was observed correlating with increasing multiplicity of infection. Higher relative infectivity of the virus was observed in tumor tissue compared with normal breast tissue. Replication of the virus was demonstrated through increases in E4 copy number at 48 and 72 hours after infection in human breast tumor slices.
Conclusions
We have shown that a tropism modified oncolytic oncolytic adenovirus can infect and replicate in breast cancer tissue slices, which may be an important preclinical indicator for its therapeutic utility.
Keywords: adenovirus, breast cancer, CD46, CAR, oncolytic, tissue slice model
INTRODUCTION
Breast cancer is the most common female malignancy in the United States. Breast cancer affects one in nine women in the United States. Each year, 46 000 women die of breast cancer despite early detection methods and advanced conventional treatments. Investigating the molecular mechanisms underlying neoplastic transformation and progression has resulted in the understanding that breast cancer can be regarded as a genetic disease, arising from accumulating a series of acquired genetic lesions. As the limits of existing treatment regimes for breast cancer are recognized, clearly novel therapies may be useful for the successful treatment of carcinoma of the breast.
Virotherapy employing oncolytic adenoviruses represents a promising targeted intervention relevant to a wide array of neoplastic diseases, including breast cancer [1]. Using oncolytic adenoviruses represents a method to achieve efficient tumor cell oncolysis and mitigate tumor cell infection limitations [2,3]. Ideally, cancer-specific replication of oncolytic adenoviruses results in viral-mediated replication and oncolysis of infected tumor tissues. Release of virus progeny results in further propagation in surrounding tumor cells but not in those of normal tissues that would be refractory to oncolytic adenovirus replication.
Relative resistance of tumor tissues to adenovirus infection have been noted in many in vivo gene therapy trials [4–6]. A relative lack of expression of the primary adenovirus receptor, the coxsackie and adenovirus receptor (CAR), in primary tumors has now been understood to be the biologic basis of this phenomenon [7]. This observation can be explained by the finding that CAR has been shown to exhibit tumor suppression activity when introduced into tumor cells [8]. Therefore, it has been proposed that gene delivery by “CAR-independent” pathways may be required to circumvent this key aspect of tumor biology [9,10].
To characterize the specificity of oncolytic adenoviral replication required for breast cancer virotherapy, it is necessary to evaluate and test them in the most stringent pre-clinical model available. Most studies simply use cell lines as the necessary and sufficient analysis medium for this validation. However, human cancer cell lines, passaged in vitro for years, may not reflect the true biology of tumors in vivo. Thus, we determined to evaluate efficiency of infection and replication ex vivo in an explant model system of primary breast tumor tissue using a tissue slice technique [11, 12]. The tissue slice model system also represents the heterogeneity of the tumor and maintains its three-dimensional structure in vitro. We have recently explored this tissue slice technology, which offers a powerful and representative ex vivo model system for pre-clinical evaluation of oncolytic adenoviruses [13,14]. Slices allow subsequent histologic evaluation as well as biochemical evaluation methods - something not possible with current in vitro cell culture methods.
In this study, we decided to explore an oncolytic adenovirus vector in which altered cancer transduction could be achieved by substituting the capsid fiber gene with one from a different serotype. In this case we used a chimeric 5/3 fiber, in which the fiber is composed of the tail and shaft domains from serotype 5 and the knob domain from serotype 3. This modification alters the receptor recognition profile of the virus containing the 5/3 fiber chimera to CD46 [15,16], which is over expressed in breast cancer, but has limited expression in liver [17]. We evaluated the replication potential and infectivity of the tropism modified oncolytic adenovirus using the tissue slice model system. Infection and replication of this modified virus was compared with a vector containing a wild-type fiber gene.
MATERIALS AND METHODS
Preparation of Solid Tumor Tissue Slices
Approval to perform this study was obtained from the Institutional Review Board (IRB) at LSUHSC-S. Tumor tissue samples were obtained at the time of resection and placed on ice in sterile RPMI (Cellgro; Mediatech, Manassas, VA) containing 10% Fetal Bovine Serum (Gemini Bio-Products, West Sacramento, CA) and 1% antibiotic/antimycotic (Invitrogen, Carlsbad, CA). Samples were washed in RPMI containing 10% FBS and 10% antibiotic/antimycotic before use. Within 2 hrs of surgical resection, tissue cores of 8 mm diameter were created using a hand-held coring tool in a sterilized biological safety cabinet. Tissue cores were placed into a Krumdieck Tissue Slicer (Alabama Research and Development, Birmingham, AL) in ice-cold PBS solution (Mediatech) and sliced at a thickness setting of 250 μ to generate slices containing approximately 1 × 106 cells. The Krumdieck tissue slicer was sterilized and used according to the manufacturer’s instructions at a blade oscillation setting of 30 rpm. Slices were collected and placed into 6-well tissue culture dishes with complete culture media (RPMI containing 10% FBS and 1% antibiotic/antimycotic). The plates were then placed on a 60 rpm shaker in a 5% CO2 incubator at 37° C for 2 h before infection.
Immunohistochemistry and Histopathology of Sample
Sections of tumor sample were fixed and embedded in paraffin for use in immunohistochemistry studies for CD46 expression and for H&E staining for tumor morphology. Immunohistochemical analysis was performed using an automated processor (Dako Autostainer Plus; Dako, Glostrup Denmark) under the following conditions: anti-CD46 (mouse anti-human CD46 monoclonal; BioLegend, San Diego, CA) at 1:300 dilution for 1 h followed by biotinylated secondary antibody (Dako) for 15 min at RT. The sections were visualized incubation for 3 min using 3,3′-diaminobenzidine tetrahydrochloride (DAB) and H2O2. Afterwards, the slides were mounted and scanned with an Ariol automated system (Genetix, San Jose, CA).
Oncolytic Adenoviruses
TA tropism modified virus (Ad5/3-CXCR4-E1A-IX-mRFP1) was constructed by homologous recombination in Escherichia coli [18]. The adenovirus serotype 5 knob domain of the fiber protein was replaced with that of serotype 3. This chimeric fiber protein allows virus entry into the host cell to be dependent on the presence of CD46 instead of the coxsackie and adenovirus receptor CAR [15]. The E1 gene region of this virus was replaced with an expression cassette containing the human chemokine receptor CXCR4 gene promoter region driving adenovirus E1A gene expression necessary for viral replication. In addition, the RFP (red fluorescent protein) sequence was fused to the carboxy terminus of the viral capsid pIX protein (IX-mRFP1) to allow for reporting of infection efficacy using fluorescence microscopy. A wild-type serotype 5 adenovirus containing a pIX-RFP fusion protein (Ad5-wt-IX-mRFP1) described previously [19] was used for comparison of tumor-specific infection and replication with the Ad5/3-CXCR4-E1A-IX-mRFP1 virus.
Infection of Solid Tumor Tissue Slices
Viral infections were carried out at 2 h after plating the tumor slices with serial dilutions (5, 50, and 500 vp/cell) of adenoviral vectors in 3 mL RPMI containing 2% FBS and 1% antibiotic/antimycotic. The tissue slices were incubated in 5% CO2 incubators at 37° C for 2 h with adenoviruses, after which the infection media was removed. The slices were washed in 2 mL sterile PBS, and placed in complete culture media and returned to a 5% CO2 incubator at 37° C until analyzed.
Determination of Infection Efficacy
Fluorescence microscopy was used to determine infection efficiency of the tissue slices at 72 h post-infection. A Nikon Eclipse TE300 inverted microscope with an epiflourescence attachment was used with a 20× objective and a 0.50 numerical aperture for visualization, and images were acquired using a Photometrics CoolSNAP fx monochrome 12-bit CCD camera using the IPLab 3.7 software (Scanalytics, Fairfax, VA). Files were saved as 8-bit TIFF images and further analysis of images was performed using ImageJ for microscopy software (National Institute of Mental Health, Bethesda, MD).
Assessment of Replication of Virus
Increase in the copy number of the E4 gene was used as a surrogate to determine the number of virions. At 24, 48, and 72 h post-infection, aliquots of culture media (300 μL) were removed and frozen at −80° C until use. Total DNA was purified using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA). E4 gene copy number was determined using specific E4 primers and Taqman probes for real-time PCR as previously described [20] with TaqMan fast reagents (Applied Biosystems, Framingham, MA) and measured against a standard curve made from known amounts of adenovirus genome.
RESULTS
Characterization of breast tumor tissue sources
Tissue samples from five patients were obtained for experiments from Surgical Oncology, and were processed within 2 h after excision according to the protocol described above. Patient demographic information as well as information about histological type is found in Table 1.
TABLE 1.
Patient demographics and histological types
| Patient | Age | Race/Gender | Histological type |
|---|---|---|---|
| 1 | 70 | AAF | infiltrating ductal |
| 2 | 38 | AAF | features of infiltrating ductal and infiltrating lobular* |
| 3 | 59 | AAF | infiltrating ductal |
| 4 | 55 | AAF | infiltrating ductal |
Patient has had previous chemotherapy
Determination of tropism modified oncolytic adenovirus infectivity in breast cancer tissue slices
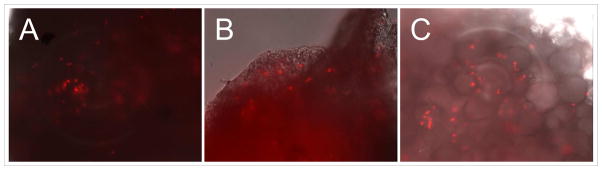
In order to demonstrate viral infection in our breast cancer tissue slices, at 72 h after infection with Ad5/3-CXCR4-E1A-IX-mRFP1 at 5, 50, and 500 MOI, the tissue slices were viewed with fluorescent microscopy; in this vector construct, the RFP (red fluorescent protein) sequence was fused to the carboxy terminus of the viral capsid pIX protein (IX-mRFP1) to allow for fluorescent reporter expression in infected cells. As shown in Fig. 1, a qualitative increase in the number of cells showing RFP expression was observed, which correlated with increasing multiplicity of infection in the breast cancer tissue slices. In a second experiment, infection of breast cancer tissue slices was directly compared with infection of normal breast tissue slices obtained from the same patient. Analysis of fluorescent reporter expression demonstrated a higher relative infectivity in tumor tissue (Fig. 2A) compared with normal breast tissue (Fig. 2B) using the tropism-modified oncolytic adenovirus (Ad5/3-CXCR4-E1A-IX-mRFP1). A similar result was obtained using a wild-type serotype 5 adenovirus containing a pIX-RFP fusion protein (Ad5-wt-IX-mRFP1): a higher relative infectivity in tumor tissue (Fig. 2C) compared with normal breast tissue (Fig. 2D).
Figure 1. Fluorescence microscopy images of breast tumor tissue slices infected with Ad5/3-CXCR4-IX-mRFP1.
Slices were infected with increasing multiplicities of infection at 5, 50, and 500 viral particles/cell and examined after 72 h.
Figure 2. Infectivity of breast cancer tissue slices compared with normal breast tissue slices obtained from the same patient.
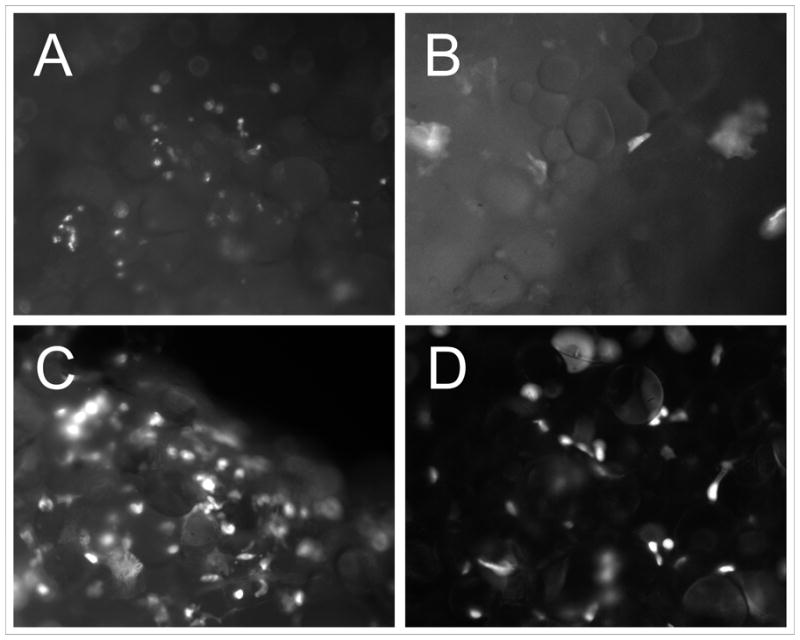
Analysis of fluorescent reporter expression in tumor tissue slices (Fig. 2A) slices and normal breast tissue slices (Fig. 2B) from the same patient infected with Ad5/3-CXCR4-E1A-IX-mRFP1 vector. Analysis of fluorescent reporter expression in tumor tissue slices (Fig. 2C) slices and normal breast tissue slices (Fig. 2D) from the same patient infected with Ad5-wt-IX-mRFP1 vector.
Evaluation of CD46 expression in breast cancer tissue slices using immunohistochemistry
In order to demonstrate CD46 expression in breast cancer tissue samples, a portion of the sample was fixed and embedded in paraffin, and used with an anti-CD46 antibody in immunohistochemistry. As shown in Fig. 3, a representative breast tumor tissue slice sample stained positively for CD46. However, there was very little staining in the no-primary antibody control indicating the specificity of the antibody.
Figure 3. Immunohistochemical analysis of CD46 expression in breast tumor tissue slices.
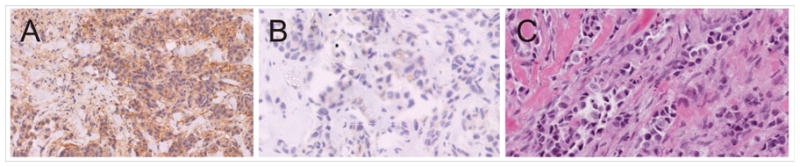
Representative staining pattern of sections using (A) an anti-CD46 monoclonal antibody or using (B) no primary antibody. An adjacent section was H&E stained for identification of histology.
Determination of oncolytic adenovirus replication in breast cancer tissue slices
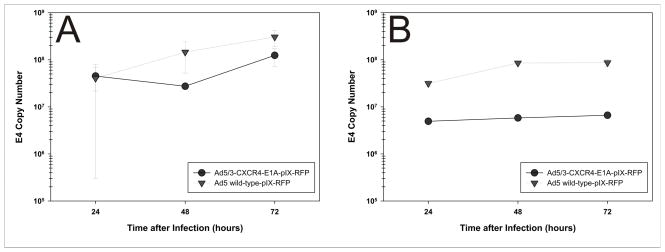
In order to determine the replicative potential of oncolytic adenoviruses, tumor tissue slices were infected with Ad5/3-CXCR4-IX-mRFP1 virus or with Ad5-wt-IX-mRFP1 virus at a multiplicity of infection at 500 viral particles/cell. At 24, 48, and 72 h after infection, media aliquots were removed and analyzed for the amount of adenoviral particles by E4 gene copy number. Both Ad5/3-CXCR4-IX-mRFP1 and Ad5-wt-IX-mRFP1 replicated efficiently in breast cancer tissue slices (Fig. 4), with comparative copy number increases in patient samples. Replication of the tropism modified virus Ad5/3-CXCR4-IX-mRFP1 was demonstrated in a set of experiments in which tumor tissue slices were infected, and E4 copy number was obtained as a surrogate marker for number of virions. Increases in viral copy number between 24 and 72 hours post-infection were indicative of viral replication. Replication of the wild-type virus was demonstrated in all patients, while replication of the Ad5/3-CXCR4-IX-mRFP1 virus was observed in three out of four patients tested (Table 2). This result indicates that the Ad5/3-CXCR4-IX-mRFP1 virus can replicate efficiently in breast tumor tissue slices.
Figure 4. Replication of oncolytic adenoviruses in breast tumor tissue slices.
Viral copy number as an indicator of viral replication in breast tumor tissue slices from (A) Patient 1 and (B) Patient 2. Growth medium was collected at 24, 48, and 72 h after infection with Ad5/3-CXCR4-E1A-IX-mRFP1 or Ad5-wt-IX-mRFP1. Points represent the mean ± SD of three replicate samples.
TABLE 2.
Viral replication in tissue slices
| Ad5/3-CXCR4-IX-mRFP1 | Ad5-wt-IX-mRFP1 | |||||
|---|---|---|---|---|---|---|
| 24 hr | 72 hr | -fold | 24 hr | 72 hr | -fold | |
| Patient 1 | 4.0 × 107 | 1.2 × 108 | 3.0 | 4.0 × 107 | 3.0 × 108 | 7.5 |
| Patient 2 | 5.0× 106 | 6.6 × 106 | 1.3 | 3.1 × 107 | 9.0 × 107 | 2.9 |
| Patient 3 | 8.0 × 108 | 9.4 × 108 | 1.2 | 4.7 × 109 | 1.0 × 1010 | 2.1 |
| Patient 4 | 9.0 × 105 | 4.6 × 106 | 5.0 | 2.4 × 107 | 9.0 × 107 | 3.8 |
DISCUSSION
Low infectivity in target cells represents a significant obstacle in adenovirus oncolytic virotherapy approaches, and is one of the mechanisms underlying the poor efficacy in previous adenoviral clinical trials. The relatively poor in vivo tumor infectivity seen in the past is ascribed to the variable expression of the coxsackie and adenovirus receptor (CAR), the major mediator of endocytosis of adenovirus virions [7]. CAR is variably expressed in human cancers, and often down regulated in more aggressive cancers, as it is thought to have tumor suppressor qualities [8]. Modifications of the fiber region of the adenovirus capsid have been demonstrated to successfully change the virus’ tropism and retarget the virus toward receptors more common in advanced human malignancies. An alteration of the knob domain of the fiber protein to that of serotype 3, resulting in a 5/3 virus, effectively changes the mediator of virus uptake to CD46 [15, 16], a surface protein involved in complement inactivation, the expression of which is up regulated in human malignancies.
In this study, an increase in infectivity of breast tissue slices using a 5/3 tropism modified oncolytic adenovirus, Ad5/3-CXCR4-IX-mRFP1, correlating with increasing multiplicity of infection was demonstrated; the Ad5/3-CXCR4-IX-mRFP1 virus showed infectivity of the breast cancer tissue slices similar to a wild-type Ad5-wt-IX-mRFP1. In one experiment, Ad5/3-CXCR4-IX-mRFP1 showed higher quality of RFP expression, and therefore infectivity in breast tumor tissue slice infection than in benign breast tissue slices. This difference in infection efficiency between tumor tissue slices and benign tissue slices was not as pronounced in those infected with Ad5-wt-IX-mRFP1, suggesting that the 5/3 fiber modification displays increased tumor selectivity for tumors. This result correlates with the higher relative expression of CD46 in breast cancer tissue than in normal tissue, and relatively equal expression of the CAR-targeted by the wild-type Ad5-wt-IX-mRFP1 virus. These results demonstrating equal expression, may be due to high levels of CAR expression in these patient samples resulting in high infectivity of the wild-type virus. Nonetheless a greater differential in expression between benign and tumor tissue samples using the Ad5/3-CXCR4-IX-mRFP1 virus would indicate a better safety profile than wild-type Ad5-wt-IX-mRFP1.
These experiments have demonstrated infection and replication of a tropism-modified oncolytic adenovirus construct in breast cancer tissue slices. Demonstrating functionality of these retargeted viruses within the ex vivo tissue slice model system may be a useful bridge between in vitro and in vivo studies. The tissue slice model system is a stringent preclinical system for evaluating the infectivity and replication potential of oncolytic adenovirus constructs [13, 14]. Importantly, the cancer cells targets of oncolytic adenoviruses have a morphology and phenotype in this model system that more closely resembles the setting in vivo in tumors than the cancer cell lines of conventional cell culture. Our results demonstrate this model system can be used to test virus function, and evaluate efficacy and cytotoxicity ex vivo as a precursor to clinical trials. Future studies will determine the value of tissue slice based experiments in predicting the effects of additional oncolytic adenovirus modifications.
Footnotes
Presented at the 5th Annual Academic Surgical Congress, February 3–5, 2010, San Antonio, TX
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rein DT, Breidenbach M, Curiel DT. Current developments in adenovirus-based cancer gene therapy. Future Oncol. 2006;2:137–143. doi: 10.2217/14796694.2.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alemany R, Balague C, Curiel DT. Replicative adenoviruses for cancer therapy. Nat Biotechnol. 2000;18:723–727. doi: 10.1038/77283. [DOI] [PubMed] [Google Scholar]
- 3.Kirn D, Martuza RL, Zwiebel J. Replication-selective virotherapy for cancer: biological principles, risk management and future directions. Nat Med. 2001;7:781–787. doi: 10.1038/89901. [DOI] [PubMed] [Google Scholar]
- 4.Rancourt C, Rogers BE, Sosnowski BA, et al. Basic fibroblast growth factor enhancement of adenovirus-mediated delivery of the herpes simplex virus thymidine kinase gene results in augmented therapeutic benefit in a murine model of ovarian cancer. Clin Cancer Res. 1998;4:2455–2461. [PubMed] [Google Scholar]
- 5.Sterman DH, Treat J, Litzky LA, et al. Adenovirus-mediated herpes simplex virus thymidine kinase/ganciclovir gene therapy in patients with localized malignancy: results of a phase I clinical trial in malignant mesothelioma. Hum Gene Ther. 1998;9:1083–1092. doi: 10.1089/hum.1998.9.7-1083. [DOI] [PubMed] [Google Scholar]
- 6.Vasey PA, Shulman LN, Campos S, et al. Phase I Trial of Intraperitoneal Injection of the E1B-55-kd-Gene–Deleted Adenovirus ONYX-015 (dl1520) Given on Days 1 Through 5 Every 3 Weeks in Patients With Recurrent/Refractory Epithelial Ovarian Cancer. J Clin Oncol. 2000;20:1562–1569. doi: 10.1200/JCO.2002.20.6.1562. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Zinn KR, Barnett BG, et al. The therapeutic efficacy of adenoviral vectors for cancer gene therapy is limited by a low level of primary adenovirus receptors on tumour cells. Eur J Cancer. 2002;38:1917–1926. doi: 10.1016/s0959-8049(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 8.Okegawa T, Li Y, Pong RC, et al. The Dual Impact of Coxsackie and Adenovirus Receptor Expression on Human Prostate Cancer Gene Therapy. Cancer Res. 2000;60:5031–5036. [PubMed] [Google Scholar]
- 9.Haviv YS, Blackwell JL, Kanerva A, et al. Adenoviral Gene Therapy for Renal Cancer Requires Retargeting to Alternative Cellular Receptors. Cancer Res. 2002;62:4273–4281. [PubMed] [Google Scholar]
- 10.Kawakami Y, Li H, Lam JT, et al. Substitution of the Adenovirus Serotype 5 Knob with a Serotype 3 Knob Enhances Multiple Steps in Virus Replication. Cancer Res. 2003;63:1262–1269. [PubMed] [Google Scholar]
- 11.Krumdieck CL. A new instrument for the rapid preparation of tissue slices. Anal Biochem. 1980;104:118 –123. doi: 10.1016/0003-2697(80)90284-5. [DOI] [PubMed] [Google Scholar]
- 12.Parrish AR, Gandolfi AJ, Brendel K. Precision-cut tissue slices: applications in pharmacology and toxicology. Life Sci. 1995;57:1887–1901. doi: 10.1016/0024-3205(95)02176-j. [DOI] [PubMed] [Google Scholar]
- 13.Kirby TO, Rivera A, Rein D, et al. A novel ex vivo model system for evaluation of conditionally replicative adenoviruses therapeutic efficacy and toxicity. Clin Cancer Res. 2004;10:8697–703. doi: 10.1158/1078-0432.CCR-04-1166. [DOI] [PubMed] [Google Scholar]
- 14.Stoff-Khalili MA, Stoff A, Rivera A, et al. Preclinical evaluation of transcriptional targeting strategies for carcinoma of the breast in a tissue slice model system. Breast Cancer Res. 2005;7:1141–1152. doi: 10.1186/bcr1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirena D, Lilienfeld B, Eisenhut M, et al. The Human Membrane Cofactor CD46 Is a Receptor for Species B Adenovirus Serotype 3. J Virol. 2004;78:4454–4462. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanerva A, Mikheeva GV, Krasnykh V, et al. Targeting Adenovirus to the Serotype 3 Receptor Increases Gene Transfer Efficiency to Ovarian Cancer Cells. Clin Cancer Res. 2002;8:275–280. [PubMed] [Google Scholar]
- 17.Madjd Z, Durrant LG, Pinder SE, et al. Do poor-prognosis breast tumours express membrane cofactor proteins (CD46)? Cancer Immunol Immunother. 2005;54:149–156. doi: 10.1007/s00262-004-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He TC, Zhou S, da Costa LT, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le LP, Le HN, Dmitriev IP, et al. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006;98:203–214. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- 20.Haviv YS, Takayama K, Glasgow JN, et al. A model system for the design of armed replicating adenoviruses using p53 as a candidate transgene. Mol Cancer Ther. 2002;1:321–328. [PubMed] [Google Scholar]